Abstract
Heterochromatin formation in the yeast Saccharomyces cerevisiae is characterized by the assembly of the Silent Information Regulator (SIR) complex, which consists of the histone deacetylase Sir2 and the structural components Sir3 and Sir4, and binds to unmodified nucleosomes to provide gene silencing. Sir3 contains an AAA+ ATPase-like domain, and mutations in an exposed loop on the surface of this domain abrogate Sir3 silencing function in vivo, as well in vitro binding to the Sir2/Sir4 subcomplex. Here, we found that the removal of a single methyl group in the C-terminal coiled-coil domain (mutation T1314S) of Sir4 was sufficient to restore silencing at the silent mating-type loci HMR and HML to a Sir3 version with a mutation in this loop. Restoration of telomeric silencing required further mutations of Sir4 (E1310V and K1325R). Significantly, these mutations in Sir4 restored in vitro complex formation between Sir3 and the Sir4 coiled-coil, indicating that the improved affinity between Sir3 and Sir4 is responsible for the restoration of silencing. Altogether, these observations highlight remarkable properties of selected amino-acid changes at the Sir3-Sir4 interface that modulate the affinity of the two proteins.
Keywords: gene silencing, repression, chromatin, Sir1
The genome of eukaryotic organisms is packaged with histone and nonhistone proteins into chromatin, which is the substrate for all processes on the genetic material, like DNA replication, transcription, DNA repair, and chromosome segregation. The chromatin architecture differs among different genomic regions, and allows the organism to implement individual gene expression programs according to cellular function, for instance, during development (Ehrenhofer-Murray 2004). Large regions assume a repressive structure termed heterochromatin, which is thought to result from a more condensed folding of the chromatin fiber, and is brought about by heterochromatin proteins that bind to the nucleosomes. In higher eukaryotes, such regions typically are found at the telomeres, where they prevent degradation and recombination, and centromeres, where they are important for proper chromosome segregation (Perrod and Gasser 2003).
An archetypal form of heterochromatin is found in the budding yeast Saccharomyces cerevisiae at the silent mating-type loci HML and HMR and at the telomeres (Rusche et al. 2003). The establishment and formation of heterochromatin at these loci is mediated by the silent information regulator (SIR) complex, which consists of the NAD+-dependent histone deacetylase (HDAC) Sir2, and the structural components Sir3 and Sir4 (Kueng et al. 2013). All three components of the complex are necessary for transcriptional gene silencing (Rine and Herskowitz 1987). The establishment and spreading of silent chromatin is a stepwise process, in which the SIR complex does not directly bind to the DNA, but is recruited via sequence-specific DNA binding proteins, and undergoes specific contacts with the histones in the nucleosomes (Oppikofer et al. 2013a). In a first step, a Sir2/Sir4 subcomplex binds to proteins/protein complexes like Rap1 (Moretti et al. 1994), the origin recognition complex (ORC) (Triolo and Sternglanz 1996), and Abf1, which themselves bind cis-acting DNA sequences (Kimmerly et al. 1988), the so-called silencer elements. At the HM silencers, this interaction is bridged by the Sir1 protein (Bose et al. 2004). Subsequently, Sir2 deacetylates the lysines in the N-termini of histones H3 and H4, including H4 lysine 16, which is essential for effective silencing (Imai et al. 2000; Landry et al. 2000). This leads to the recruitment of Sir3 to the unmodified nucleosomes (Hecht et al. 1995). This process of deacetylation of histones and SIR protein binding is repeated in multiple cycles, and allows the SIR complex to spread along the chromatin fiber (Rusche et al. 2002). The extent of SIR spreading depends on the concentration of each component, as well as on histone acetylation as the substrate for Sir2 deacetylation, and overexpression of Sir3 leads to extension of the silent region (Renauld et al. 1993; Maillet et al. 1996).
A mechanistic understanding of heterochromatin architecture requires detailed molecular insights into the interactions among the SIR proteins. Structural information is available for several homologs of Sir2 (Marmorstein 2004), as well as for S. cerevisiae Sir2 bound to a fragment of Sir4 (Sir2 interaction domain SID, aa 737–893) (Hsu et al. 2013). Sir2 consists of a Rossman fold domain, and a smaller zinc-containing regulatory domain, and the Sir4 fragment contacts the interface between the N-terminal regulatory domain, and the catalytic domain. Sir2 forms a stable heterodimer with Sir4, and this interaction strongly stimulates the HDAC activity of Sir2 (Tanny et al. 2004; Cubizolles et al. 2006; Hsu et al. 2013).
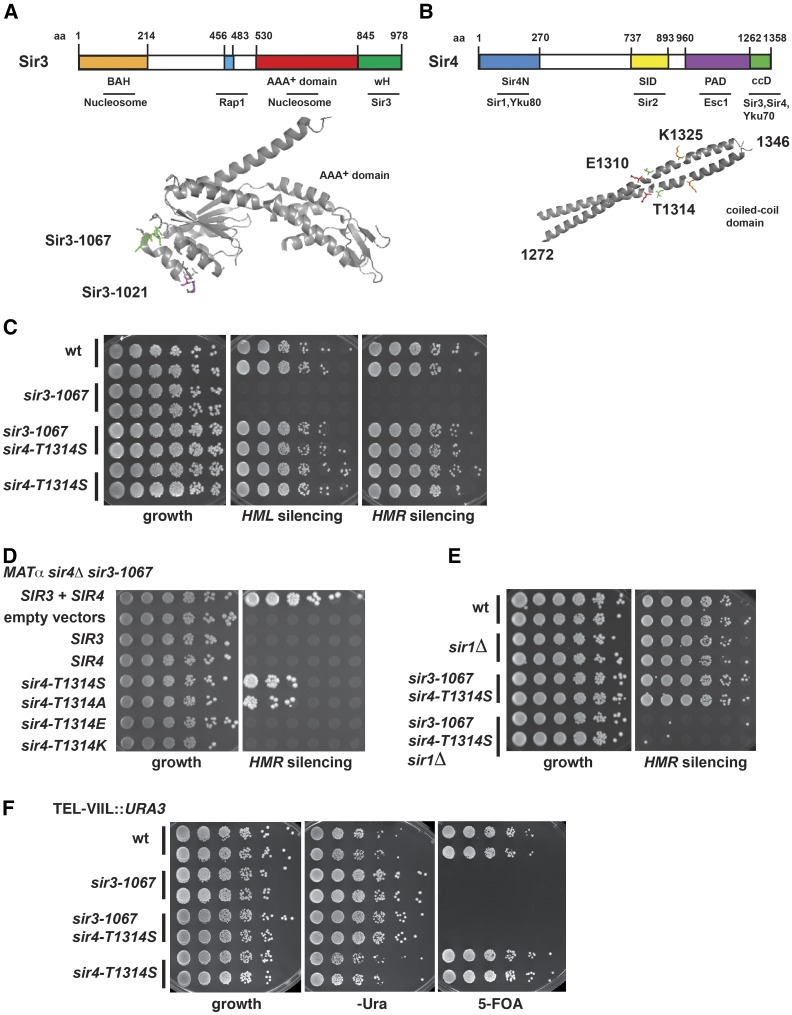
Sir3 shares its domain structure with its paralog Orc1, a component of the replication initiation complex ORC (Norris and Boeke 2010). It consists of three functional domains: the N-terminal bromo-adjacent homology (BAH) domain, the AAA+ ATPase-like domain, and the C-terminal winged helix-turn-helix (wH) domain (Figure 1A). (1) The BAH domain (aa 1–214) is a nucleosome-binding module (Onishi et al. 2007; Sampath et al. 2009), whose structure with the nucleosome shows important contacts with at least 28 histones residues, and whose binding is inhibited by acetylation of H4 K16 and methylation of H3 K79 (Armache et al. 2011; Wang et al. 2013), but enhanced by Nα-acetylation of Sir3 (Arnaudo et al. 2013; Yang et al. 2013). (2) The C-terminal 138 amino acids (aa) of Sir3 show a variant winged helix-turn-helix conformation that forms a dimer, and dimerization is essential for its silencing capacity. This domain also contributes to nucleosome binding of the SIR complex, but does not itself bind to chromatin (Oppikofer et al. 2013b). (3) Our structural analysis of the Sir3 AAA+ domain (Ehrentraut et al. 2011) revealed the typical structures of a base and a lid subdomain, as observed in other AAA+ ATPases, but an unusual topology of the domains relative to each other that disfavors nucleotide binding in the cleft between the domains. In contrast to other AAA+ ATPases, Sir3 lacks the residues required for ATP binding and hydrolysis (Bell et al. 1995). We identified several mutations in the Sir3 AAA+ domain that abrogate Sir3 silencing function (Ehrentraut et al. 2011). In particular, mutations in an extended loop of the base subdomain that connects α-helix 4 to strand 3 of the central β-sheet (K657A, K658A, and allele sir3-1067) cause a strong loss of Sir3 silencing function, and abrogates the in vitro interaction of Sir3 with Sir2/Sir4. Conversely, mutations in a loop connecting the α-helices 3 and 4 (D640A, S642L, and allele sir3-1021) abrogate HM silencing, but do not affect the Sir3–Sir4 interaction. The AAA+ domain is also able to bind nucleosomes in vitro in a H3 K79 methylation-sensitive fashion, and thus also contributes to chromatin binding of full-length Sir3. In addition to these three domains, structural information is also available for a short fragment of Sir3 that interacts with Rap1. This region (aa 456–483) lies N-terminal to the AAA+ domain, and forms a short α-helix that binds to a C-terminal region of Rap1 (Chen et al. 2011).
Figure 1.
Identification of a mutation in the coiled-coil domain of Sir4 that suppresses the HM silencing defect of a mutation in the Sir3 AAA+ loop. (A) Schematic illustration of the Sir3 protein (top), and structure of the AAA+ ATPase-like domain (below). BAH, bromo-adjacent homology domain; wH, winged-helix domain. The sir3 alleles sir3-1067 (K657A, K658A) and sir3-1021 (D640A, S642L) are mapped on the structure (PDB: 3TE6) (B) Schematic illustration of the Sir4 protein including the C-terminal coiled-coil domain (ccD, aa 1262–1358), the Sir2 interaction domain (SID), and the partitioning and anchoring domain (PAD) (top). Below, structure of the Sir4 coiled-coil domain. The amino acids E1310, T1314, and K1325 that are relevant for this study are mapped on the structure (PDB: 1PL5). (C) Mutation of Sir4-T1314 to serine (sir4-T1314S) restored the silencing defect of sir3-1067 at HMR (AEY5554) and HML (AEY5555). A semiquantitative mating assay was performed as described in Materials and Methods, and plates were incubated for 3 d at 30°. (D) Mutation of Sir4-T1314 to serine and alanine (sir4-T1314S, sir4-T1314A), but not glutamine (E) or lysine (K), restored the HMR-silencing defect of sir3-1067. Plasmids encoding the respective Sir3 and Sir4 versions were transformed into a MATα sir3-1067 sir4Δ strain (AEY5184). (E) Suppression of sir3-1067 by sir4-T1314S depended on Sir1. (F) Sir4-T1314S did not suppress the telomeric silencing defect of sir3-1067. sir3-1067 and sir4-T1314S were chromosomally integrated into a TEL-VIIL::URA3 strain, and serial dilutions were spotted on a 5-FOA containing plate to analyze their ability to silence the URA3 reporter gene at the telomere. Cells were additionally spotted on supplemented minimal medium as a growth control. The plates were incubated for 3 d at 30°.
The largest protein in the SIR complex is the Sir4 protein, which undergoes a multitude of protein–protein interactions to ensure efficient repression (Figure 1B), and thus is regarded as a scaffold protein for silencing (Kueng et al. 2013). Little structural information is available for Sir4, as its nonglobular nature has hindered its biochemical analysis. Apart from the above-mentioned dimerization with Sir2, Sir4 also interacts with the Ku heterodimer (Roy et al. 2004) as well as with Rap1 (Moretti et al. 1994; Kueng et al. 2012). The partitioning and anchoring domain of Sir4 (PAD, aa 960–1262) also mediates anchorage to the nuclear envelope via interaction with Esc1, which is associated with the nuclear envelope (Taddei et al. 2004). The N-terminus of Sir4 interacts with naked DNA and with Sir1 (Triolo and Sternglanz 1996; Martino et al. 2009; Kueng et al. 2012), which serves to recruit Sir2/Sir4 to the silencers. The only further structural information available on Sir4 is from the extreme C-terminus (aa 1272–1358), which dimerizes and assumes a parallel coiled-coil conformation (Chang et al. 2003; Murphy et al. 2003). Mutations that disrupt the dimerization activity abrogate Sir4 silencing function. Importantly, the Sir4 coiled-coil is sufficient to interact in vitro with a Sir3 fragment comprising the AAA+ domain and the wH domain (aa 464–978), and mutations were identified in a hydrophobic patch on the surface of the coiled-coil that do not interfere with dimerization, but abrogate the in vitro interaction with Sir3 (Chang et al. 2003). A previous study showed that these mutations also abrogated in vivo silencing (Rudner et al. 2005).
Altogether, the SIR complex contains two known dimerization domains, the Sir3 wH (Oppikofer et al. 2013b), and the Sir4 coiled-coil (Chang et al. 2003; Murphy et al. 2003). The SIR complex therefore possibly can form a heterohexamer with two subunits each of Sir2, Sir3, and Sir4. Each heterohexamer contains two Sir3 BAH, and two AAA+ domains, both of which bind nucleosomes, and whose binding is further supported by the Sir3 wH.
Here, we sought to characterize the defect of mutations in the interaction loop of the Sir3 AAA+ domain with Sir4 (Ehrentraut et al. 2011). We hypothesized that the Sir3 loop interacts directly with surface residues of the Sir4 C-terminus. Our work led us to identify a point mutation in the Sir4 coiled-coil domain, a mutation of threonine 1314 to serine (sir4-T1314S), that was able to suppress the silencing defect of the Sir3 loop mutation at HMR and HML. Interestingly, further mutations in the vicinity (E1310V, K1325R) were required to also restore telomeric silencing. We show that these mutations restore two-hybrid interactions with Sir3. Furthermore, the mutant Sir4 coiled-coil domains were capable of in vitro association with Sir3 versions with a defective Sir4 interaction loop. Altogether, our work shows the surprising discovery that removal of a single methyl group at position 1314 of Sir4 is sufficient to enhance binding between the Sir4 coiled-coil and Sir3, and thus to enhance heterochromatin formation in vivo.
Material and Methods
Yeast strains and plasmids
The yeast strains and plasmids used in this study are listed in Supplemental Material, Table S1 and Table S2. Yeast was grown and manipulated according to standard procedures (Sherman 1991). Yeast was grown on full medium (YPD) and selective minimal plates (YM), and plates containing 5-fluoro-orotic acid (US Biological) were used to select against URA3. Chromosomal integration of sir4 alleles was obtained by transferring them onto a yeast integrating plasmid (pRS306, URA3-marked), and introducing them into yeast strains by integrative transformation followed by loop-out on 5-FOA medium. Semiquantitative mating assays were performed by generating serial dilutions (1:10, start OD600 of one) of the respective strain in a microtiter dish. For the growth control, cells were transferred to agar plates using a replica tool. An equal volume of the mating tester strain (suspension of 10 OD600 per milliliter) was then added to the strain in the microtiter well, and a replica of this mixture was transferred to a plate selective for the growth of diploids. Plates were incubated for 2–3 d at 30°.
pGBD-C2-sir4 plasmids were generated by excising a ClaI/BlpI fragment of sir4 alleles from pAE2289 or pAE2029, and inserting it into ClaI/BlpI-cleaved pAE1355. pGAD-C2 and pGBD-C2 plasmids encoding the coiled-coil domain of Sir4 were generated by inserting Sir4 fragments using ClaI and BglII.
Random mutagenesis of SIR4
To isolate sir4 alleles that suppress the HMR silencing defect of sir3-1067, a region of SIR4 corresponding to aa 747–1358 was amplified from a URA3-SIR4 plasmid (pAE233) using mutagenic PCR conditions (van Loo et al. 2004). The plasmid was cleaved with ClaI/SmaI, and the backbone was cotransformed with the mutagenized PCR product into a MATα sir3-1067 sir4∆ strain (AEY5184) in order to generate URA3-sir4 plasmids by gap repair. Resulting transformants were tested for their ability to mate with a MATa tester strain. Among ∼15,000 transformants, one candidate was identified that restored mating ability. The URA3-sir4 plasmid was isolated from the candidate, amplified in Escherichia coli, retested in AEY5184 for restoration of mating, and subjected to sequencing. The plasmid carried 17 mutations compared to SIR4, 10 of which caused aa changes. Further investigation showed that the mutation causing the suppression phenotype lies between aa 1142 and 1358 of the mutant sir4 allele, which carries 2 aa changes (T1314S and V1351A). The sir4-T1314S mutation was constructed de novo by PCR sewing and gap repair to generate pAE2112. The equivalent procedure was carried out in a MATα sir3-1067 sir4∆ TEL-VIIL::URA3 strain (AEY5461), but using a HIS3-SIR4 plasmid (pAE2137), and a total of 37,000 candidates were screened for restoration of silencing to the telomeric URA3 reporter. This resulted in the identification of two candidate plasmids that restored telomeric silencing. One candidate carried nine mutations (four silent mutations) in SIR4, including E1310V and T1314S, and the other candidate had nine mutations (no silent mutations), including T1314S and K1325R.
Expression and purification of recombinant Sir3 and Sir4 constructs
Sir3 (464–978 aa) constructs were cloned into pET21d using NcoI/BamHI. The constructs were expressed in E. coli strain BL21 Rosetta, and protein production was induced by auto-induction (Studier 2005). The purification was performed according to Chang et al. (2003) and King et al. (2006). Briefly, cell pellets were resuspended in lysis buffer containing 50 mM Hepes (pH 7.6), 500 mM KCl, 5% glycerol, 5 mM β-mercaptoethanol, 1 mM PMSF, protease inhibitors, and lysozyme. The lysate was incubated 30 min on ice. After sonification, the insoluble material was pelleted for 60 min at 20,000 × g. The lysate was incubated with Protino Ni-IDA resin (Macherey and Nagel) for 1 hr at 4°. Bound protein was washed and eluted with 250 mM imidazole. The eluate was subsequently diluted five times in 50 mM Hepes (pH 7.6), 1 mM EDTA, 1 mM PMSF, and 1 mM DTT, and eluted with a gradient of 100–500 mM KCl on a HiTrap FF Sepharose ion exchange column (GE Healthcare). Pooled fractions were concentrated, and loaded on a Superdex 200 gel filtration column. Peak fractions were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80°. Sir4 (1217–1358 aa) constructs were cloned into pET15b using XhoI/BamHI. Sir4 plasmids were transformed in E. coli strain BL21 Rosetta, and protein production was induced with 0.5 mM IPTG at 20° overnight. The cell pellets were resuspended in lysis buffer containing 50 mM NaH2PO4, 300 mM NaCl (pH 8), 1 mM PMSF, 1 mM β-mercaptoethanol, 0.2% Tween 20 and lysozyme. After lysis, the purification of Sir4 constructs with Ni-NTA agarose (Qiagen) was performed according to the manufacturer’s instructions. Subsequently, the Sir4 containing eluates were concentrated and loaded on a Superdex 75 gel filtration column. Pooled fractions were concentrated, flash frozen in liquid nitrogen, and stored at −80°.
Analytical gel filtration
For analytical size exclusion chromatography (SEC) experiments, the proteins were mixed in equimolar ratio, and incubated for 40 min on ice. Subsequently, the SEC experiments were performed on a Superdex 200 Increase 3.2/300 column under isocratic flow conditions at 4°. The eluates were fractionated, loaded onto SDS-PAGE, and the gels were stained with Coomassie Brilliant Blue.
RNA expression analysis
Total RNA from 50 ml yeast cultures was extracted with TriFast (Peqlab), followed by an additional DNase treatment and cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Subsequently, quantitative real-time PCR was performed with Sybr Green Mastermix (Quanta) to determine the expression of the indicated subtelomeric genes. Primer sequences for quantitative real-time PCR are available upon request.
Chromatin immunoprecipitation (ChIP)
ChIP and quantititative real-time PCR were performed as previously described with the following exceptions (Samel et al. 2012). For crosslinking, the cells were incubated for 20 min with 1% formaldehyde, and the reaction was stopped with 125 mM glycine for 5 min; 5 µl of α-His antibody (Sigma, H-1029) was used for the immunoprecipitation of Sir4. The cell lysate was subsequently incubated with Dynabeads Protein G (Invitrogen) for 5 hr at 4°. Primer sequences for quantitative real-time PCR are available upon request.
Data availability
Strains and plasmids are available upon request. Table S1 contains the genotypes of all strains used in this study. Table S2 contains all plasmids used in this study.
Results
Removal of a methyl group by a threonine-to-serine mutation at Sir4 position T1314 restores HM silencing with Sir3 mutated in the AAA+ domain
Our earlier structure-function analysis of the Sir3 AAA+ domain showed that mutations in a loop of the AAA+ domain (K657A, K658A; named sir3-1067) abrogated Sir3 silencing function as well as the interaction of Sir3 with Sir4 [Figure 1A, and see Ehrentraut et al. (2011)]. Here, we sought to identify the region of Sir4, whose interaction with Sir3 is disrupted by sir3-1067 by isolating alleles of SIR4 that suppressed the silencing defect of sir3-1067. We reasoned that such mutant Sir4 proteins might restore silencing by improved binding to the mutant Sir3-1067 protein.
Since Sir3 has previously been shown to interact with the C-terminus of Sir4 (Chang et al. 2003; Murphy et al. 2003), we performed a random mutagenesis of the C-terminal region of a plasmid-encoded SIR4 corresponding to aa 747–1358 (see Materials and Methods for details), and isolated suppressors of the HMR silencing defect of a MATα sir3-1067 sir4Δ strain. Further analysis revealed that a single point mutation, threonine 1314 to serine (sir4-T1314S), was able to suppress the silencing defect of sir3-1067 at HMR (Figure S1). Interestingly, this mutation is located within the coiled-coil domain of Sir4 that has previously been shown to interact with Sir3 [Figure 1B, and see Chang et al. (2003)]. Sir4-T1314S was fully functional for Sir4 function in cells containing wild-type Sir3, since it provided wild-type silencing in a sir4Δ strain (Figure 1C and Figure S2A), showing that the substitution of threonine to serine did not interfere with normal Sir4 function.
Since Sir4 protein levels can vary when expressed from a plasmid, which might influence silencing levels (Cockell et al. 1998; Larin et al. 2015), we determined the effect of sir4-T1314S when genomically encoded from the native SIR4 locus. Sir4-T1314S was expressed at similar levels as the wild-type Sir4 protein as determined by Western blotting (Figure S2B). Importantly, genomically encoded sir4-T1314S restored silencing at HMR as well as HML to wild-type levels, as measured by the ability of sir3-1067 sir4-T1314S strains to form diploids with tester strains of opposite mating type (Figure 1C). This showed that the suppression was not due to abnormal levels of plasmid-encoded Sir4.
We next asked whether the suppression of sir3-1067 depended on the chemical nature of the substitution at position 1314 of Sir4. Interestingly, as for Sir4-T1314S, the substitution of threonine to alanine (sir4-T1314A) was able to suppress the silencing defect of sir3-1067 (Figure 1D). Conversely, mutations to lysine (K) or glutamic acid (E) showed no restoration of silencing at HMR, and these substitutions caused a loss of Sir4 function in a sir4Δ strain (Figure S2A). This indicated that a positively or negatively charged aa at position 1314 led to surface charge changes that interfere with Sir4 function, whereas a neutral substitution improved Sir4 silencing function, potentially by altering Sir4 interaction with Sir3.
Although sir4-T1314S supported HM silencing levels in sir3-1067 that were comparable to those of the wild-type Sir4 and Sir3 proteins (Figure 1C), this silencing was exquisitely sensitive to the absence of Sir1 (Figure 1E), which is in stark contrast to the subtle silencing defects at HMR in sir1∆ cells that express wild-type Sir3 and Sir4 (Rine and Herskowitz 1987). Also, sir4-T1314S was unable to suppress the silencing defect of sir3-1067 at telomeres as measured by silencing of URA3 integrated at telomere VII-L (Figure 1F). This indicated that, while sir4-T1314S can suppress some aspects of the sir3-1067 phenotype, it did not completely suppress sir3-1067, raising the question whether other amino acid changes in Sir4 might have a more penetrant phenotype, and restore silencing function at the telomeres.
Restoration of telomeric silencing in AAA+-mutated Sir3 by amino acid changes E1310V, T1314S, and K1325R in Sir4
Since sir4-T1314S was unable to suppress the telomeric silencing defects of sir3-1067, we screened for stronger alleles of SIR4 that restored telomeric silencing in sir3-1067 sir4∆ cells. Remarkably, even though we started the mutagenesis with wild-type SIR4, we isolated two sir4 suppressor alleles that both contained, in addition to other amino acid changes, the mutation T1314S that we had previously isolated as a suppressor of the HMR silencing defect. One allele also contained the mutation E1310V, whereas the other allele additionally carried K1325R. We therefore created new alleles with either mutation alone, all possible double mutants, and the triple mutant, and tested their ability to suppress the silencing defects of sir3-1067 at the telomeres, and at HMR. Importantly, none of these mutations alone supported telomeric silencing in sir3-1067 cells, and only sir4-T1314S suppressed the HMR silencing defect (Figure S3). Although sir4-E1310V, T1314S and sir4-T1314S, K1325R partially suppressed telomeric silencing in sir3-1067, sir4-E1310V, K1325R (i.e., without T1314S) did not. The strongest suppression at all loci was observed in the triple mutant sir4-E1310V, T1314S, K1325R (referred to as sir4-ETK below). Sir4-ETK function did not depend on the expression of the mutant Sir3-1067, as a plasmid-borne sir4-ETK fully suppressed HMR silencing defects in sir4∆ SIR3 cells (Figure S4).
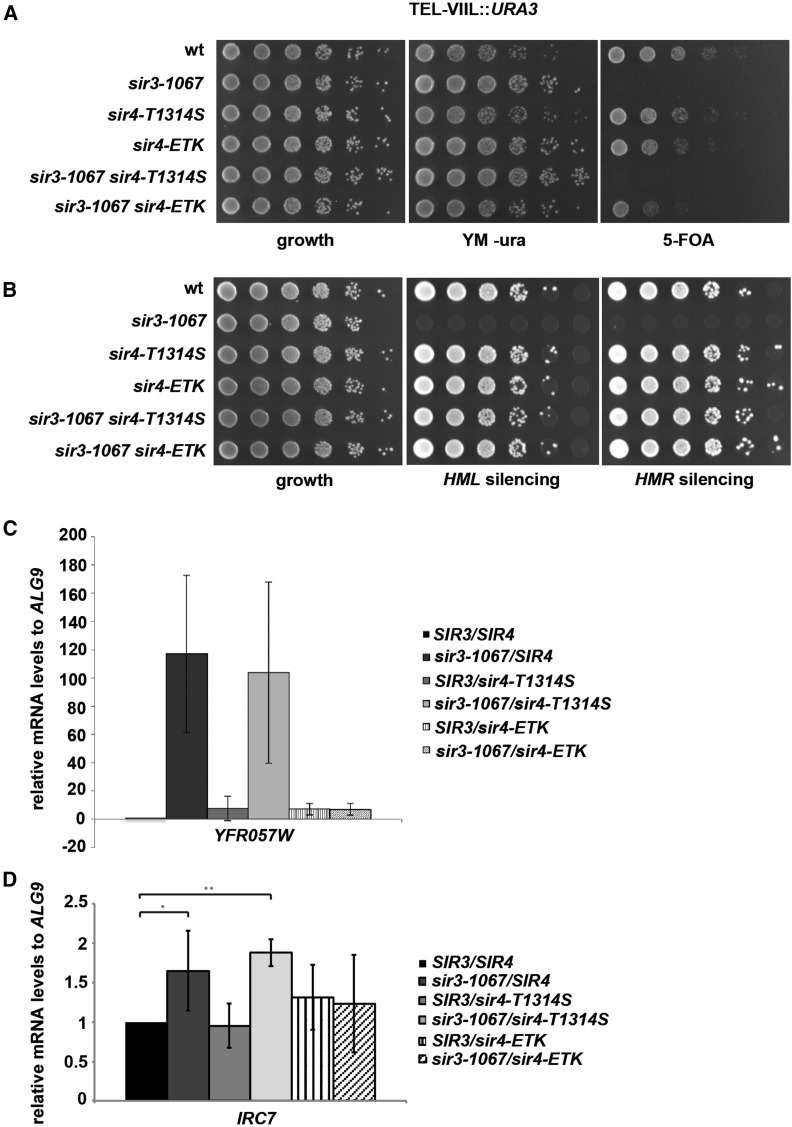
As above, to avoid potential complications of plasmid-encoded Sir4, we determined the silencing levels provided by genomically encoded sir4-ETK, which showed Sir4 protein levels that were comparable to wild-type Sir4 protein (Figure S2B). Significantly, this allele restored wild-type silencing levels to sir3-1067 at HMR and HML, and nearly wild-type silencing of the telomeric URA3 reporter (Figure 2, A and B).
Figure 2.
Sir4-E1310V, T1314S, K1325R restored sir3-1067 silencing defects at the telomeres as well as the HM loci. (A) Sir4-E1310V, T1314S, K1325R (sir4-ETK) restored telomeric silencing in sir3-1067. The silencing assay was performed as in Figure 1F. (B) Sir4-ETK suppressed the silencing defect of sir3-1067 at HML and HMR. Mating assays were performed as in Figure 1C. (C) sir4-ETK suppressed the expression of the subtelomeric gene YFR057W on chromosome VI-R in a sir3-1067 strain background. Relative mRNA levels of the subtelomeric gene YFR057W relative to ALG9 mRNA level was measured via qPCR. Errors bars give SDs of three biological triplicates. (D) sir4-ETK suppressed the expression of the subtelomeric gene IRC7 in a sir3-1067 background. Representation as in Figure 2C. The asterisks indicate significant differences, * P < 0.05, ** P value < 0.005.
We furthermore investigated the expression of native subtelomeric genes in the different mutant contexts. In agreement with the results from the telomeric URA3 reporter, sir3-1067 caused derepression of the subtelomeric genes YFR057W and IRC7, and this derepression was suppressed by sir4-ETK, but not sir4-T1314S (Figure 2, C and D).
We furthermore measured chromatin association of Sir4, Sir4-T1314S, and Sir4-ETK by ChIP. The levels of the mutant Sir4 versions were slightly reduced as compared to wild type at the telomeres as well as at the HM loci, even though the epitope-tagged versions used for ChIP were fully functional (Figure S5).
Altogether, these results suggest that the amino acid change T1314S improves interaction of Sir4 to a silencing interaction partner, possibly to Sir3, to restore HM silencing in sir3-1067, and that this interaction is further strengthened by the substitutions E1310V and K1325R.
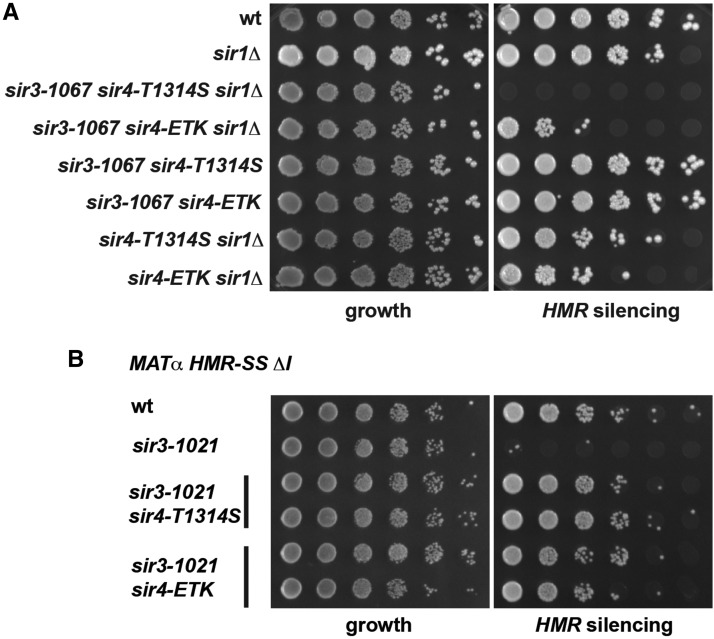
We furthermore asked whether the suppression of sir3-1067 by sir4-ETK was dependent on the presence of Sir1. Interestingly, while there was a substantial loss of HMR silencing by sir1∆ in sir3-1067 sir4-ETK (Figure 3A), this loss was not as dramatic as that of sir1∆ in sir3-1067 sir4-T1314S (Figure 1E). Thus, sir4-ETK was more resilient toward perturbations than sir4-T1314S, which was in agreement with its ability to silence at telomeres. However, sir4-ETK (in the presence of wild-type SIR3) was more susceptible to sir1∆ than wild-type SIR4 (Figure 3A), showing that this allele cannot fully replace the wild-type SIR4.
Figure 3.
Suppression of the sir3-1067 HMR silencing defect by sir4-T1314S and sir4-ETK was impaired by deletion of SIR1. (A) Semiquantitative mating was performed as in Figure 1C. (B) Sir4-T1314S and sir4-ETK suppressed the silencing defect of sir3-1021 at an HMR allele carrying a synthetic version of the HMR silencer and lacking HMR-I (HMR-SS ∆I). Mating assays were performed as above.
We furthermore asked whether sir4-T1314S and sir4-ETK were allele-specific suppressors of sir3-1067, or whether they were able to suppress another sir3 allele. We therefore tested suppression of sir3-1021, which carries the mutations D640A and S642L (Figure 1A), which do not abrogate interaction with Sir4 (Ehrentraut et al. 2011), and which causes derepression at an HMR allele carrying a synthetic HMR-E silencer and lacking HMR-I (HMR-SS ∆I). Importantly, the HMR silencing defect of sir3-1021 was strongly suppressed by sir4-T1314S and sir4-ETK (Figure 3B), showing that these sir4 alleles were able to provide silencing in another sir3 mutant background. However, these mutants were unable to suppress the silencing defects of sir3∆ cells (not shown), showing that a mutant form of Sir3 needed to be present for suppression.
Mutations in the Sir4 coiled-coil restore its in vitro interaction with mutant Sir3
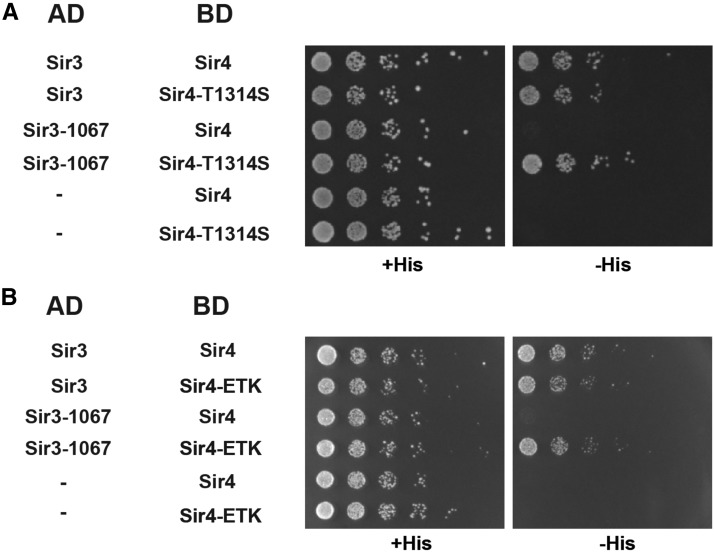
We hypothesized that the amino acid changes identified here restored silencing function of Sir4 with mutant Sir3 because they restore the physical interaction between Sir4 and Sir3. Previous work has shown an interaction between the Sir4 coiled-coil and a C-terminal fragment of Sir3 (Chang et al. 2003), and we hypothesized that this interaction might be restored by the sir4 mutants. As a first test of this hypothesis, we investigated the two-hybrid interactions of the different Sir3 and Sir4 mutant combinations. As reported earlier, Sir3 and Sir4 show a robust two-hybrid interaction, as measured by the growth of cells on medium lacking histidine as an indicator of activation of the HIS3 two-hybrid reporter. Consistent with our previous work, Sir3-1067 was unable to interact with Sir4 (Ehrentraut et al. 2011). Strikingly, however, this interaction was restored by both Sir4-T1314S and Sir4-ETK (Figure 4), thus suggesting that the improved silencing could be attributed to improved binding between the mutant Sir3 and Sir4 proteins.
Figure 4.
(A) Sir4-T1314S and (B) Sir4-E1310V, T1314S, K1325R (Sir4-ETK) restored the two-hybrid interaction to Sir3-1067. Strains [AEY3055 transformed with the respective plasmids carrying Sir3 (307–978) and Sir4 (839–1358)] were tested for activation of the two-hybrid reporter HIS3 by plating serial dilutions on minimal medium with or without histidine.
We also investigated other mutations in Sir4-T1314 for their two-hybrid interaction with wild-type and defective sir3. In agreement with the observation that sir4-T1314A suppressed sir3-1067 silencing defects, this allele also restored its interaction with Sir3-1067 (Figure S6). In contrast, sir4-T1314E and -K were both unable to restore the interaction with mutant Sir3, which was in line with their inability to suppress sir3-1067. However, neither allele abrogated the interaction with wild-type Sir3, which was unexpected because they show a loss of Sir4 silencing function in SIR3 cells, and thus are hypothesized to have lost interaction to Sir3. Thus, apparently, the in vivo silencing by Sir3 and Sir4 is more sensitive to perturbations than their two-hybrid interaction, and two-hybrid assay does not recapitulate all aspects of the physiologically relevant interactions of these proteins.
sir4-T1314S and sir4-ETK both mutate surface residues on the Sir4 coiled-coil, and we confirmed that these substitutions do not affect Sir4 dimerization as measured by a two-hybrid interaction. Neither mutant abrogated the interaction with wild-type Sir4 or to itself (Figure S7), indicating that they did not disrupt dimerization.
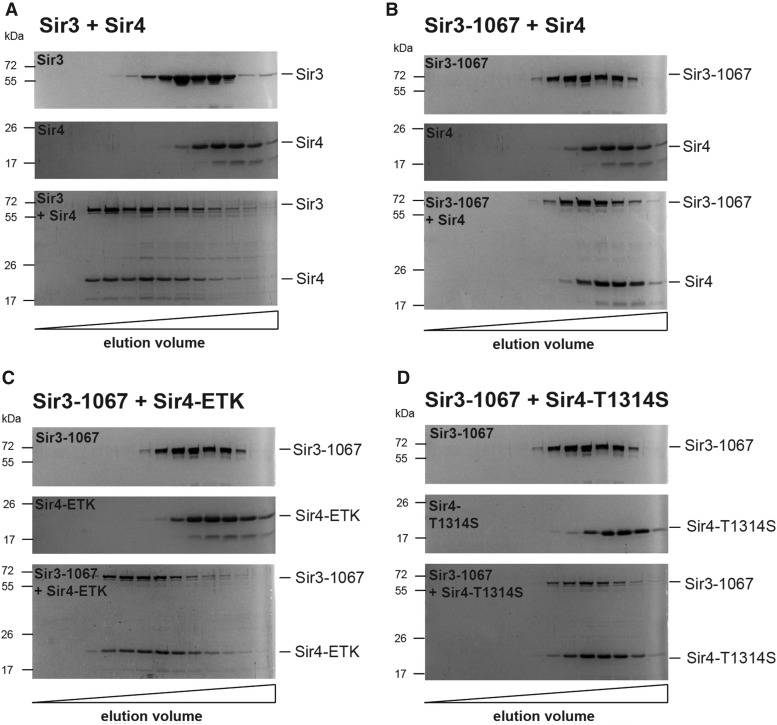
In order to directly test whether the mutations in the Sir4 coiled-coil influenced the in vitro interaction of Sir4 with Sir3, we measured the in vitro interaction between bacterially produced and purified Sir3 (aa 464–978) and the Sir4 coiled-coil domain (aa 1217–1358) by size exclusion chromatography (SEC). Both fragments have previously been shown to behave roughly as dimers during gel filtration (Chang et al. 2003). Accordingly, in our SEC analysis, Sir3 (464–978) and Sir4 (1217–1358) alone eluted as single species. As expected, prior coincubation of the two wild-type proteins resulted in elution of both proteins at a higher apparent molecular mass, indicating Sir3–Sir4 complex formation (Figure 5A). Although the Sir3-1067(464–978) fragment showed similar behavior to the wild-type fragment when analyzed alone by SEC, coincubation with Sir4 (1217–1358) did not result in the formation of a higher-molecular weight complex (Figure 5B). This was in agreement with our earlier work that showed a loss of Sir3-1067 binding to full-length Sir2/Sir4 (Ehrentraut et al. 2011). Significantly, however, the Sir3-1067(464–978) fragment was capable of forming a high molecular weight complex with a Sir4-ETK (1217–1358) coiled-coil fragment (Figure 5C). This data showed that selected amino acid changes on the surface of the Sir4 coiled-coil increased in vitro binding to Sir3, and further indicated that the restoration of this interaction was the cause for improved silencing in vivo.
Figure 5.
Sir4-ETK restored in vitro interaction to Sir3-1067. Sir4 (1217–1358) and Sir3 (464–978) as well as the respective mutant versions were expressed and purified separately from bacteria, and coincubated before separation by analytical gel filtration. (A) SDS-PAGE of SEC of wild-type Sir3 and Sir4, separately (top, middle), and after coincubation prior to SEC (bottom). Gels were stained with Coomassie Brilliant Blue. (B) SEC of Sir3-1067 with Sir4. Analysis is shown as in (A). (C) SEC of Sir3-1067 with Sir4-ETK. (D) SEC of Sir3-1067 with Sir4-T1314S.
Although sir4-T1314S suppressed some silencing defects of sir3-1067, the purified Sir4-T1314S coiled-coil was not able to interact with the mutant Sir3 fragment (Figure 5D). This was surprising given that we had observed the restoration of the two-hybrid interaction between the two mutant proteins (Figure 4), and suggests that SEC may exaggerate defects in binding between these two proteins. Altogether, this showed that selected amino acid changes on the surface of the Sir4 coiled-coil increased in vitro binding to Sir3, and further indicated that the restoration of this interaction was the cause for improved silencing in vivo.
Discussion
Multiple contacts among the Sir proteins are necessary for the assembly of a functional SIR complex that binds nucleosomes, and thus establishes heterochromatin. Here, we investigated the interaction between the Sir3 AAA+ domain and the Sir4 C-terminal coiled-coil. We made the surprising discovery that removal of a methyl group on the surface of the Sir4 coiled-coil domain (T1314S) restored silencing function in the presence of a Sir3 version with mutations in the Sir3 AAA+ domain. This site on Sir4 lies on the outer surface of the Sir4 coiled-coil [Figure 1B, and see Chang et al. (2003) and Murphy et al. (2003)]. We furthermore show that a combination of mutations on this surface restores the physical interaction between a mutant Sir3 AAA+ domain and Sir4, indicating that the restoration of this interaction is responsible for the regained silencing in vivo. We suggest that these mutations increase the affinity of Sir4 to Sir3.
Interestingly, surface residues on the Sir4 coiled-coil have previously been shown to be required for Sir3 interaction. Specifically, the changes M1307N, E1310R and I1311N in Sir4 caused a loss of in vitro interaction between Sir4 (1267–1358) and Sir3 (464–978) (Chang et al. 2003), and these three mutations disrupt the interaction between the full length proteins and silencing in vivo (Rudner et al. 2005). Remarkably, our mutation E1310V improves, rather than decreases, binding to Sir3, though only in conjunction with T1314S. It is also interesting to note that earlier studies found no effect on Sir4–Sir3 binding for the mutations Sir4-T1314N and K1325E (Chang et al. 2003; Rudner et al. 2005). We here identified other amino acid changes at these same sites that strengthened Sir4 binding to Sir3. Of note, K1325 lies within a hydrophobic surface of Sir4, via which two Sir4 coiled-coil dimers make crystal contacts (Murphy et al. 2003), and mutation of the nearby F1322 residue was shown to abrogate Sir4 silencing function. Our data suggest that this hydrophobic surface is, in fact, an interaction region with Sir3.
Based on the observation that mutations in Sir4 threonine 1314 abrogate the interaction with Sir3, one can speculate whether this residue might be regulated by post-translational modification, for instance phosphorylation. Such a modification is expected to disrupt the interaction to Sir3, since the phosphomimetic sir4-T1314E causes a silencing defect. However, phosphorylation at this site has not been observed so far (Kueng et al. 2012).
Where precisely is the contact of the Sir4 coiled-coil on Sir3? Our initial intention was to identify the site of contact with Sir4 of the loop in the Sir3 AAA+ domain by searching for site-specific suppressors. Although such mutations are expected to be allele-specific suppressors, we found suppression by the sir4 alleles of a sir3 mutation that lies one α-helix away from this loop (sir3-1021). This suggests that the Sir4 coiled-coil mutations increase binding to the Sir3 AAA+ domain, and that this enhanced binding may be sufficient to rescue other defects in Sir3. The contact therefore may be at the mentioned loop, but contact sites on other Sir3 surfaces are also possible. A further structural analysis will be required to resolve this question. We hypothesize that the Sir4 mutations increase binding not only to mutant, but also to wild-type Sir3. Interestingly, this may come at a cost for HM silencing, since HMR silencing by sir4-ETK was more sensitive to the loss of Sir1 function than silencing by wild-type SIR4 (Figure 3A).
We also note with interest that there were differences in the “suppressability” of sir3-1067 silencing defects at the HM loci compared to the telomeres, in that multiple mutations in the Sir4 surface were required to improve telomeric silencing. One interpretation of this observation is that a stronger interaction between Sir3 and Sir4 is required for telomeric silencing, because it is initiated by cis recruitment sites at the chromosome ends, and telomeric heterochromatin spreads unidirectionally toward subtelomeric regions. This is in contrast to HM silencing, where each silent locus is flanked on either side by an E and an I silencer as SIR recruitment sites, and SIR spreading proceeds in a convergent fashion. We suggest that the combined Sir4 mutations identified here increase the affinity of Sir4 to Sir3, and that this enhances SIR spreading, which is particularly important for telomeric silencing.
Another interpretation comes from the observation that the sir4-ETK is more proficient at HMR silencing in the absence of Sir1 than sir4-T1314S, which indicates that the increased affinity of Sir4-ETK to Sir3-1067 can overcome a nucleation defect at HMR caused by the absence of Sir1, whereas Sir4-T1314S cannot. Since telomeric silencing does not require Sir1 (Aparicio et al. 1991), the sir3-1067 allele may require the stronger suppressor for telomeric silencing. The notion of increased affinity improving silencing is supported by earlier work showing that silencing of a derepressed HM silencer can be restored by perinuclear tethering to increased SIR protein concentration at the nuclear periphery (Andrulis et al. 1998; Taddei et al. 2009).
Interestingly, the restored HMR silencing achieved here was exquisitely sensitive to the recruitment factor Sir1, which binds to the HM silencers, and bridges the interaction of DNA binding proteins with the SIR complex. Sir1 has previously been shown to interact with the Sir4 N-terminus (Triolo and Sternglanz 1996; Kueng et al. 2012), i.e., a region that, on the primary amino-acid sequence, is distant from the C-terminal coiled-coil. In this regard, it is interesting to note that the sir4-ETK mutant showed less HMR silencing in a sir1Δ mutant than the sir4-ETK mutant or sir1Δ mutant alone. One explanation is that the increased binding of sir4-ETK to wild-type Sir3 is detrimental for silencing in this context. How the Sir1 dependence reflects SIR binding and recruitment again will require more structural insights into the complex.
As in yeast, the formation of heterochromatin in higher eukaryotes relies on the spreading of chromatin-binding proteins like Heterochromatin Protein 1, or the Polycomb Repressive Complex 1. Learning about the basic principles of this process using the yeast SIR complex as a model will allow important insights into universal mechanisms of heritable chromatin silencing in eukaryotes.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.037739/-/DC1.
Acknowledgments
We thank Maria Bongartz, Michaela Sievers, and Nora Fresmann for early work on this project. We furthermore thank Silke Steinborn, Josta Hamann, Karolin Jänen, and Edward Schwartz for technical support, and Ekaterina Anedchenko for advice on biochemical experiments. This work was supported by the Deutsche Forschungsgemeinschaft (EH237/11-1) and Humboldt-Universität zu Berlin. The authors declare that they have no conflicts of interest.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Andrulis E. D., Neiman A. M., Zappulla D. C., Sternglanz R., 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595. [DOI] [PubMed] [Google Scholar]
- Aparicio O. M., Billington B. L., Gottschling D. E., 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in Saccharomyces cerevisiae. Cell 66: 1279–1287. [DOI] [PubMed] [Google Scholar]
- Armache K. J., Garlick J. D., Canzio D., Narlikar G. J., Kingston R. E., 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334: 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaudo N., Fernandez I. S., McLaughlin S. H., Peak-Chew S. Y., Rhodes D., et al. , 2013. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nat. Struct. Mol. Biol. 20: 1119–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Mitchell J., Leber J., Kobayashi R., Stillman B., 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA-replication and transcriptional silencing. Cell 83: 563–568. [DOI] [PubMed] [Google Scholar]
- Bose M. E., McConnell K. H., Gardner-Aukema K. A., Muller U., Weinreich M., et al. , 2004. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 24: 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. F., Hall B. E., Tanny J. C., Moazed D., Filman D., et al. , 2003. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure 11: 637–649. [DOI] [PubMed] [Google Scholar]
- Chen Y., Rai R., Zhou Z. R., Kanoh J., Ribeyre C., et al. , 2011. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat. Struct. Mol. Biol. 18: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M., Gotta M., Palladino F., Martin S. G., Gasser S. M., 1998. Targeting Sir proteins to sites of action: a general mechanism for regulated repression. Cold Spring Harb. Symp. Quant. Biol. 63: 401–412. [DOI] [PubMed] [Google Scholar]
- Cubizolles F., Martino F., Perrod S., Gasser S. M., 2006. A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol. Cell 21: 825–836. [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 271: 2335–2349. [DOI] [PubMed] [Google Scholar]
- Ehrentraut S., Hassler M., Oppikofer M., Kueng S., Weber J. M., et al. , 2011. Structural basis for the role of the Sir3 AAA+ domain in silencing: interaction with Sir4 and unmethylated histone H3K79. Genes Dev. 25: 1835–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M., 1995. Histone H3 and H4 N-termini interact with Sir3 and Sir4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592. [DOI] [PubMed] [Google Scholar]
- Hsu H. C., Wang C. L., Wang M., Yang N., Chen Z., et al. , 2013. Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding. Genes Dev. 27: 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L., 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J., 1988. Roles of 2 DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 7: 2241–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D. A., Hall B. E., Iwamoto M. A., Win K. Z., Chang J. F., et al. , 2006. Domain structure and protein interactions of the silent information regulator Sir3 revealed by screening a nested deletion library of protein fragments. J. Biol. Chem. 281: 20107–20119. [DOI] [PubMed] [Google Scholar]
- Kueng S., Tsai-Pflugfelder M., Oppikofer M., Ferreira H. C., Roberts E., et al. , 2012. Regulating repression: roles for the Sir4 N-terminus in linker DNA protection and stabilization of epigenetic states. PLoS Genet. 8: e1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S., Oppikofer M., Gasser S. M., 2013. SIR proteins and the assembly of silent chromatin in budding yeast. Annu. Rev. Genet. 47: 275–306. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., et al. , 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97: 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larin M. L., Harding K., Williams E. C., Lianga N., Dore C., et al. , 2015. Competition between heterochromatic loci allows the abundance of the silencing protein, Sir4, to regulate de novo assembly of heterochromatin. PLoS Genet. 11: e1005425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., et al. , 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and SIR protein concentration in silencer-mediated repression. Genes Dev. 10: 1796–1811. [DOI] [PubMed] [Google Scholar]
- Marmorstein R., 2004. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem. Soc. Trans. 32: 904–909. [DOI] [PubMed] [Google Scholar]
- Martino F., Kueng S., Robinson P., Tsai-Pflugfelder M., van Leeuwen F., et al. , 2009. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol. Cell 33: 323–334. [DOI] [PubMed] [Google Scholar]
- Moretti P., Freeman K., Coodly L., Shore D., 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere binding-protein RAP1. Genes Dev. 8: 2257–2269. [DOI] [PubMed] [Google Scholar]
- Murphy G. A., Spedale E. J., Powell S. T., Pillus L., Schultz S. C., et al. , 2003. The Sir4 C-terminal coiled coil is required for telomeric and mating type silencing in Saccharomyces cerevisiae. J. Mol. Biol. 334: 769–780. [DOI] [PubMed] [Google Scholar]
- Norris A., Boeke J. D., 2010. Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev. 24: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Liou G. G., Buchberger J. R., Walz T., Moazed D., 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 28: 1015–1028. [DOI] [PubMed] [Google Scholar]
- Oppikofer M., Kueng S., Gasser S. M., 2013a SIR-nucleosome interactions: structure-function relationships in yeast silent chromatin. Gene 527: 10–25. [DOI] [PubMed] [Google Scholar]
- Oppikofer M., Kueng S., Keusch J. J., Hassler M., Ladurner A. G., et al. , 2013b Dimerization of Sir3 via its C-terminal winged helix domain is essential for yeast heterochromatin formation. EMBO J. 32: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrod S., Gasser S. M., 2003. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell. Mol. Life Sci. 60: 2303–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H., Aparicio O. M., Zierath P. D., Billington B. L., Chhablani S. K., et al. , 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by Sir3 dosage. Genes Dev. 7: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Rine J., Herskowitz I., 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Meier B., McAinsh A. D., Feldmann H. M., Jackson S. P., 2004. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J. Biol. Chem. 279: 86–94. [DOI] [PubMed] [Google Scholar]
- Rudner A. D., Hall B. E., Ellenberger T., Moazed D., 2005. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 25: 4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516. [DOI] [PubMed] [Google Scholar]
- Samel A., Cuomo A., Bonaldi T., Ehrenhofer-Murray A. E., 2012. Methylation of CenH3 arginine 37 regulates kinetochore integrity and chromosome segregation. Proc. Natl. Acad. Sci. USA 109: 9029–9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath V., Yuan P., Wang I. X., Prugar E., van Leeuwen F., et al. , 2009. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol. Cell. Biol. 29: 2532–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Studier F. W., 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41: 207–234. [DOI] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F. R., Bauer C., Gasser S. M., 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23: 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Nagai S., Erb I., van Nimwegen E., et al. , 2009. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 19: 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J. C., Kirkpatrick D. S., Gerber S. A., Gygi S. P., Moazed D., 2004. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 24: 6931–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo T., Sternglanz R., 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381: 251–253. [DOI] [PubMed] [Google Scholar]
- van Loo B., Spelberg J. H. L., Kingma J., Sonke T., Wubbolts M. G., et al. , 2004. Directed evolution of epoxide hydrolase from A. radiobacter toward higher enantioselectivity by error-prone PCR and DNA shuffling. Chem. Biol. 11: 981–990. [DOI] [PubMed] [Google Scholar]
- Wang F., Li G., Altaf M., Lu C. N., Currie M. A., et al. , 2013. Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA. Proc. Natl. Acad. Sci. USA 110: 8495–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Fang Q., Wang M., Ren R., Wang H., et al. , 2013. Nα-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nat. Struct. Mol. Biol. 20: 1116–1118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. Table S1 contains the genotypes of all strains used in this study. Table S2 contains all plasmids used in this study.