Abstract
The American cranberry (Vaccinium macrocarpon Ait.) is a recently domesticated, economically important, fruit crop with limited molecular resources. New genetic resources could accelerate genetic gain in cranberry through characterization of its genomic structure and by enabling molecular-assisted breeding strategies. To increase the availability of cranberry genomic resources, genotyping-by-sequencing (GBS) was used to discover and genotype thousands of single nucleotide polymorphisms (SNPs) within three interrelated cranberry full-sib populations. Additional simple sequence repeat (SSR) loci were added to the SNP datasets and used to construct bin maps for the parents of the populations, which were then merged to create the first high-density cranberry composite map containing 6073 markers (5437 SNPs and 636 SSRs) on 12 linkage groups (LGs) spanning 1124 cM. Interestingly, higher rates of recombination were observed in maternal than paternal gametes. The large number of markers in common (mean of 57.3) and the high degree of observed collinearity (mean Pair-wise Spearman rank correlations >0.99) between the LGs of the parental maps demonstrates the utility of GBS in cranberry for identifying polymorphic SNP loci that are transferable between pedigrees and populations in future trait-association studies. Furthermore, the high-density of markers anchored within the component maps allowed identification of segregation distortion regions, placement of centromeres on each of the 12 LGs, and anchoring of genomic scaffolds. Collectively, the results represent an important contribution to the current understanding of cranberry genomic structure and to the availability of molecular tools for future genetic research and breeding efforts in cranberry.
Keywords: centromere region, genetic map, simple sequence repeat, single nucleotide polymorphism, Vaccinium
The American cranberry (Vaccinium macrocarpon Ait.) is a diploid woody perennial in the Ericaceae family, and like other Ericaceous species, it grows well in acidic, nutrient-poor soils (Vander Kloet 1983b; Kron et al. 2002; Vander Kloet and Avery 2010). Within the Ericaceae family, the cranberry and blueberry species of sections Oxycoccus (Hill) Koch and Cyanococcus A. Gray share a common base chromosome number (n = 12), and a similar karyotype with relatively uniformly sized metacentric chromosomes (Camp 1945; Hall and Galleta 1971; Vander Kloet 1983a,b; Costich et al. 1993). Both crops are growing in popularity and commercial importance because of the numerous health benefits attributed to their phytochemical constituents, and global production of cranberry and blueberry has expanded to encompass a combined 100,000+ hectares valued at >1.5 billion U.S. dollars (Howell et al. 1998; Foo et al. 2000; Duarte et al. 2006; Food and Agriculture Organization of the United Nations 2012; Feghali et al. 2012). However, cranberry genetic improvement has lagged behind its blueberry brethren for various reasons including: the lack of continued breeding efforts, the limited number of breeding programs in existence, the long breeding cycle due to lengthy establishment periods (2–4 yr) followed by long evaluation periods (4–5 yr) required to measure biennial bearing (Eaton et al. 1983; Elle 1996; Vorsa and Johnson-Cicalese 2012; Schlautman et al. 2015a), and a lack of adoption of molecular-assisted breeding strategies which could increase the rate of domestication and genetic gain in cranberry.
Prerequisites to marker-assisted breeding strategies, such as molecular-assisted seedling selection and/or genomic prediction (Ru et al. 2015; Covarrubias-Pazaran 2016), include the availability of numerous molecular markers distributed throughout the genome for haplotype estimation, knowledge of associations between markers and traits of interest, available populations with characterized genetic and phenotypic diversity, and high-throughput marker genotyping methodologies that justify the costs associated with molecular-assisted breeding compared to classical breeding methods. Large-scale exploration and development of genetic and genomic resources in cranberry has only begun recently with the advent of next generation sequencing technologies (NGS). For example, NGS technologies such as pyrosequencing (i.e., 454), sequencing by oligonucleotide ligation and detection, and Illumina have been useful in assembling both the plastid and mitochondrial genomes (Fajardo et al. 2013, 2014), a transcriptome (Polashock et al. 2014), and a draft nuclear genome assembly (Polashock et al. 2014). Simple sequence repeat (SSR) mining of those sequence resources resulted in the development of >900 informative (polymorphic) SSR markers (Georgi et al. 2011, 2013; Zhu et al. 2012; Schlautman et al. 2015b, 2016), with 136 and 541 of those SSRs placed in cranberry SSR linkage maps by Georgi et al. (2013) and Schlautman et al. (2015b), respectively. However, means for gathering SSR genotype information in an efficient, cost-effective manner at a breeding program scale have not been achieved in cranberry, and uncertainty remains about whether the available set of cranberry SSRs is large enough to saturate the genome.
Covarrubias-Pazaran et al. (2016) recently demonstrated the potential for genotyping-by-sequencing (GBS) in cranberry to serve as a high-throughput platform that integrates single nucleotide polymorphism (SNP) marker discovery and genotyping into a single procedure (Elshire et al. 2011). The Covarrubias-Pazaran et al. (2016) study increased the availability of cranberry DNA markers 10-fold and used them to construct the first high-density SNP cranberry linkage map. Therefore, to further saturate the cranberry genome with SNP markers and to test the utility of GBS to detect SNPs that are polymorphic across multiple populations, an experiment was designed to develop a cranberry composite map using GBS for multi-pedigree linkage mapping. In this study, six parental bin maps were constructed from three interrelated cranberry populations, whose ancestry trace back to seven historically important cranberry wild selections. The high density of SNPs identified herein were placed in a composite map that allowed characterization of important chromosome structural aspects including identification of segregation distortion regions, centromere placement, and anchoring of cranberry nuclear scaffolds containing predicted coding DNA sequences.
Materials and Methods
Plant material and DNA extraction
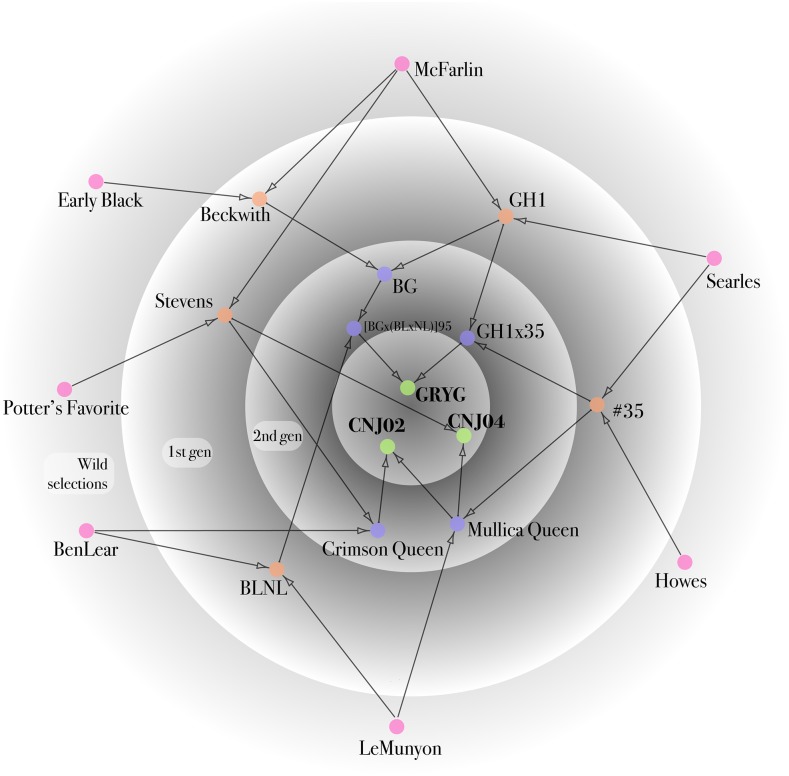
Three full-sib linkage mapping populations (i.e., CNJ02, CNJ04, and GRYG) were derived from crosses between five interrelated cranberry parental genotypes (Figure 1). The CNJ02 population included 168 progeny from a cross between maternal parent, CNJ97-105 (Mullica Queen), and paternal parent, NJS98-23 (Crimson Queen); the CNJ04 population included 67 progeny from a reciprocal cross between CNJ97-105 (Mullica Queen) and Stevens; and the GRYG population included 352 progeny from a cross between the maternal parent, [BGx(BLxNL)]95, and the paternal parent, GH1x35 (Figure 1). The CNJ02 and CNJ04 populations were generated and are maintained at the Rutgers University P.E. Marucci Center in Chatsworth, NJ and were planted in separate unreplicated, completely randomized designs. The GRYG population was generated and is maintained by the Valley Corporation in Tomah, WI. Genomic DNA from all parents and progeny was extracted from flash frozen newly emerged leaves using a Macherey-Nagel (MN) Plant II kit (Düren, Germany) following the manufacturer’s instructions.
Figure 1.
Description of the pedigrees of the three mapping populations (green), CNJ02, CNJ04, and GRYG, derived from crosses between five interrelated cranberry parental genotypes (blue), CNJ97-105 (Mullica Queen), NJS98-23 (Crimson Queen), Stevens, [BGx(BLxNL)]95, and GH1x35. All pedigrees trace to “The Big Seven” native cranberry selections (red), which have played important roles in cranberry production and breeding history. The pedigree contains five first generation removed from the wild genotypes resulting from crosses between two native selections (orange), and additional later generation genotypes (blue).
SSR and SNP genotyping and data processing
SSR marker data generated for the CNJ02 (541 SSRs) and GRYG (189 SSRs) populations in previous linkage mapping studies were incorporated into the current study (Schlautman et al. 2015a; Covarrubias-Pazaran et al. 2016). A subset of these previously mapped and additional unmapped SSR markers were used to genotype the CNJ04 progeny and its parents in this study (Schlautman et al. 2015a,b). In addition, SSRs previously developed and mapped in blueberry (Rowland et al. 2014) and newly developed SSR primer pairs were used to genotype the CNJ02 progeny. Multiplex (3×) polymerase chain reactions (PCR) and fragment analysis for SSR genotyping were performed according to Schlautman et al. (2015a).
EcoT221-associated DNA fragments for the progeny and parents of the CNJ02 and CNJ04 mapping populations were generated and sequenced according to the genotyping-by-sequencing (GBS) approach described by Elshire et al. (2011). During library construction, adapters containing unique barcodes were ligated to restriction-digested DNA for the 235 progeny from the CNJ02 and CNJ04 populations and their three parental genotypes and then were pooled into three 96-plex sequencing libraries. To guarantee higher sequencing coverage in the parental genotypes, the parents for the CNJ02 and CNJ04 populations were represented by six samples each, and the resulting libraries were sequenced (single-end) on the Illumina HiSeq 2000 platform in the Cornell University Biotechnology Resource Center Genomics Facility. Library preparation and sequencing for the GRYG population (352 progeny) was performed previously following the same methodology described herein, except the two parental genotypes were represented by four samples each. This sequence data, used in a previous publication (Covarrubias-Pazaran et al. 2016), was incorporated into the current study to increase the density of the final composite map and reprocessed to ensure that the linkage maps for all three populations were constructed according to the same standard set of parameters.
The reference-based Tassel v3.0.166 GBS analysis pipeline was used to filter and process the resulting sequence reads, align and merge sequence tags by genotype, and to call SNPs in the resulting datasets for the CNJ02 and CNJ04 populations using the parameters outlined in Glaubitz et al. (2014) (Supplemental Material, File S1). The reference genome used during SNP discovery and genotyping with the Tassel pipeline was created by concatenating the 229,745 scaffold cranberry genome assembly (N50 = 4237 bp) produced by Polashock et al. (2014). After generating the filtered HapMap, the SNP marker data were further processed by removing SNPs separately from each of the three populations which had >20% missing data, had a minor allele frequency (MAF) of <10%, or had severely distorted segregation ratios.
Parental component map construction
Linkage analysis was performed with JoinMap v4.1 using the pseudo-testcross method. Markers were formatted and separated into two categories: (a) uniparental markers heterozygous in only a single parent (lm × ll and nn × np); and (b) biparental markers heterozygous in both parents (hk × hk, ef × eg, and ab × cd) (Van Ooijen 2006, 2011). Linkage groups (LGs) were determined with a logarithm base 10 of odds (LOD) threshold >5.0 (Van Ooijen 2006), and marker order was determined using the maximum likelihood (ML) algorithm (Van Ooijen 2011). A strict approach was used to determine and remove markers causing errors in estimation of marker ordering or inflation of map distances. First, markers which obviously caused problems and/or were placed far from the ends of the LGs were identified visually when observing the phased genotype data. Additionally, iterative rounds of mapping were performed to remove markers that caused a nearest neighbor fit (centimorgans) >2 cM, a nearest neighbor stress >0.035, or a nearest neighbor stress (centimorgans) >3.5 cM. Genetic distances among loci were then recalculated with the regression approach and the Kosambi mapping function using the fixed marker order determined by ML to facilitate map comparisons between the current maps and previously published maps.
Linkage mapping proceeded by first constructing a maternal and paternal component map for each population using only the uniparental markers (i.e., lm × ll or nn × np) and removing all markers causing problems in map order and distance per the guidelines outlined above. Using the resulting marker positional information, linkage-informed imputation was performed using the linkim package in R to impute missing marker information in the parental datasets (R Core Team 2015; Xu and Wu 2015). The multiple spanning tree (MST) algorithm implemented in the ASMap R package was then used to detect remaining genotyping errors and to perform bin mapping (determining bins of identical markers) for each of the parents (Wu et al. 2008; Taylor and Butler 2015).
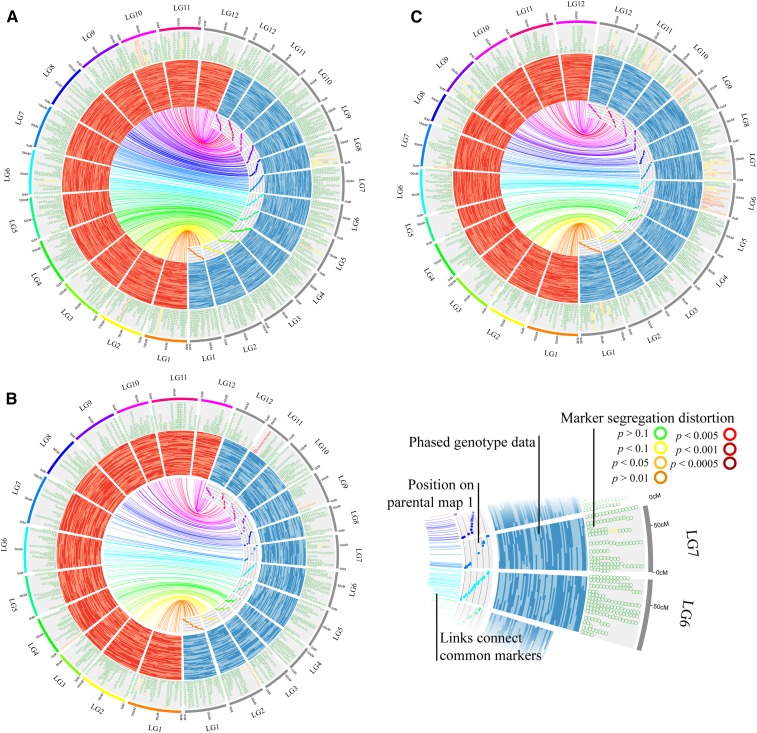
Next, biparental markers were added to each parental map. For each population, a single uniparental marker from each bin of each parental bin map was added to the remaining biparental maker set (i.e., hk × hk, ef × e.g., and ab × cd markers) and linkage mapping was performed to construct integrated maps for the three populations using the before-mentioned guidelines with the cp approach in JoinMap v4.1. The resulting marker positional and phase information was used to convert the hk × hk biparental SNP markers to ab × cd maker format by linkage-informed imputation of the hk genotypes using functions from the sommer R package (Covarrubias-Pazaran 2016). Each biparental marker was then split into two uniparental markers (either lm × ll for alleles from the maternal parent or nn × np for alleles from the paternal parent) to create a new dataset for each parent containing all possible markers. The MST algorithm was used once again to detect genotyping errors within the biparental marker sets and to create bins for each of the parental maps using the Kosambi mapping function to estimate map distances (Wu et al. 2008; Taylor and Butler 2015). Pair-wise Spearman rank correlations comparing marker order in the LGs of the six parental component bin maps were estimated to ensure that they were syntenic and collinear and could be used in composite map construction. Additionally, collinearity between LGs was visually assessed in Circos circular ideograms generated in the R package SOFIA by plotting the links between homologous markers in the LGs of the parental bin maps in each of the three populations (Krzywinski et al. 2009).
Composite map construction and map comparisons
A synthetic composite map was constructed from the six parental maps with the LPmerge package in R, which uses linear programming to minimize the mean absolute error between marker intervals in the parental maps and the composite map (Endelman and Plomion 2014). During composite mapping, iterations of the maximum interval size (k) ranging from k = 1 to k =10 were tested for each LG, and the k which minimized the root mean square error (RMSE) when comparing the position of makers in the composite LG to the six parental LGs was chosen for construction of the final composite LG. Spearman rank correlations and visual assessment in Circos were used to determine the collinearity of the composite map with each of the six component bin maps, with the previous SSR map constructed for the CNJ02 population (Schlautman et al. 2015a), and with the first GBS-based SNP map developed previously for the GRYG population (Covarrubias-Pazaran et al. 2016). Cranberry scaffolds from Polashock et al. (2014) containing predicted coding DNA sequences (CDS) and SSRs or SNPs mapped in this study were anchored using the markers’ positions in the composite map.
Genome-wide segregation distortion and centromere placement
Segregation distortion for each mapped marker in parental bin maps was analyzed using chi-square tests (χ2) with one degree of freedom for codominant markers as implemented in JoinMap v4.1. The χ2 P-value for each locus in each parental component map was then plotted to examine patterns of segregation distortion and to determine if segregation distortion regions existed in any LGs. Markers with a χ2 P-value < 0.1 were considered distorted.
Centromeric regions in the cranberry LGs of the component bin maps were explored and identified following the methodology of Limborg et al. (2016). Recombination frequencies (RFM) were estimated from phased genotype data by recording the proportion of offspring with an observed recombination (i.e., change of phase) in each interval between the terminal marker (m0) and every subsequent marker (mn) in both directions along the 12 cranberry LGs. For metacentric LGs, centromeric regions in the LGs were defined as the region from the point of intersection between the RFM estimates made from each terminal marker extending outwards until reaching the first marker interval with an RFM = 0.45 in both directions (Limborg et al. 2016).
Data availability
File S2 contains marker positions in the composite linkage map and the six parental component bin maps. File S3 contains all SNP genotype data for the six parental component bin maps.
Results
SSR and SNP genotyping
Differing sizes of SSR marker datasets were available for mapping in each of the three populations. The CNJ02 population, which was used in the first high-density SSR linkage mapping study, had the largest amount of SSR data (629 SSRs) available for linkage mapping (Table 1 and Table S1) (Schlautman et al. 2015a). The GRYG population, which was used in the first cranberry SNP genotyping study (Covarrubias-Pazaran et al. 2016), had a similar amount of SSR data compared to the CNJ04 population with 189 and 221 SSR markers, respectively (Table 1 and Table S1). The CNJ04 population has never been used in a peer-reviewed study, and all SSR and SNP marker data for that population was generated herein using multiplex (3×) PCR reactions.
Table 1. Summary of SSR and SNP marker availability and genotyping results for linkage mapping in the GRYG, CNJ02, and CNJ04 cranberry populations following marker filtering steps and merging with previous marker datasets.
| Population | # of SNPs in HapMap | # SNPs After Filtering for Missing Dataa | # SNPs After Filtering for MAFb | # SNPs After Filtering for SDc | # SSRs | Total # of Mappable Markers |
|---|---|---|---|---|---|---|
| GRYG | 18,499d | 18,374 | 8345 | 5150 | 189d | 5339 |
| CNJ02 | 15,197 | 15,028 | 8721 | 5564 | 629e | 6193 |
| CNJ04 | 15,224 | 14,818 | 8566 | 4566 | 221 | 4787 |
SNP loci with >20% missing data were excluded.
SNP loci with a MAF <10% were excluded.
SNP loci with extreme segregation distortion were excluded.
Generated in Covarrubias-Pazaran et al. (2016).
Generated in Schlautman (2015a).
After using the Tassel v3.0.166 GBS analysis pipeline to filter and process the sequence reads, a total of 15,197 and 15,224 SNPs with good sequence tag coverage were detected that were polymorphic in the CNJ02 and CNJ04 populations, respectively; and 18,499 SNPs for the GRYG population were available from the previous study (Table 1). Further filtering was performed to exclude SNP loci from each population with too much missing data (<20%), a MAF <10%, or extreme segregation distortion with χ2 P-values <0.00001. The 5150, 5564, and 4566 remaining SNPs were combined with the SSR marker datasets so that 5339, 6193, and 4787 markers were available for linkage mapping in the GRYG, CNJ02, and CNJ04 populations, respectively (Table 1).
Parental component bin map construction
The number of markers mapped in each of the six parental component bin maps ranged from 1774 markers for MQ (CNJ04 maternal parent) to 2487 markers for CQ (CNJ02 paternal parent) (Table S2, Table S3, and Table S4), with an average of 2080 markers per parental component map. The number of SNPs mapped in the three populations was very similar (CNJ04 = 2915, CNJ02 = 3326, GRYG = 3158); however, the CNJ02 population had more total markers mapped because of its larger SSR dataset (Table S5). Twelve LGs, corresponding to the expected haploid chromosome number in cranberry (2n = 2x = 24) (Hall and Galleta 1971), were retrieved for each of the cranberry parents, and total length of those 12 LGs varied from 845.2 cM for ST (CNJ04 maternal parent) to 1296.5 cM for MQ (CNJ02 maternal parent) (Table S2, Table S3, and Table S4).
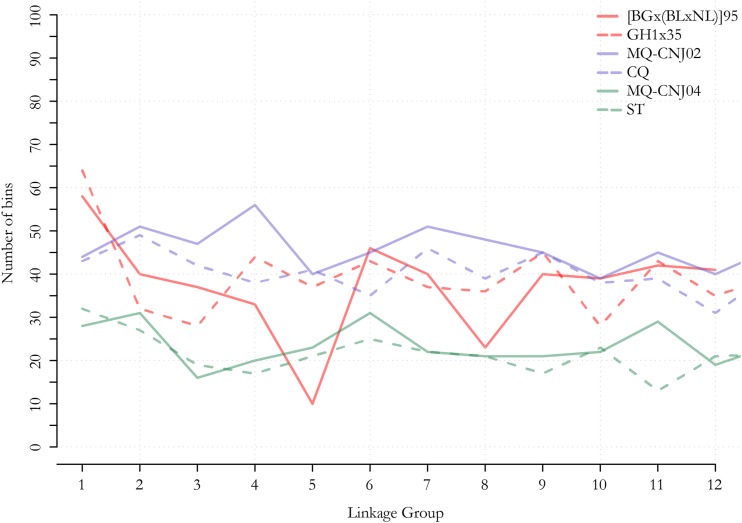
On average, the LGs in the six parental bin maps spanned 89.4 cM and contained 173.3 markers placed in 34.7 unique bins (Table S2, Table S3, and Table S4); however, notable exceptions were present in the component maps. For example, linkage group 5 from the [BGx(BLxNL)]95 component map only contained 34 markers placed in 10 marker bins spanning 61 cM (Table S2). Population size appeared to have some effect on the average number of bins per LG, a reflection of the number of observed recombination events per LG in the population of parental gametes that fused to form the progeny, such that fewer marker bins were present in the LGs of parents from the CNJ04 population, which was approximately one third and one fifth the size of the CNJ02 and GRYG populations, respectively (Figure 2). However, there was no obvious difference in number of bins per LG in the CNJ02 and GRYG parents despite the CNJ02 population containing only half as many progeny (Figure 2).
Figure 2.
Line graphs representing the distribution of the number of recombination bins, estimated using the multiple spanning tree algorithm implemented in ASMap (Wu et al. 2008; Taylor and Butler 2015), in each linkage group (LG) of the six parental component bin maps constructed for three cranberry full-sib populations: GRYG {i.e., [BGx(BLxNL)]95 × GH1x35}, CNJ02 (i.e., MQ × CQ), and CNJ04 (i.e., MQ × ST). The GRYG population (red) included 352 progeny; the CNJ02 population (blue) included 168 progeny, and the CNJ04 population (green) included 67 progeny.
In each parental map, the mean number of recombination events per progeny per LG ranged from 0.6 to 1.1 (Table S2, Table S3, and Table S4). Comparing the number of recombination events between the 36 maternal vs. paternal LG pairs, we found that the maternal LGs had a significantly greater number of recombination events per progeny per LG (P < 1.0−7 according to a paired Student’s t-test). The number of recombination events was greater for the maternal LG or equal to the paternal LG in 92% of the 36 pair-wise comparisons, and on average 0.18 more recombination events per progeny per LG were observed in the maternal LGs. Exceptions to this trend were observed in GRYG LG 8 and CNJ04 LG 11 where there were ∼0.1 more recombination events per progeny per LG in the paternal vs. maternal parent and in GRYG LG 5 where there were 0.3 more recombination events per progeny per LG in the paternal vs. maternal parent (Table S2, Table S3, and Table S4).
Pair-wise Spearman rank correlations comparing marker order revealed exceptionally high levels of synteny and collinearity between the LGs in the six parental component bin maps (Table 2). An average of 57.3 markers were used in each of the 180 pair-wise comparisons of LGs, and a mean Spearman rank correlation of 0.993 was observed across all comparisons (Table 2). The minimum Spearman rank correlation (r = 0.92) was observed in comparison of LG 9 between parents [BGx(BLxNL)]95 and ST (Table 2). Most differences in marker order between LGs were due to markers being in a single marker bin in one parent while being in adjacent marker bins in the other parent. The high degree of synteny and collinearity among LGs in the component maps was also visually observed in alignments of the linkage maps of the parents in each of the three cranberry populations in Circos and in scatterplots of the relative position of the common markers in each map on different axes (Figure 3).
Table 2. Pair-wise Spearman rank correlations between the LGs of the six parental component maps constructed for the parents (P) of the GRYG {i.e., P1 = [BGx(BLxNL)]95 and P2 = GH1x35}, CNJ02 (P3 = MQ and P4 = CQ), and CNJ04 (P5 = MQ and P6 = ST) full-sib cranberry populations.
| LG | P1xP2 | P1xP3 | P1xP4 | P1xP5 | P1xP6 | P2xP3 | P2xP4 | P2xP5 | P2xP6 | P3xP4 | P3xP5 | P3xP6 | P4xP5 | P4xP6 | P5xP6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.00 (91) | 1.00 (78) | 1.00 (100) | 1.00 (56) | 1.00 (75) | 1.00 (95) | 1.00 (85) | 1.00 (81) | 0.99 (71) | 1.00 (96) | 1.00 (119) | 1.00 (59) | 1.00 (52) | 0.99 (69) | 1.00 (87) |
| 2 | 1.00 (42) | 1.00 (50) | 1.00 (100) | 1.00 (46) | 1.00 (43) | 1.00 (37) | 1.00 (50) | 1.00 (29) | 1.00 (30) | 1.00 (89) | 1.00 (135) | 1.00 (32) | 1.00 (49) | 1.00 (55) | 1.00 (51) |
| 3 | 1.00 (41) | 0.99 (42) | 1.00 (56) | 0.97 (27) | 0.98 (24) | 1.00 (91) | 1.00 (37) | 0.97 (56) | 0.98 (32) | 1.00 (77) | 0.99 (97) | 0.99 (37) | 0.98 (25) | 0.99 (64) | 0.99 (44) |
| 4 | 1.00 (49) | 1.00 (66) | 1.00 (47) | 0.99 (39) | 0.99 (32) | 1.00 (68) | 1.00 (46) | 1.00 (42) | 1.00 (37) | 1.00 (96) | 1.00 (102) | 0.99 (23) | 0.99 (31) | 0.99 (62) | 0.99 (29) |
| 5 | 0.99 (15) | 0.98 (16) | 0.98 (19) | 0.95 (11) | 0.93 (8) | 1.00 (85) | 1.00 (86) | 1.00 (48) | 0.99 (34) | 1.00 (85) | 1.00 (93) | 1.00 (15) | 0.99 (35) | 1.00 (56) | 1.00 (19) |
| 6 | 1.00 (70) | 1.00 (57) | 1.00 (86) | 0.99 (43) | 1.00 (49) | 1.00 (96) | 1.00 (51) | 0.99 (83) | 0.99 (42) | 1.00 (80) | 0.99 (143) | 0.99 (49) | 1.00 (52) | 1.00 (89) | 0.99 (55) |
| 7 | 1.00 (45) | 1.00 (54) | 1.00 (50) | 0.99 (37) | 0.99 (22) | 1.00 (99) | 1.00 (44) | 0.99 (68) | 0.99 (14) | 1.00 (91) | 1.00 (122) | 0.98 (29) | 0.99 (42) | 1.00 (48) | 0.98 (34) |
| 8 | 0.99 (32) | 0.99 (29) | 0.99 (41) | 0.99 (26) | 0.99 (24) | 1.00 (35) | 1.00 (36) | 1.00 (31) | 1.00 (15) | 1.00 (83) | 1.00 (97) | 1.00 (21) | 0.99 (46) | 0.99 (55) | 1.00 (25) |
| 9 | 1.00 (84) | 1.00 (67) | 1.00 (70) | 0.99 (49) | 0.92 (54) | 1.00 (83) | 1.00 (57) | 0.99 (56) | 0.98 (37) | 1.00 (93) | 0.99 (141) | 0.97 (31) | 1.00 (39) | 0.98 (46) | 0.98 (36) |
| 10 | 1.00 (48) | 1.00 (34) | 1.00 (46) | 1.00 (31) | 0.99 (69) | 1.00 (79) | 0.98 (53) | 1.00 (59) | 0.99 (33) | 1.00 (82) | 1.00 (100) | 0.98 (27) | 0.99 (29) | 0.99 (64) | 0.99 (31) |
| 11 | 1.00 (56) | 1.00 (66) | 1.00 (77) | 0.99 (49) | 0.99 (32) | 1.00 (114) | 1.00 (69) | 0.99 (94) | 0.99 (28) | 1.00 (97) | 1.00 (159) | 0.99 (34) | 1.00 (61) | 0.99 (47) | 0.98 (43) |
| 12 | 1.00 (66) | 0.99 (48) | 1.00 (77) | 0.98 (40) | 0.98 (46) | 0.99 (85) | 1.00 (57) | 0.99 (64) | 0.97 (37) | 0.99 (77) | 0.99 (127) | 0.98 (32) | 0.99 (39) | 0.97 (75) | 0.98 (45) |
| CoAa | 0.094 | 0.078 | 0.094 | 0.078 | 0.063 | 0.156 | 0.031 | 0.156 | 0.063 | 0.000 | 0.500c | 0.000 | 0.000 | 0.250 | 0.000 |
| Mean | 1.00 (53.3) | 1.00 (50.6) | 1.00 (64.1) | 0.99 (37.8) | 0.98 (39.8) | 1.00 (80.6) | 1.00 (55.9) | 0.99 (59.3) | 0.99 (34.2) | 1.00 (87.2) | 1.00 (119.6) | 0.99 (32.4) | 0.99 (41.7) | 0.99 (60.8) | 0.99 (41.6) |
| Totalb | 639 | 607 | 769 | 454 | 478 | 967 | 671 | 711 | 410 | 1046 | 1435 | 389 | 500 | 730 | 499 |
Coefficients of coancestry (CoA), or kinship coefficients, were calculated for the parents involved in each pair-wise comparison. The number of common markers available and used in each comparison is listed in parentheses.
Coefficient of coancestry calculated using pedigree information.
Total markers in common between the compared parental bin maps.
Self-coancestry is 0.5 assuming no prior inbreeding.
Figure 3.
Comparisons of the LGs of the parental linkage maps for the (A) GRYG {[BGx(BLxNL)]95 × GH1x35}, (B) CNJ02 (Mullica Queen × Crimson Queen), and (C) CNJ04 (Mullica Queen × Stevens) full-sib cranberry mapping populations. LGs from the maternal maps are on the left side of each circular ideogram while the paternal LGs are on the right. Links are drawn between common markers in the LGs of the two parental maps in each population. Scatterplots of the position (centimorgans) of the common markers in the paternal map plotted on the x-axis and position in the maternal map plotted on the y-axis display collinearity of marker order. Bars display phased genotype data which show position (centimorgans) of phase changes (the gametic recombination) which occurred in both parents for a random subset of 60 progeny from each population. Outer ring displays the position (centimorgans) of markers in the parental maps colored by the χ2 P-value obtained from the tests for distortion from expected Mendelian segregation ratios. Marker colors range from dark green for markers not showing distortion (χ2 P > 0.1) to dark red for markers showing highly significant segregation distortion (χ2 P < 0.0005).
Composite map construction
The high degree of collinearity between the component maps allowed for the construction of a cranberry composite map with LPmerge by retaining the synthetic composite map for each LG computed with the maximum interval size, k, which minimized the RMSE (Endelman and Plomion 2014). The composite map contained 6073 markers (636 SSRs and 5437 SNPs) spanning 1124.3 cM (Table 3, Table S6, and File S2). The 12 LGs of the composite map ranged from 84.1 to 115.9 cM in length, and each LG contained an average of 506 (453 SNPs and 53 SSRs) markers (Table 3). The 6073 markers in the composite map corresponded to 1560 unique marker positions (bins), with an average gap of 0.7 cM between unique marker positions (Table 3). The largest gap in the composite map was on LG10 and spanned 6.6 cM; however, there were only three gaps >5 cM and nine gaps >4 cM in the entire composite map (Table S7). The composite map anchored a total of 3989 cranberry scaffolds from the Polashock et al. (2014) assembly totaling 21.8 Mb (4.6%) of the predicted 470-Mb cranberry genome (Table S8). Of the 3989 anchored scaffolds, 1654 contained predicted CDS (Table S8).
Table 3. Features of the cranberry composite map, constructed using the six parental component bin maps for the parents of the CNJ02, CNJ04, and GRYG populations, including the length of the LGs, the total number of SSR and SNP markers mapped, the number of unique marker positions, the number of unique marker positions containing an SSR, and the mean gap distance in centimorgans (cM) between unique positions.
| LG | Length (cM) | # SNPs | # SSRs | Total # Markers | # Unique Marker Positions | # Positions with an SSR | Mean Gap (cM) |
|---|---|---|---|---|---|---|---|
| LG1 | 115.9 | 588 | 50 | 638 | 158 | 37 | 0.74 |
| LG2 | 100.8 | 416 | 63 | 479 | 152 | 45 | 0.67 |
| LG3 | 92.4 | 407 | 54 | 461 | 99 | 34 | 0.94 |
| LG4 | 86.4 | 423 | 61 | 484 | 110 | 36 | 0.79 |
| LG5 | 93.1 | 405 | 35 | 440 | 110 | 24 | 0.85 |
| LG6 | 93.8 | 473 | 50 | 523 | 157 | 37 | 0.6 |
| LG7 | 97.0 | 418 | 59 | 477 | 158 | 47 | 0.62 |
| LG8 | 85.2 | 370 | 54 | 424 | 128 | 35 | 0.67 |
| LG9 | 89.9 | 532 | 54 | 586 | 112 | 33 | 0.81 |
| LG10 | 84.1 | 378 | 51 | 429 | 115 | 32 | 0.74 |
| LG11 | 95.3 | 499 | 42 | 541 | 152 | 30 | 0.63 |
| LG12 | 90.6 | 528 | 63 | 591 | 109 | 32 | 0.84 |
| Mean | 93.7 | 453 | 53 | 506 | 130 | 35 | 0.74 |
| Total | 1124.3 | 5437 | 636 | 6073 | 1560 | 422 |
Collinearity between the cranberry composite map and the six component parental bin maps was high; the mean Spearman correlation across the 72 pairwise comparisons of marker order was r = 0.997 (Table 4). Collinearity between the parents of the two larger populations (i.e., CNJ02 and GRYG) and the composite map were slightly higher (i.e., r ≥ 0.999) than the CNJ04 parents (i.e., r = 0.995 and 0.991) (Table 4). Marker order variation between the composite and six component maps was highest in LG12, with perfect correlation observed only between the [BGx(BLxNL)]95 component map and the composite map (Table 4). Spearman rank correlations comparing the LGs of the composite map to the 12 LGs from the Covarrubias-Pazaran et al. (2016) SNP-SSR map and the Schlautman et al. (2015a) SSR map were also high (i.e., r = 0.976 and r = 0.996, respectively) (Table S9).
Table 4. Pair-wise Spearman rank correlations between the LGs of the six parental component maps constructed for the parents of the GRYG {[BGx(BLxNL)]95 and GH1x35}, CNJ02 (MQ and CQ), and CNJ04 (MQ and ST) full-sib cranberry populations compared to the LGs of the cranberry composite map.
| LG | [BGx(BLxNL)]95 | GH1x35 | MQ-CNJ02 | CQ | MQ-CNJ04 | ST |
|---|---|---|---|---|---|---|
| LG1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| LG2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| LG3 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 |
| LG4 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 |
| LG5 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.97 |
| LG6 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| LG7 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| LG8 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| LG9 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.97 |
| LG10 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| LG11 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 |
| LG12 | 1.00 | 0.99 | 0.99 | 0.99 | 0.96 | 0.98 |
| Mean | 1.000 | 0.999 | 0.999 | 0.999 | 0.995 | 0.991 |
Genome-wide segregation distortion and centromere placement
Approximately 8% of the markers in the six parental component bin maps displayed significant segregation distortion (SD) at the χ2 P < 0.1 level (Figure 3 and Table S10). SD was not randomly distributed across the LGs or populations. The GH1x35 parental map contained a much higher proportion of markers displaying SD (i.e., 26%) compared to the other five parents, with as many as 81% of all markers in LG 6 displaying distortion (Figure 3 and Table S10). Segregation distortion regions (SDRs) were observed in LGs of each of the six parental bin maps, and some SDRs, such as the SDR on LG 9, appeared to be present in multiple parental genomes (Figure 3 and Table S10).
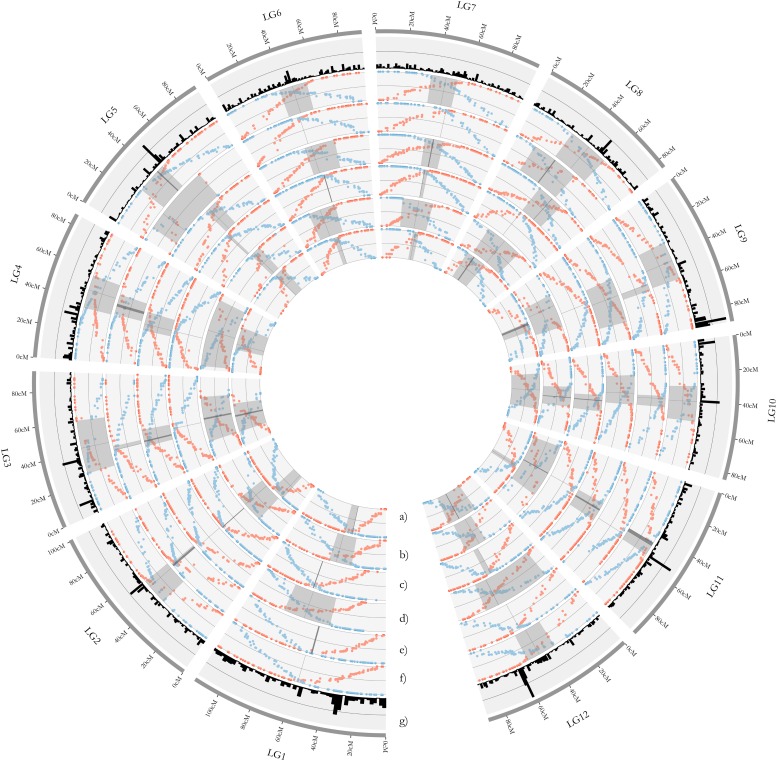
Phasing the genotype data and estimating RFM for each marker interval from the terminal markers in both directions allowed for centromere placement on each of the 12 LGs of the component maps using the method developed in Limborg et al. (2016) (Figure 4 and Table 5). The recombination phasing method allows for distinguishing between metacentric and acrocentric LGs. All cranberry LGs appeared to be metacentric, which is consistent with the karyotype for cranberry and diploid Vacciniums observed in Hall and Galleta (1971), and centromere spans averaged 18.6 cM across the 12 LGs of the 6 component maps (Figure 4 and Table 5).
Figure 4.
Plots of recombination frequency (RFM) estimated from phased genotype data by starting at the terminal markers at the beginning (red points) and end (blue points) of each linkage and recording the proportion of offspring with an observed recombination (i.e., change of phase) in the interval between the terminal marker (m0) and each subsequent marker (mn) for the 12 cranberry LGs. Centromere spans (gray regions) were placed on the 12 LGs of the cranberry parental component bin maps constructed for the parents of the CNJ04 population, Mullica Queen (a) and Stevens (b); the CNJ02 population, Mullica Queen (c) and Crimson Queen (d); and the GRYG population, [BGx(BLxNL)]95 (e) and GH1x35 (f) using the method developed in Limborg et al. (2016). Centromeric spans in the LGs were defined as the range (centimorgans) extending from the intersection (dark lines) of the recombination frequency (RFM) estimates made from both ends of the LG outwards until reaching the first marker with an RFM = 0.45 in both directions. (g) Marker density in the cranberry composite map is shown to explore the relationship between marker density and centromere position.
Table 5. Centromere spans placed on the 12 LGs of the cranberry parental component bin maps constructed for the parents of the CNJ04 population, Mullica Queen (MQ) and Stevens (ST); the CNJ02 population, MQ and Crimson Queen (CQ); and the GRYG population, [BGx(BLxNL)]95 (BGBLNL95) and GH1x35.
| Parent | LG | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| MQ-CNJ04 | 44.3–55.0 (44.3) | 24.2–42.1 (28.2) | 20.8–58.6 (41.1) | 29.9–58.7 (36.9) | 47.1–58.5 (47.7) | 38.1–43.8 (43.5) | 42.3–49.6 (45.4) | 13.9–41.1 (23.9) | 43.8–55.3 (44.6) | 15.0–61.5 (36.6) | 46.9–53.0 (50.2) | 24.9–64.7 (49.9) |
| ST | 38.0–62.8 (55) | 28.2–59.2 (50.2) | 20.8–69.9 (45.5) | 4.2–79.0 (43.9) | 44.7–56.1 (48.3) | 21.0–49.8 (34.2) | 24.3–57.6 (42.3) | 20.0–62.4 (41.7) | 25.9–62.7 (38.1) | 31.7–52.8 (43.4) | 6.6–66.3 (38.1) | 35.4–60.1 (43.7) |
| MQ-CNJ02 | 68.8–68.8 (68.8) | 47.8–47.8 (47.8) | 42.9–42.9 (42.9) | 33.9–33.9 (33.9) | 42.4–44.7 (42.4) | 48.9–48.9 (48.9) | 42.3–45.4 (42.3) | 36.2–36.2 (36.2) | 49.3–49.3 (49.3) | 39.0–54.2 (44.0) | 38.7–38.7 (38.7) | 48.0–52.9 (48) |
| CQ | 46.1–85.2 (67.8) | 49.3–49.3 (49.3) | 33.4–47.0 (37.1) | 28.7–43.9 (33.9) | 44.7–56.1 (48.3) | 40.4–65.4 (45.9) | 38.3–53.5 (44.8) | 36.2–58.8 (49.4) | 38.1–73.2 (52.5) | 21.2–45.3 (36) | 38.1–53.6 (43.5) | 9.8–66.4 (55.3) |
| BGBLNL95 | 53.9–53.9 (53.9) | 50.0–52.5 (50.3) | 36.5–39.3 (36.5) | 35.6–50.1 (41.5) | 9.8–61.0 (22.4) | 44.1–44.1 (44.1) | 49.9–49.9 (49.9) | 14.7–50.2 (32.1) | 52.5–57.2 (52.5) | 37.1–44.4 (39.0) | 50.2–50.2 (50.2) | 49.6–49.6 (49.6) |
| GH1x35 | 58.1–58.1 (58.1) | 37.4–54.5 (50.2) | 26.0–61.0 (42.9) | 34.9–55.9 (46.7) | 26.4–46.7 (41.8) | 37.2–55.3 (45.9) | 35.7–52.9 (45.4) | 30.9–58.1 (44.7) | 31.0–56.3 (43.8) | 29.8–52.7 (39.9) | 44.6–53.6 (44.6) | 34.9–54.7 (46.3) |
| Mean | 51.5–64.0 (58.0) | 39.5–50.9 (46.0) | 30–53.1 (41.0) | 27.9–53.6 (39.4) | 35.9–53.9 (41.8) | 38.3–51.2 (43.7) | 38.8–51.5 (45.0) | 25.3–51.1 (38.0) | 40.1–59 (46.8) | 29.0–51.8 (39.8) | 37.5–52.6 (44.2) | 33.8–58.1 (48.8) |
Method developed in Limborg et al. (2016) was used. Centromeric spans (x cM − y cM) in the LGs were defined as the range from the intersection between the recombination frequency (RFM) estimates between each marker and the terminal marker on both ends of the LG extending outwards until reaching the first marker with an RFM = 0.45 in both directions. The intersection point (centimorgans) of RFM estimates in each LG is provided in parentheses.
Discussion
Genotyping-by-sequencing is an effective strategy for simultaneous SNP discovery and genotyping in cranberry, and was used to identify thousands of polymorphic SNP loci which were transferable between interrelated full-sib mapping populations. A composite map was developed by merging the six bin maps constructed for the parents of each full-sib family, which allowed for exploration of SDRs and centromere placement. The map will serve as a framework for future genomics studies to identify quantitative trait loci (QTL) regions of interest and for the continued investigation of genome organization and evolution in the genus Vaccinium. Furthermore, the cranberry genomic scaffolds and predicted CDS anchored in the composite map demonstrate its utility in future efforts to anchor physical maps and assist in the assembly of cranberry and Vaccinium genome sequences.
Map construction
The use of three full-sibling populations of diverse genetic backgrounds facilitated an increase in the number of mapped markers (SNPs and SSRs) and resulting genome coverage in the cranberry composite map that has not been possible in past studies using single populations (Georgi et al. 2013; Schlautman et al. 2015a; Covarrubias-Pazaran et al. 2016). More importantly, the pedigrees of the three full-sibling populations all trace their ancestry to the seven wild cranberry selections, “The Big Seven,” made in the 19th and 20th centuries, which collectively account for the entire genetic base of the modern cranberry industry (Figure 1) (Eck 1990; Clark and Finn 2010; Fajardo et al. 2012). Therefore, the composite cranberry map affords the ideal opportunity for exploring and connecting the genetic diversity to the phenotypic diversity that has been responsible for the historical success of cranberry production. A key use for the composite map will be to allow for concurrent identification and integration of QTL and marker trait loci (MTL) which colocalize across populations, across environments, and across phenotypes within and among future linkage mapping, QTL mapping, and genome-wide association studies (GWAS).
The composite map will be most useful for studies within section Oxycoccus, and it may aid in future targeted introgression of genomic regions involved in the expression of unique metabolic pathways or disease resistance genes from the small-fruited cranberry, V. oxycoccos, to the American cranberry, V. macrocarpon (Vorsa and Polashock 2005). However, due to the high level of synteny and collinearity detected between cranberry and blueberry using cross-transferable SSRs (Schlautman 2016), information from the composite map should also be applicable to many other commercially important Vacciniums including blueberry (Vaccinium section Cyanococcus) and closely related sections lingonberry (Vaccinium section Vitis-Idaea) and sparkle berry (Vaccinium section Batodendron) (Lyrene 2011; Schlautman et al. 2016).
Two methods are commonly employed for integration of independent linkage maps. The first method uses pooled segregation and recombination information from independent maps; however, this method is computationally unfeasible for integration of high-density linkage maps containing thousands of loci (Van Ooijen 2011; Endelman and Plomion 2014; Bodénès et al. 2016). The second method, which was used in the present study and other recent composite mapping studies in pine and oak (Chancerel et al. 2013; Bodénès et al. 2016), performs map integration based on marker position rather than observed recombination such as is proposed and implemented in the LPmerge package (Endelman and Plomion 2014). Using LPmerge, the six parental component bin maps, each containing an average of 2080 markers (Table S2, Table S3, and Table S4), were merged to obtain a synthetic composite map that included 6073 loci (1560 unique marker positions) covering 1124 cM on 12 LGs, making it the highest-density map in the Vaccinium genus and the entire Ericaceae family (File S2 and Table 3). This map achieved considerably higher marker saturation (i.e., 0.7 cM mean interval between unique marker positions) than previous published cranberry linkage maps based on SSR and SNP markers (Georgi et al. 2013; Schlautman et al. 2015a; Covarrubias-Pazaran et al. 2016), while decreasing the total map length by 50 cM compared to the highest-density cranberry SSR map and increasing total map length by only 13 cM compared to the first cranberry SNP map (Table 3) (Schlautman et al. 2015a; Covarrubias-Pazaran et al. 2016).
The similarity in total map lengths between the current composite map and former maps reflects the robust marker-ordering and marker distance estimation achieved in construction of the six individual component maps using stringent parameters during mapping, imputing missing data and correcting potential genotyping errors, and the bin mapping strategy. Accuracy of the cranberry parental linkage maps is supported by the near perfect collinearity of the parental bin maps as quantified by Spearman rank correlations [e.g., Spearman rank correlations (r) exceeded 0.99 for 58 (80%) of the 72 pair-wise comparisons of marker order in the LGs of the composite map compared to the LGs of each of the component parental bin maps and exceeded 0.95 for all 72 pair-wise comparisons], and is especially apparent when observing the recombination events as phase changes in the phased genotype data of each of the parental bin maps (Figure 3, Table 2, and Table 4). Spearman rank correlations comparing the LGs of the composite map to LGs from the Covarrubias-Pazaran et al. (2016) SNP-SSR map and the Schlautman et al. (2015a) SSR map were also high (i.e., r = 0.976 and r = 0.996, respectively) and provided further evidence of the composite map quality (Table S9).
Previous high-density linkage mapping studies in outcrossing populations have often excluded the biparental markers (i.e., hk × hk segregation) because of difficulties they cause during mapping and because of assumptions that they are less informative for linkage mapping (Ward et al. 2013; Bodénès et al. 2016); however, including the biparental SNPs in this study greatly increased the number of total markers mapped and the number of markers in common between component maps. All genetic maps (i.e., composite and component maps) in this study could be directly compared because of the high frequency of transferability of SNP markers across parents and populations. A total of 2921 (54%) of the SNPs in the composite map were mapped in two or more populations and 1040 (19%) of the SNPs were mapped in all three populations (Table S5). The number of markers in common between LGs in the parental maps appeared to be correlated (r = 0.74) with the degree of shared ancestry between the parents. For example, there were, on average, 80.1 markers in common per LG for parents with a coefficient of coancestry >0.1, while parents with a coefficient of coancestry <0.1 had an average of 49 markers in common per LG (Figure 1 and Table 2). A high degree of synteny and collinearity was visually observed in alignments of the parental maps in each population (Figure 3), and the mean Spearman rank correlation of r = 0.993 was remarkably high in comparisons of marker order, based on an average of 57.3 markers per LG, across all 180 pair-wise comparisons of LGs in the parental component maps (Table 2). The observed transferability of SNP markers across the five cranberry parents from three populations suggests that GBS will be an effective marker discovery and genotyping platform for exploring cranberry genetic diversity and population structure and, aided by the placement of >5000 SNPs in the composite map, will be useful in estimating linkage disequilibrium in cranberry diversity panels for GWAS.
In addition to the >5000 SNP markers mined from GBS data and mapped in this study, the composite map also included 636 SSRs, which represents the largest collection of mapped SSR markers available for cranberry and an important genetic resource for future cranberry breeding efforts (Table 3 and Table S1). Of the 1540 unique marker positions in the composite map, 27% include one or more SSR markers (Table 3). Within the Mullica Queen and Crimson Queen parental bin maps for the CNJ02 population, an average of 53% of the unique marker bins include an SSR marker suggesting that a larger number of marker positions in the composite map could have included one or more SSR markers if more SSR data had been available for the other two full-sib populations (Table S3). The large number of positioned SSRs, and their apparent distribution throughout the entire cranberry genome, should allow them to be useful in marker-assisted seedling selection of MTL in cranberry using multiplexing panels like those developed in Schlautman (2016). Despite the obvious benefits of simultaneous marker discovery and genotyping afforded by GBS, the high cost per sample does not yet justify its use for large-scale genotyping of thousands of seedlings for selection of a few MTL in a specialty crop such as cranberry. A better way to allocate resources will be to utilize the SSRs, which are likely to be polymorphic in much more diverse backgrounds than SNPs (Hamblin et al. 2007), to select for must-have MTL in large populations for simple traits, and then use GBS to further explore the genetic diversity or perform genomic selection in the MTL selected individuals (Dekkers 2007; Collard et al. 2008; Singh and Singh 2015).
Recombination rate variation
Comparisons across the six parental linkage maps developed in this study highlighted interesting differences that are related to the rates of recombination in each parent and may have important implications for cranberry breeding and genetic studies. First, the average number of marker bins per linkage group, which reflects the number of unique recombination events in the parental gametes, was lower in the parents of the CNJ04 population (23 bins per parent per LG), than in the GRYG and CNJ02 populations (38 bins and 43.5 bins per parent per LG, respectively) (Figure 2, Table S2, Table S3, and Table S4). The fewer bins observed in the parents of the CNJ04 population was expected and is likely due to its small size (67 progeny), suggesting that breeders may need less genetic data (DNA markers) to capture all the recombination history in small biparental breeding populations. Unexpectedly, there were similar numbers of marker bins per LG in the CNJ02 and GRYG populations, although the GRYG population (352 progeny) was much larger than the CNJ02 (168 progeny) population. This observation indicates that cranberry genotypes can exhibit different rates of recombination or different numbers of recombination hot and cold spots. Differential rates of recombination frequencies between cranberry genotypes were also observed in Schlautman et al. (2015a) and Covarrubias-Pazaran et al. (2016), and both genotypic and environmental effects on genetic recombination were observed in tetrad analysis of cranberry reciprocal translocation heterozygotes (Ortiz and Vorsa 2004). Finally, the smaller number of bins observed in the CNJ04 population suggests that increasing cranberry breeding population size from ∼70 progeny to ∼150 progeny can increase the number of unique recombination events observed. However, the similar number of bins observed in the large (GRYG) and medium size (CNJ02) populations suggests that cranberry breeders could better allocate their resources by planting many medium size populations (∼150 progeny) rather than a few large populations (∼350 progeny) when detecting slightly more unique recombination events is not as valuable as saving field space and observing additional breeding populations.
The second interesting observation is that the paternal LGs were consistently shorter than the maternal LGs, and there were fewer phase changes (i.e., recombination events) per progeny per LG in the paternal gametes than in the maternal gametes (Figure 3, Table S2, Table S3, and Table S4). Sex-specific differences in recombination rates have previously been observed in cranberry (Schlautman et al. 2015a; Covarrubias-Pazaran et al. 2016), blueberry (Schlautman 2016), apple (Maliepaard et al. 1998), and olive (Sadok et al. 2013). It is uncertain whether these observed differences in recombination rates between sexes are more than coincidental differences in patterns and rates of recombination in the five cranberry parents. However, we speculate that the differential recombination rates in male and female gametes observed in Vaccinium linkage mapping studies may be a true consequence resulting from a combination of common practices employed by cranberry and blueberry breeders during population development, differences between microsporogenesis and megasporogenesis, and the unique pollen morphology of the Vaccinium genus.
During megasporogenesis, three of the four megaspores disintegrate, and only one megaspore survives; conversely, all four microspores survive during microsporogenesis (Fehr 1991). Previous studies in Vaccinium species including cranberry revealed that pollen, rather than being shed as single grains, which is common in flowering plants, are shed as groups of four pollen grains (i.e., the four microspores) called tetrads, which are derived from the same meiotic division (Eck 1990; Roper and Vorsa 1997). This has important consequences considering that, during meiosis, chiasma almost always occur between only two of the four chromatids on each side of the centromere (Roeder 1997). Therefore, assuming Vaccinium chromosomes are metacentric (Hall and Galleta 1971), for any single chromosome in a Vaccinium pollen tetrad (i.e., a total of eight chromosome arms), four of the eight chromosome arms in the four haploid gametes (i.e., four microspores) represent meiotic recombinants, but in reality, there are only two unique positions of recombination because the reflection of each recombinant chromosome arm exists as one of the three remaining recombinant chromosome arms in the pollen tetrad.
In addition, pollen germination studies in both cranberry and blueberry have revealed that all four pollen grains are viable in the majority of Vaccinium pollen tetrads (Huang and Johnson 1996; Cane 2009), and that fruit set does not increase after loading >8 tetrads on a cranberry stigma which contain an average of 32 ± 4 ovules (Sarracino and Vorsa 1991; Cane and Schiffhauer 2003). Consequently, it is highly likely that any seed within a Vaccinium fruit shares a paternal meiotic history with one or more seeds in the same fruit; however, because of the disintegration of three of the four megaspores, no seed in a fruit shares a maternal meiotic history with any other seed in the same fruit. The result of this phenomenon (high Vaccinium microspore fertility with pollen shed as tetrads), combined with the fact that Vaccinium breeders and geneticists generally only make a few crosses (i.e., harvest seeds from a few fruits) to generate breeding and/or linkage mapping populations, may explain the reduced rate of recombination observed in the paternal vs. maternal gametes in this study. Specifically, in a population of 40 cranberry full-sib progeny with a meiotic history tracing to 10 cranberry pollen tetrads (i.e., 40 haploid paternal gametes), assuming strong cross-over interference, a maximum of 20 unique recombinant chromosome arms out of 80 could be observed for any single paternal metacentric chromosome in the full-sib progeny; conversely, a maximum of 80 out of 80 unique recombinant chromosome arms could be observed for the same maternal metacentric chromosome in the full-sib progeny. Although future studies will be needed to confirm this hypothesis, these results suggest that Vaccinium breeders and geneticists should consider harvesting seeds from as many crosses as possible (i.e., more fruits) during population development to ensure equal representation of recombination in both the maternal and paternal genomes. Although pollen tetrad formation could be a challenge in Vaccinium breeding and genetics, pollen tetrad analysis, which is not feasible in most higher Eukaryotes, has been highly advantageous and is used extensively in genetic studies of Arabidopsis mutants to detect every genetic change between chromatids, to simplify genetic map construction, to define centromere positions, and to quantify cross-over and chromatid interference (Preuss et al. 1994; Copenhaver et al. 2002; Brieuc et al. 2014). Furthermore, it has already been useful in detecting chromosomal rearrangements, specifically reciprocal translocations, in cranberry cultivars (Ortiz and Vorsa 2004).
Genome characterization
SNP and SSR loci present within nuclear genome scaffolds from the Polashock et al. (2014) assembly were used to anchor 3989 nuclear scaffolds containing 21.8 Mb (4.6%) of the predicted 470 Mb cranberry genome in the composite map (Table S8). Although the present composite map anchored 1500 more nuclear scaffolds than the previous SNP linkage map, those 1500 scaffolds only represented a 1.9% increase in the total megabases of cranberry genome anchored (Covarrubias-Pazaran et al. 2016). This is reflective of the sheer number of scaffolds (i.e., 229,745 scaffolds) and their size (i.e., N50 = 4237 bp), and suggests the need for a higher quality cranberry genome assembly (Polashock et al. 2014).
Segregation distortion affects accuracy of linkage map construction by introducing errors in map distance estimation and marker order, and thus, could affect mapping QTL (Zhang et al. 2010). Deviations from expected Mendelian inheritance were widespread throughout the cranberry genome, and ∼8% of the markers positioned in the parental component maps displayed significant SD (i.e., P < 0.1) according to χ2 tests with one degree of freedom (Figure 3 and Table S10). Previous high-density linkage mapping studies have sometimes observed that distorted markers are not always randomly distributed, but rather, are grouped together in SDRs (Yin et al. 2004; Wang et al. 2012a; Bodénès et al. 2016; Covarrubias-Pazaran et al. 2016). Likewise, apparent SDRs were observed in the present study in all six of the parental component maps (Figure 3 and Table S10). Many of the SDRs are unique to each parental cranberry genome, as was previously observed in oak and palm (Ting et al. 2014; Bodénès et al. 2016). However, there is apparent overlapping between some SDRs in the six parents {e.g., SDR in LG9 for the [BGx(BLxNL)]95 and Stevens parents, and SDR in LG8 for the [BGx(BLxNL)]95 and Crimson Queen parents}, which could represent biologically significant phenomena such as gametic competition, gametophytic selection, or sterility (Wang et al. 2005; Bloom and Holland 2012; Xu et al. 2013). The identification of SDR regions in this study provides a framework to further study the cause of this phenomenon in cranberry and to improve experimental designs for association mapping studies and marker-assisted selection strategies.
There have been minimal efforts to characterize chromosomal structures, such as centromeres, in cranberry, and modern cytogenetic approaches such as fluorescent in situ hybridization have not yet been attempted. Centromeres are central components of chromosome architecture that play fundamental roles in the regulation and cross-over formation during meiosis (Roeder 1997; Zickler and Kleckner 1999); therefore, knowledge of centromere location is critically important in attempts to manage the occurrence of meiotic recombination in plant breeding (Wijnker and de Jong 2008). However, many high-density linkage mapping studies in cranberry and other commercially important crops have not attempted or failed to place centromeres onto the LGs (Wang et al. 2012b; Castro et al. 2013; Ting et al. 2014; Bodénès et al. 2016; Covarrubias-Pazaran et al. 2016), which could limit their future applicability in interpretations of genome organization, genome divergence, and of the genomic architecture of adaptive or economically important traits (Limborg et al. 2016). Therefore, we utilized the recombination phasing method outlined in Limborg et al. (2016) and effectively applied in Mckinney et al. (2016) to define centromere regions in the cranberry LGs (Figure 4 and Table 5).
In general, the Limborg et al. (2016) recombination phasing method appeared to work well in cranberry. Centromeres were placed on all 12 cranberry LGs in each of the 6 parental component maps (Figure 4 and Table 5). All LGs were identified as metacentric, consistent with a previous cranberry karyotype presented in Hall and Galleta (1971). The centromere regions in the parental LGs, although sometimes large (i.e., an average of 18.6 cM across LGs), were similar in size to those observed in Mckinney et al. (2016). The centromere regions appeared to have the same general location across LGs in the parental maps, providing further confidence in the estimation of their positions. Interestingly, observed marker densities were often higher in centromere regions and had reduced recombination rates (i.e., recombination “cold spots”) compared to the rest of the respective LGs (Figure 4). The knowledge of centromere location generated herein should facilitate future meiotic studies in cranberry exploring cross-over interference and recombination “hot spots” and “cold spots,” and could potentially be useful for developing cranberry crop improvement strategies for managing meiotic recombination and for overcoming barriers to recombination in cranberry chromosomes (Martinez-Perez and Moore 2008).
In conclusion, GBS has been shown to be a highly efficient means for SNP marker discovery and genotyping in cranberry. A large proportion of the GBS-based SNP loci were polymorphic and transferable between three full-sib cranberry populations, which allowed for the construction of the first high-density composite linkage map in cranberry composed of both SNP and SSR markers. The stringent parameters used during component map construction and the remarkable collinearity observed between the six component maps and the composite map suggest that estimation of marker position and distance was performed in an accurate, reproducible manner. The large number of cross-transferable mapped markers allowed for characterization of cranberry genomic architecture such as the detection of centromeric regions, and we foresee that the composite map and the marker data will be used extensively for future QTL detection, GWAS, and development of molecular-assisted breeding strategies in cranberry.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.037556/-/DC1.
Acknowledgments
J.Z. and B.S. wish to express their gratitude through 1 Cor 10:31. This project was supported by USDA-SRCI under grant 2008-51180-04878; USDA-NIFA-AFRI Competitive grant USDA-NIFA-2013-67013-21107; USDA-ARS (project no. 3655-21220-001-00 provided to J.Z.); WI-DATCP (SCBG Project #14-002); National Science Foundation (DBI-1228280); Ocean Spray Cranberries, Inc.; NJ Cranberry and Blueberry Research Council; Wisconsin Cranberry Growers Association; and the Cranberry Institute. B.S. was supported by the Frank B. Koller Cranberry Fellowship Fund for Graduate Students; G.C.-P. and L.D.-G. were supported by the Consejo Nacional de Ciencia and Tecnologia (CONACYT, Mexico). M.I. was supported by the USDA National Institute of Food and Agriculture, Hatch project 1008691.
Author contributions: BS, GC-P, NV, JP, and JZ conceived the study. JZ, MI, NV, and JP supervised the research. BS and GC-P prepared samples and conducted DNA genotyping. BS and GC-P formatted genotypic data, performed imputations, and constructed the linkage and composite maps. BS performed all statistical analyses. BS and LD-G identified putative centromere regions. EG, NV, JP, and MI provided intellectual contributions and/or germplasm resources. BS and JZ wrote the manuscript. BS and LD-G prepared the final figures. The manuscript was read, edited, and approved by all authors.
Footnotes
Communicating editor: S. A. Jackson
Literature Cited
- Bloom J. C., Holland J. B., 2012. Genomic localization of the maize cross-incompatibility gene, Gametophyte factor 1 (ga1). Maydica 56: 379–387. [Google Scholar]
- Bodénès C., Chancerel E., Ehrenmann F., Kremer A., Ploimon C., 2016. High-density linkage mapping and distribution of segregation distortion regions in the oak genome. DNA Res. 23(2): 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieuc M. S. O., Waters C. D., Seeb J. E., Naish K. A., 2014. A dense linkage map for Chinook salmon (Oncorhynchus tshawytscha) reveals variable chromosomal divergence after an ancestral whole genome duplication event. G3 (Bethesda) 4: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp W., 1945. The North American blueberries with notes on other groups of Vacciniaceae. Brittonia 5: 203–275. [Google Scholar]
- Cane J. H., 2009. Pollen viability and pollen tube attrition in cranberry (Vaccinium macrocarpon Aiton). Acta Hortic. 810(2): 563–566. [Google Scholar]
- Cane J. H., Schiffhauer D., 2003. Dose-response relationships between pollination and fruiting refine pollinator comparisons for cranberry (Vaccinium macrocarpon [Ericaceae]). Am. J. Bot. 90: 1425–1432. [DOI] [PubMed] [Google Scholar]
- Castro P., Stafne E. T., Clark J. R., Lewers K. S., 2013. Genetic map of the primocane-fruiting and thornless traits of tetraploid blackberry. Theor. Appl. Genet. 126: 1–12. [DOI] [PubMed] [Google Scholar]
- Chancerel E., Lamy J. B., Lesur I., Noirot C., Klopp C., et al. , 2013. High-density linkage mapping in a pine tree reveals a genomic region associated with inbreeding depression and provides clues to the extent and distribution of meiotic recombination. BMC Biol. 11: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. R., Finn C. E., 2010. Register of new fruit and nut cultivars list 45. HortScience 45: 716–756. [Google Scholar]
- Collard B. C., Mackill D. J., 2008. Marker-assisted selection : an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363: 557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver G. P., Housworth E. A., Stahl F. W., 2002. Crossover interference in Arabidopsis. Genetics 160: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costich D. E., Ortiz R., Meagher T. R., Bruederle L. P., Vorsa N., 1993. Determination of ploidy level and nuclear DNA content in blueberry by flow cytometry. Theor. Appl. Genet. 86: 1001–1006. [DOI] [PubMed] [Google Scholar]
- Covarrubias-Pazaran G., 2016. Genome-assisted prediction of quantitative traits using the R package sommer. PLoS One 11: e0156744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias-Pazaran G., Diaz-Garcia L., Schlautman B., Deutsch J., Salazar W., et al. , 2016. Exploiting genotyping by sequencing to characterize the genomic structure of the American cranberry through high-density linkage mapping. BMC Genomics 17: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J. C. M., 2007. Prediction of response to marker-assisted and genomic selection using selection index theory. J. Anim. Breed. Genet. 124: 331–341. [DOI] [PubMed] [Google Scholar]
- Duarte S., Gregoire S., Singh A. P., Vorsa N., Schaich K., et al. , 2006. Inhibitory effects of cranberry polyphenols on formation and acidogenicity of Streptococcus mutans biofilms. FEMS Microbiol. Lett. 257: 50–56. [DOI] [PubMed] [Google Scholar]
- Eaton G. W., Shawa A. Y., Bowen P. a., 1983. Productivity of individual cranberry uprights in Washington and British Columbia. Sci. Hortic. (Amsterdam) 20: 179–184. [Google Scholar]
- Eck P., 1990. The American Cranberry. Rutgers University Press, New Brunswick, NJ. [Google Scholar]
- Elle E., 1996. Reproductive trade-offs in genetically distinct clones of Vaccinium macrocarpon, the American cranberry. Oecologia 107: 61–70. [DOI] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6(1): e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., Plomion C., 2014. LPmerge: an R package for merging genetic maps by linear programming. Bioinformatics 30: 1623–1624. [DOI] [PubMed] [Google Scholar]
- Fajardo D., Morales J., Zhu H., Steffan S. A., Harbut R., et al. , 2012. Discrimination of American cranberry cultivars and assessment of clonal heterogeneity using microsatellite markers. Plant Mol. Biol. Report. 31: 264–271. [Google Scholar]
- Fajardo D., Senalik D., Ames M., Zhu H., Steffan S. a., et al. , 2013. Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genet. Genomes 9: 489–498. [Google Scholar]
- Fajardo D., Schlautman B., Steffan S., Polashock J., Vorsa N., et al. , 2014. The American cranberry mitochondrial genome reveals the presence of selenocysteine (tRNA-Sec and SECIS) insertion machinery in land plants. Gene 536: 336–343. [DOI] [PubMed] [Google Scholar]
- Feghali K., Feldman M., La V. D., Santos J., Grenier D., 2012. Cranberry proanthocyanidins: natural weapons against periodontal diseases. J. Agric. Food Chem. 60: 5728–5735. [DOI] [PubMed] [Google Scholar]
- Fehr W., 1991. Principles of Cultivar Development. Iowa State University, Ames, IA. [Google Scholar]
- Foo L. Y., Lu Y., Howell A. B., Vorsa N., 2000. A-type proanthocyanidin trimers from cranberry that inhibit adherence of uropathogenic P-fimbriated Escherichia coli. J. Nat. Prod. 63: 1225–1228. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations, 2012 Food and agriculture commodities production. Available at: http://faostat3.fao.org/browse/Q/QC/E. Accessed: October 10, 2016.
- Georgi L., Herai R. H., Vidal R., Carazzolle M. F., Pereira G. G., et al. , 2011. Cranberry microsatellite marker development from assembled next-generation genomic sequence. Mol. Breed. 30: 227–237. [Google Scholar]
- Georgi L., Johnson-Cicalese J., Honig J., Das S. P., Rajah V. D., et al. , 2013. The first genetic map of the American cranberry: exploration of synteny conservation and quantitative trait loci. Theor. Appl. Genet. 126: 673–692. [DOI] [PubMed] [Google Scholar]
- Glaubitz J. C., Casstevens T. M., Lu F., Harriman J., Elshire R. J., et al. , 2014. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9(2): e90346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S., Galleta G., 1971. Comparative chromosome morphology of diploid Vaccinium species. Amer. Soc. Hort. Sci. 96: 289–292. [Google Scholar]
- Hamblin M. T., Warburton M. L., Buckler E. S., 2007. Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS One 2: e1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A. B., Vorsa N., Der Marderosian A., Foo L. Y., 1998. Inhibition of the adherence of P-fimbriated Escherichia coli to uroepithelial-cell surfaces by proanthocyanidin extracts from cranberries. N. Engl. J. Med. 339: 1085–1086. [DOI] [PubMed] [Google Scholar]
- Huang Y. H., Johnson C. E., 1996. A convenient and reliable method to evaluate blueberry pollen viability. HortScience 31: 1235. [Google Scholar]
- Kron K. A., Judd W. S., Stevens P. F., Crayn D. M., Anderberg A. A., et al. , 2002. Phylogenetic classification of Ericaceae: molecular and morphological evidence. Bot. Rev. 68: 335–423. [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limborg M. T., Mckinney G. J., Seeb L. W., Seeb J. E., 2016. Recombination patterns reveal information about centromere location on linkage maps. Mol. Ecol. Resour. 16: 655–661. [DOI] [PubMed] [Google Scholar]
- Lyrene P. M., 2011. First report of Vaccinium arboreum hybrids with cultivated highbush blueberry. HortSci 46: 563–566. [Google Scholar]
- Maliepaard C., Alston F. H., Van Arkel G., Brown L. M., Chevreau E., et al. , 1998. Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theor. Appl. Genet. 97: 60–73. [Google Scholar]
- Martinez-Perez E., Moore G., 2008. To check or not to check? The application of meiotic studies to plant breeding. Curr. Opin. Plant Biol. 11: 222–227. [DOI] [PubMed] [Google Scholar]
- Mckinney G. J., Seeb L. W., Larson W. A., Gomez-Uchida D., Limborg M. T., et al. , 2016. An integrated linkage map reveals candidate genes underlying adaptive variation in Chinook salmon (Oncorhynchus tshawytscha). Mol. Ecol. Resour. 16: 769–783. [DOI] [PubMed] [Google Scholar]
- Ortiz R., Vorsa N., 2004. Transmission of a cyclical translocation in two cranberry cultivars. Hereditas 140: 81–86. [DOI] [PubMed] [Google Scholar]
- Polashock J., Zelzion E., Fajardo D., Zalapa J., Georgi L., et al. , 2014. The American cranberry: first insights into the whole genome of a species adapted to bog habitat. BMC Plant Biol. 14(1): 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S. Y., Davis R. W., 1994. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2015. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Roeder G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621. [DOI] [PubMed] [Google Scholar]
- Roper T. R., Vorsa N., 1997. Cranberry: botany and horticulture, pp. 215–249 in Horticultural Reviews, Vol. 21, edited by Janick J. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- Rowland L. J., Ogden E. L., Bassil N., Buck E. J., McCallum S., et al. , 2014. Construction of a genetic linkage map of an interspecific diploid blueberry population and identification of QTL for chilling requirement and cold hardiness. Mol. Breed. 34: 2033–2048. [Google Scholar]
- Ru S., Main D., Evans K., Peace C., 2015. Current applications, challenges, and perspectives of marker-assisted seedling selection in Rosaceae tree fruit breeding. Tree Genet. Genomes 11(1): 1–12. [Google Scholar]
- Sadok B. I., Celton J. M., Essalouh L., El Aabidine A. Z., Garcia G., et al. , 2013. QTL mapping of flowering and fruiting traits in olive. PLoS One 8(5): e62831 (erratum: PLoS One 9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarracino J. M., Vorsa N., 1991. Self and cross fertility in cranberry. Euphytica 58: 129–136. [Google Scholar]
- Schlautman, B., 2016 Accelerating domestication and genetic gain in the American cranberry (Vaccinium macrocarpon Ait.): new genetic and genomic resources for the cranberry breeders’ toolbox. Ph.D. Thesis, University of Wisconsin-Madison, WI. [Google Scholar]
- Schlautman B., Covarrubias-Pazaran G., Diaz-Garcia L., Johnson-Cicalese J., Iorrizo M., et al. , 2015a Development of a high-density cranberry SSR linkage map for comparative genetic analysis and trait detection. Mol. Breed. 35: 177. [Google Scholar]
- Schlautman B., Fajardo D., Bougie T., Wiesman E., Polashock J., et al. , 2015b Development and validation of 697 novel polymorphic genomic and EST-SSR markers in the American cranberry (Vaccinium macrocarpon Ait.). Molecules 20: 2001–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlautman B., Covarrubias-Pazaran G., Fajardo D., Steffan S., Zalapa J., 2016. Discriminating power of microsatellites in cranberry organelles for taxonomic studies in Vaccinium and Ericaceae. Genet. Resour. Crop Evol. 64: 451–466. [Google Scholar]
- Singh D., Singh K., 2015. Marker-Assisted Plant Breeding: Principles and Practices. Springer, New Delhi, India. [Google Scholar]
- Taylor, J., and D. Butler, 2015 ASMap: linkage map construction using the MSTmap algorithm. R package version 0.4–5. https://CRAN.R-project.org/package=ASMap
- Ting N.-C., Jansen J., Mayes S., Massawe F., Sambanthamurthi R., et al. , 2014. High density SNP and SSR-based genetic maps of two independent oil palm hybrids. BMC Genomics 15: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Kloet S., 1983a The taxonomy of Vaccinium section Cyanococcus: a summation. Can. J. Bot. 61: 256–266. [Google Scholar]
- Vander Kloet S., 1983b The taxonomy of Vaccinium section Oxycoccus. Rhodora 85: 1–43. [Google Scholar]
- Vander Kloet S. P., Avery T. S., 2010. Vaccinium on the edge. Edinb. J. Bot. 67: 7–24. [Google Scholar]
- Van Ooijen J. W., 2006. JoinMap 4: software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen, Netherlands.
- Van Ooijen J. W., 2011. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res. 93: 343–349. [DOI] [PubMed] [Google Scholar]
- Vorsa N., Johnson-Ccicalese J., 2012. Fruit Breeding, edited by Badenes M. L., Byrne D. H. Springer US, Boston, MA. [Google Scholar]
- Vorsa N., Polashock J., 2005. Alteration of anthocyanin glycosylation in cranberry through interspecific hybridization. J. Am. Soc. Hortic. Sci. 130: 711–715. [Google Scholar]
- Wang C., Zhu C., Zhai H., Wan J., 2005. Mapping segregation distortion loci and quantitative trait loci for spikelet sterility in rice (Oryza sativa L.). Genet. Res. 86: 97–106. [DOI] [PubMed] [Google Scholar]
- Wang G., He Q. Q., Xu Z. K., Song R. T., 2012a High segregation distortion in maize B73 x teosinte crosses. Genet. Mol. Res. 11: 693–706. [DOI] [PubMed] [Google Scholar]
- Wang N., Fang L., Xin H., Wang L., Li S., 2012b Construction of a high-density genetic map for grape using next generation restriction-site associated DNA sequencing. BMC Plant Biol. 12: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. A., Bhangoo J., Fernández-Fernández F., Moore P., Swanson J. D., et al. , 2013. Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genomics 14: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E., de Jong H., 2008. Managing meiotic recombination in plant breeding. Trends Plant Sci. 13: 640–646. [DOI] [PubMed] [Google Scholar]
- Wu Y., Bhat P. R., Close T. J., Lonardi S., 2008. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4: e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Li L., Dong X., Jin W., Melchinger A. E., et al. , 2013. Gametophytic and zygotic selection leads to segregation distortion through in vivo induction of a maternal haploid in maize. J. Exp. Bot. 64: 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Wu J., 2015. linkim: linkage information based on genotype imputation method. R package version 0.1. https://CRAN.R-project.org/package=linkim
- Yin T. M., DiFazio S. P., Gunter L. E., Riemenschneider D., Tuskan G. A., 2004. Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor. Appl. Genet. 109: 451–463. [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang S., Li H., Deng Q., Zheng A., et al. , 2010. Effects of missing marker and segregation distortion on QTL mapping in F2 populations. Theor. Appl. Genet. 121: 1071–1082. [DOI] [PubMed] [Google Scholar]
- Zhu H., Senalik D., McCown B. H., Zeldin E. L., Speers J., et al. , 2012. Mining and validation of pyrosequenced simple sequence repeats (SSRs) from American cranberry (Vaccinium macrocarpon Ait.). Theor. Appl. Genet. 124: 87–96. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N., 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S2 contains marker positions in the composite linkage map and the six parental component bin maps. File S3 contains all SNP genotype data for the six parental component bin maps.