Abstract
Investigation of human immunodeficiency virus type 1 (HIV-1) in the genital tract of women is crucial to the development of vaccines and therapies. Previous analyses of HIV-1 in various anatomic sites have documented compartmentalization, with viral sequences from each location that were distinct yet phylogenetically related. Full-length RNA genomes derived from different compartments in the same individual, however, have not yet been studied. Furthermore, although there is evidence that intrapatient recombination may occur frequently, recombinants comprising viruses from different sites within one individual have rarely been documented. We compared full-length HIV-1 RNA sequences in the plasma and female genital tract, focusing on a woman with high HIV-1 RNA loads in each compartment who had been infected heterosexually and then transmitted HIV-1 by the same route. We cloned and sequenced 10 full-length HIV-1 RNA genomes from her genital tract and 10 from her plasma. We also compared viral genomes from the genital tract and plasma of four additional heterosexually infected women, sequencing 164 env and gag clones obtained from the two sites. Four of five women, including the one whose complete viral sequences were determined, displayed compartmentalized HIV-1 genomes. Analyses of full-length, compartmentalized sequences made it possible to document complex intrapatient HIV-1 recombinants that were composed of alternating viral sequences characteristic of each site. These findings demonstrate that the genital tract and blood harbor genetically distinct populations of replicating HIV-1 and provide evidence that recombination between strains from the two compartments contributes to rapid evolution of viral sequence variation in infected individuals.
Worldwide, a large fraction of human immunodeficiency virus type 1 (HIV-1) infections have been transmitted through exposure to virus in the female genital tract. Heterosexual transmission is the major route of HIV-1 infection in most countries (22), with female-to-male transmission occurring as frequently as male to female (47). The female genital tract is also a major site of exposure in perinatal transmission (40). Relatively little is known about HIV-1 in the female genital tract, however, because the vast majority of molecular and detailed virologic analyses to date have focused on virus in the peripheral blood (30). To develop new vaccines and therapeutics, it is necessary to fully characterize HIV-1 derived from the female genital tract as well as the blood (34, 37).
HIV-1 genomes in infected individuals demonstrate marked heterogeneity (5, 19, 27, 29, 54, 59, 62). Even within a single individual, the virus exists as a population of highly related but genetically distinct variants called quasispecies (35). Although much HIV-1 variation can be attributed to the error-prone nature of reverse transcriptase, viral recombination provides the opportunity for evolutionary leaps, with genetic consequences far greater than those resulting from the steady accumulation of individual mutations (35). Recent work suggests that intrapatient HIV-1 recombination may occur very frequently (13, 23, 24, 32), but recombination between quasispecies from different anatomic sites within the same individual has not been studied extensively.
Previous analyses comparing HIV-1 sequences from different tissues documented compartmentalization, the occurrence of distinct yet phylogenetically related HIV-1 genotypes within different anatomic sites. Compartmentalization has been rigorously documented in various different tissues, including the brain, blood and tissue lymphocytes, and plasma (5, 27, 41, 59, 61). HIV-1 sequence heterogeneity has also been observed in several pioneering studies of blood and genital secretions of infected women (18, 24, 39, 45, 51), but complete RNA genomes from the genital tract have not been fully characterized on a molecular level. Study of HIV-1 in the female genital tract has been hampered by the difficulty of cultivating virus from this biologically critical site (28).
We aimed to characterize HIV-1 genomes from the genital tract and plasma of well-characterized women with a spectrum of disease. Because viral RNA represents the currently replicating virus pool more accurately than cell-associated proviral DNA (6, 53), we previously developed an efficient method to clone full-length HIV-1 genomes from viral RNA (10). We used this method to determine complete viral sequences from a woman with advanced disease who had been infected heterosexually and subsequently transmitted HIV-1 by the same route, selecting a subject in whom HIV-1 derived from the genital tract was likely to have played a role in transmission and pathogenesis. By comparing multiple complete HIV-1 RNA genomes from the genital tract and plasma of this subject as well as numerous partial viral genomes from the two sites in several other heterosexually infected women, we focused on HIV-1 variation and recombination within the same individual. Analyses of full-length, compartmentalized sequences made it possible to document complex intrapatient HIV-1 recombinants.
MATERIALS AND METHODS
Study population.
We analyzed HIV-1 sequences from five infected women participating in longitudinal studies of HIV-1 disease. Patients were interviewed and examined, and both blood and gynecologic specimens, including cervicovaginal lavage (CVL), were obtained. The first patient, WC10, was followed at Montreal General Hospital in Montreal, Canada. Her only risk for HIV-1 infection was heterosexual exposure, with no history of intravenous drug use or transfusion. In 1991, she transmitted HIV-1 heterosexually to a male partner. The specimens from WC10 were obtained in October 1996, when she had a history of cutaneous Kaposi's sarcoma affecting the limbs and multiple opportunistic infections, including Pneumocystis carinii pneumonia, cytomegalovirus retinitis, and disseminated Mycobacterium avium complex infection. Kaposi's sarcoma did not involve the genital tract, nor was there evidence of other genital tract infections or inflammation. She had not received antiretroviral therapy at the time that the specimens were collected.
Four additional patients were participants in the Bronx-Manhattan site of the Women's Interagency HIV Study, a multicenter study of the natural history of HIV-1 infection in women (1). All four Women's Interagency HIV Study participants were clinically asymptomatic, and none of them displayed infection or inflammation of the lower genital tract.
The institutional review boards at each clinical site and the New York State Department of Health approved the investigation, and each woman provided informed consent.
Sample preparation and analysis.
Peripheral blood was separated into plasma and cell components (1). CVL was collected by spraying 10 ml of sterile nonbacteriostatic saline against the cervical os and endocervix and aspirating it from the posterior vaginal fornix (28). CVL specimens were examined for the presence of blood with a DiaScreen dipstick to detect hemoglobin and red blood cells (Chronimed, Minneapolis, Minn.) and tested for semen with the SEMA test (Humagen Fertility Diagnostics, Charlottesville, Va.) or the OneStep ABAcard p30 test (Abacus Diagnostics, Inc., West Hills, Calif.). Specimens contaminated by blood or semen were not studied. We quantitated HIV-1 RNA in plasma and CVL with NucliSens (bioMérieux, Durham, N.C.), with a lower limit of quantitation of ≈80 copies/ml. Human genotypes for the CCR5 coreceptor were determined as described previously (43).
Cloning of full-length HIV-1 RNA genomes.
Virion-derived RNA was extracted from plasma and genital tract specimens. We then employed an efficient method to clone full-length HIV-1 genomes directly by reverse transcription and long PCR amplification of genomes as described previously (10, 11, 12). To minimize the likelihood of recombination between molecules during PCR, limiting-dilution reaction conditions were employed (11). In addition, we performed multiple PCRs on each cDNA preparation and then cloned and sequenced a single molecule from each positive reaction to achieve an accurate picture of the predominant genotypes present in the viral population (10, 11, 12, 31). Samples from different patients and from different compartments were also processed, amplified, and sequenced separately to minimize potential cross-contamination or mislabeling.
Full-length genomic sequences were assembled from overlapping 5-kbp clones of the 5′ and 3′ halves of the genome. Assembly required analyzing the sequence of the 700-bp overlap and pairing those clones with identical overlapping sequences. The large overlap makes it likely that the assembled 5′ and 3′ sequences represent the same viral species (10).
Cloning HIV-1 gag and env genes.
Reverse transcription (RT)-PCR was also used to clone the HIV-1 gag and env genes. After amplifying the HIV-1 gag gene by RT-PCR with primers gag651F (5′-GACTTGAAAGCGAAAGTAAAGCC-3′) and gag2335R (5′-GCTCCTGTATCTAATAGAGCTTCC-3′), we constructed panels of full-length gag clones, each approximately 1.7 kbp in size. Panels of full-length env clones were constructed by the method of Singh and Collman (55). To determine HIV-1 coreceptor usage by performing a fusion assay, amplified env genes were also cloned into a mammalian expression vector with the pTarget mammalian vector expression system (Promega). As with amplification and cloning of full-length constructs, measures to ensure high fidelity and representative HIV-1 gag and env clones were performed.
Fusion assay for coreceptor utilization.
HIV-1 coreceptor usage was determined via a cell fusion assay that employs HL3T1 and GHOST cells. The HL3T1 cell line was derived by stable transfection of parental HeLa cells with a chloramphenicol acetyltransferase (CAT) reporter construct (14). The CAT gene is linked to the HIV-1 long terminal repeat promoter so that these cells produce CAT protein only upon introduction of active HIV-1 Tat. HL3T1 cells were transfected with a cloned env gene derived from virion-associated HIV-1 RNA from the patient of interest. The parental GHOST line is a human osteogenic sarcoma cell line stably expressing high levels of CD4 (36). GHOST cells transfected with genes encoding either CCR5 or CXCR4 in addition to CD4 (cell lines GHOST.CCR5 and GHOST.CXCR4, respectively) serve as indicator lines for coreceptor usage (36). GHOST.CCR5 and GHOST.CXCR4 cells were transfected with pSV2tat72, a construct expressing high levels of HIV-1 Tat under the control of the simian virus 40 early promoter.
To initiate fusion, transfected HL3T1 and GHOST.CCR5 or GHOST.CXCR4 cells were mixed in six-well plates at 37°C and allowed to fuse for 48 h. To quantitate fusion, the cells were lysed with 0.5% NP-40, and aliquots of the cell lysates were monitored for CAT production with a commercially available kit (CAT-ELISA; Boehringer Mannheim). All assays were performed in duplicate. HIV-1 JR-FL- and LAV/HTLV-IIIB-derived envelope clones were assayed in parallel as CCR5- and CXCR4-specific positive controls, respectively. Cells that were not transfected were used as negative controls. To control for fusion due to low levels of endogenous coreceptor expression, parental GHOST cells were also assayed in parallel. Experimental results were discarded if any parental GHOST culture yielded fusion or any JR-FL- or LAV/HTLV-IIIB-positive control was negative. If the variance in CAT production level between duplicate assays was greater than 25%, the coreceptor utilization assay for that particular envelope isolate was repeated.
Sequence analyses.
HIV-1 genomes were sequenced by automated fluorescent sequencing in the Wadsworth Center Molecular Genetics Core. Reference sequences were obtained from the HIV sequence database of the Los Alamos National Laboratory (http://hiv-web.lanl.gov).
Computational analyses.
We performed sequence editing and alignment by using the program Bioedit, version 5.0.6, obtained from Tom Hall (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The Phylip 3.5 suite of sequence analysis programs was used for detailed phylogenetic analyses (http://evolution.genetics.washington.edu/phylip.html). Sequences were initially aligned with Clustal W (56) and then checked manually. Gaps and ambiguous sites within the sequence alignment were excluded from all analyses.
Pairwise evolutionary distances were calculated with the Kimura two-parameter model for estimation of distances (25). Phylogenetic trees were constructed by the neighbor-joining method (49). The reliability of each tree was assessed by bootstrap resampling with a minimum of 1,000 replicates (15); interior branch testing (48); and comparison against phylogenetic relationships constructed by the maximum-likelihood method with randomized input order (38). HIV-1 subtypes were determined by phylogenetic analysis, distance plotting, and bootscanning, the latter two with the help of SimPlot v2.5 (33; http://www.med.jhu.edu/deptmed/sray/download). Clones suspected of being recombinants based on phylogenetic analyses were also investigated with SimPlot v2.5, with a window size of 600 bp and a 20-bp sliding step. Each putative recombinant clone was chosen as a query sequence. Representative sequences from the plasma and CVL of the same patient as well as reference sequence were analyzed to determine degree of similarity to the query sequence. We also defined the recombination breakpoints by informative-site analysis as described previously (17, 33).
Aligned CVL- and plasma-derived amino acid sequences were analyzed for distinctive signature sequence variations with VESPA (Viral Epidemiology Signature Pattern Analysis), obtained from Bette Korber of the Los Alamos HIV Sequence Database (26). N-linked glycosylation sites were identified and counted with the Glycosite tool from the Los Alamos HIV sequence database (http://hiv-web.lanl.gov).
Statistical analysis and diversity measurement.
Based upon pairwise analyses and construction of phylogenetic trees supported by bootstrap resampling, the patterns of sequence variation between the genital tract and plasma genomes were classified as follows. Compartmentalization denotes quasispecies from different anatomic sites that are clearly distinct but sufficiently related phylogenetically to suggest evolution from a single ancestor, and intermingled quasispecies are phylogenetically related and distributed in both sites. Bootstrap values greater than 95% were considered significant.
Diversity measurements were determined with the distance matrix constructed for neighbor-joining analysis with Kimura's two-parameter estimation of distances (25), as implemented in the Phylip package with a transition/transversion rate ratio of 2.0. For each patient, the mean pairwise distances within and between compartments were computed and compared with Student's t test.
To perform the informative-site analysis, each gene region in the test sequence was compared to the same gene region in the representative plasma- or CVL-like genome. Exact P values for the differences in the number of sites that clustered with the plasma-like or CVL-like sequences were assessed with Fisher's exact test, a nonparametric test that is powerful in small sample sizes.
Nucleotide sequence accession number.
The sequences described here have been deposited in GenBank under accession numbers AY314044 through AY314188.
RESULTS
Study patients.
To compare HIV-1 replicating in the female genital tract and plasma, we cloned viral RNA genomes obtained from each site contemporaneously and analyzed viral genotypes and phenotypes in each compartment. Plasma and CVL samples were obtained from five HIV-1-infected women whose virologic, immunologic, and clinical characteristics are shown in Table 1. Although the women displayed a broad spectrum of CD4+ cell counts, they were selected because each provided evidence of viral replication in the genital tract. For all five subjects, the viral loads in each site exceeded 50,000 copies/ml, and in most of the women, they were considerably higher (Table 1). All five women reported that their only risk for HIV-1 infection was heterosexual contact. These patients carried the homozygous wild-type genotype for the coreceptor CCR5 (43).
TABLE 1.
Virologic, immunologic, and clinical characteristics of the subjects
| Patient | Race | Age (yr) | Viral level (RNA copies/ml)
|
CD4+ cell count (cells/mm3) | Antiviral therapya | |
|---|---|---|---|---|---|---|
| Plasma | CVL | |||||
| WC10 | Caucasian | 37 | 180,000 | 2,500,000 | 6 | None |
| WC11 | Latina | 31 | 448,415 | 849,825 | 571 | AZT |
| WC12 | Latina | 35 | 426,181 | 53,777 | 221 | AZT |
| WC13 | Latina | 32 | 272,847 | 91,419 | 396 | AZT |
| WC14 | Latina | 39 | 71,618 | 76,507 | 293 | AZT |
AZT, zidovudine.
Phylogenetic and diversity analyses.
We first analyzed viral RNA sequences from a highly instructive patient, WC10. Because WC10 had been infected heterosexually and subsequently transmitted the infection to a male partner, it is likely that HIV-1 derived from her genital tract played a role in HIV-1 transmission and pathogenesis. We cloned and sequenced 10 full-length HIV-1 RNA genomes from her plasma and 10 from her CVL. Detailed computational analyses of sequences from this and other patients focused in particular on genomic variation and coreceptor usage in the two compartments. Initial analyses of open reading frames revealed no significant deletions, alterations, or nonsense mutations. Extensive Blast searches also revealed no evidence of viral contamination of sequences from any of the five patients.
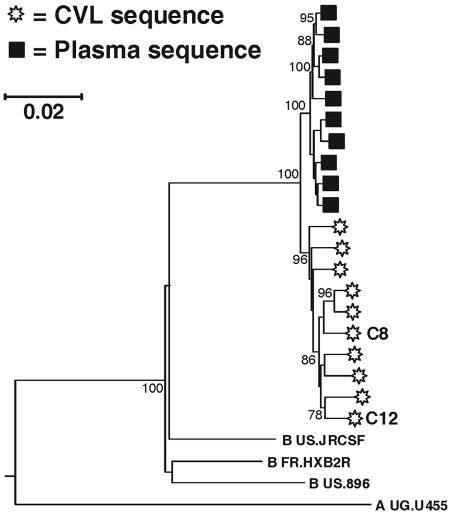
Phylogenetic trees were constructed by using three methods: the neighbor-joining method (depicted in the figures), maximum-likelihood analysis, and the interior branch length test of minimum evolution. All three methods produced similar results in computational analyses of sequences from all five patients examined here (data not shown). Phylogenetic trees revealed that the 20 full-length sequences from WC10 clustered in a single branch when compared to reference sequences (Fig. 1). Analyses also revealed that all 10 genital tract-derived variants formed a tightly clustered group of sequences that were clearly distinct from those found in the plasma. Thus, WC10's HIV-1 sequences were highly compartmentalized; quasispecies from each site were clearly distinct but sufficiently related phylogenetically to suggest evolution from a single ancestor.
FIG. 1.
Phylogenetic tree of complete HIV-1 genomic clones obtained from the blood and genital tract of patient WC10. A consensus tree constructed with the neighbor-joining method is shown, with evolutionary distance calculated with the Kimura two-parameter algorithm (25). Values at key branch nodes indicate the percentage of bootstrap replicates in which the cluster to the right was found (1,000 data sets).
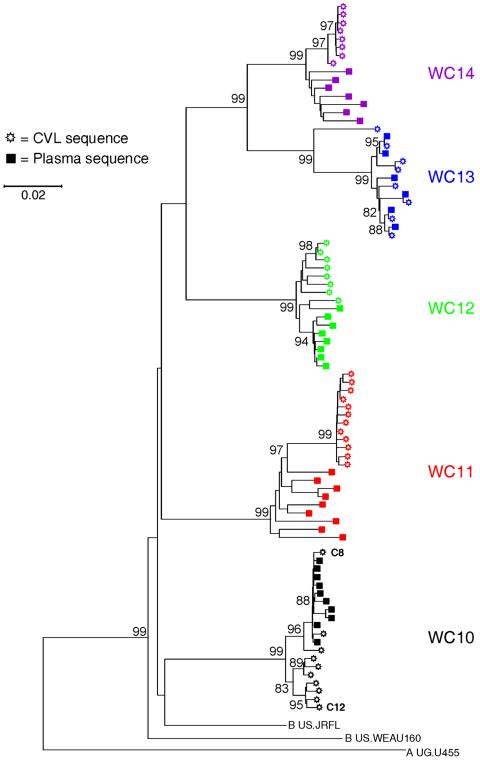
We extended the analyses of viral sequence variation in the two compartments by studying four subjects in addition to patient WC10 (Table 1). Focusing on a 1,250-bp region encompassing the first through fifth variable portions of the gp120 envelope gene (V1 to V5), we compared HIV-1 genomes from the CVL and plasma samples of five patients, including WC10. All 164 env genes were identified as subtype B, and each set of sequences from a single patient clustered as a monophyletic group when compared with those from other women in this study or with the control sequences (Fig. 2).
FIG. 2.
Phylogenetic tree of gp120 envelope clones derived from the blood and genital tract of all patients, constructed with the neighbor-joining method. Bootstrap values of key branch nodes are indicated (1,000 data sets). Different colors are used to distinguish sequences derived from each patient.
Phylogenetic analyses of env sequences from four patients (WC10, WC11, WC12, and WC14) demonstrated compartmentalization (Fig. 2). Sequences from WC14 and WC11 were highly compartmentalized, with all variants derived from the CVL clustering separately on the evolutionary tree from those from plasma. env sequences from patients WC10 and WC12 were compartmentalized to a slightly lesser degree (Fig. 2). gp120 sequences from patient WC13, by contrast, were intermingled; they were phylogenetically related and displayed a uniformly distributed pattern of viral variants between CVL and plasma (Fig. 2).
In the four patients with compartmentalized HIV-1 genomes (WC10, WC11, WC12, and WC14), not only did phylogenetic trees reveal distinct clustering of sequences from CVL and plasma, but the levels of sequence diversity in the env genes from each site differed significantly as well (P < 0.05) (Fig. 2 and Table 2). A comparatively greater level of diversity was detected in either CVL or plasma, depending on which patient was studied. In two women, WC10 and WC12, the CVL sequences displayed higher levels of diversity than those from plasma. In WC11 and WC14, the plasma sequences were significantly more diverse. The full-length sequences derived from WC10's CVL were also significantly more diverse than complete genomes from her plasma (Table 2). The levels of sequence diversity in the two compartments from WC13, the patient with intermingled HIV-1 genomes, by contrast, were very similar. These data suggest that differences in the level of HIV-1 RNA sequence diversity provide additional evidence for compartmentalization of viral replication in each anatomic site.
TABLE 2.
HIV-1 sequence diversity in CVL and plasma
| Sequence | Patient | Pattern of HIV-1 sequence variation | Mean (range) sequence diversitya (%)
|
P | |
|---|---|---|---|---|---|
| Plasma | CVL | ||||
| gp120env | WC10 | Compartmentalized | 0.8 (0.1-1.6) | 1.8 (0.2-2.5) | 0.046b |
| WC11 | Compartmentalized | 3.1 (1.7-4.6) | 0.3 (0.1-0.6) | <0.001b | |
| WC12 | Compartmentalized | 0.8 (0.2-1.8) | 1.6 (0.2-2.6) | <0.01b | |
| WC13 | Intermingled | 1.6 (0.4-2.7) | 1.7 (0.3-2.8) | 0.11 | |
| WC14 | Compartmentalized | 2.8 (0.6-4.9) | 0.3 (0.1-0.5) | <0.001b | |
| Full length | WC10 | Compartmentalized | 0.6 (0.3-0.9) | 1.3 (0.6-2.0) | 0.04b |
Level of diversity among sequences derived from each compartment.
Differences in level of diversity among sequences derived from plasma as compared to those from CVL were statistically significant (P ≤ 0.05).
Although compartmentalization of viral genomes between the plasma and genital tract was seen in four of the five patients, we found no distinctive sequences distinguishing the strains from each anatomic site. Comparison of plasma- and CVL-derived envelope sequences with the aid of VESPA (26) failed to identify a signature pattern characteristic of blood or genital tract strains.
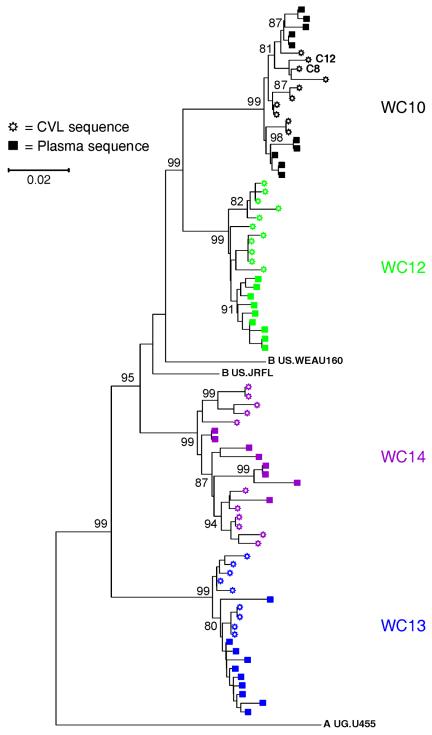
Independent phylogenetic analyses of HIV-1 gag sequences from CVL and plasma were also performed. Figure 3 displays phylogenetic trees constructed of ≈400-bp sequences encompassing the entire open reading frame of p17gag. The gag sequences from patient WC11 were fragmentary and therefore excluded from these analyses. Each set of patient-derived sequences clustered as a monophyletic group. Sequences from the plasma and CVL of patients WC10, WC12, and WC14 demonstrated viral compartmentalization. Although p17 sequences from patient WC13 clustered to a limited degree, extensive computational analyses, including maximum-likelihood analysis, interior branch length testing of minimum evolution trees (data not shown), and statistical analysis of inter- and intracompartment pairwise differences, failed to demonstrate compartmentalized viral populations. Thus, in the patients whose env and gag genes were both analyzed, the same pattern of sequence variation was displayed in both genes: compartmentalization of sequences from patients WC10, WC12, and WC14 and intermingling of sequences from WC13.
FIG. 3.
Phylogenetic analysis of HIV-1 p17gag sequences cloned from the blood and genital tract of patients WC10, WC12, WC13, and WC14. Bootstrap values for key topological clusters are shown (1,000 data sets). Different colors are used to distinguish sequences derived from each patient.
Traits encoded by gp120: HIV-1 coreceptor usage and N-linked glycosylation.
Having documented compartmentalization of env sequences in four patients, we asked whether the traits encoded by gp120 were also compartmentalized. We first investigated whether viral coreceptor usage, a phenotypic trait encoded primarily by the HIV-1 V3 loop, differed in each compartment. Two methods were employed to determine whether an HIV-1 strain used CCR5 (R5 strains) or CXCR4 (X4 strains): prediction of coreceptor usage for each clone based upon the presence of basic or acidic residues at positions 275 and 287 in the V3 portion of the env gene (2, 20) and a phenotypic assay. All 164 V3 loop sequences isolated from the five patients were predicted to use CCR5. Phenotypic analysis of functional gp160 envelope clones derived independently confirmed these predictions. A total of 79 full-length envelope clones were obtained, 5 to 10 from each anatomical site. All clones were biologically functional in a fusion assay to determine coreceptor utilization, and all used CCR5. None of the clones was found to be dual tropic in this phenotypic assay.
Another trait encoded by the env gene, N-linked glycosylation of gp120, has been reported to play a role in viral evolution and persistence by creating a “glycan shield” that helps HIV-1 escape neutralization (60). We therefore compared predicted patterns of N-linked glycosylation of envelope sequences obtained from each anatomical site. No statistically significant differences were seen in any of the patients.
Recombinant HIV-1 genomes.
Because of recent reports suggesting that intrapatient HIV-1 recombination may be frequent in infected individuals (23, 24, 32), we searched for evidence of recombination between strains from different body sites in the same person. Compartmentalization of HIV-1 quasispecies between plasma and CVL made it possible to distinguish sequences derived from each site. To maximize the opportunity of detecting chimeric genomes, we focused on full-length sequences derived from patient WC10, thereby identifying intrapatient recombination between HIV-1 genomes from contemporaneous CVL and plasma samples from the same individual.
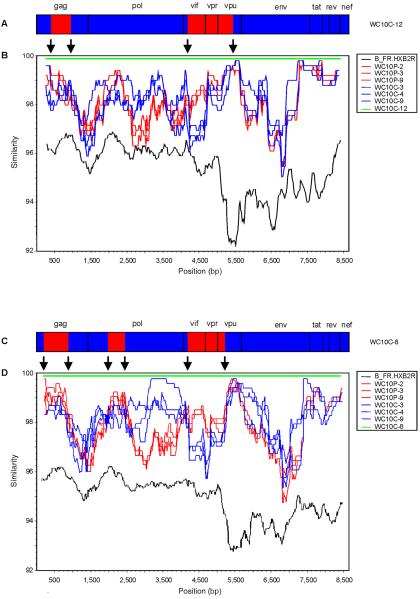
We began our search for hybrid molecules by performing multiple Simplot (33) analyses comparing the 20 complete sequences from WC10's CVL and plasma samples. These analyses identified two putative recombinant sequences, WC10C-8 and WC10C-12, both of which displayed evidence of the occurrence of multiple recombinatorial events involving various HIV-1 genes (Fig. 1 and 4). To map the recombination breakpoints, we next used informative-site analysis. This computational method examines the linear distribution of individual nucleotides, called phylogenetically informative sites, indicating whether each portion of the putative “hybrid” sequence clusters phylogenetically with one of the “parental” subtypes or the other (17, 33). By comparing representative viral genomes from both sites to the sequences of the putative recombinants, informative-site analysis suggested that both WC10C-8 and WC10C-12 were composed of alternating viral sequences characteristic of each site (Table 3).
FIG. 4.
(A) Intrapatient recombination. Open reading frames of the HIV-1 genome relative to the WC10C-12 sequence. Genital tract-like regions are depicted in blue, plasma-like sequences in red. (B) Simplot comparison of clone WC10C-12 with contemporaneous clones derived from CVL and plasma samples from the same patient. Each curve is a comparison of recombinant genome WC10C-12, derived from CVL, to nonrecombinant reference sequences derived from the plasma (red) and genital tract (blue) of the same patient. Similarity plots comparing WC10C-12 to strain HXB2 (black) and itself (green) are included for reference. Recombination breakpoints confirmed by informative-site analysis are indicated with black arrows. (C) Open reading frames of the HIV-1 genome relative to the WC10C-8 sequence. (D) Simplot comparison of clone WC10C-8 with clones derived from the genital tract and plasma of the same patient. Similarity plots comparing WC10C-8 to strain HXB2 and itself are also included.
TABLE 3.
Informative-site analysis of clones derived from CVLa
| Isolate | Regionb (HXB2 numbering) | No. of informative sites
|
|
|---|---|---|---|
| Plasma-like | CVL-like | ||
| WC10C-3 | gag (793-2296) | 1 | 9 |
| (nonrecombinant) | pol (2088-5099) | 3 | 23 |
| Accessory (5044-6302) | 0 | 12 | |
| env (6230-8805) | 0 | 9 | |
| WC10C-8 | gag (793-2296) | 3 | 3 |
| (recombinant) | pol (2088-5099) | 8 | 13 |
| Accessory (5044-6302) | 10 | 8 | |
| env (6230-8805) | 0 | 7 | |
| WC10C-12 | gag (793-2296) | 6 | 1 |
| (recombinant) | pol (2088-5099) | 10 | 28 |
| Accessory (5044-6302) | 9 | 4 | |
| env (6230-8805) | 0 | 9 | |
Distribution of phylogenetically informative sites between plasma and CVL, indicating whether each region of the sequence under analysis clusters phylogenetically with representative sequences from each compartment. The distribution indicates how many informative sites of these clones, all derived from the CVL of WC10, cluster with characteristic genomic sequences obtained from the genital tract and plasma of the same patient.
Accessory refers to the region of the HIV-1 genome encompassing the overlapping rev, tat, vif, vpr, and vpu open reading frames.
Although recombinant clone WC10C-12 was obtained from the CVL, informative-site analysis suggested that the sequence resembled representative CVL strains most closely in the env region, whereas the gag and accessory genes appeared to be derived from the predominant plasma strains (Table 3). To map the recombination breakpoints more precisely, we analyzed the informative sites further, scoring them to look for clear stretches indicative of sequences from each site. When the relative abundance and pattern of these sites were taken into account, this analysis confirmed the pattern seen in the Simplot. As illustrated by the recombinant breakpoints and genomic diagram in Fig. 4A and B, the gag and accessory genes of this chimeric HIV-1 variant were characteristic of contemporaneous plasma strains, although the remaining regions of the genome most resembled strains from the CVL. A phylogenetic analysis of WC10C-8 revealed that it too was a hybrid clone, displaying evidence of multiple recombinational events and a pattern of breakpoints that was similar but not identical to that of WC10C-12 (Fig. 4C and D). Regions of gag, the 5′ portion of pol, and the accessory genes were most characteristic of plasma sequences, with the rest of the viral genome resembling the strains obtained from the CVL samples. Examination of phylogenetically informative sites of the other full-length RNA sequences from patient WC10 did not identify any additional recombinants.
DISCUSSION
Worldwide, a large proportion of HIV-1 infections have been transmitted through exposure to virus in the female genital tract (22), yet the vast majority of detailed analyses of HIV-1 pathogenesis have focused on the blood of men (30). To understand the pathogenesis of HIV-1 and halt its spread both heterosexually and perinatally, it is important to examine the virus in the genital tract of women.
We therefore focused on five women who acquired the infection heterosexually and displayed high levels of HIV-1 RNA in the CVL, with the aim of studying individuals in whom viral replication in the genital tract played a significant role in the initiation and course of infection. One of these women, WC10, not only was infected heterosexually but also transmitted HIV-1 by the same route. To increase our understanding of both HIV-1 compartmentalization and infection of the female genital tract, we compared HIV-1 RNA sequences from the plasma and CVL, focusing on complete viral genomes from patient WC10 and large portions of the viral genome from the others.
Previous studies examining portions of the HIV-1 genome documented viral variants in the genital tract that differed from those in the blood (8, 9, 18, 19, 24, 39, 45, 46). The present study went on to examine 10 full-length viral genomes derived from each anatomic site and found compartmentalization of all the complete sequences. Although the full-length HIV-1 genomes from WC10 were phylogenetically related, all 10 sequences from the CVL constituted a separate branch on an evolutionary tree from those derived from the plasma (Fig. 1). Compartmentalization was also observed in the env and gag genes of four of the five women (Fig. 2 and 3).
Phylogenetic analyses demonstrated not only compartmentalization but also high heterogeneity among the sequences from each patient, with the env genes differing significantly in the two compartments (Table 2). These data provide strong evidence for the presence of distinct populations of HIV-1 replicating in each site. Although we did not discern sequences that conferred different phenotypes on the viruses in each compartment, such differences have been observed (8, 9, 24, 44, 54). The detailed documentation of HIV-1 compartmentalization presented here underscores the potential for distinct, clinically relevant characteristics such as antiretroviral resistance or coreceptor usage in separate, biologically significant compartments.
It is noteworthy that despite the striking compartmentalization of HIV-1 genomes in four of five patients, there was no evidence of particular signature sequences that identified a strain as originating in one compartment or the other. Although the compartmentalized sequences displayed a significantly different level of diversity in each compartment, the plasma sequences were more diverse in two women, whereas the CVL sequences displayed more diversity in the other two. Furthermore, sequences from the two compartments in each patient resembled each other more than they resembled sequences from the same compartment of different individuals.
The forces leading to compartmentalization are not well delineated. Compartmentalization may stem from a founder effect, with seeding of various tissues during primary infection followed by local evolution of viral strains. Selective pressures may also shape distinct viral populations, and previous studies suggested that immune pressures are likely to play a role. One study of sequence variation in different anatomic sites during acute and chronic infection demonstrated that compartmentalization emerged only after infection became chronic, providing evidence that homogeneous viral strains disseminate during primary infection and then diversify under selective pressure in the separate compartments (61). Another study, which examined women with a spectrum of immune states, demonstrated an association of higher CD4+ cell counts and compartmentalization of HIV-1 env sequences, providing evidence that the immune response influences the development of viral genotypes in each compartment (24).
Immunologic studies have revealed significant differences in the frequency of cytotoxic T-lymphocyte epitope variants among HIV-1 quasispecies in different compartments, consistent with the existence of distinct immune pressures in different sites (7, 16, 50, 59). Together these findings support the idea that the compartmentalization of HIV-1 quasispecies in the genital tract versus plasma reflects localized disparities in immune pressure. The next step in this line of research is to determine experimentally the cellular and humoral responses in both the genital tract and blood of patients with various patterns of compartmentalization. Examination of the immune responses in each compartment that target autologous HIV-1 strains is likely to increase our understanding of the containment of HIV-1 infection and may help to identify viral epitopes that elicit protective responses in the genital tract and blood.
Local conditions affecting HIV-1 replication in each site, including cell type, cytokine profile, and inflammation, may exert pressures leading to viral compartmentalization as well (28, 41). We did not, however, detect evidence for local selective pressures such as sexually transmitted infections or differences in HIV-1 coreceptor usage between the two compartments in the patients whom we studied. Antiretroviral therapy may also constitute a powerful selective pressure for compartmentalization (3, 4), but it is unlikely to have played a role in the pattern of sequence variation seen here for a number of reasons: the only treatment was zidovudine, used by four patients; therapy did not correlate with viral compartmentalization; viremia was not suppressed in either compartment; and we observed compartmentalized sequences encoding the gp120 envelope and p17gag proteins, which were not the targets of the therapeutic agent. Antiretroviral therapy, however, is likely to serve as a potent selective pressure for HIV-1 compartmentalization in other patients because treatment has become more complex and widespread. Compartmentalization of drug resistance therefore merits additional examination.
Our examination of HIV-1 coreceptor usage by employing functional env clones did not detect compartmentalization of this phenotypic trait; all clones used CCR5 exclusively. The lack of X4 variants is consistent with the immune status of the patients whom we studied. HIV-1 strains transmitted in vivo generally use CCR5, but later, X4 strains emerge in a proportion of infected individuals (52), often heralding CD4+ cell depletion. X4 viruses are therefore usually seen in patients with low CD4+ cell counts. Four of the five patients studied here, however, had CD4+ cell counts of >200 cells/mm3, so that exclusive R5 coreceptor usage is not surprising in these patients.
This study demonstrated striking genomic compartmentalization in four of five women, but the forces maintaining the distinct viral populations were not wholly successful in containing the distinct viruses in each site. Two of the four women displayed env and gag sequences that were largely but not completely compartmentalized. In addition, we documented two intrapatient recombinants composed of sequences characteristic of each site. These hybrid strains could only have arisen via migration of virus from one compartment to another and coinfection of one cell by viruses originating in the two sites. These findings suggest that the selective pressures maintaining compartmentalization are frequently balanced by those leading to mixing of sequences from different sites. There is evidence that as the immune system loses control of HIV-1 replication in the later stages of HIV-1 infection, it may also lose the ability to influence the evolution of viral strains in the different compartments, permitting intermingling of viral quasispecies (24).
Characterization of highly compartmentalized, full-length sequences made it possible to document intrapatient HIV-1 recombinants composed of alternating sequences from viruses from each anatomic site. Recombination among genetically distinct subtypes of HIV-1 has been recognized with increasing frequency worldwide (13, 17, 33, 42, 57). Elegant molecular studies have demonstrated that intrapatient recombination is very common (23, 32), but it is difficult to demonstrate this phenomenon in infected individuals because most quasispecies within a single person are too similar to allow detection of such hybrid strains. The compartmentalization of multiple viral genes and the large number of full-length sequences derived from patient WC10, however, permitted the identification of two intrapatient recombinants obtained from the CVL.
Although some degree of recombination during both reverse transcription and PCR is inevitable given currently available technology (11), the likelihood that the hybrid clones described here reflect in vivo recombination is high for a number of reasons. The strongest piece of evidence supporting in vivo recombination is the similar pattern of breakpoints for each of the two recombinant clones, as indicated by Simplot and informative-site analyses (Fig. 4 and Table 3). Although the clones were the products of independent PCR amplifications, they displayed breakpoints occurring at nearly the same sites in the viral genome, suggesting that they represent phylogenetically related but not identical quasispecies obtained from patient WC10. Furthermore, a series of control measures were taken to minimize PCR-mediated recombination and contamination, particularly contamination of materials from different compartments (10, 11, 12). These procedures include endpoint dilution of DNA before amplification, controlled reaction conditions, and handling of specimens from different compartments separately during every stage of laboratory analysis. Both the composition of the hybrid molecules and the control measures in the laboratory make it likely that the recombinant clones reflect in vivo recombination.
A recent study of the dynamics of HIV-1 recombination in vitro by Levy et al. made several observations about recombination (32) that were exemplified in vivo by the strains characterized here. First, we identified 2 recombinant HIV-1 genomes out of 20 from WC10, consistent with the finding that recombinant genomes are extremely common in cultured immune cells and the SCID-hu system. Production of recombinants in the model systems was proportional to the infection rate of cells, suggesting that high viral load increases the risk of recombination and viral diversification (32). This finding was also confirmed by studying WC10, who displayed high viral burdens in both compartments, particularly the source of the recombinant clones, the CVL, where the HIV-1 RNA load was 2,500,000 copies/ml. Finally, Levy et al. documented that recombination usually entails multiple recombinational strand transfer events, leading to hybrid viral genomes with multiple breakpoints and an alternating pattern of sequences derived from the two parental strains. The two recombinant strains characterized in this study displayed multiple recombinant breakpoints in different viral genes, leading to alternating stretches of sequence derived from viruses from the CVL and plasma (Fig. 4).
Recombination has tremendous potential to accelerate the formation and dissemination of viral variants within infected individuals (35). Because recombination results in evolutionary leaps, it can rapidly lead to the accumulation of numerous genetic traits that confer a selective advantage on the new viral strain. Viruses bearing multidrug resistance mutations are predicted to arise through recombinatorial events as well as strains that escape the host immune response. The increased risk of recombination as a consequence of high viral load provides an additional reason to suppress HIV-1 replication in infected individuals. To assess the clinical and virologic effects of recombination in vivo, however, it will be necessary to examine the recombinant strains derived from infected individuals for their ability to evade the selective pressures of antiretroviral drugs and immune response.
Acknowledgments
We thank T. Moran, M. Shudt, and the Wadsworth Center Molecular Genetics Core for oligonucleotide synthesis and DNA sequence analysis; D. Bopst, M. Bradley, L. Hirsch, and W. A. Meyer III for technical help and specimen procurement; S. Back, R. Castro, M. Vasco, and W. Gao for obtaining patient data; S. Beck for help preparing the manuscript; and the study patients for their participation. The NIH AIDS Research and Reference Reagent Program provided the GHOST and HL3T1 cell lines, the clones of HIV-1 LAV/HTLVIII and JR-FL, and the pSV2tat72 plasmid construct.
This work was supported by NIH grants U01-AI-35004, R01-AI-42555, and R01AI52015 and a National Research Service Award (1 F32 HD08478-01) to S. Philpott.
REFERENCES
- 1.Anastos, K., S. J. Gange, B. Lau, B. Weiser, R. Detels, J. V. Giorgi, J. Margolick, M. Cohen, J. Phair, S. Melnick, C. R. Rinaldo, A. Kovacs, A. Levine, S. Landesman, M. Young, A. Munoz, and R. M. Greenblatt. 2000. The association of race and gender with HIV-1 RNA levels and immunologic progression. J. Acquired Immune Defic. Syndr. Hum. Retroviruses 24:218-226. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya, D., B. R. Brooks, and L. Callahan. 1996. Positioning of positively charged residues in the V3 loop correlates with HIV type 1 syncytium-inducing phenotype. AIDS Res. Hum. Retroviruses. 12:83-90. [DOI] [PubMed] [Google Scholar]
- 3.Byrnes, V. W., V. V. Sardana, W. A. Schleif, J. H. Condra, J. A. Waterbury, J. A. Wolfgang, W. J. Long, C. L. Schneider, A. J. Schlabach, and B. S. Wolanski. 1993. Comprehensive mutant enzyme and viral variant assessment of human immunodeficiency virus type 1 reverse transcriptase resistance to nonnucleoside inhibitors. Antimicrob. Agents Chemother. 37:1576-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavert, W., D. Notermans, K. Staskus, S. Wietgrefe, M. Zupancic, K. Gebhard, K. Henry, Z. Zhang, R. Mills, H. McDade, C. M. Schuwirth, J. Goudsmit, S. A. Danner, and A. T. Haase. 1997. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science 276:960-964. [DOI] [PubMed] [Google Scholar]
- 5.Cheng-Mayer, C., C. Weiss, D. Seto, and J. A. Levy. 1989. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc. Natl. Acad. Sci. USA 86:8575-8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 7.Crotty, S., B. L. Lohman, F. X. Lu, S. Tang, C. J. Miller, and R. Andino. 1999. Mucosal immunization of cynomolgus macaques with two serotypes of live poliovirus vectors expressing simian immunodeficiency virus antigens: Stimulation of humoral, mucosal, and cellular immunity. J. Virol. 73:9485-9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delwart, E., J. Mullins, P. Gupta, G. Learn Jr., M. Holodniy, D. Katzenstein, B. Walker, and M. Singh. 1998. Human immunodeficiency virus type 1 populations in blood and semen. J. Virol. 72:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellerbrock, T., J. Lennox, K. Clancy, R. Schinazi, T. Wright, M. Pratt-Palmore, T. Evans-Strickfaden, C. Schnell, R. Pai, L. Conley, E. E. Parrish-Kohler, T. J. Bush, K. Tatti, and C. E. Hart. 2001. Cellular replication of human immunodeficiency virus type 1 occurs in vaginal secretions. J. Infect. Dis. 184:28-36. [DOI] [PubMed] [Google Scholar]
- 10.Fang, G., B. Weiser, A. A. Visosky, L. Townsend, and H. Burger. 1996. Molecular cloning of full-length HIV-1 genomes directly from plasma viral RNA. J. Acquired Immune Defic. Syndr. 12:352-357. [DOI] [PubMed] [Google Scholar]
- 11.Fang, G., G. Zhu, H. Burger, J. S. Keithly, and B. Weiser. 1998. Minimizing DNA recombination during long RT-PCR. J. Virol. Methods 76:139-148. [DOI] [PubMed] [Google Scholar]
- 12.Fang, G., H. Burger, C. Chappey, S. Rowland-Jones, A. Visosky, C.-H. Chen, T. Moran, L. Townsend, M. Murray, and B. Weiser. 2001. Analysis of transition from long-term nonprogressive to progressive infection identifies sequences that may attenuate HIV type 1. AIDS Res. Hum. Retroviruses 17:1395-1404. [DOI] [PubMed] [Google Scholar]
- 13.Fang, G., B. Weiser, C. Kuiken, S. M. Philpott, S. Rowland-Jones, F. Plummer, J. Kimani, B. Shi, R. Kaul, J. Bwayo, O. Anzala, and H. Burger. 2004. Recombination following superinfection by HIV-1. AIDS 18:153-159. [DOI] [PubMed] [Google Scholar]
- 14.Felber, B. K., and G. N. Pavlakis. 1988. A quantitative bioassay for HIV-1 based on trans-activation. Science 239:184-187. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 16.Gallichan, W. S., and K. L. Rosenthal. 1996. Long-lived cytotoxic T lymphocyte memory in mucosal tissues after mucosal but not systemic immunization. J. Exp. Med. 184:1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goulston, C., E. Stevens, D. Gallo, J. I. Mullins, C. V. Hanson, and D. Katzenstein. 1996. Human immunodeficiency virus in plasma and genital secretions during the menstrual cycle. J. Infect. Dis. 174:858-861. [DOI] [PubMed] [Google Scholar]
- 19.Gupta, P., C. Leroux, B. K. Patterson, L. Kingsley, C. Rinaldo, M. Ding, Y. Chen, K. Kulka, W. Buchanan, B. McKeon, and R. Montelaro. 2000. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral quasispecies between blood and semen. J. Infect. Dis. 182:79-87. [DOI] [PubMed] [Google Scholar]
- 20.Hung, C.-S., N. Vander Heyden, and L. Ratner. 1999. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J. Virol. 73:8216-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joint United Nations Programme on HIV/AIDS and World Health Organization. 2002. The report on global HIV/AIDS epidemic, “The Barcelona Report.” Joint United Nations Programme on HIV/AIDS, Geneva, Switzerland.
- 23.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 24.Kemal, K. S., B. Foley, H. Burger, K. Anastos, H. Minkoff, C. Kitchen, S. M. Philpott, W. Gao, E. Robison, S. Holman, C. Dehner, S. Beck, W. A. Meyer 3rd, A. Landay, A. Kovacs, J. Bremer, and B. Weiser. 2003. HIV-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc. Natl. Acad. Sci. USA 100:12972-12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 26.Korber, B., and G. Myers. 1992. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res. Hum. Retroviruses 8:1549-1560. [DOI] [PubMed] [Google Scholar]
- 27.Korber, B. T., K. J. Kunstman, B. K. Patterson, M. Furtado, M. M. McEvilly, R. Levy, and S. M. Wolinsky. 1994. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 68:7467-7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovacs, A., S. S. Wasserman, D. Burns, D. J. Wright, J. Cohn, A. Landay, K. Weber, M. Cohen, A. Levine, H. Minkoff, P. Miotti, J. Palefsky, M. Young, and P. Reichelderfer. 2001. Determinants of HIV-1 shedding in the genital tract of women. Lancet 358:1593-1601. [DOI] [PubMed] [Google Scholar]
- 29.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 30.Kuiken, C., B. Foley, E. Freed, B. Hahn, B. Korber, P. A. Marx, F. McCutchan, J. W. Mellors, and S. Wolinksy (ed.). 2002. HIV sequence compendium. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 31.Learn, G. H., Jr., B. T. M. Korber, B. Foley, B. H. Hahn, S. M. Wolinsky, and J. I. Mullins. 1996. Maintaining the integrity of human immunodeficiency virus databases. J. Virol. 70:5720-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy, D. N., G. M. Aldrovandi, O. Kutsch, and G. M. Shaw. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. USA 101:4204-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length Human Immunodeficiency Virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 35.Malim, M. H., and M. Emerman. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469-472. [DOI] [PubMed] [Google Scholar]
- 36.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mostad, S. B., and J. K. Kreiss. 1996. Shedding of HIV-1 in the genital tract. AIDS 10:1305-1315. [DOI] [PubMed] [Google Scholar]
- 38.Olsen, G. J., H. Matsuda, R. Hagstrom, R., and R. Overbeek. 1994. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 39.Overbaugh, J., R. J. Anderson, J. O. Ndinya-Achola, and J. K. Kreiss. 1996. Distinct but related HIV-1 variant populations in genital secretions and blood. AIDS Res. Hum. Retroviruses 12:107-115. [DOI] [PubMed] [Google Scholar]
- 40.Panther, L. A., L. Tucker, C. Xu, R. Tuomala, J. I. Mullins, and D. J. Anderson. 2001. Genital tract human immunodeficiency virus type 1 (HIV-1) shedding and inflammation and HIV-1 env diversity in perinatal HIV-1 transmission. J. Infect. Dis. 181:555-563. [DOI] [PubMed] [Google Scholar]
- 41.Patterson, B., A. Landay, J. Andersson, C. Brown, H. Behbahani, D. Jiyamapa, Z. Burki, D. Stanislawski, M. Czerniewski, and P. Garcia. 1998. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am. J. Pathol. 153:481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peeters, M. 2000. Recombinant HIV sequences: their role in the global epidemic, p. I39-154. In C. L. Kuiken, B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinksy (ed.), HIV sequence compendium 2000. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 43.Philpott, S., B. Weiser, P. Tarwater, S. H. Vermund, C. A. Kleeberger, S. J. Gange, K. Anastos, M. Cohen, R. M. Greenblatt, A. Kovacs, H. Minkoff, M. A. Young, P. Miotti, M. Dupuis, C.-H. Chen, and H. Burger. 2003. CC chemokine receptor 5 genotype and susceptibility to transmission of human immunodeficiency virus type 1 in women. J. Infect. Dis. 187:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ping, L., M. Cohen, I. Hoffman, P. Vernazza, F. Moiseiwitsch, H. Chakraborty, P. Kazembe, D. Zimba, M. Maida, S. Fiscus, J. J. Eron, R. Swanstrom, and J. A. Nelson. 2000. Effects of genital tract inflammation on human immunodeficiency virus type 1 V3 populations in blood and semen. J. Virol. 74:8946-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poss, M., H. L. Martin, J. K. Kreiss, L. Granville, B. Chohan, P. Nyange, K. Mandaliya, and J. Overbaugh. 1995. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with HIV-1. J. Virol. 69:8118-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poss, M., A. Rodrigo, J. Gosink, G. Learn, D. de Vange Panteleeff, H. Martin Jr., J. Bwayo, J. Kreiss, and J. Overbaugh. 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J. Virol. 72:8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinn, T. C., M. J. Wawer, N. Sewankambo, D. Serwadda, C. Li, F. Wabwire-Mangen, M. O. Meehan, T. Lutalo, and R. H. Gray. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 342:921-929. [DOI] [PubMed] [Google Scholar]
- 48.Rzhetsky, A., and M. Nei. 1994. METREE: a program package for inferring and testing minimum-evolution trees. Comput. Appl. Biosci. 10:409-412. [DOI] [PubMed] [Google Scholar]
- 49.Saitou, N., and M. Nei. 1987. The neighbor-joining method: A new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 50.Shacklett, B., S. Cu-Uvin, T. Beadle, C. Pace, N. Fast, S. Donahue, A. Caliendo, T. Flanigan, C. Carpenter, and D. Nixon. 2000. Quantification of HIV-1-specific T-cell responses at the mucosal cervicovaginal surface. AIDS 14:1911-1915. [DOI] [PubMed] [Google Scholar]
- 51.Shaheen, F., A. V. Sison, L. McIntosh, M. Mukhtar, and R. H. Pomerantz. 1999. Analysis of HIV-1 in the cervicovaginal secretions and blood of pregnant and nonpregnant women. J. Hum. Virol. 2:154-166. [PubMed] [Google Scholar]
- 52.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmonds, P., L. Zhang, F. McOmish, P. Balfe, C. Ludlam, and A. J. Brown. 1991. Discontinuous sequence change of human immunodeficiency virus (HIV) type 1 env sequences in plasma viral and lymphocyte-associated proviral populations in vivo: implications for models of HIV pathogenesis. J. Virol. 65:6266-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, A., G. Besson, A. Mobasher, and R. G. Collman. 1999. Patterns of chemokine receptor fusion cofactor utilization by human immunodeficiency virus type 1 variants from the lungs and blood. J. Virol. 73:6680-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh, A., and R. G. Collman. 2000. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J. Virol. 74:10229-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W-improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomson, M. M., L. Perez-Alvarez, and R. Najera. 2002. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect. Dis. 2:461-467. [DOI] [PubMed] [Google Scholar]
- 58.Wolinsky, S. M., B. T. M. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, J. T. Safrit, and R. A. Koup. 1996. Adaptive evolution of HIV-1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 59.Wong, J. K., C. C. Ignacio, F. Torriani, D. Havlir, N. J. Fitch, and D. D. Richman. 1997. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J. Virol. 71:2059-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiping, W., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, L., L. Rowe, T. He, C. Chung, J. Yu, W. Yu, A. Talal, M. Markowitz, and D. Ho. 2002. Compartmentalization of surface envelope glycoprotein of human immunodeficiency virus type 1 during acute and chronic infection. J. Virol. 76:9465-9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, D. A. Cooper, and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zorr, B., A. P. A. Schafer, I. Dilger, K.-O. Habermahl, and M. Kosh. 1994. HIV-1 detection in endocervical swabs and mode of HIV infection. Lancet 343:852. [PubMed] [Google Scholar]