Abstract
Through a process known as RNA interference (RNAi), double-stranded short interfering RNAs (siRNAs) silence gene expression in a sequence-specific manner. Recently, several viral proteins, including the nonstructural protein NSs of tomato spotted wilt virus (a plant-infecting bunyavirus), the interferon antagonist protein NS1 of influenza virus, and the E3L protein of vaccinia virus, have been shown to function as suppressors of RNAi, presumably as a counterdefense against cellular mechanisms that decrease viral production. La Crosse virus (LACV), a member of the California serogroup of orthobunyaviruses, has a trisegmented negative-stranded genome comprised of large (L), medium (M), and small (S) segments. To develop a strategy for segment-specific inhibition of transcription, we designed 13 synthetic siRNAs targeting specific RNA segments of the LACV genome that decreased LACV replication and antigen expression in mammalian (293T) and insect (C6/36) cells. Furthermore, NSs, a LACV nonstructural protein, markedly inhibited RNAi directed both against an LACV M segment construct and against a host gene (glyeraldehyde-3-phosphate dehydrogenase), suggesting a possible role for this viral protein in the suppression of RNA silencing. Segment-specific siRNAs will be useful as a tool to analyze LACV transcription and replication and to obtain recombinant viruses. Additionally, NSs will help us to identify molecular pathways involved in RNAi and further define its role in the innate immune system.
The Bunyaviridae are a large and diverse family of enveloped, negative-stranded RNA viruses divided into the following five genera: Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus (57). The orthobunyaviruses, the first members of the family that were identified, are maintained in nature in a transmission and amplification cycle that alternates between arthropod vectors, predominantly culcine and anopheline mosquitoes, and small mammals. Within the genus Orthobunyavirus, the California serogroup consists of about 14 viruses that are antigenically related to its original member, California encephalitis virus (7), which while described in association with a human infection, has since been seldom isolated, and rarely in relationship to central nervous system disease (20). Human infections by several other members of the California serogroup have been well documented (2, 32, 51, 54, 55, 60), including La Crosse virus (LACV) and Tahyna virus (TAHV), which have been studied most extensively. LACV is an important cause of pediatric encephalitis and aseptic meningitis in the Midwestern United States, where its principal vector, Aedes triseriatus, resides (33, 44, 54). TAHV is associated with an influenza-like illness in central Europe (16, 55).
The bunyavirus genome is composed of three negative-sense RNA segments designated by their size, as follows: large (L), medium (M), and small (S). The L segment encodes an RNA-dependent RNA polymerase (6, 23); the M segment encodes a polyprotein precursor that is posttranslationally cleaved into two envelope glycoproteins, G1 and G2, and a third polypeptide, NSm, of unknown function (24). All three segments of the bunyavirus genome are encapsidated by the S segment-encoded nucleocapsid (N) protein; this segment also encodes the 12-kDa NSs protein in an overlapping reading frame (21, 22, 28). Interestingly, the NSs protein of Bunyamwera virus, the prototypic member of the family and of the genus Orthobunyavirus, has been reported to decrease RNA synthesis in a mini-replicon system (62), to act as an interferon antagonist (3, 38, 52, 61), and to play an important role in viral pathogenesis (3). Although a similar function for the NSs proteins of the California serogroup viruses has not been described to date, a recent report demonstrating sequence homology between Bunyavirus NSs proteins and Reaper, a proapoptotic protein identified in Drosophila melanogaster, suggests a role for LACV NSs in promoting neuronal apoptosis (12), a function that has been previously described for the whole virus (47). Like Reaper, NSs can induce mitochondrial cytochrome c release and caspase activation, further suggesting that NSs and Reaper may be involved in a common mechanism of cell death (12).
Double-stranded short interfering RNAs (siRNAs) have been demonstrated to silence gene expression in a sequence-specific manner through a process known as RNA interference (RNAi). Naturally occurring RNAi is initiated by the double-stranded RNA (dsRNA)-specific endonuclease/helicase Dicer-RDE-1, which cleaves long dsRNA species into 21- to 25-nucleotide fragments called siRNAs (18). siRNAs are incorporated into a protein complex known as the RNA-induced silencing complex, which recognizes and cleaves target mRNAs (18). First described for plants, in which it represents an important mechanism for virus resistance (58), RNAi has been studied intensively in invertebrates and has been shown to function in antiviral defense and development (8, 25, 36, 56). Overall, the building blocks of the RNAi-mediated gene silencing pathway have remarkable similarities in otherwise disparate organisms (11, 37, 58), suggesting an ancient origin of gene silencing in pathogen resistance and organismal development. To counteract the RNA silencing mechanism of their host, plant viruses have developed ways to evade or neutralize RNAi (4, 48, 53, 58). Recently, NSs of the Tomato spotted wilt virus (TSWV), a plant-infecting Bunyavirus of the genus Tospovirus, was shown to suppress RNAi, suggesting a role for this protein in viral pathogenesis (5, 53). While the NSs protein of TSWV is significantly longer than LACV NSs, these proteins share 33.33% identity and 66.7% similarity within a 27-amino-acid overlap according to the Smith-Waterman algorithm for protein sequence and structural similarity.
While the role of RNAi in plants and invertebrates has been related to an innate response to pathogens, its role in mammalian cells is not entirely clear. Nevertheless, siRNAs have been shown to decrease viral replication in human immunodeficiency virus (HIV), influenza virus, hepatitis C virus, and several other viral infections (27, 34, 35, 43). Here we describe 13 siRNAs that target individual segments of the tripartite LACV genome and that inhibit virus replication in both human and insect cells. These siRNAs may be used as a tool to study the mechanism of orthobunyavirus replication, as a novel method for creating reassortants or pseudotypes, or potentially to develop single-segment reverse genetics. Furthermore, we describe a role for the LACV NSs protein in the suppression of RNAi, a function shared by several viral proteins, including NS1 of influenza virus, the E3L protein of vaccinia virus, NSs of TSWV, and B2 of flock house virus (5, 40, 41, 53). This study reinforces the potential physiologic role of siRNAs in viral resistance in animal cells and suggests that NSs may be important in bunyavirus pathogenesis by, among other effects, counteracting the cellular response to viral transcription.
MATERIALS AND METHODS
siRNA transfection.
LACV is a negative-strand RNA virus in which the viral genome (− strand) is transcribed into a positive-strand mRNA that serves as a template for the synthesis of viral protein and a cRNA that is the template for the generation of additional viral RNAs. siRNAs were designed for either the genomic RNA (negative strand) or the antigenomic RNA (positive strand) of LACV by use of the Dharmacon (Lafayette, Colo.) siRNA design center, which selects optimal siRNA sequences based on the Tuschl rules, an empirical algorithm that predicts siRNA sequences that are likely to be effective for gene silencing (19). While either the negative- or positive-strand sequence was used to design effective siRNAs, it is unclear which strand is targeted in this system because of the complementarity of the siRNA duplex (27). All LACV siRNAs were supplied in a 2′-deprotected, annealed, and desalted form with 3′ dTdT overhangs (Dharmacon). LACV siRNAs (Table 1) were transfected into a human embryonic kidney cell line, 293T, and an epithelial cell-like mosquito cell line, C6/36. The 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Grand Island, N.Y.) enriched with 10% heat-inactivated fetal calf serum (FCS) (Atlanta Biologicals, Norcross, Ga.) and 2 mM l-glutamine (Gibco). The C6/36 cells were maintained at 28°C in 5% CO2 in minimal essential medium (Gibco) supplemented with nonessential amino acids (Gibco), 1.5 g of sodium bicarbonate/liter, 1 mM sodium pyruvate (BioWhittaker, Walkersville, Md.), and 10% heat-inactivated FCS (Atlanta Biologicals). Antibiotic-free medium was used for the duration of the siRNA experiments.
TABLE 1.
Change in LACV PFU/ml in siRNA-versus mock-transfected 293T cells
| siRNAb | Target sequence | Target segment | Fold change at 48 ha | Strand used for siRNA design |
|---|---|---|---|---|
| L2436* | AAGACACAACCACUUGCGGA | L | −15 | + |
| L4782* | AAUAUACGGGAGUCCUCAACG | L | −40.5 | + |
| L1949* | AAUCAUGUAGCGUGCUGGCU | L | −58.6 | − |
| L785 | AAAUCCACUAGCCAGGAAA | L | −1.8 | − |
| M209* | AAUGGCUAGUCUCUGAUUG | M (G2) | −38.9 | + |
| M459* | UCAAACUUGCGAACACCUU | M (G2) | −48.9 | + |
| M312* | AAUCUGCGCUGCAAACAUAU | M (G2) | −50.7 | + |
| M941* | UUACUGCGGUACUGGUCUU | M (G2) | −53 | + |
| M2843* | AAGCACCCGCAAUAUCAUCU | M (G1) | −47.9 | − |
| M2860* | AACUUGCCGCACAUCAAACC | M (G1) | −142 | − |
| M1566* | AAUUUCCGUACUAGAUGUCCC | M (G1) | −51.5 | + |
| S272* | AAAUUUGGAGAGUGGCAGGUGG | S | −53.1 | + |
| S103* | UCCUAACUGCAGCAAGGUU | S | −6,800 | − |
| S528* | AAGGCAACGCUAUGGCACUCU | S | −30.9 | + |
| S528c | AAGGCAACGGUAUGGCACUCU | S | 1.8 | + |
| S652 | AACCUAUAACUGCUUAGUGU | S | 2.3 | − |
(LACV MOI = 0.0001).
*, statistically significant decrease in virus titer compared to mock-transfected cells (P < 0.01; Student's t test).
Approximately 24 h before transfection, 8 × 104 293T cells or C6/36 cells were plated in a 24-well plate in a volume of 500 μl/well. Chemically synthesized siRNAs (Dharmacon) were mixed with the amine siPORT (293T) or lipid siPORT (C6/36) siRNA transfection reagent (Ambion, Austin, Tex.) and Opti-MEM (Gibco) per the kit's instructions for 30 min before being added to the cells at a concentration of 100 nM siRNA per well. Forty-eight hours after siRNA transfection, the cells were infected for 1 h with either the LACV/original (54) or TAHV/Bardos 92 (42) strain at a multiplicity of infection (MOI) of 0.01 to 0.0001 PFU/cell in DMEM supplemented with 3% FCS, after which the virus was removed and the cells were resuspended in growth medium. Cells and cell culture supernatants were harvested at 12, 24, 48, and 72 h postinfection. All experiments were performed in triplicate.
Plaque assays.
Plaque assays for LACV and TAHV were performed in Vero cells, an African green monkey kidney cell line. Vero cells were plated overnight at 3 × 105 cells/well in a six-well plate, and serial dilutions of cell culture supernatants (10−2 to 10−6) and uninfected control supernatants were applied in a volume of 500 μl for 1 h. The diluted culture supernatants were then removed, and a 1:1 mixture of 2% sea plaque agarose (Cambrex, Rockland, Maine) and 2× DMEM (Gibco) containing 4% FCS was added. Vero cells were incubated for 72 h at 32°C in 10% CO2 before the agarose overlay was removed. Plaques were then visualized by staining with 0.1% crystal violet.
Flow cytometry.
293T and C6/36 cells were harvested and fixed for 30 min in a 2% paraformaldehyde solution. Nonspecific binding was reduced by exposure to 10% goat serum (Sigma, St. Louis, Mo.) for 30 min and three washes with fluorescence-activated cell sorter (FACS) staining buffer (phosphate-buffered saline with 1% FCS and 0.1% sodium azide) prior to incubation with a 1:500 dilution of mouse immunoglobulin G (IgG) specific for either LACV G1 (807.31, 807.33, 813.13, or 807.35) or TAHV G1 (813.48) (31) for 30 min at room temperature. For each experiment, an aliquot of cells was also treated with isotype-matched control antibodies to establish background staining. The cells were then washed three times with FACS staining buffer and incubated at room temperature with a 1:100 dilution of fluorescein isothiocyanate-conjugated anti-mouse IgG (Sigma) for 30 min. After incubation with the fluorescein isothiocyanate-conjugated secondary antibody, the cells were washed three times with FACS staining buffer and resuspended in 400 μl of FACS staining buffer before acquisition and analysis on a FACScalibur instrument (Becton Dickinson, Sunnyvale, Calif.) using CellQuest software (Becton Dickinson) (University of Pennsylvania Cancer Center).
Total RNA extraction and sequencing of virus harvested at 72 h postinfection of siRNA-pretreated cells.
Total RNAs were isolated from 293T cells according to the instructions in the Rneasy handbook (Qiagen, Santa Clarita, Calif.). Approximately 1 μg of total RNA was reverse transcribed to cDNA by the use of random hexamers and the SuperScript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, Calif.) per the manufacturer's instructions. The resulting cDNA was used for sequencing (Cell Center, DNA Sequencing Facility, Department of Genetics, University of Pennsylvania).
Molecular cloning of LACV NSs.
The LACV S segment was PCR amplified from a pRB322 vector containing a complete double-stranded DNA copy of the LACV/original virus S segment (pLAC 4C-26) (6) and was then inserted into pGEM7Z by the use of Vent DNA polymerase. Briefly, specific primers for the 5′ and 3′ ends of the S segment (XhoI-LACS [GCGCTCGAGATTAGTGTATCCACTTGAATACTTTGA] and HindIII-LACS [GCGAAGCTTAGTAGTGTGCTCCACTGAATACATTTTA ], respectively; underlining indicates restriction sites) were used to amplify the S segment. The resulting fragment was digested with both XhoI and HindIII, inserted into a linearized pGEM7Z vector (Promega, Madison, Wis.), and subsequently sequenced by use of the following primers: LACS79F (5′-GTGATGTCGGATTTGGTG), LACS270R (5′-AGGGTTAGCCTTCCTCTCTGG), LACS370F (5′-GGGTATTTAGCCAGATGG), and LACS652F (5′-GCAGATAAGTGGATGTCAC). La Crosse NSs clones were amplified by PCRs using rTth DNA polymerase XL (PE Biosystems, Foster City, Calif.), with the pGEM-S segment as a template and with specific primers to introduce restriction enzyme cut sites at the 5′ and 3′ ends of NSs. Briefly, NSs was amplified by PCRs with thermal cycling conditions consisting of a 2-min denaturing step at 94°C followed by 30 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min and an extension step of 72°C for 10 min. The forward primer NSsBgl II-F was complementary to the 5′ end of NSs and contained a BglII restriction site (underlined) (GGAAGATCTTCCGATTTGGTGTTTTATGATGTCGCAT). The reverse primer NSsEcoRI-R corresponded to the 3′ end and contained an EcoRI restriction site (underlined) (GGGGAATTCGGGATCTGGCCAAATACCCAGATA). The resulting fragment was digested with both BglII and EcoRI and inserted into linearized pIRES2-EGFP (BD Biosciences, San Diego, Calif.) that had been digested with BglII and EcoRI. In addition to cloning NSs into pIRES2-EGFP in the correct orientation, we also cloned it into pIRES2-EGFP in the reverse orientation by PCR amplification with the forward primer NSsEcoRI-F, corresponding to the 5′ end of NSs and containing an EcoRI restriction site (underlined) (GGGGAATTCGGGGATTTGGTGTTTTATGATGTCGCAT), and the reverse primer NssBglII-R, corresponding to the 3′ end of NSs and containing a BglII restriction site (underlined) (GGAAGATCTTCCCATTCTCGTTATACTGATCAAGGACCC).
To detect NSs expression in the absence of a LACV NSs-specific antibody, we engineered a vector to express an NSs-FLAG fusion protein. The LACV NSs was cloned into p3XFLAG-CMV-13 (Sigma) in frame with the 3× FLAG sequence by PCR amplification with primers corresponding to the 5′ and 3′ ends of NSs and containing restriction sites for EcoRI and BglII. Two primer sets were used to clone NSs into two different sites in the multiple cloning region of p3XFLAG-CMV-13. Primer set A consisted of NSsEcoRI-F (described above) and NSsBglII-R2 (GGAAGATCTTCCATCTGGCGCAATACCCAGATA). NSsBglII-R2 was used to eliminate the NSs stop codon and to add an alanine residue (double underline), allowing the translation of NSs and the FLAG epitope. Successful transformation of the 3X-FLAG vector with the LACV NSs was confirmed by sequencing.
Molecular cloning of the LACV M segment (G1/G2) into pCAGGS.
The LACV M segment open reading frame (ORF) was subcloned from pcMORF (44) into the expression vector pCAGGS (kindly provided by Andrew Pekosz). The LACV M ORF was amplified by PCR from pcMORF (pcDNA3.1-LACV M) by use of a primer set that introduced restriction cut sites at both the 5′ and 3′ ends. Briefly, the LACV M ORF was amplified by the use of rTth DNA polymerase XL (PE Biosystems). The PCR cycle consisted of a 2-min denaturing step at 93°C followed by 35 cycles of 93°C for 30 s, 48°C for 1 min, and 72°C for 6 min and an extension step of 72°C for 10 min. The forward primer LAC-ClaIF was complementary to the 5′ end and contained a ClaI restriction site (CCATCGATGGCCAAAGATGATTTGTATATTGGTGC) (underlined), and the reverse primer LAC-XhoIR corresponded to the 3′ end and contained an XhoI restriction site (CCGCTCGAGCGGCTATCTAATTTTCATCTCTCTCTTATATGC) (underlined). The resulting fragment was digested with both ClaI and XhoI and inserted into the linearized pCAGGS expression vector digested with ClaI and XhoI. Transient cotransfection of NSs-FLAG or bacterial alkaline phosphatase (BAP)-FLAG (Sigma) and the LACV M segment mRNA-derived construct was achieved by calcium phosphate transfection (Promega) of 1 μg of DNA per well of a 24-well plate.
Western blotting.
Western blot analysis was performed after the cells were harvested by centrifugation at 600 × g for 10 min. The cell supernatants were discarded, and the pellets were lysed in 150 μl of immunoprecipitation assay buffer (50 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 50 mM Tris [pH 7.4], 1 mM EGTA, 10 mM sodium orthovanadate, 10 mM NaF, and a protease inhibitor cocktail [one tablet per 50 ml]) for 10 min at 4°C. Each sample was then centrifuged at 14,000 × g for 10 min at 4°C. After a rapid freeze (−80°C)-thaw cycle, protein levels in the whole-cell lysates were determined (DC protein assay; Bio-Rad, Hercules, Calif.). The lysates were then mixed with an equal amount of 2× sample buffer (0.125 M Tris, 4% sodium dodecyl sulfate, 20% [vol/vol] glycerol, 0.01% [wt/vol] bromophenol blue, 10% 2-mercaptoethanol) and boiled for 5 min. Ten micrograms of each cell lysate was resolved by gel electrophoresis with an 8 to 16% Tris-glycine gel (Cambrex) and then transferred to a 0.2-μm-pore-size nitrocellulose membrane (Bio-Rad). Western blot analysis for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed with anti-GAPDH (1:3,000) rabbit IgG (Abcam, Cambridge, United Kingdom), and the protein was visualized by incubation with horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG (Amersham Biosciences, Piscataway, N.J.) at a 1:10,000 dilution and subsequent detection by an enhanced chemiluminescence assay (SuperSignalWest Pico chemiluminescence substrate; Pierce, Rockford, Ill.). Western blot analysis for β-actin was performed with anti-β-actin (1:3,000) goat IgG (Santa Cruz Biotechnology, Santa Cruz, Calif.), and the protein was visualized with HRP-conjugated mouse anti-goat IgG at a dilution of 1:10,000 (Santa Cruz Biotechnology). Western blot analysis for FLAG was performed with 0.1 μg of anti-FLAG antibody (Sigma)/ml, and the protein was visualized by incubation with HRP-conjugated rabbit anti-mouse IgG (Sigma) at a 1:10,000 dilution and detection by an enhanced chemiluminescence assay.
NSs inhibition of RNAi.
NSs and control constructs were transfected into 293T cells (1.0 μg of DNA/well) 24 h prior to treatment with either a GAPDH or LACV M siRNA. As controls, 293T cells were also (i) mock DNA transfected, (ii) transfected with the 3X-FLAG vector alone, and (iii) transfected with a control FLAG fusion protein, bacterial alkaline phosphatase (BAP-FLAG). The silencing of GAPDH was assessed by Western blotting and TaqMan quantitative PCR. LACV glycoprotein expression was measured in cells that were cotransfected with the LACV M segment construct by FACS analysis with a mixture of LACV G1-specific antibodies (807.31, 807.33, 813.13, and 807.35).
Real-time PCR detection of GAPDH.
RNAs were extracted from NSs-FLAG-, BAP-FLAG-, and mock-transfected 293T cells 72 h after GAPDH siRNA treatment. Total RNAs were isolated from cells by use of an Rneasy kit (Qiagen) and reverse transcribed to cDNA by the use of random hexamers and the SuperScript first-strand synthesis system (Invitrogen). Quantitative real-time PCRs were performed on an ABI Prism 770 sequence detection system (Perkin-Elmer, Foster City, Calif.). The amplification protocol followed the instructions of a TaqMan Gold RT-PCR kit. A normalization experiment was performed simultaneously for each sample with 18S RNA primers and probe. CT (threshold) values were calculated for the target (GAPDH CT) and the standard (18S RNA CT) by determining the points at which the fluorescence exceeded a threshold limit (10 times the standard deviation of the baseline). The results are expressed as ΔCT (mean GAPDH CT − mean 18S RNA CT) (9).
RESULTS
Inhibition of LACV infection by use of siRNAs.
RNAi can be induced by the introduction of synthetic ≈21- to 23-nucleotide siRNA duplexes into cells, bypassing the requirement for processing of a long dsRNA mediated by Dicer-RDE-1 (45). These siRNAs confer transient interference of gene expression in a sequence-specific manner and are generally thought to be too short to induce an alpha/beta interferon response in mammalian cells (39, 45). 293T cells were pretreated with siRNAs targeting the LACV L, M, and S segments (Table 1). Of 15 siRNAs synthesized, 13 reduced LACV replication (>35% inhibition, as measured by the virus titer, 48 h after infection at an MOI of 0.0001 PFU/cell) (Table 1). Twelve of the siRNAs were specific for LACV; one siRNA (M2860) also inhibited replication of the closely related TAHV (data not shown).
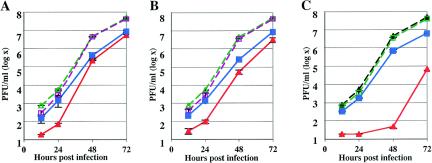
LACV and TAHV growth curves were generated for mammalian and insect cell lines (293T and C6/36 cells, respectively), with and without prior siRNA transfections. siRNAs targeting the LACV L, M, and S segments were transfected into cells prior to infection with LACV or TAHV, and the efficacies of the siRNAs were assessed by a plaque assay 12 to 72 h after infection (Fig. 1A and B). S103, an siRNA targeting the S segment and the most effective siRNA of the panel generated, inhibited LACV up to 6,800-fold in both 293T and C6/36 cells. All siRNAs that inhibited LACV were effective in both 293T and C6/36 cells, indicating that siRNA inhibition of LACV is equally potent in mammalian and insect cells. Moreover, LACV-directed siRNAs decreased LACV replication regardless of whether the L, M, or S segment was targeted.
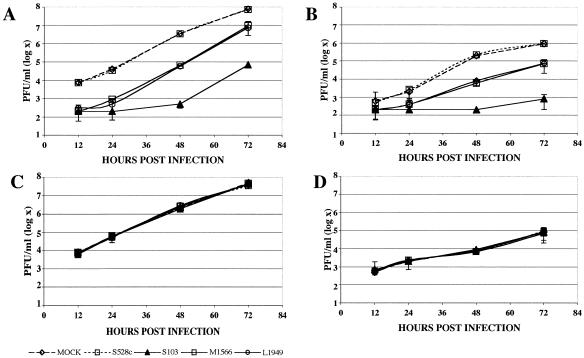
FIG. 1.
Specific inhibition of LACV replication. 293T and C6/36 cells were pretreated with LACV L, S, and M segment siRNAs. LACV (A and B) and TAHV (C and D) replication was measured in cell culture supernatants obtained from 293T (A and C) and C6/36 (B and D) cells 12, 24, 48, and 72 h after infection with LACV or TAHV at an MOI of 0.0001 PFU/cell. This representative experiment demonstrates virus replication in mock-transfected cells, cells that were pretreated with LAC528c (a control for the LACV S segment), and cells that were treated with LACS103, LACM1566, and LACL1949 siRNAs, designed for the S, M, and L segments of LACV, respectively. Error bars represent the variabilities (standard errors of the means [SEM]) between triplicate samples for each time point. TAHV replication was not inhibited by pretreatment with LACV siRNAs, except for LACM2860, which inhibited TAHV replication 138-fold at 48 h postinfection of 293T cells (data not shown).
For most of the siRNAs, maximal inhibition occurred at 48 h, with a trend toward a rebound of virus replication at 72 h (Fig. 1). No inhibition of LACV infection was observed with a control siRNA designed to target the LACV S segment (LACS528c) that had a 1-bp nucleotide substitution (Fig. 1A and B) or with an siRNA targeting GAPDH (data not shown). With one exception, TAHV replication was not inhibited by pretreatment with LACV siRNAs in either 293T or C6/36 cells. The outlier siRNA, LACM2860, inhibited TAHV replication ≈138-fold 48 h after infection in 293T cells and ≈110-fold in C6/36 cells, although its sequence is not present in the TAHV strain used for these experiments.
To determine the effect of different MOIs on inhibition by siRNAs, we also performed plaque assays with supernatants from cultures that were infected with LACV at MOIs ranging from 0.01 to 0.0001. Whereas the pretreatment of 293T cells with LACV siRNAs resulted in 99% or more inhibition of virus replication at a low MOI (0.0001), the efficiency decreased markedly when the transfected cells were challenged with increasingly higher concentrations of virus (Fig. 2). To determine if siRNAs could clear the virus when they were delivered after virus infection and viral gene expression, we infected 293T and C6/36 cells with LACV at an MOI of 0.0001 PFU/cell and transfected them with LACV L1949, S103, and M1566 segment siRNAs or control siRNAs (LAC528c and LACM1566c) at 24 h postinfection. While these siRNAs were highly effective at decreasing LACV replication when they were used before virus infection, the ability of these siRNAs to inhibit LACV replication was significantly impaired when they were delivered after virus infection (Fig. 3). The best-performing siRNAs for the LACV S and M segments decreased virus replication when they were applied after virus infection and viral gene expression, albeit to a far lesser extent than when they were transfected prior to LACV infection. The LACV L segment siRNA that we tested did not significantly decrease virus replication when it was transfected after LACV infection (Fig. 3).
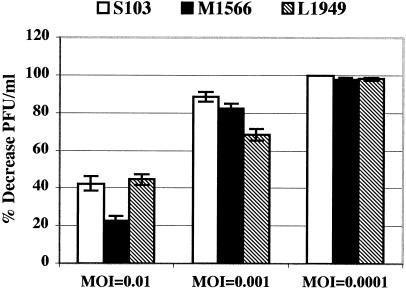
FIG. 2.
siRNA-mediated inhibition of LACV replication. The percent decrease in LACV titer in LACV siRNA-treated cells was compared to that in mock-transfected cells at 48 h for infections with various MOIs (0.01, 0.001, and 0.0001 PFU/cell). The pretreatment of 293T cells with LACV siRNAs resulted in an up to 99% inhibition of virus replication when the cells were challenged with LACV at a low MOI (0.0001). The efficiency of LACV siRNAs decreased as cells were challenged with increasing MOIs (0.001 and 0.01). Error bars represent variabilities (SEM) between triplicate samples for each time point.
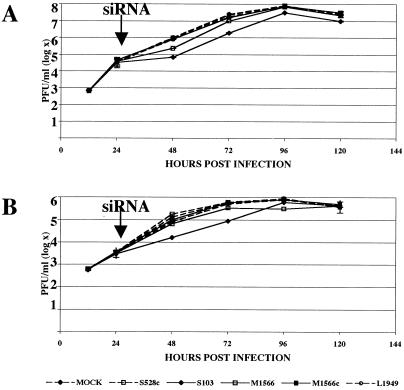
FIG. 3.
Inhibition of LACV replication is reduced when siRNAs are delivered after virus infection. 293T (A) and C6/36 cells (B) were infected with LACV at an MOI of 0.0001 PFU/cell and were transfected 24 h later (arrow) with LACV L1949, S103, and M1566 segment siRNAs or control siRNAs (LAC528c and LACM1566c). Error bars represent variabilities (SEM) between triplicate samples for each time point.
To confirm that the inhibition of viral replication in siRNA-pretreated cells was due to a decrease in gene expression, we measured antigen accumulation in 293T and C6/36 cells by FACS with a mixture of LACV G1-specific monoclonal antibodies (807.31, 807.33, 813.13, and 807.35) 12, 24, and 48 h after siRNA transfection (31). Glycoprotein expression was significantly reduced in both 293T and C6/36 cells that were pretreated with 13 of the 15 siRNAs designed for the LACV L, M, and S segments. Figure 4 shows the results for two siRNAs (LACM1566 and LACS103). The control siRNA 528c did not affect glycoprotein expression, and with the exception of M2860, the LACV siRNAs did not decrease TAHV glycoprotein expression in either 293T or C6/36 cells (data not shown). LACV siRNAs also did not inhibit GAPDH expression (data not shown), indicating that the effects of LACV siRNAs are indeed highly specific.
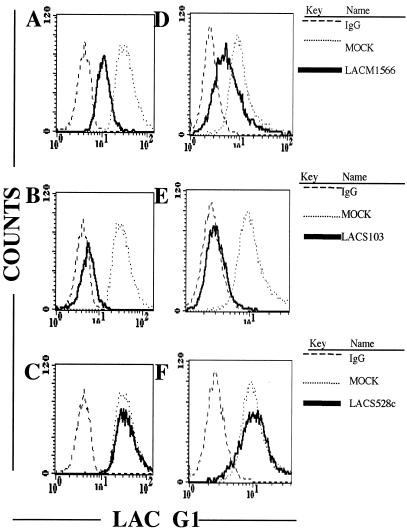
FIG. 4.
LACV glycoprotein expression is decreased in 293T cells that are pretreated with LACV siRNAs. LACV antigen accumulation in 293T (A to C) and C6/36 (D to F) cells was measured by FACS with a panel of LACV G1-specific antibodies 48 h after siRNA transfection. As shown in these representative plots, LACV-specific siRNAs, including M1627 (A and D) and S356 (B and E), inhibited LACV G1 expression. The LACV control siRNA 528c (C and F) did not affect LACV glycoprotein expression in 293T cells. LACV siRNAs did not decrease TAHV glycoprotein expression in either 293T or C6/36 cells, with the exception of M2860 (data not shown).
siRNA-resistant viruses.
Others have reported that RNAi-resistant viruses can emerge in siRNA-treated cells by either mutation or deletion of the siRNA-targeted sequence (14, 29, 30). To determine whether RNAi-resistant viruses emerged at points after infection when inhibition was no longer evident, we used supernatants harvested 72 h after LACV infection of 293T cells that had been pretreated with LACV siRNAs to infect a second round of cells that were pretreated with the same LACV siRNAs (Fig. 5). Assays were performed after infection with either LACV alone or LACV cultured for 72 h in 293T cells that were pretreated with the LACM1566 (Fig. 5A), LACL1949 (Fig. 5B), or LACS103 (Fig. 5C) siRNA. A slight increase (approximately 1 log) in LACV titer was observed for viruses that were treated with a siRNA and had been previously exposed to the same inhibitor (LACM1566 [Fig. 5A] and LACL1949 [Fig. 5B]) compared with wild-type LACV (Fig. 5A and B). However, the differences were negligible at later time points. In contrast, the virus that was previously exposed to LACS103 demonstrated a highly significant (Student's t test; P < 0.01) increase in titer, particularly at later time points (Fig. 5C). When the M and S segments from virus supernatants harvested 72 h after LACM1566, LACS103, or mock transfection were sequenced, both wild-type LACV and viruses with single or double nucleotide changes were observed (Table 2). At least 30 cDNA clones were sequenced for each group. Significantly fewer (χ2 = 10.8; P = 0.01) wild-type S segment sequences (26.6%) were observed for S segment clones obtained from siRNA LACS103-treated cells than for wild-type S segment clones (66.6%) derived from mock-transfected cells (Table 2). While fewer wild-type LACV M segment clones were obtained from the siRNA LACM1566-pretreated progeny virus (38.7%) than from mock-transfected cells (61.3%), this trend did not reach statistical significance (χ2 = 3.84; P = 0.1).
FIG. 5.
RNAi resistance of LACV. Supernatants harvested 72 h after LACV infection of 293T cells that had been pretreated with LACV siRNAs were used to infect a second round of 293T cells that were pretreated with the same LACV siRNAs (LACM1566 [A], LACL1949 [B], and LACS103 [C]). Plaque assays were performed 12, 24, 48, and 72 h after infection with either LACV or LACV cultured for 72 h in 293T cells that were pretreated with LACM1566. In all instances, green lines represent mock siRNA-treated wild-type LACV while red lines represent LACV siRNA-treated wild-type LACV (A), LACL1949 (B), or LACS103 (C). (A) Green, mock siRNA-treated wild-type LACV; purple, mock siRNA-treated, LACM1566-pretreated LACV; red, LACM1566 siRNA-treated wild-type LACV; blue, LACM1566 siRNA-treated, LACM1566-pretreated LACV. (B) Green, mock siRNA-treated LACV; purple, mock siRNA-treated, LACL1949-pretreated LACV; red, LACL1949 siRNA-treated LACV; blue, LACL1949 siRNA-treated, LACL1949-pretreated LACV. (C) Green, mock siRNA-treated LACV; purple, mock siRNA-treated, LACS103-pretreated LACV; red, LACS103 siRNA-treated LACV; blue, LACS103 siRNA-treated, LACS103-pretreated LACV. A significant (P < 0.01 by Student's t test) increase in titer was observed at 12 to 72 h for viruses that were previously exposed to LACS103 compared to naïve, wild-type LACV.
TABLE 2.
LACV clones recovered at 72 hours postinfection of LACV siRNA-pretreated 293T cells
| siRNA | mRNA target (+ strand)a | Mutation | No. of wild-type or mutant clones/total no. of clones sequenced (%) |
|---|---|---|---|
| S103 | TCACTCAACCTTGCTGCAGTTAGGATCTTCTT | LAC wild type | 8/30 (26.6)b |
| TCACTCAACTTTGCTGCAGTTAGGATCTTCTT | Thr→Thr (NSs), Leu→Phe (N) | 5/30 (16.6) | |
| TCACTCAACCTTGCTGCAATTAGGATCTTCTT | Gln→Gln (NSs), Val→Ile (N) | 6/30 (20) | |
| TCACTCAACCTTACTGCAATTAGGATCTTCTT | Leu→Leu (NSs), Aln→Thr (N) | 1/30 (3.3) | |
| TCACTCAACCTTGCAGCAGTTAGGATCTTCTT | Leu→Gln (NSs), Aln→Aln(N) | 8/30 (26.6) | |
| TCACTCAGCCTTGCTGCAGTTAGGATCTTCTT | Thr→Ala (NSs), Asn→Ser | 1/30 (3.3) | |
| TCACTCAACCTGTCTGCAGGTTAGTCTTCTT | Frame shift (NSs, N) | 1/30 (3.3) | |
| Mock | TCACTCAACCTTGCTGCAGTTAGGATCTTCTT | LAC wild type | 22/33 (66.6) |
| TCACTCAACCTTGCAGCAGTTAGGATCTTCTT | Leu→Gln (NSs), Aln→Aln(N) | 7/33 (21.2) | |
| TCACTCAACCTTGCTGCAATTAGGATCTTCTT | Gln→Gln (NSs), Val→Ile (N) | 4/33 (12.1) | |
| M1566 | AAGGCAATTTCCGTACTAGATGTCCCTATAA | LAC wild type | 12/31 (38.7)c |
| AAGGCAATTACCGTACTAGATGTCCCTATAA | Ser→Thr (G1) | 2/31 (6.5) | |
| AAGGCAATTTCCGTGCTAGATGTCCCTATAA | Val→Val (G1) | 5/31 (16.1) | |
| AAGGCAATTTCCGTACTGGATGTCCCTATAA | Leu→Leu (G1) | 6/31 (19.4) | |
| AAGGCAATTTCCGTACTGGATGTTCCTATAA | Leu→Leu and Val→Val (G1) | 3/31 (9.6) | |
| AAGGCAATTTCCGTACTAGATGAACCTATAA | Val→Glu (G1) | 2/31 (6.5) | |
| AAGGCGGTTTCCGTACTAGATGTCCCTATAA | Ala→Ala and ile→Val (G1) | 1/31 (3.2) | |
| Mock | AAGGCAATTTCCGTACTAGATGTCCCTATAA | LAC wild type | 19/31 (61.3) |
| M1566 | AAGGCAATTTCCGTGCTAGATGTCCCTATAA | Val→Val (G1) | 5/31 (16.1) |
| AAGGCAATTTCCGTACTGGATGTCCCTATAA | Leu→Leu (G1) | 4/31 (12.9) | |
| AAGGCAATTACCGTACTAGATGTCCCTATAA | Ser→Thr (G1) | 2/31 (6.5) | |
| AAGGCAATTTCCGTACTAGATGAACCTATAA | Val→Glu (G1) | 1/31 (3.2) |
Bold letters indicate the target (positions 103 to 122 for the S segment and positions 1627 to 1646 for the M segment). Underlining indicates mutations.
Significantly fewer wild-type LACV clones were obtained from LAC S103 siRNA-pretreated cells than from mock-transfected cells (χ2 = 10.8, P = 0.01).
Fewer wild-type LACV clones were obtained from LACM1566 siRNA-pretreated cells than from mock-transfected cells. However, this trend did not reach statistical significance (χ2 = 3.84, P = 0.10).
NSs inhibition of RNAi.
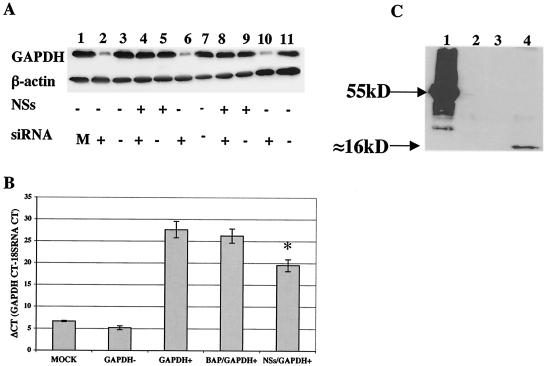
The NSs protein of TSWV, a Bunyavirus of the genus Tospovirus, was shown to suppress RNA silencing (5, 53). To determine whether a similar function could be associated with LACV NSs, we cloned the gene for this protein into a pIRES2-EGFP vector in both the correct (NSs-CO) and the opposite (NSs-WO) orientation and into a 3X-FLAG vector in the correct orientation (NSs-FLAG). These NSs constructs were transfected into 293T cells (1.0 μg of DNA/well), and interference with silencing of the endogenous gene GAPDH was determined. As controls, 293T cells were also (i) mock transfected, (ii) transfected with the 3X-FLAG vector alone, and (iii) transfected with a control FLAG fusion protein, BAP-FLAG. Silencing of GAPDH was observed for 293T cells that were transfected with a GAPDH siRNA (GAPDH+) compared with mock-transfected cells or cells that were transfected with a negative control for the GAPDH siRNA (GAPDH−) (Fig. 6). GAPDH silencing was observed for 293T cells that were transfected with a vector containing NSs in the incorrect (opposite) orientation (NSs-WO) or with the 3X-FLAG vector. In contrast, GAPDH expression was not decreased in 293T cells that were transfected with NSs in the correct orientation (NSs-CO) or with the NSs-FLAG fusion protein. Similar results were observed with NIH 3T3 cells (data not shown). The failure of the GAPDH+ siRNA to decrease the expression of GAPDH in the presence of NSs suggests that the NSs protein of LACV may function in part as an RNA silencing suppressor.
FIG. 6.
NSs inhibition of GAPDH RNAi. (A) Expression of NSs FLAG in 293T cells and inhibition of RNA silencing. LACV NSs-FLAG (1.0 μg of DNA/well) was transfected into 293T cells by use of the ProFection mammalian calcium phosphate transfection system (Promega) 24 h prior to GAPDH and control siRNA treatment. GAPDH expression in 293T cells was assessed 72 h after treatment with GAPDH siRNA (+) and controls, as measured by Western blotting (see Materials and Methods). Effective silencing of GAPDH was observed for 293T cells that were transfected with a GAPDH siRNA (+) (Ambion) (lane 2) compared to mock-transfected cells (lane 1) or cells that were transfected with a negative control for GAPDH siRNA (−) (lane 3) (Ambion). A similar inhibition of GAPDH expression was observed for 293T cells that were transfected with a vector containing NSs in the wrong orientation (NSs-WO; lane 6) or with the BAP-FLAG vector (lane 10). GAPDH expression was not silenced in 293T cells that were transfected with NSs in the correct orientation (NSs-Co; lane 4) or with the NSs-FLAG fusion protein (lane 8). (B) Quantitative real-time TaqMan PCR was used to examine the effects of GAPDH siRNA on GAPDH expression. RNAs were extracted from NSs-FLAG-, BAP-FLAG-, and mock-transfected 293T cells 72 h after GAPDH siRNA treatment. Quantitative real-time PCRs for GAPDH were performed on an ABI Prism 770 sequence detection system (Perkin-Elmer). A normalization 18S RNA experiment was performed simultaneously for each sample by the use of 18S RNA primers and probe. CT values were calculated for the target (GAPDH CT) and the standard (18S RNA CT), and the results are expressed as ΔCTs (mean GAPDH CT − mean 18S RNA CT). High CT values are equated with low copy numbers. Therefore, low ΔCTs correlate with high GAPDH expression levels and high ΔCTs correlate with low GAPDH expression levels. GAPDH mRNA expression was significantly reduced in 293T cells that were transfected with the GAPDH siRNA (GAPDH+) compared to mock-transfected cells and cells that were transfected with a GAPDH siRNA negative control (GAPDH−). In addition, GAPDH mRNA was significantly increased in GAPDH+ siRNA-transfected 293T cells that were cotransfected with NSs-FLAG compared to mock- and BAP-FLAG-transfected 293T cells (asterisk; P < 0.01 by ANOVA). (C) BAP-FLAG and NSs-FLAG expression. As controls, 293T cells were mock transfected (lane 2), transfected with the 3X-FLAG vector alone (lane 3), and transfected with a control FLAG fusion protein, BAP-FLAG (lane 1). With an anti-FLAG monoclonal antibody, NSs FLAG was detected (lane 4) at approximately 16 kDa, the predicted size for the NSs-FLAG fusion protein.
To confirm the results observed at the protein level, we used quantitative real-time TaqMan PCR to examine the effects of the GAPDH siRNA on GAPDH mRNA expression. RNAs were extracted from NSs-FLAG-, BAP-FLAG-, and mock-transfected 293T cells 72 h after GAPDH siRNA treatment. Quantitative real-time PCRs for GAPDH were performed in conjunction with normalization for 18S RNA. Cycle threshold (CT) values were calculated for the target (GAPDH CT) and the standard (18S RNA CT), and the results were analyzed as ΔCTs (mean GAPDH CT − mean 18S RNA CT). Because high CT values are equated with low copy numbers, low ΔCTs correlate with high GAPDH expression levels and high ΔCTs correlate with low GAPDH expression levels. GAPDH mRNA expression was significantly reduced in 293T cells that were transfected with the GAPDH siRNA (GAPDH+) compared to mock-transfected cells and cells transfected with a GAPDH siRNA negative control (GAPDH−). In addition, the GAPDH mRNA level was significantly increased in GAPDH+ siRNA-transfected 293T cells that were cotransfected with NSs-FLAG compared to mock- and BAP-FLAG-transfected 293T cells (P < 0.01 by analysis of variance [ANOVA]). The increased GAPDH mRNA expression in GAPDH siRNA-treated cells in the presence of NSs-FLAG supports a role for LACV NSs as a suppressor of RNAi.
We next examined the ability of NSs to inhibit RNAi of the LACV G1 glycoprotein. 293T cells that had been pretreated with the LACM1566 siRNA or a scrambled LACM1566 control siRNA (AACCGTTTCATTAGAGTTCCC) were cotransfected with the LACV M segment (G1/G2) and either NSs-FLAG or the BAP-FLAG control vector (Fig. 7A). LACV glycoprotein expression was measured by FACS with a mixture of LACV G1-specific antibodies (807.31, 807.33, 813.13, and 807.35). Transfection with NSs-FLAG and BAP-FLAG did not affect LACV glycoprotein expression in LACM (G1/G2)-cotransfected cells (Fig. 7A). LACV glycoprotein expression was also not inhibited significantly in 293T cells that were pretreated with the M segment control siRNA (Fig. 7B). As expected, LACV glycoprotein expression was downregulated in mock-transfected or BAP-FLAG-cotransfected 293T cells that were pretreated with the LACM1566 siRNA (Fig. 7C). However, LACV M expression was partially recovered in 293T cells that were cotransfected with NSs (Fig. 7C). This experiment was performed on three separate occasions, and the data were analyzed as percentages of maximum expression of the mean fluorescence intensities (Fig. 7D). LACV glycoprotein expression was significantly increased in 293T cells that were pretreated with the LACM1566 siRNA and transfected with NSs-FLAG compared with BAP-FLAG-transfected 293T cells or cells that were transfected only with the siRNA (ANOVA; P < 0.05 compared to BAP-FLAG- and mock-transfected cells) (Fig. 7D). The inability of the LACM1566 siRNA to maximally inhibit LACV glycoprotein expression in the presence of NSs supports the potential role of NSs as an RNA silencing suppressor. However, NSs-FLAG did not affect siRNA inhibition of LACV infection itself when NSs was transfected into cells prior to siRNA transfection and LACV infection (data not shown), indicating that the maximal NSs effect may occur in infected cells that express this protein.
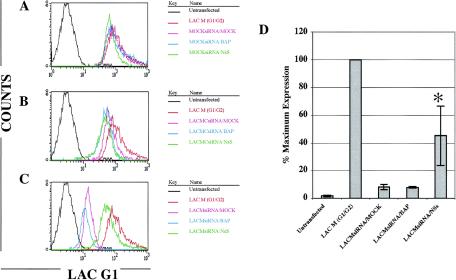
FIG. 7.
Inhibition of LACV M segment RNAi by NSs. 293T cells were transfected with either NSs-FLAG or BAP-FLAG, were treated 24 h later with the LACM1566 siRNA, and were cotransfected with the LACV M segment (G1/G2). LACV glycoprotein expression was measured by FACS with a cocktail of LACV G1-specific antibodies (807.31, 807.33, 813.13, and 807.35). Representative FACS plots from one of three experiments are shown (A to C). (A) Transfection with NSs-FLAG and BAP-FLAG did not affect LACV glycoprotein expression in LACM (G1/G2)-cotransfected cells. (B) LACV glycoprotein expression was not inhibited significantly in 293T cells that were pretreated with an M segment control siRNA. (C) LACV glycoprotein expression was downregulated in mock-transfected or BAP-FLAG-cotransfected 293T cells that were pretreated with the LACM1566 siRNA. Importantly, LACM expression was partially recovered in 293T cells that were cotransfected with NSs. (D) This experiment was performed on three separate occasions, and the data were analyzed as percentages of maximum expression of the mean fluorescence intensities. LACV glycoprotein expression was significantly increased in NSs-FLAG-transfected 293T cells that were treated with the LACM1566 siRNA compared to mock- and BAP-FLAG-transfected 293T cells (asterisk; P < 0.05 by ANOVA).
DISCUSSION
We have demonstrated that siRNAs targeting the L, M, and S segments of LACV inhibit virus replication for up to 72 h after infection. A siRNA treatment directed against the M segment was associated with diminished expression of the G1 glycoprotein, reinforcing the proposed mechanism for its effects. Collectively, these results indicate that siRNAs targeting the LACV genome may be used to confer intracellular immunization. More importantly, these findings suggest that siRNAs may be a useful tool for studying the mechanisms of LACV replication and pathogenesis and, at least theoretically, for the creation of orthobunyavirus segment-specific genome reassortants. Moreover, the ability to induce RNAi of LACV replication in both human and insect cells indicates that these LACV siRNAs may be exploited to study arthropod-borne transmission and may potentially be used for prophylaxis and therapy of LACV infections. The reduced susceptibility of LACV to a second round of siRNA treatment and the emergence of RNAi-resistant viral isolates after only 72 h in culture may have resulted from mutant viruses present in the initial swarm inoculum or from the acquisition of mutations during siRNA treatment (14, 30) and will make any therapeutic applications challenging. However, when similar patterns of RNAi-resistant viruses were described for poliovirus and for HIV, it was suggested that a successful approach to siRNA-mediated antiviral therapies for RNA viruses will need to include multiple siRNAs, a strategy that is theoretically possible given our results (14, 29, 30, 34).
We propose that LACV segment-specific siRNAs may be adapted for protocols for reverse genetics, a strategy used to engineer specific mutations into viral genomes and for the generation of recombinant viruses. If successful, the use of siRNAs in reverse genetics for the genus Orthobunyavirus will be less cumbersome than the approaches that are currently available (26, 46). Moreover, the use of siRNAs in reverse genetics can be applied to other viruses with segmented RNA genomes, such as influenza virus.
Antiviral RNA silencing has been demonstrated for both plants and insects as a natural defense mechanism for antiviral protection (1, 10, 13, 40). Plant viruses have been shown to counteract RNA silencing defense systems (5, 17, 48, 50, 53, 59). Tospoviruses have life cycles in both plants and arthropod vectors (thrips) (63), and thus these bunyaviruses confront RNA silencing defense mechanisms of both plant and insect hosts (4, 49). Likewise, arboviruses that replicate in both vertebrates and invertebrates, such as the orthobunyaviruses, may need to compensate for both the complex immune systems of the vertebrate host and the RNA silencing defense mechanisms of the insect vector. Here we describe the apparent diminution of siRNA-mediated silencing of GAPDH and LACM (G1/G2) by LACV NSs. The ability to inhibit suppression of both a host (GAPDH) and a viral (LACM) gene in 293T cells suggests that the NSs of LACV may function in part as an RNA silencing suppressor.
Recently, it was demonstrated that the interferon antagonist proteins of influenza virus and vaccinia virus, named NS1 and E3L, respectively, are also suppressors of RNAi (40, 41). These suppressors of RNAi may function by binding and sequestering dsRNA. A similar mechanism has been implicated for the p19 protein of tombusvirus, which also functions as an RNA silencing suppressor (50). There is little structural or sequence similarity between these proteins (5, 15, 41). However, the suppression of RNAi by both E3L and NS1 requires the N-terminal dsRNA binding domain, which is also required for the inhibition of innate antiviral immunity (64). Therefore, it cannot be ruled out that the inhibition of RNAi is unrelated to the targeting of innate antiviral immunity. Accordingly, LACV NSs-mediated inhibition of silencing may be due to a direct effect or to indirect effects related to its putative function as an interferon antagonist, a function demonstrated for the NSs protein of the closely related Bunyamwera virus (3, 38, 52, 61). The ability of LACV NSs to suppress RNAi in mammalian cells adds to the growing body of evidence suggesting that RNAi also plays an antiviral role in mammalian cells, but its capacity to bind and sequester dsRNA and to act as an interferon antagonist has yet to be determined. Future studies are required to determine the mechanisms of suppression of RNAi by LACV NSs and whether the interferon system is involved.
Acknowledgments
This work was supported by Public Health Service grants NS-30606 and NS-07180 (training grant for S.S.S.). M.K.M. was funded by NIH grant T32 GM-07229.
We thank Julio Martin-Garcia, Robin Vos, and other members of the González-Scarano laboratory for their many helpful comments and suggestions. In addition, we thank Karen Stachelek and Thomas Harvey for their technical help.
REFERENCES
- 1.Al-Kaff, N. S., S. N. Covey, M. M. Kreike, A. M. Page, R. Pinder, and P. J. Dale. 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113-2115. [DOI] [PubMed] [Google Scholar]
- 2.Arunagiri, C. K., L. P. Perera, S. B. Abeykoon, and J. S. Peiris. 1991. A serologic study of California serogroup bunyaviruses in Sri Lanka. Am. J. Trop. Med. Hyg. 45:377-382. [DOI] [PubMed] [Google Scholar]
- 3.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Bucher, E., T. Sijen, P. De Haan, R. Goldbach, and M. Prins. 2003. Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J. Virol. 77:1329-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabradilla, C. D., Jr., B. P. Holloway, and J. F. Obijeski. 1983. Molecular cloning and sequencing of the La Crosse virus S RNA. Virology 128:463-468. [DOI] [PubMed] [Google Scholar]
- 7.Calisher, C. H. 1996. History, classification, and taxonomy of viruses in the family Bunyaviridae, p. 1-17. In R. M. Elliot (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 8.Caplen, N. J., Z. Zheng, B. Falgout, and R. A. Morgan. 2002. Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference. Mol. Ther. 6:243-251. [DOI] [PubMed] [Google Scholar]
- 9.Carcelain, G., R. Tubiana, A. Samri, V. Calvez, C. Delaugerre, H. Agut, C. Katlama, and B. Autran. 2001. Transient mobilization of human immunodeficiency virus (HIV)-specific CD4 T-helper cells fails to control virus rebounds during intermittent antiretroviral therapy in chronic HIV type 1 infection. J. Virol. 75:234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrington, J. C., and S. A. Whitham. 1998. Viral invasion and host defense: strategies and counter-strategies. Curr. Opin. Plant Biol. 1:336-341. [DOI] [PubMed] [Google Scholar]
- 11.Cogoni, C., and G. Macino. 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10:638-643. [DOI] [PubMed] [Google Scholar]
- 12.Colon-Ramos, D. A., P. M. Irusta, E. C. Gan, M. R. Olson, J. Song, R. I. Morimoto, R. M. Elliott, M. Lombard, R. Hollingsworth, J. M. Hardwick, G. K. Smith, and S. Kornbluth. 2003. Inhibition of translation and induction of apoptosis by bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol. Biol. Cell 14:4162-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covey, S. N., and N. S. Al-Kaff. 2000. Plant DNA viruses and gene silencing. Plant Mol. Biol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 14.Das, A. T., T. R. Brummelkamp, E. M. Westerhout, M. Vink, M. Madiredjo, R. Bernards, and B. Berkhout. 2004. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 78:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delgadillo, M. O., P. Saenz, B. Salvador, J. A. Garcia, and C. Simon-Mateo. 2004. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J. Gen. Virol. 85:993-999. [DOI] [PubMed] [Google Scholar]
- 16.Demikhov, V. G., V. G. Chaitsev, A. M. Butenko, M. S. Nedyalkova, and T. N. Morozova. 1991. California serogroup virus infections in the Ryazan region of the USSR. Am. J. Trop. Med. Hyg. 45:371-376. [DOI] [PubMed] [Google Scholar]
- 17.Dunoyer, P., S. Pfeffer, C. Fritsch, O. Hemmer, O. Voinnet, and K. E. Richards. 2002. Identification, subcellular localization and some properties of a cysteine-rich suppressor of gene silencing encoded by peanut clump virus. Plant J. 29:555-567. [DOI] [PubMed] [Google Scholar]
- 18.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir, S. M., J. Martinez, A. Patkaniowska, W. Lendeckel, and T. Tuschl. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eldridge, B. F., C. Glaser, R. E. Pedrin, and R. E. Chiles. 2001. The first reported case of California encephalitis in more than 50 years. Emerg. Infect. Dis. 7:451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott, R. M. 1989. Nucleotide sequence analysis of the small (S) RNA segment of Bunyamwera virus, the prototype of the family Bunyaviridae. J. Gen. Virol. 70:1281-1285. [DOI] [PubMed] [Google Scholar]
- 22.Elliott, R. M., and A. McGregor. 1989. Nucleotide sequence and expression of the small (S) RNA segment of Maguari bunyavirus. Virology 171:516-524. [DOI] [PubMed] [Google Scholar]
- 23.Endres, M. J., D. R. Jacoby, R. S. Janssen, F. Gonzalez-Scarano, and N. Nathanson. 1989. The large viral RNA segment of California serogroup bunyaviruses encodes the large viral protein. J. Gen. Virol. 70:223-228. [DOI] [PubMed] [Google Scholar]
- 24.Fazakerley, J. K., F. Gonzalez-Scarano, J. Strickler, B. Dietzschold, F. Karush, and N. Nathanson. 1988. Organization of the middle RNA segment of snowshoe hare Bunyavirus. Virology 167:422-432. [PubMed] [Google Scholar]
- 25.Fjose, A., S. Ellingsen, A. Wargelius, and H. C. Seo. 2001. RNA interference: mechanisms and applications. Biotechnol. Annu. Rev. 7:31-57. [DOI] [PubMed] [Google Scholar]
- 26.Flick, R., K. Flick, H. Feldmann, and F. Elgh. 2003. Reverse genetics for Crimean-Congo hemorrhagic fever virus. J. Virol. 77:5997-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge, Q., M. T. McManus, T. Nguyen, C. H. Shen, P. A. Sharp, H. N. Eisen, and J. Chen. 2003. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. USA 100:2718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentsch, J. R., and D. H. Bishop. 1978. Small viral RNA segment of bunyaviruses codes for viral nucleocapsid protein. J. Virol. 28:417-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gitlin, L., and R. Andino. 2003. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J. Virol. 77:7159-7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Scarano, F., R. E. Shope, C. E. Calisher, and N. Nathanson. 1982. Characterization of monoclonal antibodies against the G1 and N proteins of LaCrosse and Tahyna, two California serogroup bunyaviruses. Virology 120:42-53. [DOI] [PubMed] [Google Scholar]
- 32.Grimstad, P. R., C. L. Shabino, C. H. Calisher, and R. J. Waldman. 1982. A case of encephalitis in a human associated with a serologic rise to Jamestown Canyon virus. Am. J. Trop. Med. Hyg. 31:1238-1244. [DOI] [PubMed] [Google Scholar]
- 33.Griot, C., F. Gonzalez-Scarano, and N. Nathanson. 1993. Molecular determinants of the virulence and infectivity of California serogroup bunyaviruses. Annu. Rev. Microbiol. 47:117-138. [DOI] [PubMed] [Google Scholar]
- 34.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapadia, S. B., A. Brideau-Andersen, and F. V. Chisari. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA 100:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennerdell, J. R., and R. W. Carthew. 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 18:896-898. [DOI] [PubMed] [Google Scholar]
- 37.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohl, A., R. F. Clayton, F. Weber, A. Bridgen, R. E. Randall, and R. M. Elliott. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77:7999-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar, M., and G. G. Carmichael. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62:1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 41.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malkova, D. 1971. Virulence of Tahyna virus in mice and its relation to thermosensitivity and character of plaque population markers. Acta Virol. 15:473-478. [PubMed] [Google Scholar]
- 43.McCown, M., M. S. Diamond, and A. Pekosz. 2003. The utility of siRNA transcripts produced by RNA polymerase in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology 313:514-524. [DOI] [PubMed] [Google Scholar]
- 44.McJunkin, J. E., E. C. de los Reyes, J. E. Irazuzta, M. J. Caceres, R. R. Khan, L. L. Minnich, K. D. Fu, G. D. Lovett, T. Tsai, and A. Thompson. 2001. La Crosse encephalitis in children. N. Engl. J. Med. 344:801-807. [DOI] [PubMed] [Google Scholar]
- 45.McManus, M. T., and P. A. Sharp. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3:737-747. [DOI] [PubMed] [Google Scholar]
- 46.Pekosz, A., B. He, and R. A. Lamb. 1999. Reverse genetics of negative-strand RNA viruses: closing the circle. Proc. Natl. Acad. Sci. USA 96:8804-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pekosz, A., J. Phillips, D. Pleasure, D. Merry, and F. Gonzalez-Scarano. 1996. Induction of apoptosis by La Crosse virus infection and role of neuronal differentiation and human bcl-2 expression in its prevention. J. Virol. 70:5329-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfeffer, S., P. Dunoyer, F. Heim, K. E. Richards, G. Jonard, and V. Ziegler-Graff. 2002. P0 of beet Western yellows virus is a suppressor of posttranscriptional gene silencing. J. Virol. 76:6815-6824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Pruss, G., X. Ge, X. M. Shi, J. C. Carrington, and V. Bowman Vance. 1997. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srihongse, S., M. A. Grayson, and R. Deibel. 1984. California serogroup viruses in New York State: the role of subtypes in human infections. Am. J. Trop. Med. Hyg. 33:1218-1227. [DOI] [PubMed] [Google Scholar]
- 52.Streitenfeld, H., A. Boyd, J. K. Fazakerley, A. Bridgen, R. M. Elliott, and F. Weber. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 77:5507-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeda, A., K. Sugiyama, H. Nagano, M. Mori, M. Kaido, K. Mise, S. Tsuda, and T. Okuno. 2002. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 532:75-79. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, W. H., B. Kalfayan, and R. O. Anslow. 1965. Isolation of California encephalitis group virus from a fatal human illness. Am. J. Epidemiol. 81:245-253. [DOI] [PubMed] [Google Scholar]
- 55.Traavik, T., R. Mehl, and R. Wiger. 1978. California encephalitis group viruses isolated from mosquitoes collected in Southern and Arctic Norway. Acta Pathol. Microbiol. Scand. B 86:335-341. [DOI] [PubMed] [Google Scholar]
- 56.Tuschl, T., P. D. Zamore, R. Lehmann, D. P. Bartel, and P. A. Sharp. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 13:3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Family Bunyaviridae, p. 35-43. In R. B. Wickner (ed.), Virus taxonomy: 7th report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 58.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 59.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 60.Watts, D. M., J. W. LeDuc, C. L. Bailey, J. M. Dalrymple, and T. P. Gargan II. 1982. Serologic evidence of Jamestown Canyon and Keystone virus infection in vertebrates of the DelMarVa Peninsula. Am. J. Trop. Med. Hyg. 31:1245-1251. [DOI] [PubMed] [Google Scholar]
- 61.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber, F., E. F. Dunn, A. Bridgen, and R. M. Elliott. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67-74. [DOI] [PubMed] [Google Scholar]
- 63.Wijkamp, I., J. van Lent, R. Kormelink, R. Goldbach, and D. Peters. 1993. Multiplication of tomato spotted wilt virus in its insect vector, Frankliniella occidentalis. J. Gen. Virol. 74:341-349. [DOI] [PubMed] [Google Scholar]
- 64.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]