Abstract
Nitrogen is one of the most important mineral elements for plant growth. We studied the functional roles of Oryza sativa DNA BINDING WITH ONE FINGER 18 (OsDOF18) in controlling ammonium uptake. The growth of null mutants of OsDOF18 was retarded in a medium containing ammonium as the sole nitrogen source. In contrast, those mutants grew normally in a medium with nitrate as the sole nitrogen source. The gene expression was induced by ammonium but not by nitrate. Uptake of ammonium was lower in osdof18 mutants than in the wild type, while that of nitrate was not affected by the mutation. This indicated that OsDOF18 is involved in regulating ammonium transport. Among the 10 ammonium transporter genes examined here, expression of OsAMT1;1, OsAMT1;3, OsAMT2;1, and OsAMT4;1 was reduced in osdof18 mutants, demonstrating that the ammonium transporter genes function downstream of OsDOF18. Genes for nitrogen assimilation were also affected in the mutants. These results provide evidence that OsDOF18 mediates ammonium transport and nitrogen distribution, which then affects nitrogen use efficiency.
Keywords: ammonium, ammonium transporters, nitrate, rice, transcription factor DOF
INTRODUCTION
Nitrogen (N) is an essential component for plant growth and development (Sonoda et al., 2003; Tabuchi et al., 2007). The major sources of inorganic N ions are ammonium (NH4+) and nitrate (NO3−), which can be absorbed and used by paddy crops in submerged soil (Tabuchi et al., 2007). As a N source, ammonium is preferable to nitrate for root uptake because less energy is needed for assimilation in the plants (Bloom et al., 1992; Gu et al., 2013; Masumoto et al., 2010).
Ammonium is mobilized by ammonium transporter (AMT). Rice (Oryza sativa) contains 10 members of the AMT family: OsAMT1;1, OsAMT1;2, OsAMT1;3, OsAMT2;1, OsAMT2;2, OsAMT2;3, OsAMT3;1, OsAMT3;2, OsAMT3;3, and OsAMT4;1. Whereas the OsAMT1 members are characterized as high-affinity transporters, the other three family members are considered low-affinity transporters (Loqué and von Wirén, 2004; Sonoda et al., 2003; Suenaga et al., 2003). Three OsAMT1 genes show distinct expression patterns: OsAMT1;1 is constitutively expressed in shoots while OsAMT1;2 and OsAMT1;3 are expressed specifically in roots (Sonoda et al., 2003). OsAMT1;1 and OsAMT1;2 are up-regulated in plants following exposure to ammonium, whereas OsAMT1;3 is up-regulated by N-deprivation (Kumar et al., 2003; Sonoda et al., 2003; Suenaga et al., 2003; Xuan et al., 2013). Overexpression of OsAMT1;1 results in higher production of biomass with increased amounts of ammonium and glutamine (Ranathunge et al., 2014). However, overexpression of OsAMT1;3 causes growth retardation (Bao et al., 2015). OsAMT2;1 shares only 20 to 25% sequence identity with proteins in the AMT1 family, and is more closely related to the yeast METHYLAMINE PERMEASE (MEP) transporter sequence (Suenaga et al., 2003).
Up to 40% of the total N is taken up in the form of nitrate by NITRATE TRANSPORTER (NRT) in paddy (Kirk and Kronzucker 2005). In rice, there are four high affinity NTR2; Os-NRT2;1, OsNRT2;2, OsNRT2;3, and OsNRT2;4 (Feng et al., 2011). The coding region sequences of OsNRT2.1 and Os-NRT2.2 are identical although their untranscribed regions are different. These NRT2 genes are highly homologous to other monocotyledons, while OsNRT2.3 and OsNRT2.4 are closely related to Arabidopsis NRT2 (Cai et al. 2008).
Ammonium is first assimilated by glutamine synthetase (GS) to yield the amino group of glutamine that serves as a major nitrogen source transported from root to shoot in rice (Kiyomiya et al. 2001). Glutamine synthetase is coupled with glutamate synthase (GOGAT) in the GS/GOGAT cycle. Os-NADH-GOGAT1 is predominantly expressed at root tips, leaves and seeds, while OsNADH-GOGAT2 is highly expressed in mature leaves (Tamura et al., 2011). Phosphoenolpyruvate carboxylase (PEPC) plays an important role in carboxylation of phosphoenolpyruvate to form oxaloacetate. NADP-malate dehydrogenase (MDH) converts the reaction between oxaloacetate and malate. OsPEPC4 and OsMDH play crucial roles in ammonium assimilation (Kurai et al., 2011; Masumoto et al., 2010).
DNA-binding with one finger (DOF) transcription factors participate in various biological processes, including tissue differentiation and hormone signaling (Noguero et al., 2013). Zea mays DOF1 (ZmDOF1) enhances the C4 pathway genes PEPC and cytosolic orthophosphate dikinase (cyPPDK) by binding to their promoters. Zea mays DOF2 (ZmDOF2) represses the promoter activity of ZmPEPC and ZmPPDK by blocking transactivation of ZmDOF1 (Yanagisawa and Izui, 1993; Zhang et al., 1995). Overexpression of ZmDOF1 increases the nitrogen content in transgenic Arabidopsis plants (Yanagisawa et al., 2004) and results in better growth of transgenic rice plants under low-N conditions (Kurai et al., 2011).
Rice OsDOF18, also named OsDOF24 or OsDOF25, is most homologous to ZmDOF1 (Kushwaha et al., 2010; Lijavetzky et al., 2003). Its heterologous expression in Arabidopsis alters carbon and nitrogen metabolism (Santos et al., 2012). In addition, OsDOF18 appears to have a function in carbohydrate metabolism by controlling OsPPDK (Zhang et al., 2015). Here, we demonstrated that OsDOF18 modulates ammonium uptake by inducing ammonium transporter genes.
MATERIALS AND METHODS
Plant materials and growth conditions
Japonica rice (Oryza sativa cv. Dongjin) plants were grown in controlled environment rooms as previously described (Ryu et al., 2009). Seeds were germinated either on an MS medium containing 3% sucrose or in soil, as previously reported (Yi and An, 2013). The T2 progeny of osdof18 knockout mutants were grown on a 1/2 MS medium containing 50 μg mL−1 hygromycin. For ammonium uptake experiment, plants were grown hydroponically on Yoshida medium containing 1.44 mM NH4NO3, 0.3 mM NaH2PO4, 0.5 mM K2SO4, 1.0 mM CaCl2, 1.6 mM MgSO4, 0.075 μM (NH4)6Mo7O24, 18.8μM H3BO3, 9.5 μM MnCl2, 0.16 μM CuSO4, 0.15 μM ZnSO4, 35.6 μM FeCl3, and 74.4 μM citric acid, pH5.5 (Yoshida et al., 1976).
Isolation of osdof18 mutants
A T-DNA-tagged osdof18-1 mutant (Line number 3A-16330) was identified from the rice T-DNA insertion sequence database (An et al., 2005a; 2005b; Jeong et al., 2006). Homozygous mutants were confirmed by PCR, using genomic DNA. The Ds-tagged osdof18-2 mutant (Line number Ds-17925), generated in ‘Dongjin’, was obtained from the Rice Division of Yeongnam Agricultural Research Institute, National Institute of Crop Science, Korea. Gene-specific primers are listed in Supplemental Table S1.
Analyses of nitrogen induction
Plants were grown in a chamber under the following conditions: 200 μmol m−2s−1 photosynthetic photon flux, 12-h photoperiod, 70% relative humidity, and 28°C/24°C (day/night). Seeds were first treated with 2% sodium hypo-chlorite for 30 min, then washed with distilled water three to five times, and placed in a glass bottle containing distilled water. At 14 days after germination (DAG), the seedlings were transferred to a glass tube containing 4 ml of Yoshida medium (Yoshida et al., 1976) that contains both ammonium and nitrate as the N source. Medium was harvested at 2-d intervals during the nitrogen uptake experiments, and ammonium and nitrate levels were determined with a UV-1800 spectrometer (Shimadzu, Japan) at OD625 and OD220, respectively (Martín-Rodríguez et al., 2015; Weatherb, 1967; Wu et al., 2015).
RT-PCR analyses
Total RNA was isolated from seedling roots at 4 DAG using RNAiso Plus (TaKaRa, Shiga, Japan; http://www.takarabio. com). The cDNAs were synthesized and quantitative real-time RT-PCR was performed as previously described (Cho et al., 2016; Ryu et al., 2009; Yang et al., 2013; 2014). Expression levels were normalized with rice UBQ5 (LOC_Os01g 22490). All experiments were conducted at least three times, with three or more samples taken at each point. To ensure primer specificity, we performed the experiments when the melting curve showed a single sharp peak. The PCR products were sequenced to verify the specificity of the reaction (Ryu et al., 2009; Yang et al., 2013; 2014). All primers are listed in Supplemental Table S1.
RESULTS
Identification of knockout mutants in OsDOF18
The T-DNA insertion mutant osdof18-1 was isolated from our T-DNA tagging population (An et al., 2003; 2005a; Jeon et al., 2000; Jeong et al., 2002; 2006; Ryu et al., 2004) and another allele, osdof18-2, was identified from Ds insertion lines (Chin et al., 1999; Kim et al., 2004) (Fig. 1A). In both lines, insertions occurred within the coding region, and OsDOF18 transcript was not detectable in the plants (Fig. 1B). This indicated that both are null mutants.
Fig. 1. Structure of OsDOF18 and position of T-DNA/Ds insertion.
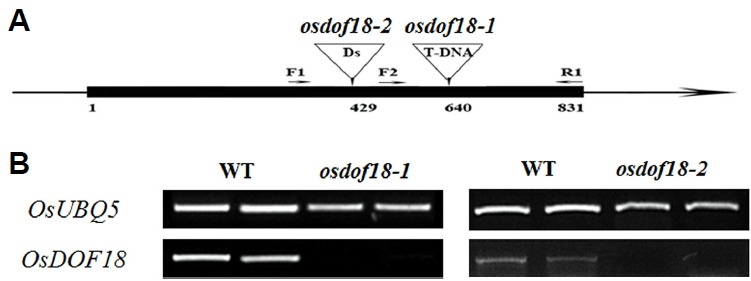
(A) Positions of insertion mutants. Black boxes, exons; lines between boxes, introns. Arrows indicate primers for genotyping. (B) RT-PCR analyses of OsDOF18 expression levels.
Mutations in OsDOF18 cause growth retardation when ammonium is the sole N source
The osdof18 mutants grew normally in Yoshida medium containing both ammonium and nitrate (Figs. 2A and 2D). Shoot growth was slightly reduced in the mutant compared to WT when they were grown on Yoshida medium with nitrate as the sole N source (Figs. 2B and 2E). However, when placed on the medium where ammonium was the sole N source, growth of the mutants was significantly retarded (Figs. 2C and 2F). At 4 DAG, primary root lengths from the mutant seedlings were 60% of that measured from the WT. Dry weights for roots and shoots from the mutants at 7 DAG were 60% and 73%, respectively, of values recorded for the WT (Figs. 2G and 2H). The defect was likely due to their diminished capability in taking up or assimilating ammonium.
Fig. 2. Phenotypes of osdof18 mutants.
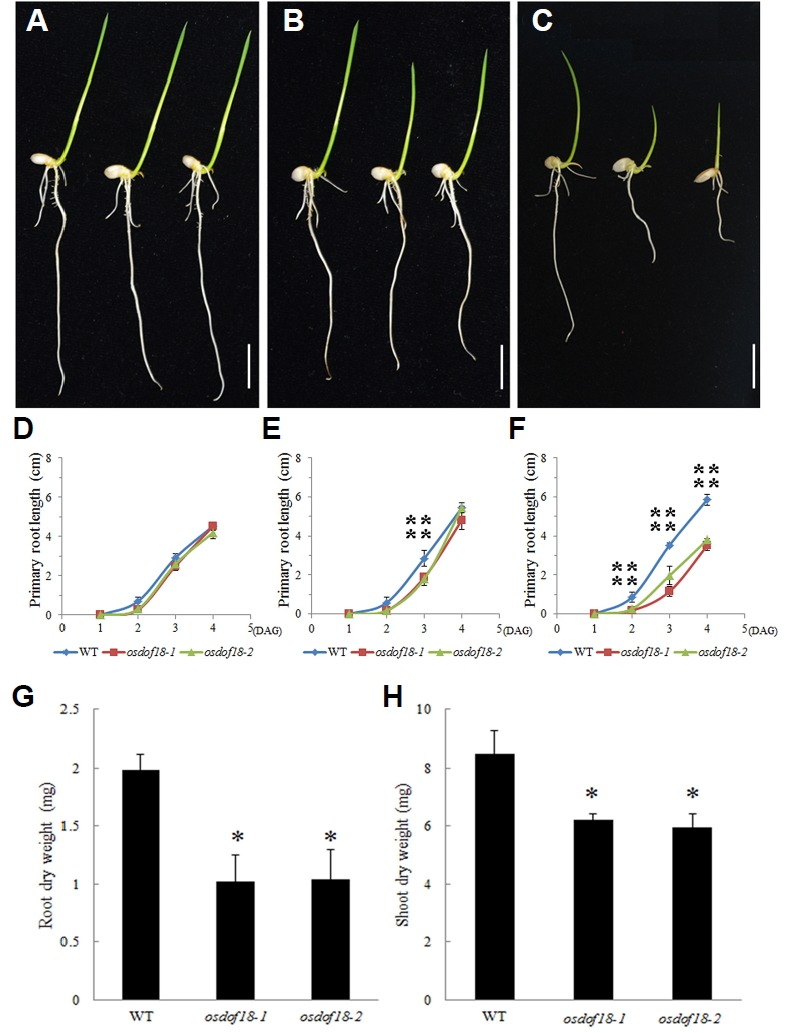
(A-C) Plant phenotypes in Yoshida medium (A), Yo-shida medium with nitrate as sole N source (B), or Yoshida medium with ammonium as sole N source (C). (D–F) Primary root lengths in Yoshida medium (D), Yoshida medium with nitrate as sole N source (E), or Yoshida medium with ammonium as sole N source (F). Blue line, WT; red line, osdof18-1; green line, osdof18-2. Root (G) and shoot (H) dry weights of osdof18 mutants and WT at 7 DAG grown in Yoshida medium with ammonium as a sole N source. Error bars represent standard error (SE) for at least 5 samples. *, P < 0.05; **P, < 0.01.
Ammonium uptake level is slow in osdof18 mutants
To study ammonium uptake level, we grew mutant and WT seedlings in water until 14 DAG to deplete the N stored in the seeds. The plants were then transferred to the Yoshida medium and uptake levels were estimated by measuring the amount of ammonium that remained in the medium. The initial concentration of ammonium was 1.44 mM. For WT plants, that concentration was rapidly reduced in the first 2 d, with approximately 69.3% of the ammonium being removed (Fig. 3A). The concentration was further reduced as the plants grew, with only 9.1% of the ammonium remaining at Day 6. By comparison, the amount of ammonium in the medium where osdof18 mutants were grown was reduced by only 13.5% at 2 d and 36.8% at 6 d (Fig. 3A).
Fig. 3. Uptake activity of ammonium (A–C) and nitrate (D–F) in osdof18 mutants and WT.
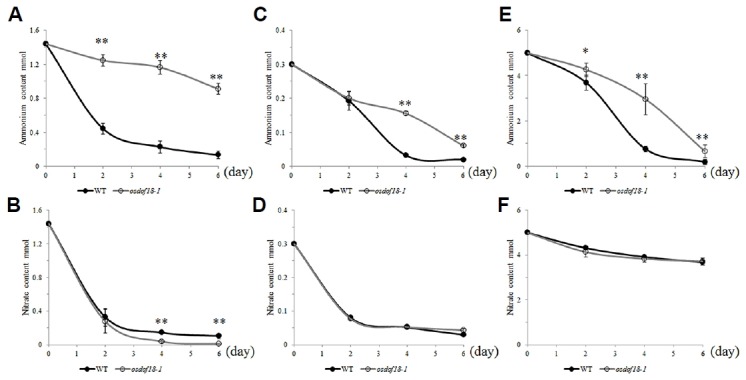
A–B; Plants were grown on Yoshida medium with 1.44 mM NH4NO3 as sole N source; C–D; Plants were grown on Yoshida medium with 0.3 mM NH4NO3 as sole N source; E–F; Plants were grown on Yoshida medium with 5 mM NH4NO3 as sole N source. Closed circles, WT; open circles, osdof18-1. Error bars represent SE for at least 3 plants. *P < 0.05; **P, < 0.01.
We also examined the ammonium uptake level at 5 mM and 0.3 mM concentrations. The uptake level in the mutants was slower compared to WT at both reduced (Fig. 3C) and increased (Fig. 3E) concentrations of ammonium. These experiments indicate that OsDOF18 affects long term ammonium uptake at both low and high concentration. In contrast, nitrate uptake levels were similar between the WT and mutant plants (Figs. 3B, 3D and 3F). These experiments supported our conclusion that OsDOF18 functions in the uptake of ammonium but not nitrate into the roots.
Expression of OsDOF18 is induced by ammonium
Seedlings were grown in water for 14 d to deplete stored N in the seeds. After they were transferred to Yoshida medium with ammonium as the sole N source, the level of OsDOF18 expression increased within 30 min ammonium and peaked at 1 h (Fig. 4A). However, the gene expression was not increased during the 12 h treatment in Yoshida medium with nitrate as the sole N source. Using OsAMT1;1 and OsNRT2;1 as controls (Figs. 4B and 4C), we found that the former responded more significantly to ammonium supply than to nitrate while the latter responded mainly to a nitrate supply. These results suggested that OsDOF18 functions in controlling OsAMT1;1.
Fig. 4. Expression patterns of OsDOF18, OsAMT1;1, and OsNRT2;1 after plants were supplied with ammonium or nitrate.
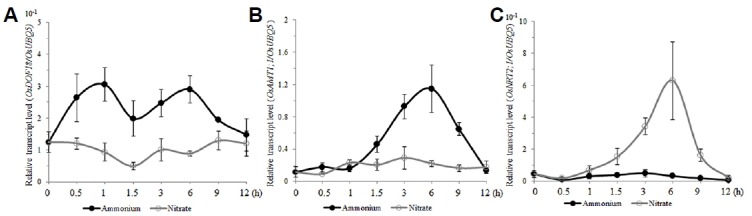
Quantitative RT-PCR analyses of transcript levels of OsDOF18 (A), OsAMT1;1 (B), and OsNRT2;1 (C) after ammonium or nitrate was supplied to 14 DAG seedlings grown in water. Expression levels were normalized to OsUBQ5. Closed circles, ammonium treatment; open circles, nitrate treatment. Error bars represent SE for at least 3 samples.
Mutations in OsDOF18 affect expression of ammonium transporter genes
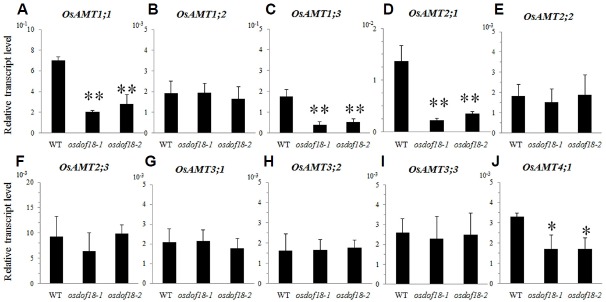
Transcript levels of 10 ammonium transporter genes were compared between osdof18 mutants and WT in roots from 4 DAG seedlings grown on the ammonium-supplemented Yoshida medium. When compared with the WT, expression of OsAMT1;1, OsAMT1;3, OsAMT2;1, and OsAMT4;1 was low in the mutants while that of OsAMT1;2, OsAMT2;2, OsAMT2;3, OsAMT3;1, OsAMT3;2, and OsAMT3;3 was not significantly affected by the mutation (Fig. 5). Transcript levels of the nitrate transporter genes, OsNRT2;1, OsNRT2;2 and OsNRT2;3, were not changed. We also observed no alteration of the transcript level of OsIDD10 that acts as an inducer of OsAMT1;2 (Figs. 6A–6D).
Fig. 5. Expression levels of OsAMT genes.
Quantitative RT-PCR analyses of transcript levels of OsAMT1;1 (A), OsAMT1;2 (B), OsAMT1;3 (C), OsAMT2;1 (D), OsAMT2;2 (E), OsAMT2;3 (F), OsAMT3;1 (G), OsAMT3;2 (H), OsAMT3;3 (I), and OsAMT4;1 (J) in WT and osdof18 mutants. RNAs were isolated from roots of 4 DAG plants grown in Yoshida medium with ammonium as sole N source. Expression levels were normalized to OsUBQ5. Error bars represent SE for at least 3 samples. *P < 0.05; **P < 0.01.
Fig. 6. Expression levels of OsNRTs, OsIDD10, OsGOGAT1, OsGOGAT2, OsPEPC4, and OsMDH genes.
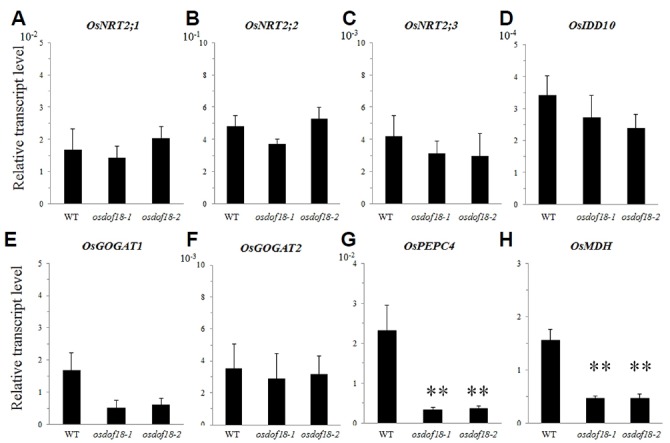
<Quantitative RT-PCR analyses of transcript levels of OsNRT2;1 (A), OsNRT2;2 (B), OsNRT2;3 (C) and OsIDD10 (D), OsGOGAT1 (E), OsGOGAT2 (F), OsPEPC4 (G), and OsMDH (H) in WT and osdof18 mutants. RNAs were isolated from roots of 4 DAG plants grown in Yoshida medium with ammonium as sole N source. Expression levels were normalized to OsUBQ5. Error bars represent SE for at least 3 samples. *P < 0.05; **P < 0.01.
Expression of OsGOGAT1, OsPEPC4, and OsNADPH-MDH was also decreased in the mutants while that of Os-GOGAT2 was unchanged (Figs. 6E–6H). These results suggested that OsDOF18 functions to affect ammonium uptake as well as the expression of genes involved in ammonium assimilation.
DISCUSSION
Transcription factors containing a zinc finger motif control ammonium transport
Mutations in OsDOF18 caused growth retardation when ammonium was the sole N source. The level of ammonium uptake was significantly lower in osdof18 mutants than in the WT. This ammonium-dependent phenotype was similar to that described for osidd10, which displays delayed germination and slower primary root growth when the medium is supplemented only with ammonium (Xuan et al., 2013). However, the molecular mechanisms involved in ammonium uptake are apparently different between OsIDD10 and OsDOF18. Whereas the former enhances the expression of AMT1;2 by binding to its promoter region, the latter does not affect AMT1;2 expression. The influence of OsIDD10 on AMT1;1 is not clear because gene expression is moderately reduced in both osidd10 knockout mutants and OsIDD10-overexpressing plants. The relationship between OsIDD10 and other AMT genes has not yet been examined. Therefore, further study is needed to determine whether OsAMT2;2, OsAMT2;3, OsAMT3;1, OsAMT3;2, and OsAMT3;3, which are not targets of OsDOF18, are controlled by OsIDD10. Although OsDOF18 and OsIDD10 belong to different gene families, they have zinc finger motifs in common, with the former containing one zinc finger and the latter having four such motifs.
OsDOF18 expression is induced by ammonium
Transcription levels of OsDOF18 were induced when ammonium was supplied, but were not significantly altered by nitrate supplementation. Expression of OsAMT1;1 was also ammonium-dependent, thereby indicating that both genes are associated with ammonium transport. However, OsDOF18 was induced in response to ammonium, peaking within the first 60 min after application whereas OsAMT1;1 was slowly induced, reaching the highest level only after 6 h. These differential induction rates demonstrated that OsDOF18 functions upstream of OsAMT1;1.
OsDOF18 regulates ammonium transporter genes
Multiple AMTs function in ammonium uptake by the roots (Kumar et al., 2003; Sonoda et al., 2003; Suenaga et al., 2003). We observed that four AMT genes were affected in osdof18 mutants. Of these, OsAMT1;1 and OsAMT1;3 encode high-affinity ammonium transporters while OsAMT2;1 and OsAMT4;1 encode low-affinity transporters. This indicates that OsDOF18 controls both types. A reduction in OsAMT1;3 levels in osdof18 was unexpected because the AMT gene is not induced but, instead, is repressed when plants are supplemented with ammonium (Suenaga et al., 2003). This may result when ammonium accumulates in the growth medium because the mutants are defective in their uptake capacity.
Heterogeneous expression of OsDOF18 in Arabidopsis enhances ammonium uptake by increasing expression of At-AMT1.1 and AtAMT2.1, and promotes ammonium assimilation by elevating transcription of AtPK1, AtPK2, AtPEPC1, AtPEPC2, AtGS1.1, AtGS1.2, AtGS1.3, and AtGS2 (Santos et al., 2012). This indicates that functional role of OsDOF18 is conserved between Arabidopsis and rice. Overexpression of ZmDOF1, a maize ortholog of OsDOF18, promotes PEPC and PPDK expression (Yanagisawa et al., 2004). Chromatin immunoprecipitation assays show enrichment of OsDOF18 on the promoter region of OsPPDK chromatin (Zhang et al., 2015), suggesting that OsDOF18 may also function in photosynthesis.
Supplementary data
ACKNOWLEDGMENTS
We thank Kyungsook An for generating the T-DNA insertional lines and managing the transgenic seeds, and Priscilla Licht for her critical proofreading of the manuscript. This work was supported in part by grants from the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01108001), Rural Development Administration, Republic of Korea; the Republic of Korea Basic Research Promotion Fund (Grant No. NRF-2007-0093862); and Kyung Hee University (20130214) to G.A.
Footnotes
Note: Supplementary Information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- An S.Y., Park S., Jeong D.H., Lee D.Y., Kang H.G., Yu J.H., Hur J., Kim S.R., Kim Y.H., Lee M., et al. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 2003;133:2040–2047. doi: 10.1104/pp.103.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G., Jeong D.H., Jung K.H., Lee S. Reverse genetic approaches for functional genomics of rice. Plant Mol Biol. 2005a;59:111–123. doi: 10.1007/s11103-004-4037-y. [DOI] [PubMed] [Google Scholar]
- An G., Lee S., Kim S.H., Kim S.R. Molecular genetics using T-DNA in rice. Plant Cell Physiol. 2005b;46:14–22. doi: 10.1093/pcp/pci502. [DOI] [PubMed] [Google Scholar]
- Bao A., Liang Z., Zhao Z., Cai H. Overexpressing of OsAMT1-3, a high affinity ammonium transporter gene, modifies rice growth and carbon-nitrogen metabolic status. Int J Mol Sci. 2015;16:9037–9063. doi: 10.3390/ijms16059037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom A.J., Sukrapanna S.S., Warner R.L. Root respiration associated with ammonium and nitrate absorption and assimilation by barley. Plant Physiol. 1992;99:1294–1301. doi: 10.1104/pp.99.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.H., Wang J.Y., Zhu Y.G., Shen Q.R., Li B., Tong Y.P., Li Z.S. Gene structure and expression of high-affinity nitrate transport system in rice roots. J Int Plant Biol. 2008;50:443–451. doi: 10.1111/j.1744-7909.2008.00642.x. [DOI] [PubMed] [Google Scholar]
- Chin H.G., Choe M.S., Lee S.H., Park S.H., Park S.H., Koo J.C., Kim N.Y., Lee J.J., Oh B.G., Yi G.H., et al. Molecular analysis of rice plants harboring an Ac/Ds transposable element-mediated gene trapping system. Plant J. 1999;19:615–624. doi: 10.1046/j.1365-313x.1999.00561.x. [DOI] [PubMed] [Google Scholar]
- Cho L.H., Yoon J., Pasriga R., An G. Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 2016;170:2159–2171. doi: 10.1104/pp.15.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H.M., Yan M., Fan X.R., Li B.Z., Shen Q.R., Miller A.J., Xu G.H. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot. 2011;62:2319–2332. doi: 10.1093/jxb/erq403. [DOI] [PubMed] [Google Scholar]
- Gu R., Duan F.Y., An X., Zhang F.S., von Wirén N., Yuan L.X. Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays L.) Plant Cell Physiol. 2013;54:1515–1524. doi: 10.1093/pcp/pct099. [DOI] [PubMed] [Google Scholar]
- Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Yang K., Nam J., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeong D.H., An S.Y., Kang H.G., Moon S., Han J.J., Park S., Lee H.S., An K.S., An G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002;130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., An S., Park S., Kang H.G., Park G.G., Kim S.R., Sim J., Kim Y.O., Kim M.K., Kim S.R., et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Kim C.M., Piao H.L., Park S.J., Chon N.S., Je B.I., Sun B.Y., Park S.H., Park J.Y., Lee E.J., Kim M.J., et al. Rapid, large-scale generation of Ds transposant lines and analysis of the Ds insertion sites in rice. Plant J. 2004;39:252–263. doi: 10.1111/j.1365-313X.2004.02116.x. [DOI] [PubMed] [Google Scholar]
- Kirk G.J.D., Kronzucker H.J. The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: a modelling study. Annals Bot. 2005;96:639–646. doi: 10.1093/aob/mci216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomiya S., Nakanishi H., Uchida H., Tsuji A., Nishiyama S., Futatsubashi M., Tsukuda H., Ishioka N.S., Watanabe S., Ito T., et al. Real time visualization of 13N-translocation in rice under different environment conditions using position emitting tracer imaging system. Plant Physiol. 2001;125:1743–1754. doi: 10.1104/pp.125.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Silim S.N., Okamoto M., Siddiqi M.Y., Glass A.D.M. Differential expression of three members of the AMT1 gene family encoding putative high affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant Cell Environ. 2003;26:907–914. doi: 10.1046/j.1365-3040.2003.01023.x. [DOI] [PubMed] [Google Scholar]
- Kurai T., Wakayama M., Abiko T., Yanagisawa S., Aoki N., Ohsugi R. Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol J. 2011;9:826–837. doi: 10.1111/j.1467-7652.2011.00592.x. [DOI] [PubMed] [Google Scholar]
- Kushwaha H., Gupta S., Singh V.K., Rastogi S., Yadav D. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol Biol Rep. 2010;38:5037–5053. doi: 10.1007/s11033-010-0650-9. [DOI] [PubMed] [Google Scholar]
- Lijavetzky D., Carbonero P., Vicente-Carbajosa J. Genomewide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol. 2003;23:3–17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D., von Wirén N. Regulatory levels for the transport of ammonium in plant roots. J Exp Bot. 2004;55:1293–1305. doi: 10.1093/jxb/erh147. [DOI] [PubMed] [Google Scholar]
- Masumoto C., Miyazawa S., Ohkawa H., Fukuda T., Taniguchi Y., Murayama S., Kusano M., Saito K., Fukayama H., Miyao M. Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc Natl Acad Sci USA. 2010;107:5226–5231. doi: 10.1073/pnas.0913127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Rodríguez A.J., Babarro J.M., Lahoz F., Sansón M., Martín V.S., Norte M., Fernández J.J. From broad-spectrum biocides to quorum sensing disruptors and mussel repellents: antifouling profile of alkyl triphenylphosphonium salts. PLoS One. 2015;10:e0123652. doi: 10.1371/journal.pone.0123652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguero M., Atif R.M., Ochatt S., Thompson R.D. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013;209:32–45. doi: 10.1016/j.plantsci.2013.03.016. [DOI] [PubMed] [Google Scholar]
- Ranathunge K., El-Kereamy A., Gidda S., Bi Y.M., Rothstein S.J. AMT1;1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J Exp Bot. 2014;65:965–979. doi: 10.1093/jxb/ert458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C.H., You J.H., Kang H.G., Hur J.H., Kim Y.H., Han M.J., An K.S., Chung B.C., Lee C.H., An G. Generation of T-DNA tagging lines with a bidirectional gene trap vector and the establishment of an insertion-site database. Plant Mol Biol. 2004;54:489–502. doi: 10.1023/B:PLAN.0000038257.93381.05. [DOI] [PubMed] [Google Scholar]
- Ryu C.H., Lee S., Cho L.H., Kim S.L., Lee Y.S., Choi S.C., Jeong H.J., Yi J., Park S.J., Han C.D., An G. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009;32:1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- Santos L.A., de Souza S.R., Fernandes M.S. OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnol Rep. 2012;6:327–337. [Google Scholar]
- Sonoda Y., Ikeda A., Saiki S., von Wiren N., Yamaya T., Yamaguchi J. Distinct expression and function of three ammonium transporter genes (OsAMT1;1–1;3) Plant Cell Physiol. 2003;44:726–734. doi: 10.1093/pcp/pcg083. [DOI] [PubMed] [Google Scholar]
- Suenaga A., Moriya K., Sonoda Y., Ikeda A., von Wirén N., Hayakawa T., Yamaguchi J., Yamaya T. Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 2003;44:206–211. doi: 10.1093/pcp/pcg017. [DOI] [PubMed] [Google Scholar]
- Tabuchi M., Abiko T., Yamaya T. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.) J Exp Bot. 2007;58:2319–2327. doi: 10.1093/jxb/erm016. [DOI] [PubMed] [Google Scholar]
- Tamura W., Kojima S., Toyokawa A., Watanabe H., Tabuchi-Kobayashi M., Hayakawa T., Yamaya T. Disruption of a novel NADH-glutamate synthase2 gene caused marked reduction in spikelet number of rice. Front Plant Sci. 2011;2:57. doi: 10.3389/fpls.2011.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherb M.W. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- Wu Z., Akter R., Arirob W., Juntawong N., Ma C., Deangmanee P. Effects of light intensity and the remaining nitrate concentration on the beta-carotene accumulation of a wild Dunaliella salina strain isolated from the saline soil. Microbiology. 2015;6:6233. [Google Scholar]
- Xuan Y.H., Priatama R.A., Huang J., Je B.I., Liu J.M., Park S.J., Piao H.L., Son D.Y., Lee J.J., Park S.H., et al. Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots. New Phytol. 2013;197:791–804. doi: 10.1111/nph.12075. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S., Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem. 1993;268:16028–16036. [PubMed] [Google Scholar]
- Yanagisawa S., Sheen J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell. 1998;10:75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S., Akiyama A., Kisaka H., Uchimiya H., Miwa T. Metabolic engineering with Dof1 transcription factor in plants: Improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA. 2004;101:7833–7838. doi: 10.1073/pnas.0402267101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S., Hang R., Kim S.R., Lee Y.S., Cao X., Amasino R., An G. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 2013;73:566–578. doi: 10.1111/tpj.12057. [DOI] [PubMed] [Google Scholar]
- Yi J., An G. Utilization of T-DNA tagging lines in rice. J Plant Biol. 2013;56:85–90. [Google Scholar]
- Yoon J., Cho L.H., Kim S.L., Choi H., Koh H.J., An G. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 2014;79:717–728. doi: 10.1111/tpj.12581. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Forno D.A., Cock J.H., Gomez K.A. Laboratory Manual for Physiological Studies of Rice The third edition. International Rice Research Institute; 1976. pp. 61–64. [Google Scholar]
- Zhang B., Chen W., Foley R.C., Buttner M., Singh K.B. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 1995;7:2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Verhoeff N.I., Chen Z., Chen S., Wang M., Zhu Z., Ouwerkerk P.B. Functions of OsDof25 in regulation of OsC4PPDK. Plant Mol Biol. 2015;89:229–242. doi: 10.1007/s11103-015-0357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.