Abstract
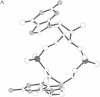
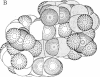
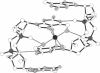
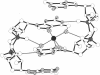
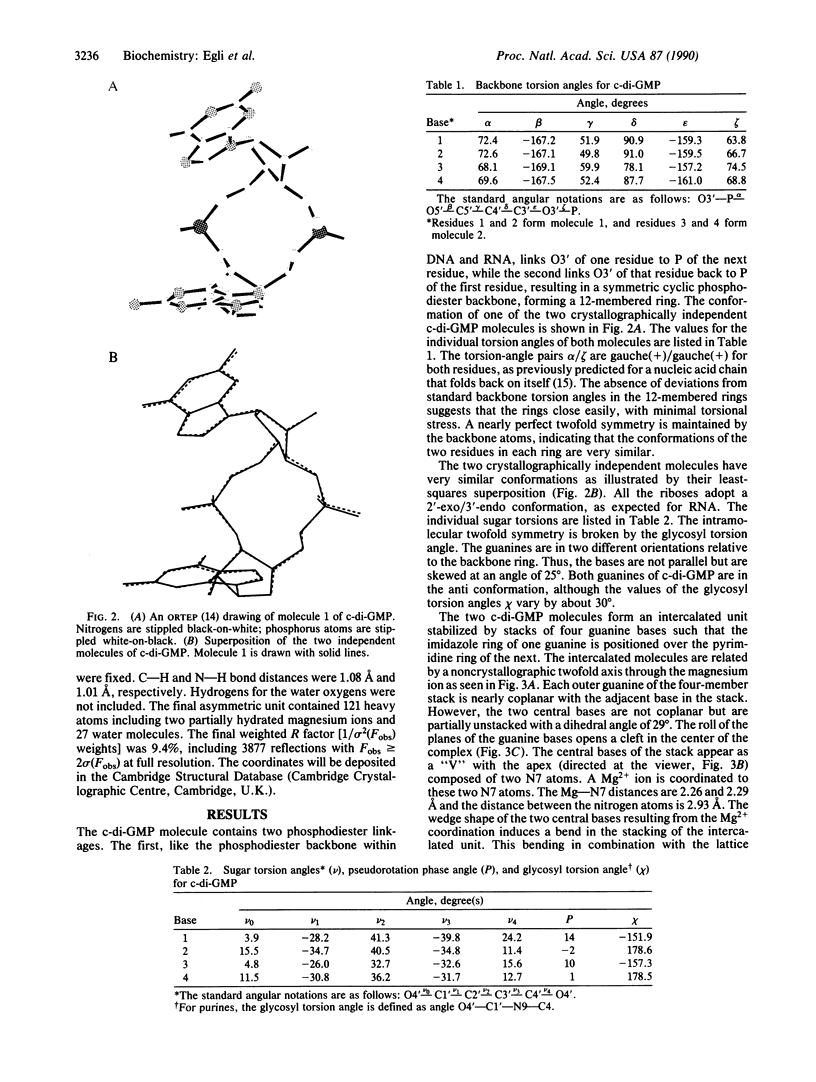
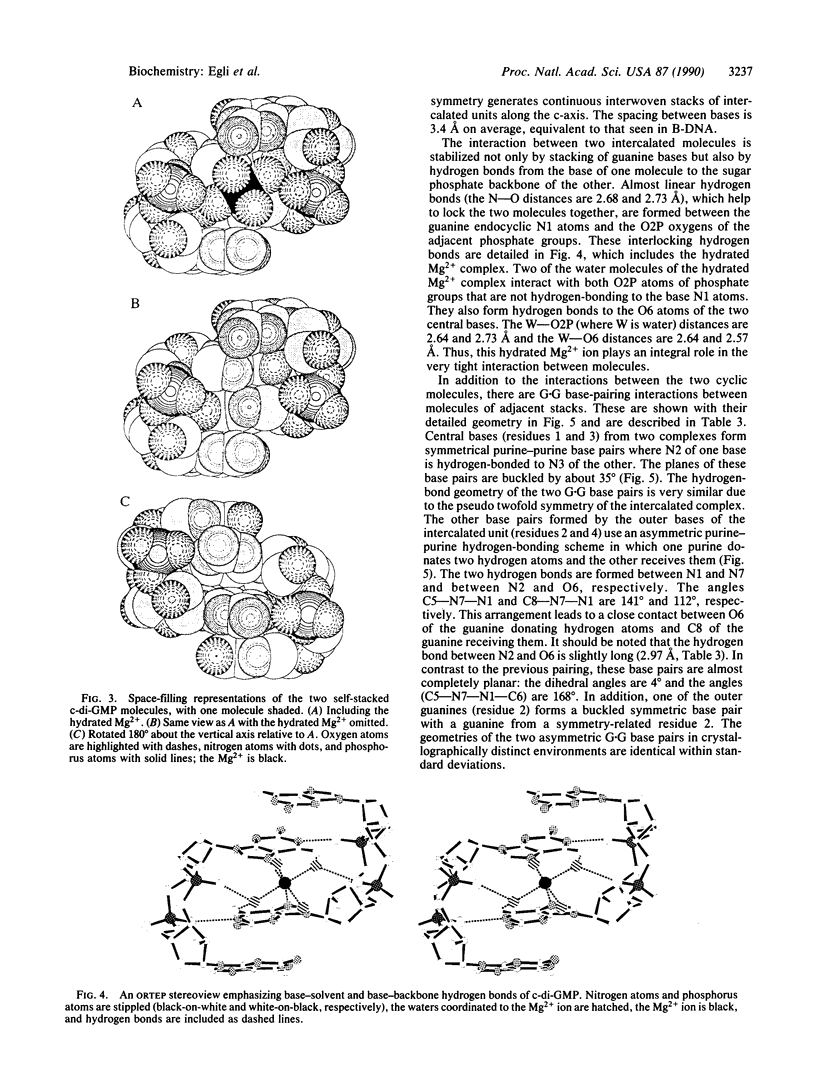
Cyclic diguanylic acid acts as a regulator for cellulose synthase activity in the bacterium Acetobacter xylinum. We report the x-ray crystal structure of the regulator at atomic resolution. The structure contains two independent molecules that adopt almost identical conformations. The two molecules form self-intercalated units that are stacked on each other. Two different G.G base-pairing modes occur between the stacks. The more stable one has two or possibly three hydrogen bonds between two guanines and is related to the type of hydrogen bonding that is believed to exist between G-rich strands at the ends of chromosomes.
Full text
PDF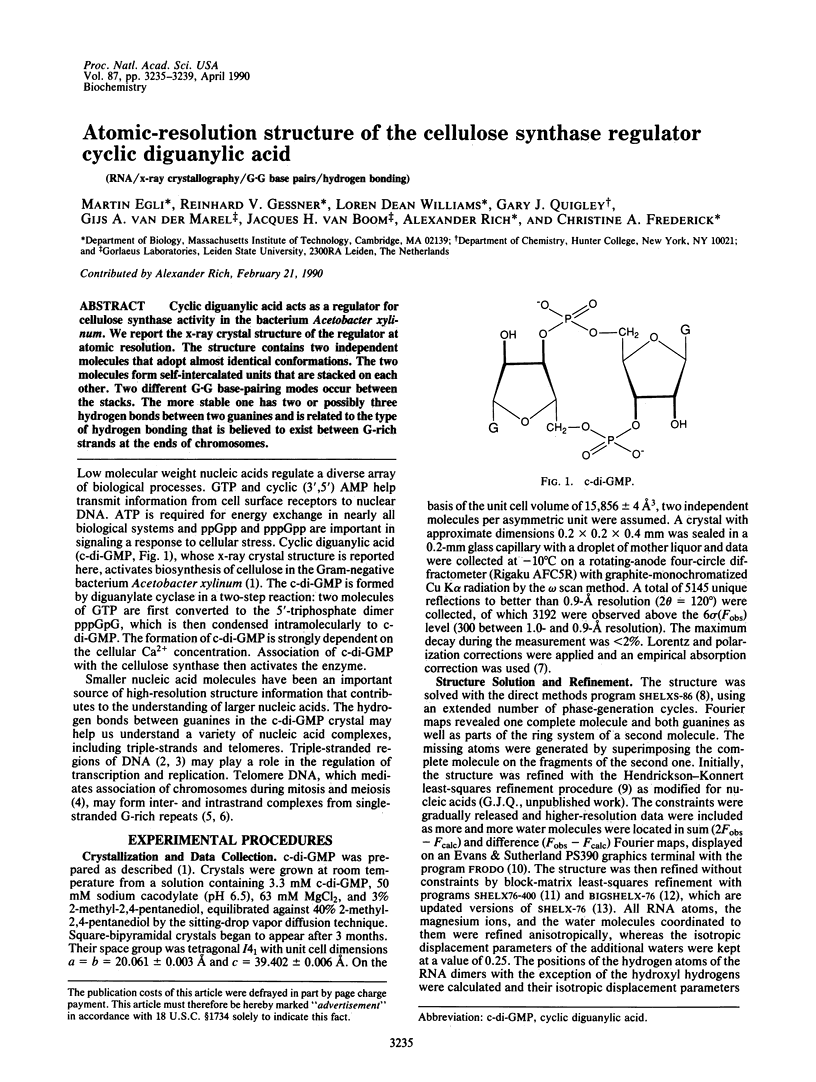


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blackburn E. H. The molecular structure of centromeres and telomeres. Annu Rev Biochem. 1984;53:163–194. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- Bugg C. E., Thewalt U. T., Marsh R. E. Base stacking in nucleic acid components: the crystal structures of guanine, guanosine and iosine. Biochem Biophys Res Commun. 1968 Nov 8;33(3):436–440. doi: 10.1016/0006-291x(68)90591-3. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Coll M., van der Marel G. A., van Boom J. H., Wang A. H. Molecular structure of cyclic deoxydiadenylic acid at atomic resolution. Biochemistry. 1988 Nov 1;27(22):8350–8361. doi: 10.1021/bi00422a010. [DOI] [PubMed] [Google Scholar]
- GELLERT M., LIPSETT M. N., DAVIES D. R. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962 Dec 15;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson E. R., Blackburn E. H. An overhanging 3' terminus is a conserved feature of telomeres. Mol Cell Biol. 1989 Jan;9(1):345–348. doi: 10.1128/mcb.9.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Kohwi Y., Kohwi-Shigematsu T. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3781–3785. doi: 10.1073/pnas.85.11.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Sasisekharan V., Zimmerman S., Davies D. R. The structure of helical 5'-guanosine monophosphate. J Mol Biol. 1975 Feb 25;92(2):171–179. doi: 10.1016/0022-2836(75)90221-1. [DOI] [PubMed] [Google Scholar]
- Sherman S. E., Gibson D., Wang A. H., Lippard S. J. X-ray structure of the major adduct of the anticancer drug cisplatin with DNA: cis-[Pt(NH3)2(d(pGpG))]. Science. 1985 Oct 25;230(4724):412–417. doi: 10.1126/science.4048939. [DOI] [PubMed] [Google Scholar]
- Sundquist W. I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989 Dec 14;342(6251):825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Thewalt U., Bugg C. E., Marsh R. E. The crystal structure of guanosine dihydrate and inosine dihydrate. Acta Crystallogr B. 1970 Aug 15;26(8):1089–1101. doi: 10.1107/s0567740870003667. [DOI] [PubMed] [Google Scholar]
- Williamson J. R., Raghuraman M. K., Cech T. R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989 Dec 1;59(5):871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984 May 11;224(4649):574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]