Abstract
Purpose
We investigated the prevalence of BRCA1/2 small mutations and large genomic rearrangements in high risk breast cancer patients who attended a genetic counseling clinic.
Methods
In total 478 patients were assessed for BRCA1/2 mutations by direct sequencing, of whom, 306 were identified as non-carriers of BRCA1/2 mutation and assessed for large rearrangement mutations by multiplex ligation-dependent probe amplification. Family history and clinicopathological characteristics of patients were evaluated.
Results
Sixty-three mutation carriers (13.2%) were identified with BRCA1 mutations (6.3%) and BRCA2 mutations (6.9%), respectively. Mutation frequency was affected by familial and personal factors. Breast cancer patients with family history of breast and ovarian cancer showed the highest prevalence of BRCA1/2 mutations (67%), and triple-negative breast cancer (TNBC) patients showed high BRCA1 mutation prevalence (25%). The three probands of BRCA1 deletion (1%) represented both familial risk and personal or clinicopathological risk factors as two with TNBC and one with bilateral ovarian cancer.
Discussion
This is the largest study assessing large genomic rearrangement prevalence in Korea and BRCA1 deletion frequency was low as 1% in patients without BRCA1/2 small mutations. For clinical utility of large genomic rearrangement testing needs further study.
Keywords: BRCA1/2 mutation, Large genomic rearrangements, Breast cancer, Genetic counseling, Family counseling
Introduction
Germline mutations in the BRCA1/2 genes are the most important cause of hereditary breast cancer [1]. The average cumulative risk of breast cancer in female carriers above 70 years is estimated to be 57–65 and 45–49% for BRCA1 and BRCA2 mutation carriers, respectively [2, 3].
Risk of hereditary cancers is assessed by taking into account familial and personal factors or clinicopathological characteristics of cancers such as triple-negative breast cancers (TNBC), which do not express the genes for estrogen receptor (ER), progesterone receptor (PR), or HER2 receptor [4]. With this information, women with a high risk of developing hereditary cancers are recommended to be tested for mutations in BRCA1/2 genes [5, 6]. Given that this test targets a high-risk population and the further management plans are largely dependent on the results of BRCA1/2 mutation testing are important.
Direct Sanger sequencing, conformation-sensitive gel electrophoresis, and denaturing high-performance liquid chromatography have been used to identify BRCA1/2 mutations. However, these techniques cannot identify large genomic rearrangements, which are often reported in patients with negative results from conventional direct sequencing [7, 8]. This may lead to an underestimation of mutation prevalence and provide false-negative information to patients and their families. Previous studies showed varying results with the prevalence of large genomic rearrangements ranging from 0 to 30% [9–14]. This wide range might be caused by different genetic backgrounds and the different inclusion criteria of various studies [11]. Thus, we investigated the prevalence of BRCA1/2 large genomic rearrangements using multiplex ligation-dependent probe amplification in high-risk breast cancer patients with negative results for BRCA1/2 mutation by direct sequencing.
Materials and methods
Study process
All the patients who were referred to a genetic counseling clinic and screened for BRCA1/2 mutation, at the National Cancer Center between April 2008 and 2015, were initially considered for this study. Among the total 523 patients screened for BRCA1/2 mutations, we excluded 28 family members of known BRCA1/2 mutation carriers and 17 patients with other types of cancers. There were 478 probands with breast cancer included in this study. The referred criteria for breast cancer were: (1) breast cancer patients with a family history of breast or ovarian cancer; (2) breast cancer patients who were 40 years or younger during diagnosis, who had bilateral breast cancer or breast cancer with other primary malignancy, or male breast cancer patients, in accordance with the standard of National Medical Insurance Reimbursement in Korea. In the genetic counseling process, all patients’ mutation probabilities were estimated by CaGene5.0 software [15] using pedigree information up to second-degree relatives and considering estimates by BRCAPRO [16] and Myriad [17]. Clinicopathological characteristics of cancer, such as the stage and the hormone receptor and human epidermal growth factor receptor 2 (HER2) status, were evaluated by reviewing medical records.
The initial BRCA1/2 mutation test was performed by PCR amplification and direct sequencing covering all exons and flanking intronic sequences. A subset (73%) of the 418 cases designated as non-carriers by direct sequencing, agreed to participate in the study. Written informed consent was obtained from 306 patients, and they were screened for the presence of large genomic rearrangements using a multiplex ligation-dependent probe amplification (MLPA) assay. This study protocol was approved by the institutional review board of the National Cancer Center (NCC2015-0177). The selection of patients and the test process is presented in the Fig. 1.
Fig. 1.
The study flowchart outlining the number of subjects and the genetic testing approach used in the study. A total of 478 breast cancer patients were included and multiple ligation-dependent probe amplification (MLPA) analysis was performed for 306 patients who did not have small mutations in the BRCA1 and BRCA2 genes and agreed for this study
Direct sequencing for mutation detection of BRCA1 and BRCA2, and nomenclature
Genomic DNA was extracted from peripheral blood using the Chemagic DNA Blood 200 Kit (Chemagen, Baesweiler, Germany). Amplified products were sequenced on an ABI 3500xl analyzer (Applied Biosystems, Foster City, CA, USA), using the Bigdye Terminator v3.1 Cycle Sequencing Kit. Sequences were analyzed using Sequencher v4.10.1 software. The clinical significance of each sequence variation was determined according to the Breast Cancer Information Core database (BIC: http://research.nhgri.nih.gov/bic/) and the recommendations of the American College of Medical Genetics [18]. All the mutations are described according to HUGO-approved systematic nomenclature (http://www.hgvs.org/mutnomen/). GenBank accession sequences NM_007294.3 and NM_000059.3 were used as reference sequences for BRCA1 and BRCA2, respectively. Traditional mutation nomenclatures of the BIC were used for description.
Multiplex ligation-dependent probe amplification
MLPA was performed to detect large genomic rearrangements using the SALSA P002-D1 BRCA1 Kit (MRC Holland, Amsterdam, Holland) for BRCA1 and P045-B3 BRCA2 Kit (MRC Holland, Amsterdam, Holland) for BRCA2. PCR products were analyzed using an ABI 3500xl analyzer with GeneMarker v2.4.0 demonstration program (Softgenetics, State College, PA, USA). Peak heights were normalized, and a deletion or duplication was identified when the normalized peak ratio value was below 0.75 or above 1.30, respectively.
Risk factors and statistical analysis
The subjects were classified according to familial and personal factors. Familial factors taken into account included family history of breast cancer, number of family members with breast cancer, closest degree of family members with breast cancer, and family history of ovarian cancer. Forty-two of the 306 patients had a family history of ovarian cancer. Personal factors taken into account included early onset of breast cancer which is defined as the development of breast cancer before the age of 40, bilateral breast cancer irrespective of age at onset, both breast and ovarian cancer irrespective of age at onset, multiple organ cancers defined as breast cancer patients with other primary organ cancer except ovarian cancer, and male cancer. The clinicopathological factors considered were age at diagnosis of breast cancer, stage, and the hormone receptor (including estrogen and PR) and HER2 status.
The frequencies of mutations were presented according to familial and personal factors and clinicopathological characteristics. All analyses were conducted using SAS 9.2 software (SAS Institute, Cary, NC).
Results
Mutational status according to subjects’ characteristics
Pathogenic mutations in BRCA1/2 genes, including large genomic rearrangements, were detected in 63 of 478 (13.2%) patients. In total, 30 BRCA1 mutation carriers (6.3%) and 33 BRCA2 mutation carriers (6.9%) were identified. Table 1 shows the frequency of BRCA1/2 mutations according to familial and personal factors, and clinicopathological characteristics. BRCA1/2 mutation prevalence in familial breast cancer cases was 15.9%, which was significantly higher in non-familial breast cancer cases (6.0%, P = 0.004). When personal factors were considered, BRCA1/2 mutations were observed in 11.6% of early-onset breast cancer patients, 14.9% of bilateral breast cancer patients, and 66.7% of patients who were diagnosed with both breast and ovarian cancer. The prevalence of BRCA1/2 mutations also differed significantly according to hormone receptor and HER2 status (P = 0.003 and <0.001, respectively). The prevalence of BRCA1/2 mutation according to the combinations of familial and personal factors with denominators and numerators is described in Appendix Table 4.
Table 1.
The frequencies of BRCA1 and BRCA2 mutationsa in high-risk breast cancer patients according to familial and personal risk factors (N = 478)
| Risk category | Total | BRCA1 mutation | BRCA2 mutation | BRCA1/2 mutation |
|---|---|---|---|---|
| N (%d) | N (%e) | N (%e) | N (%e) | |
| Family history | ||||
| Breast cancer family only (without ovarian cancer)§ | 303 (63.4) | 14 (4.6) | 29 (9.6) | 43 (14.2) |
| 1 breast cancer family | 253 (52.9) | 11 (4.4) | 17 (6.7) | 28 (11.1) |
| 2 ≤breast cancer families*,§ | 50 (10.5) | 3 (6.0) | 12 (24.0) | 15 (30.0) |
| Breast cancer families in 1st degree relativesb | 217 (45.4) | 13 (6.0) | 20 (9.2) | 33 (15.2) |
| Breast cancer families in second/third degree relativesb,§ | 86 (18.0) | 1 (1.2) | 9 (10.5) | 10 (11.6) |
| Ovarian cancer familyb | ||||
| Without breast cancer§ | 29 (6.1) | 6 (20.7) | 0 (0.0) | 6 (20.7) |
| With breast cancer* | 13 (2.7) | 4 (30.8) | 2 (15.4) | 6 (46.2) |
| Any of breast/ovarian cancer familiesc,* | 345 (72.2) | 24 (7.0) | 31 (9.0) | 55 (15.9) |
| No family history* | 133 (27.8) | 6 (4.5) | 2 (1.5) | 8 (6.0) |
| Personal history | ||||
| Early-onset breast cancer (age < 40) | 199 (41.6) | 12 (6.0) | 11 (5.5) | 23 (11.6) |
| Bilateral breast cancer | 47 (9.8) | 2 (4.3) | 5 (10.6) | 7 (14.9) |
| Multiple organ cancersf | 27 (5.6) | 0 (0.0) | 2 (7.4) | 2 (7.4) |
| Both breast and ovarian cancer* | 6 (1.3) | 3 (50.0) | 1 (16.7) | 4 (66.7) |
| Male breast cancer | 4 (0.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Clinicopathological factor | ||||
| Age at diagnosis | ||||
| <40 | 186 (38.9) | 12 (6.5) | 11 (6.8) | 23 (14.3) |
| 40–49 | 172 (36.0) | 10 (5.8) | 15 (8.7) | 25 (14.5) |
| 50–59 | 89 (18.6) | 7 (7.9) | 6 (6.7) | 13 (14.6) |
| 60–79 | 31 (6.5) | 1 (3.3) | 1 (3.3) | 2 (6.5) |
| Stage* | ||||
| 0 | 55 (11.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| I | 173 (36.2) | 12 (6.9) | 12 (6.9) | 24 (13.9) |
| II | 159 (33.3) | 12 (7.6) | 18 (11.3) | 30 (18.9) |
| III+ | 88 (18.4) | 5 (7.4) | 2 (2.9) | 7 (10.3) |
| Unknown | 3 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Hormone receptor status*,§ | ||||
| ER+ & PR+ | 307 (64.2) | 5 (1.6) | 25 (8.1) | 20 (9.8) |
| ER+ & PR− | 44 (9.2) | 1 (2.3) | 4 (9.1) | 5 (11.4) |
| ER− & PR+ | 7 (1.5) | 1 (14.3) | 0 (0.0) | 1 (14.3) |
| ER− & PR− | 115 (24.1) | 23 (20.0) | 4 (3.5) | 27 (23.5) |
| Unknown | 5 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Subtype according to hormone receptor and HER2 status*,§ | ||||
| HR+ & HER2− | 252 (52.7) | 6 (2.4) | 19 (7.5) | 25 (9.9) |
| HR− & HER2+ | 28 (5.9) | 1 (3.6) | 0 (0.0) | 1 (3.6) |
| HR+ & HER2+ | 37 (7.7) | 0 (0.0) | 2 (5.4) | 2 (5.4) |
| Triple-negative | 76 (15.9) | 19 (25.0) | 3 (4.0) | 22 (29.0) |
| Unclassifiable | 85 (17.8) | 4 (4.7) | 9 (10.6) | 13 (15.3) |
| Total | 478 (100.0) | 30 (6.3) | 33 (6.9) | 63 (13.2) |
HR hormone receptor, HER2 human epidermal growth factor receptor 2
* P value <0.05 for BRCA1/2 mutation prevalence between those included in each category and those not
§ P value <0.05 between BRCA1 and BRCA2 ratio in carriers
aIncluding three patients with large genomic rearrangements in BRCA1 gene
bAmong 42 patients who had family history of ovarian cancer, 40 had one family member with ovarian cancer history and 2 had two family members with ovarian cancer history
cClosest degree of relatives with breast cancer
dPercent among all subjects (column percent)
ePercent among subjects with each risk category (row percent)
fMultiple organ cancer was defined as breast cancer patients with other primary organ cancer except ovarian cancer
Table 4.
The frequencies of BRCA1 and BRCA2 mutations according to combined family and personal characteristics
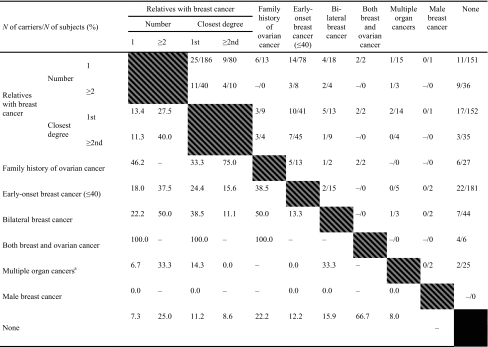
|
aMultiple organ cancer was defined as breast cancer patients with other primary organ cancer except ovarian cancer
Patients with large genomic rearrangements
Three BRCA1 deletion carriers were identified by MLPA from 306 patients BRCA1/2 mutation negative by standard sequencing. These deletion mutations account for 10% of all BRCA1 mutation carriers (Fig. 1). The characteristics of each patient are summarized in Table 2 and detailed explanation is as following. Patient A, diagnosed with ductal carcinoma in situ with TNBC at the age of 51, carried exons 5–8 deletion (Fig. 2a). She had a second- and a third-degree relative with breast cancer. Estimated mutation probabilities for BRCA1/2 before mutation test were 0.8% by BRCAPRO and 5.3% by Myriad. Following genetic counseling about the BRCA1 deletion, genetic testing was performed on her daughter and three sisters without breast cancer and her daughter was found to have the same deletion.
Table 2.
Characteristics of the probands with BRCA1 large genomic rearrangements
| Personal factor | Familial factor | Mutation carrier risk | ||||||
|---|---|---|---|---|---|---|---|---|
| Pt | Early-onset BC (age at diagnosis) | Bilateral BC | Both BC and OC | TNBC | Family history of BC (number, closest degree) | Family history of OC (number, closest degree) | Other cancer | BRCAPRO/Myriad |
| A | No (51) | No | No | Yes | Yes (2, second degree) | No | Liver Cervix Stomach Thyroid Colon |
0.8/5.3 |
| B | Yes (35) | No | No | Yes | Yes (1, second degree) | Yes (1, 1st degree) | Thyroid | 57.2/39.2 |
| C | Yes (33) | Yes | Yes | No | Yes (1, first degree) | No | Lung | 51.2/15.8 |
Fig. 2.
The 3 BRCA1 LGRs identified in the study using MLPA screening. The MLPA analysis demonstrates a exons 5–8 deletion, b exons 22–24 deletion, and c exons 1–14 deletion. Exons having a reduced peak ratio are denoted with the arrows
Patient B, diagnosed with stage III TNBC at the age of 35, carried exons 22–24 deletion (Fig. 2b). The patient had a first-degree relative with ovarian cancer and a second-degree relative with breast cancer. Estimated mutation probabilities for BRCA1/2 mutation test were 57.2% by BRCAPRO and 39.2% by Myriad. Genetic testing was performed on her four sisters without breast cancer and of them, two sisters were found to have the deletion.
Patient C, was diagnosed with bilateral stage I breast cancer for the first time at age 33 and for the second time at age 46. She was not a TNBC case, and carried a BRCA1 gene lacking exons 1–14 (Fig. 2c). The estimated mutation probabilities for BRCA1/2 mutation test were 51.2% by BRCAPRO and 15.8% by Myriad. After enrollment in this study, she was newly diagnosed with ovarian cancer. Genetic testing was performed on her two sisters and one of her sisters, diagnosed before with bilateral breast cancer, carried a BRCA1 gene with the same large genomic rearrangements.
Pathogenic variants of BRCA1/2 genes found in this study
The pathogenic variants of BRCA1/2 genes found in this study are presented in Table 3. Overall, 24 pathogenic variants in BRCA1 gene and 19 in BRCA2 gene were found in 63 index cases mutations. Many of these mutations have been reported in the BIC or previous studies in Korea. The most frequent mutation was c.7480C>T in BRCA2 and it was found in nine patients (14.3%). The next most frequent mutations were c.1399A>T in BRCA2 gene and c.390C>A in BRCA1 gene, which were found in four and three patients, respectively. The large genomic rearrangements found through MLPA were located in BRCA1 gene, including an exon 1–14 deletion, exon 5–8 deletion, and exon 22–24 deletion.
Table 3.
Frequency of pathogenic variants of BRCA1 and BRCA2 genes in patients
| Gene | Exon/intron | BIC nomenclature | HGVS cDNA | HGVS protein | N |
|---|---|---|---|---|---|
| BRCA1 | IVS5 | IVS5+1G>A | c.212+1G>A | – | 1 |
| 7 | 509C>A | c.390C>A | p.Tyr130* | 3 | |
| 11 | 1041_1042delAG | c.922_923delAG | p.Ser308Glufs* | 1 | |
| 11 | c.922_924delAGCinsT | p.Ser308* | 1 | ||
| 11 | 1137delG | c.1018delG | p.Val340Glyfs* | 1 | |
| 11 | 1599C>T | c.1480C>T | p.Gln494* | 1 | |
| 11 | c.14923_1494delTC | p.Leu498Hisfs* | 1 | ||
| 11 | 1630dupG | c.1511dupG | p.Lys505* | 1 | |
| 11 | c.1516delA | p.Arg506Glysfs* | 2 | ||
| 11 | c.2354T>A | p.Leu785* | 1 | ||
| 11 | 3415delC | c.3296delC | p.Pro1099Leufs* | 1 | |
| 11 | 3746dupA | c.3627dupA | p.Leu1210Glufs* | 1 | |
| 11 | 3819del5 | c.3700_3704delGTAAA | p.Val1210Aspfs* | 1 | |
| c.4110C>T | p.Glu1331* | 1 | |||
| 16 | 5100G>T | c.4981G>T | p.Glu1661* | 2 | |
| 20 | 5379G>T | c.5260G>T | p.Glu1754* | 2 | |
| IVS21 | c.5332+4delA | – | 1 | ||
| 23 | c.5445G>A | p.Trp1815* | 2 | ||
| IVS23 | IVS23+1G | c.5467+1G>A | – | 1 | |
| 24 | 5602delG | c.5483delG | p.Cys1828Leufs* | 1 | |
| 24 | 5615del11insA | c.5496_5506delGGTGACCCGAGinsA | p.Val1833Serfs* | 1 | |
| 1–14 | Exon 1–14 deletion | 1 | |||
| 5–8 | Exon 5–8 deletion | 1 | |||
| 22–24 | Exon 22–24 deletion | 1 | |||
| BRCA2 | 7 | 173G>T | c.518G>T | p.Gly173Val | 1 |
| 9 | 983del4 | c.755_758delACAG | p.Asp252Serfs* | 2 | |
| 10 | 1222delA | c.994delA | p.Ile332Phefs* | 1 | |
| 10 | 1627A>T | c.1399A>T | p.Lys467* | 4 | |
| 11 | 3026delCA | c.2798_2799delCA | p.Thr933Argfs* | 1 | |
| 11 | c.3096_3110delAGATATTGAAGAAC | p.Asp1033Ilefs* | 1 | ||
| 11 | 3972del4 | c.3744_3747delTGAC | p.Ser1248Glufs* | 3 | |
| 11 | 6019C>T | c.5791C>T | p.Gln1931* | 1 | |
| 11 | 6781delG | c.6553delG | p.Ala2185Leufs* | 1 | |
| 14 | c.7258G>T | p.Glu2420* | 2 | ||
| 15 | 7708C>T | c.7480C>T | p.Arg2494* | 9 | |
| 15 | c.7486G>T | p.Glu2420* | 1 | ||
| 18 | c.8300_8301insAC | p.Pro2767Hisfs* | 1 | ||
| 22 | 9179C>G | c.8951C>G | p.Ser2984* | 1 | |
| 23 | 9304C>T | c.9076C>T | p.Gln3026* | 1 | |
| 24 | c.9253delA | p.Thr3085Glnfs* | 1 | ||
| 25 | 9503del2 | c.9275_9276delAT | p.Tyr3092Phefs* | 1 | |
| 25 | 9641T>G | c.9413T>G | p.Leu3138* | 1 |
Discussion
Here, we detected BRCA1/2 mutations in 13.2% of high-risk breast cancer patients who were referred to a genetic counseling center. Of the BRCA1/2 carriers, 5% (3 out of 63) were identified by MLPA after negative direct sequencing results. All the large genomic rearrangements were found in BRCA1 gene. Therefore, 10% of BRCA1 carriers (3 out of 30) would have not been identified if MLPA had not been conducted. In Korea, BRCA1/2 mutation screening has been covered by National Health Insurance since 2012 for those who meet certain criteria. Because more than 95% of this study population was recruited after 2012, our results could provide a more current measure of the prevalence of BRCA1/2 mutations.
The prevalence in familial breast cancer cases was 15.9% in this study, which was slightly lower than previous results from Korea, which ranged 19.4–30.0% [19–22], and results from Western countries [23]. Mutation prevalence according to each personal factor in this study was comparable with previous Korean studies [19, 21, 22, 24, 25]. TNBC is an important factor used to select breast cancer patients for BRCA1/2 mutation testing [5] and this was confirmed in our study. Considering that the mutation prevalence varies according to overlapped risk factors (Appendix Table 4), multiple combinations of familial or personal factors need to be considered for a more detailed risk assessment.
The prevalence of large genomic rearrangement after negative direct sequencing results in previous studies targeting potential hereditary cancer subjects in diverse ethnic groups ranged 0–5%, and was particularly low in Asian countries [12, 26–33] (Appendix Table 5). With the exception of two studies, most large genomic rearrangements have been identified in the BRCA1 gene [12, 28], and all of the large genomic rearrangements identified in Korea [9, 10], including in this study, were in the BRCA1 gene. The common characteristics of seven BRCA1 large genomic rearrangement cases were that they all have a family history of breast and/or ovarian cancer with at least one additional personal factor. These personal factors included bilateral breast cancer, young age at onset (≤40 years old), both breast and ovarian cancer, and TNBC. Therefore, at least for BRCA1, the MLPA test should be considered for breast cancer patients with a family history of breast and/or ovarian cancer and additional personal factors such as bilateral breast cancer, young age at onset, and TNBC during the genetic counseling process. In our study population with large genomic rearrangements, the compliance with the prophylactic strategies was quite good (Appendix Table 6).
Table 5.
The prevalence of BRCA1/2 mutation and large genomic rearrangements in subjects without small mutation from direct sequencing
| Country | Number of subject | Total prevalence (%) | Prevalence among subjects with negative result of direct sequencing (%) | ||
|---|---|---|---|---|---|
| BRCA1 | BRCA2 | LRG in BRCA1 | LRG in BRCA2 | ||
| Hong Kong [24] | 1236 | 4.6 | 5.1 | 0.4 | 0.3 |
| Poland [27] | 281 | 28.8 | – | 1.5 | – |
| Lebanese Arab [22] | 250 | 2.8 | 2.8 | 0.0 | 0.0 |
| Mexico [29] | 188 | 18.0 | 3.2 | 2.0 | 0.0 |
| Slovakia [23] | 585 | 14.5 | 11.5 | 5.0 | 0.0 |
| Malaysia [11] | 324 | 7.4 | 7.1 | 0.6a | 0.3a |
| Malaysia [28] | 100 | – | – | 2.0a | 0.0 |
| Poland [26] | 64 | 67 | 4.6 | 0.0 | |
| Singapore [25] | 100 | – | – | 2.0 | 1.0 |
| Korea [8] | 221 | 35.3 | 2.1 | 0.0 | |
| Korea [9] | 122 | – | – | 0.8 | 0.0 |
| Korea—this study | 478 | 6.3 | 6.9 | 1.0 | 0.0 |
aNot certain whether the prevalence is from whole subject or from limited subjects with BRCA1/2 mutation negative
Table 6.
Follow-up status of the family members with multiple ligation-dependent probe amplification (MLPA) positive result from family with BRCA1 large genomic rearrangements
| Pt | Relationship (age) | Interval between proband identification and first genetic counseling | Known comorbidities at first genetic counseling | Interval between MLPA (+) to first preventive screening | Result of initial preventive screening for ovarian cancer | Result of initial preventive screening for breast cancer | Conducted prophylactic therapy and family care | Future plan for prophylaxis and family care |
|---|---|---|---|---|---|---|---|---|
| A | Daughter (23) |
5 months | None | 4 days | None | None | Oral contraceptive | Annual breast cancer screening until the age 25 Ovarian cancer screening per 3 months Recommended family screening for brother (age 25) |
| B | Sister 1 (39) | 4 months | Thyroid disease | 13 days | None | None | BSO (3 months after identifying carrier) | |
| Sister 2 (45) | 4 months | None | 13 days | None | None | BSO (3 months after identifying carrier) | Recommended family screening for 2 sons in the future (age 19 and 16) | |
| C | Sister (44) | 1 months | Left breast cancer (TNBC, age 40) Right breast cancer (HR positive, age 44) |
1 day | None | Known breast cancer | – | BSO Recommended family screening for 2 daughters in the future (age 15 and 13) |
BSO bilateral salpingo-oophorectomy, TNBC triple-negative breast cancer, HR hormone receptor
Several limitations of this study should be mentioned. Firstly, this study was performed by the patients of a single institute. Secondly, we could not conduct MLPA test for all BRCA1/2 small mutation non-carriers due to patient non-participation, suggesting possible participation bias. However, this study was designed to reflect common clinical settings, following the patients in the process of genetic counseling. Thirdly, the detected large genomic rearrangements through MLPA test were not confirmed by different MLPA probes or other platforms. However, the three detected large genomic rearrangements were multi-exon deletions and detected in multiple family members, showing apparent inheritance patterns, suggesting minimum probabilities of false positives. Fourth, the family history was obtained by proband recollection and we did not consider the validity and reliability of the information.
In conclusion, the prevalence of BRCA1/2 mutations was dependent on familial and personal factors. Subjects with both familial and personal factors had a much higher risk of carrying BRCA1/2 mutations. The MLPA test for BRCA1 mutation could be recommended for breast cancer patients with a family member with breast and/or ovarian cancer and additional personal factors, and who tested negative for BRCA1/2 small mutations in initial testing.
Acknowledgments
Funding
This study was funded by the National Cancer Center, Korea (Grant Numbers 1410691-3 and 1710172-1).
Appendix
Compliance with ethical standards
Conflict of interest
All of the authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study protocol was approved by the institutional review board of the National Cancer Center (NCC2015-0177).
Informed consent
Among the 418 cases designated as non-carriers by direct sequencing, written informed consent was obtained from 306 patients, and they were screened for the presence of large genomic rearrangements using MLPA assay. From 112 patients from whom the informed consent was not obtained, informed consent was exempted because this study was based on clinical chart review retrospectively. This study was approved by the institutional review board of National Cancer Center (NCC2015-0177).
Footnotes
Boyoung Park, Ji Yeon Sohn, Sun-Young Kong, and Eun Sook Lee have contributed equally to this work.
Contributor Information
Sun-Young Kong, Phone: +82-31-920-1735, Email: ksy@ncc.re.kr.
Eun Sook Lee, Phone: +82-31-920-1503, Email: eslee@ncc.re.kr.
References
- 1.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62(3):676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25(11):1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 5.NCCN: Genetic/Familial High-Risk Assessment: Breast and Ovarian. 2015, Version 1
- 6.Moyer VA. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;160(4):271–281. doi: 10.7326/M13-2747. [DOI] [PubMed] [Google Scholar]
- 7.Judkins T, Rosenthal E, Arnell C, Burbidge LA, Geary W, Barrus T, Schoenberger J, Trost J, Wenstrup RJ, Roa BB. Clinical significance of large rearrangements in BRCA1 and BRCA2. Cancer. 2012;118(21):5210–5216. doi: 10.1002/cncr.27556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011;125(2):325–349. doi: 10.1007/s10549-010-0817-z. [DOI] [PubMed] [Google Scholar]
- 9.Seong MW, Cho SI, Kim KH, Chung IY, Kang E, Lee JW, Park SK, Lee MH, Choi DH, Yom CK, et al. A multi-institutional study of the prevalence of BRCA1 and BRCA2 large genomic rearrangements in familial breast cancer patients. BMC Cancer. 2014;14:645. doi: 10.1186/1471-2407-14-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seong MW, Cho SI, Noh DY, Han W, Kim SW, Park CM, Park HW, Kim SY, Kim JY, Park SS. Low contribution of BRCA1/2 genomic rearrangement to high-risk breast cancer in the Korean population. Fam Cancer. 2009;8(4):505–508. doi: 10.1007/s10689-009-9279-z. [DOI] [PubMed] [Google Scholar]
- 11.Ewald IP, Ribeiro PL, Palmero EI, Cossio SL, Giugliani R, Ashton-Prolla P. Genomic rearrangements in BRCA1 and BRCA2: a literature review. Genet Mol Biol. 2009;32(3):437–446. doi: 10.1590/S1415-47572009005000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang P, Mariapun S, Phuah SY, Lim LS, Liu J, Yoon SY, Thong MK, Mohd Taib NA, Yip CH, Teo SH. Large BRCA1 and BRCA2 genomic rearrangements in Malaysian high risk breast-ovarian cancer families. Breast Cancer Res Treat. 2010;124(2):579–584. doi: 10.1007/s10549-010-1018-5. [DOI] [PubMed] [Google Scholar]
- 13.Montagna M, Dalla Palma M, Menin C, Agata S, De Nicolo A, Chieco-Bianchi L, D’Andrea E. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet. 2003;12(9):1055–1061. doi: 10.1093/hmg/ddg120. [DOI] [PubMed] [Google Scholar]
- 14.Palma MD, Domchek SM, Stopfer J, Erlichman J, Siegfried JD, Tigges-Cardwell J, Mason BA, Rebbeck TR, Nathanson KL. The relative contribution of point mutations and genomic rearrangements in BRCA1 and BRCA2 in high-risk breast cancer families. Cancer Res. 2008;68(17):7006–7014. doi: 10.1158/0008-5472.CAN-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. BayesMendel: an R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol. 2004;3(1):1063. doi: 10.2202/1544-6115.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parmigiani G, Berry DA, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62(1):145–158. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20(6):1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 18.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. Committee MSotALQA: ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10(4):294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 19.Ahn SH, Son BH, Yoon KS, Noh DY, Han W, Kim SW, Lee ES, Park HL, Hong YJ, Choi JJ, et al. BRCA1 and BRCA2 germline mutations in Korean breast cancer patients at high risk of carrying mutations. Cancer Lett. 2007;245(1–2):90–95. doi: 10.1016/j.canlet.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 20.Han SA, Kim SW, Kang E, Park SK, Ahn SH, Lee MH, Nam SJ, Han W, Bae YT, Kim HA, et al. The prevalence of BRCA mutations among familial breast cancer patients in Korea: results of the Korean Hereditary Breast Cancer study. Fam Cancer. 2013;12(1):75–81. doi: 10.1007/s10689-012-9578-7. [DOI] [PubMed] [Google Scholar]
- 21.Kang E, Seong MW, Park SK, Lee JW, Lee J, Kim LS, Lee JE, Kim SY, Jeong J, Han SA, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast Cancer Res Treat. 2015;151(1):157–168. doi: 10.1007/s10549-015-3377-4. [DOI] [PubMed] [Google Scholar]
- 22.Seong MW, Cho S, Noh DY, Han W, Kim SW, Park CM, Park HW, Kim SY, Kim JY, Park SS. Comprehensive mutational analysis of BRCA1/BRCA2 for Korean breast cancer patients: evidence of a founder mutation. Clin Genet. 2009;76(2):152–160. doi: 10.1111/j.1399-0004.2009.01202.x. [DOI] [PubMed] [Google Scholar]
- 23.Szabo CI, King M-C. Population genetics of BRCA1 and BRCA2. Am J Hum Genet. 1997;60(5):1013. [PMC free article] [PubMed] [Google Scholar]
- 24.Son BH, Ahn SH, Kim SW, Kang E, Park SK, Lee MH, Noh WC, Kim LS, Jung Y, Kim KS, et al. Prevalence of BRCA1 and BRCA2 mutations in non-familial breast cancer patients with high risks in Korea: the Korean Hereditary Breast Cancer (KOHBRA) Study. Breast Cancer Res Treat. 2012;133(3):1143–1152. doi: 10.1007/s10549-012-2001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi DH, Lee MH, Bale AE, Carter D, Haffty BG. Incidence of BRCA1 and BRCA2 mutations in young Korean breast cancer patients. J Clin Oncol. 2004;22(9):1638–1645. doi: 10.1200/JCO.2004.04.179. [DOI] [PubMed] [Google Scholar]
- 26.El Saghir NS, Zgheib NK, Assi HA, Khoury KE, Bidet Y, Jaber SM, Charara RN, Farhat RA, Kreidieh FY, Decousus S. BRCA1 and BRCA2 mutations in ethnic Lebanese Arab women with high hereditary risk breast cancer. Oncologist. 2015;20(4):357–364. doi: 10.1634/theoncologist.2014-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konecny M, Milly M, Zavodna K, Weismanova E, Gregorova J, Mlkva I, Ilencikova D, Kausitz J, Bartosova Z. Comprehensive genetic characterization of hereditary breast/ovarian cancer families from Slovakia. Breast Cancer Res Treat. 2011;126(1):119–130. doi: 10.1007/s10549-010-1325-x. [DOI] [PubMed] [Google Scholar]
- 28.Kwong A, Chen J, Shin VY, Ho JC, Law FB, Au CH, Chan T-L, Ma ES, Ford JM. The importance of analysis of long-range rearrangement of BRCA1 and BRCA2 in genetic diagnosis of familial breast cancer. Cancer Genet. 2015;208(9):448–454. doi: 10.1016/j.cancergen.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 29.Lim Y, Iau P, Ali A, Lee S, Wong JL, Putti T, Sng JH. Identification of novel BRCA large genomic rearrangements in Singapore Asian breast and ovarian patients with cancer. Clin Genet. 2007;71(4):331–342. doi: 10.1111/j.1399-0004.2007.00773.x. [DOI] [PubMed] [Google Scholar]
- 30.Ratajska M, Brozek I, Senkus-Konefka E, Jassem J, Stepnowska M, Palomba G, Pisano M, Casula M, Palmieri G, Borg A. BRCA1 and BRCA2 point mutations and large rearrangements in breast and ovarian cancer families in Northern Poland. Oncol Rep. 2008;19(1):263. [PubMed] [Google Scholar]
- 31.Rudnicka H, Debniak T, Cybulski C, Huzarski T, Gronwald J, Lubinski J, Gorski B. Large BRCA1 and BRCA2 genomic rearrangements in Polish high-risk breast and ovarian cancer families. Mol Biol Rep. 2013;40(12):6619–6623. doi: 10.1007/s11033-013-2775-0. [DOI] [PubMed] [Google Scholar]
- 32.Sharifah NA, Nurismah MI, Lee HC, Aisyah AN, Clarence-Ko CH, Naqiyah I, Rohaizak M, Fuad I, Jamal ARA, Zarina AL. Identification of novel large genomic rearrangements at the BRCA1 locus in Malaysian women with breast cancer. Cancer Epidemiol. 2010;34(4):442–447. doi: 10.1016/j.canep.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Villarreal-Garza C, Alvarez-Gómez RM, Pérez-Plasencia C, Herrera LA, Herzog J, Castillo D, Mohar A, Castro C, Gallardo LN, Gallardo D. Significant clinical impact of recurrent BRCA1 and BRCA2 mutations in Mexico. Cancer. 2015;121(3):372–378. doi: 10.1002/cncr.29058. [DOI] [PMC free article] [PubMed] [Google Scholar]





