Abstract
Cyclophilin A (CypA) is a peptidyl-prolyl isomerase that binds to the capsid protein (CA) of human immunodeficiency virus type 1 (HIV-1) and by doing so facilitates HIV-1 replication. Although CypA is incorporated into HIV-1 virions by virtue of CypA-Gag interactions that occur during virion assembly, in this study we show that the CypA-CA interaction that occurs following the entry of the viral capsid into target cells is the major determinant of CypA's effects on HIV-1 replication. Specifically, by using normal and CypA-deficient Jurkat cells, we demonstrate that the presence of CypA in the target and not the virus-producing cell enhances HIV-1 infectivity. Moreover, disruption of the CypA-CA interaction with cyclosporine A (CsA) inhibits HIV-1 infectivity only if the target cell expresses CypA. The effect of CsA on HIV-1 infection of human cells varies according to which particular cell line is used as a target, and CA mutations that confer CsA resistance and dependence exert their effects only if target cells, and not if virus-producing cells, are treated with CsA. The differential effects of CsA on HIV-1 infection in different human cells appear not to be caused by polymorphisms in the recently described retrovirus restriction factor TRIM5α. We speculate that CypA and/or CypA-related proteins affect the fate of incoming HIV-1 capsid either directly or by modulating interactions with unidentified host cell factors.
Cyclophilin A (CypA) is the most abundant member of a ubiquitous family of peptidyl-prolyl isomerases and binds to the capsid (CA) domain of human immunodeficiency virus type 1 (HIV-1) and SIVcpz Gag proteins (26). Consequently, it is quite efficiently packaged into HIV-1 and SIVcpz virions, with about 1 molecule of CypA incorporated per 10 Gag molecules (15, 38). The CypA-Gag interaction can be blocked by an immunosuppressive drug, cyclosporine A (CsA), and its analogues (7, 14, 26), and this manipulation inhibits the replication of most HIV-1 strains in most cells in vitro (5, 7, 10, 14, 15, 21, 33, 38, 41, 42). In addition, mutation of residues G89 or P90 of HIV-1 CA, which constitute the core of the CypA binding site (16, 46), both disrupt CypA binding and confer a substantial replication defect in human cells (2, 9, 12, 15), consistent with the notion that the CypA-CA interaction is important for HIV-1 replication.
These observations led to the proposal of a number of models incorporating attachment, entry, or postentry steps of the viral life cycle to explain why HIV-1 replication is uniquely dependent on it ability to incorporate CypA into virions (31, 34, 35). The most reasonable and popular suggestion was that, by virtue of its direct interaction with CA or its peptidyl-prolyl isomerase activity, CypA affected capsid uncoating or some other step in the subsequent and poorly characterized postentry phases of the HIV-1 life cycle (17, 23, 25, 43).
A recently proposed model to explain why HIV-1 CA uniquely binds to CypA invokes a requirement for CypA expressed by the target cell, rather than virion-encapsidated CypA, and is based on the fact that the CypA binding site is coincident with a critical determinant for recognition by CA-specific endogenous restriction factors (24, 40). These inhibitors, originally termed Lv1 and Ref1 and now known to be encoded by the α-spliced variant of the TRIM5 gene (20, 22, 30, 37, 44), target the capsid of widely divergent incoming retroviruses (6, 13, 18, 28, 39, 40). Mutations in the CypA binding site of HIV-1 CA or treatment of target cells with CsA allows HIV-1 capsids to saturate TRIM5α/Ref1, presumably by binding to it (19, 40). Thus, acquisition of CypA by incoming HIV-1 capsids may be a defense against restriction factors in human cells. In support of this notion, treatment of certain human target cells with CsA modestly inhibits (two- to fivefold) infection in single-cycle HIV-1 infectivity assays (24, 40). Remarkably, however, the CypA binding property of HIV-1 CA dramatically inhibits infection of cells of owl monkey origin (40), and infection by HIV-1 is strongly dependent on the pharmacological or genetic inhibition of CypA binding activity. Recently this was shown to be due to the presence of a novel restriction factor, TRIM-CyP, generated in owl monkeys as a consequence of transposition of a CypA pseudogene into the TRIM5 locus (36). These studies showed that functionally relevant cyclophilin-CA interactions can, in principle, occur in target cells rather than virus-producing cells or virions.
Other studies have shown that replication of HIV-1 in human cells in the presence of CsA selects for two mutations in CA, A92E and G94D, that obviate the requirement for CypA, i.e., they confer on HIV-1 the ability to replicate in the presence of CsA (1). While these mutations do not themselves appear to affect CypA-CA binding (8), they do reverse the infectivity defect observed in CypA binding site mutant HIV-1 capsids (8, 45). Remarkably, when present in isolation, A92E and G94D render HIV-1 replication dependent on CsA or CypA binding site mutations in some human cell lines, such as HeLa-CD4 and H9 (1, 45). In other cell lines, such as Jurkat, the mutations confer CsA resistance but not dependence (8, 45).
In this study we show that manipulations which prevent the CypA-CA interaction during HIV-1 assembly or production generally have little or no effect on HIV-1 infectivity. Moreover, CA mutations A92E and G94D, which confer CsA resistance or dependence, do not manifest their effects during virus production but do affect the progression of early phases of the HIV-1 life cycle and do so in a target cell type-dependent manner. Thus, cyclophilin-CA interactions that occur in human target cells, rather than during virus production, are major determinants of CsA and cyclophilin effects on HIV-1 replication. Moreover, the phenotype of HIV-1 mutants that require CsA for infection of human cells is reminiscent of that of wild-type HIV-1 in owl monkey cells (40). These facts, and the finding that the A92E mutation reverses the ability of CypA binding site mutant HIV-1 CA to saturate Ref1/TRIM5α in human TE671 cells (19), are consistent with the notion that sequences in and around the CypA binding site influence recognition by species-specific restriction factors (19, 29). Nevertheless, human and rhesus monkey versions of TRIM5α do not discriminate between wild-type and CsA-resistant or -dependent HIV-1.
MATERIALS AND METHODS
Virus and vector constructs.
CA mutations (A92E and G94D) were introduced into an HIV-1 NL4-3-based clone by using PCR-based methods as previously described (47). Each mutant Gag was transferred to the previously described HIV-1/GFP construct (13) as a BssHII-SalI fragment, and the presence of the CA mutation in the final HIV-1/GFP construct was verified by DNA sequencing. For most experiments, an env− HIV-1/GFP construct was used; however, for some experiments (see Fig. 1 and 4, below) otherwise identical constructs containing full-length env from HXB3 were used. To generate HIV-1 Gag-Pol expression plasmids, wild-type or mutant Gag-Pol-encoding sequences were amplified by PCR using primers that incorporated 5′ EcoRI and 3′ NotI sites and were inserted into the previously described expression vector, pCRV1 (18).
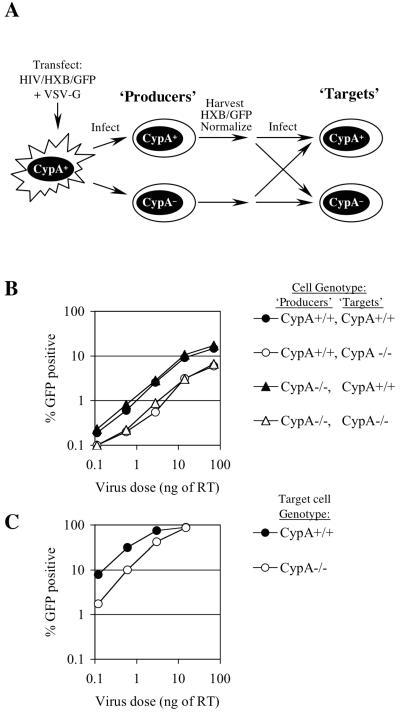
FIG. 1.
Effects of CypA expression in HIV-1-producing and target cells on HIV-1 infection during a single cycle of replication. (A) Experimental scheme. 293T cells were transfected with a full-length HIV-1 reporter virus (HIV-1/HXB/GFP) and VSV-G, and the resulting pseudotyped virus stock was used to infect normal or CypA-null Jurkat cells (producers) at high multiplicity. Virus stocks produced by these cells were normalized for reverse transcriptase content and titrated on normal or CypA-null Jurkat cells (targets). (B) Titration of HIV/HXB/GFP derived from normal and CypA-null Jurkat producers on normal and CypA-null Jurkat targets, as indicated. The percentage of infected GFP-positive cells is plotted as a function of input virus dose, in nanograms of reverse transcriptase (RT). (C) Titration of VSV-G-pseudotyped HIV-1/GFP in normal and CypA-null Jurkat target cells.
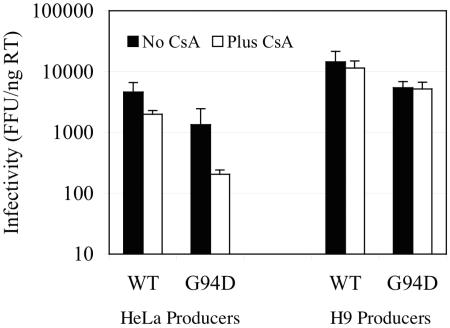
FIG. 4.
Effects of CsA treatment of HIV-1-producing cells on the infectivity of progeny virions. 293T cells were transfected with a full-length wild-type (WT) or G94D mutant HIV-1 reporter virus (HIV/HXB/GFP) and VSV-G, and the resulting pseudotyped virus stock was used to infect HeLa and H9 cells at high multiplicity. Infected cells were split in two, and one half was treated with 5 μM CsA. Virus stocks produced by these cells were normalized for reverse transcriptase content and titrated on TE671-CD4 cells. The number of GFP-positive foci (FFU) generated per nanogram of RT is plotted for cultures infected with HIV-1/HXB/GFP derived from HeLa or H9 cells treated with carrier alone or with CsA.
Cell lines and viruses.
Adherent human (TE671, HOS, HeLa, and 293T) and murine (MDTF) cells were maintained in Dulbecco's modified Eagle's medium supplemented with fetal calf serum and antibiotics. Human T-cell lines (wild type and CypA−/− Jurkat [11], H9, and CEMx174 cells) were maintained in RPMI supplemented with fetal calf serum and antibiotics. Wild-type or mutant GFP-reporter virus stocks were made by cotransfecting 293T cells with a proviral plasmid and a vesicular stomatitis virus G protein (VSV-G) expression plasmid as previously described (13, 18). HIV-1 vector stocks for infection of MDTF cells (which lack a cyclinT1 that can support HIV-1 Tat function) were made in the same way except that HIV-1 Gag-Pol expression plasmids and an HIV-1-based vector (CSGW) encoding a green fluorescent protein (GFP) reporter driven by a spleen necrosis virus promoter (4) were used in place of the proviral plasmid. All reporter virus stocks were quantified using a colorimetric reverse transcription assay (Cavidi Tech). N-tropic murine leukemia virus (N-MLV) reporter virus stocks were generated by cotransfecting 293T cells with an N-MLV Gag Pol, a GFP-expressing MLV retroviral vector, and a VSV-G expression plasmid, as previously described (18, 39). N-MLV stocks were quantified by infectivity titration on Fv1-null MDTF cells.
Infection assays.
Adherent target cells were seeded at 2 × 104 cells/well in 24-well trays the day before infection. T-cell targets were seeded at 4 × 104/well in 48-well trays the day of infection. Cells were inoculated with fivefold-serially diluted GFP reporter virus stocks in the presence of 5 μg of Polybrene/ml. In some experiments, CsA was included in the medium at 5 μM at the time of infection. Virus and CsA were replaced with fresh medium 24 h after infection, and infected cells were enumerated 48 h postinfection using a FACSCalibur instrument and CellQuest software (Becton-Dickinson).
Generation of viruses in the presence and absence of CsA or CypA and measurements of their infectivity.
To generate matched CypA-containing and CypA-free virus stocks, bearing a “natural” HIV-1 envelope, the procedure outlined in Fig. 1A was followed. An HIV-1/HXB/GFP stock pseudotyped with VSV-G was first generated by transfection of 293T cells. This stock was then used to infect CypA+/+ and CypA−/− Jurkat cells at high multiplicity, such that all, or nearly all, cells became synchronously infected (as monitored by GFP expression). The cells were then washed thoroughly (four times) 24 h after infection. Forty-eight hours postinfection, virus produced by these cells was harvested, normalized for reverse transcriptase activity, and used to infect fresh CypA+/+ and CypA−/− Jurkat cells, as described above.
A similar strategy was used to generate virus stocks bearing an HIV-1 envelope in HeLa and H9 cells. In this case, each cell line was infected with VSV-G-pseudotyped wild-type or G94D mutant HIV-1/HXB/GFP stock and split into two, 24 h later. One replicate was cultured in the presence of 5 μM CsA, and the other was cultured with carrier alone. After a further 24 h, virus stocks were harvested from each culture and normalized for reverse transcriptase activity. To measure infectivity of virus stock produced in the presence and absence of CsA, foci of GFP-positive cells were counted microscopically at limiting dilution (20 to 100 foci per well) 48 h after infection of TE671-CD4 cells. This was done to dilute CsA (>100-fold) and therefore remove its influence on target cells during infection.
TRIM5α-expressing cells.
TRIM5α was amplified by PCR from TE671, HOS, Jurkat HeLa, and H9 cells using primers directed to the 5′ and 3′ ends of the coding sequences and incorporating XhoI and SalI restriction sites. The PCR products were digested with XhoI and SalI and inserted into retroviral expression vector LNCX2 (Clontech) and pCR3.1/HA (27). To generate TRIM5α-expressing murine MDTF cells, 293T cells were transfected with LNCX2-based retroviral vectors containing a TRIM5α variant along with MLV Gag-Pol and VSV-G expression plasmids. Vector stocks produced by these cells were used to transduce MDTF cells. The transduced cells were selected in 1 mg of G418/ml for 7 to 10 days and then used as a pool of target cells. These cells were seeded in 24-well plates at 2 × 104 cells per well and inoculated with wild-type and G94D mutant HIV-1 and N-MLV vector stocks in the presence of 5 μg of Polybrene/ml. Each virus dose was selected so as to infect 20 to 50% of unmodified cells, and GFP-positive cells were enumerated as described above.
RNA interference.
Synthetic small interfering RNA (siRNA) oligonucleotide duplexes were targeted to sequences within human TRIM5 (GCUCAGGGAGGUCAAGUUG ). HeLa cells were mock transfected or transfected with 60 pmol of the TRIM5α-specific RNA duplex by using Lipofectamine 2000 and were replated 24 h later. Forty-eight hours after transfection, cells were inoculated with GFP-expressing HIV-1- or N-MLV-based vectors. The vector dose was selected to give a low but measurable number of GFP-positive cells (<10% positive), such that any enhancement could be readily measured. Infected cells were enumerated as described above. To confirm that the siRNA duplexes could silence TRIM5α expression, they were cotransfected with plasmids expressing HA-TRIM5α and GFP in HeLa cells. The abundance of these proteins was assessed by Western blotting 48 h later and showed that HA-TRIM5α expression was specifically reduced by >90% (data not shown).
Western blot analysis.
Human cells were washed three times with phosphate-buffered saline, counted, and lysed. Protein concentration was determined using the Bradford assay. Extracts were normalized for cell numbers or protein concentrations, and aliquots were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. The blots were probed with an anti-human CypA antibody (Affinity Bioreagents) and a peroxidase-conjugated secondary antibody and were developed with chemiluminescence detection reagents (Pierce). Blots were exposed to film for a minimal amount of time to avoid saturation, and signal intensities were quantitated using NIH Image.
RESULTS
CypA is not required for the production of fully infectious HIV-1 particles, but it does enhance infection when present in target cells.
Previously, several groups reported that depletion of CypA from virions, by CsA treatment of virus-producing cells, caused those virions to be less infectious (3, 5, 7, 14, 15, 33, 38). In addition, HIV-1 replicated poorly in a Jurkat-derived cell line where both alleles of the CypA gene had been inactivated (11). HIV-1 particles harvested during a spreading infection of CypA-null cells were also reported to be intrinsically less infectious (11).
Our previous findings implicated target cell CypA rather than virus-producing cell CypA as playing the major role in virus replication (40). However, those studies were done using VSV-G-pseudotyped HIV-1, which others have previously shown to be resistant to the effects of CsA when it is present during virus production (3). We reexamined this question by a somewhat different approach, shown schematically in Fig. 1A. HIV-1 stocks bearing a natural gp120 envelope from the HXB3 strain were generated by synchronously infecting normal or CypA-null Jurkat cells with a replication-competent, VSV-G-pseudotyped HIV-1/HXB/GFP reporter virus at high multiplicity and harvesting virus 48 h after inoculation. Thus, these virus stocks should be identical to each other except that virions derived from CypA-null Jurkat cells would lack CypA. These virus stocks were normalized for reverse transcriptase activity, and infectivity was measured by using normal and CypA-null Jurkat cells as target cells. As can be seen in Fig. 1B, the infectivity of HIV-1/HXB/GFP derived from CypA-null Jurkat cells was identical to that derived from normal Jurkat cells. However, when used as target cells in a single-cycle infection assay, normal Jurkat cells were approximately three- to fivefold more sensitive to infection by HIV-1/HXB/GFP than were CypA-null Jurkat cells (Fig. 1B). This was not due to differences in receptor expression levels; VSV-G-pseudotyped HIV-1/GFP derived from 293T cells exhibited approximately the same three- to fivefold reduction in infectivity in CypA-null Jurkat cells as in normal Jurkat cells (Fig. 1C), although VSV-G-pseudotyped HIV-1/GFP was much more infectious on both cell lines than was HIV-1/HXB/GFP. Moreover, we have previously shown that VSV-pseudotyped SIVmac and HIV-1 bearing an inactivating mutation in the CypA binding site exhibit the same infectivity in normal compared to CypA-null Jurkat cells (40), unlike wild-type HIV-1. We concluded from these studies that CypA is not required in virus-producing cells for the assembly of fully infectious HIV-1 particles. Rather, the presence of CypA in Jurkat target cells enhances HIV-1 infection, regardless of whether HIV-1 entry is mediated by VSV-G or HIV-1 envelope proteins.
Target cell-dependent effects of CsA and mutations conferring CsA resistance or dependence.
The ability of CsA to inhibit HIV-1 replication in cell culture and the deleterious effect of CypA binding site mutations on HIV-1 infectivity were thought to be due to the fact that both manipulations prevent the incorporation of CypA into virions. However, recent findings suggested that CsA can modestly inhibit infection of human cells when applied during infection rather than during production (24, 40). This implies that CypA recognition of the incoming viral capsid is important for HIV-1 infectivity and that the CypA that is carried by particles is less important. However, if CsA inhibits HIV-1 infection by preventing recognition of incoming HIV-1 particles by the CypA that is present in the target cells, then any inhibitory effect of CsA on HIV-1 infection should be dependent on the presence of CypA in target cells, even if virions are derived from a cell that contains CypA.
To test this notion, we infected normal and CypA-null Jurkat cells with VSV-G-pseudotyped HIV-1/GFP produced by 293T cells (and therefore containing CypA) in the presence and absence of CsA. As can be seen in Fig. 2A, CsA inhibited infection of normal Jurkat cells by four- to fivefold. In contrast, infection of CypA-null Jurkat cells was less efficient than that of normal Jurkat cells and was not further reduced by CsA (Fig. 2A). This observation indicates that the CypA which constitutes the target for inhibition of HIV-1 infection by CsA is provided by the target cell and not by the incoming virion particle.
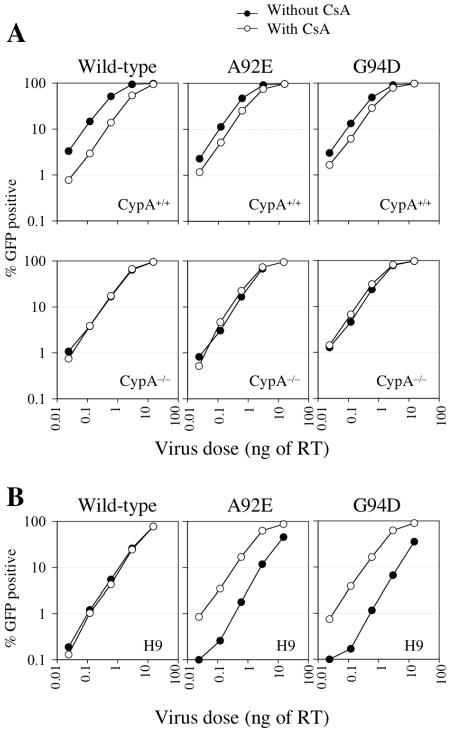
FIG. 2.
Effects of CsA and CypA on the early steps of wild-type and mutant HIV-1 infection in T-cell lines. (A) Titration of VSV-G-pseudotyped HIV-1/GFP in normal and CypA-null Jurkat target cells, as indicated, in the presence (open symbols) or absence (filled symbols) of 5 μM CsA. The percentage of infected GFP-positive cells is plotted as a function of input virus dose, in nanograms of reverse transcriptase (RT). (B) Titration of VSV-G-pseudotyped HIV-1/GFP in H9 cells in the presence (open symbols) or absence (filled symbols) of 5 μM CsA.
A further prediction of the notion that CypA affects HIV-1 infection by recognition of incoming HIV-1 capsids is that the effects of mutations that induce CsA resistance and dependence should be manifested in these single-cycle infection assays (where target cells and not virus-producing cells are treated with CsA). As can be seen in Fig. 2 and 3, this indeed proved to be the case. Specifically, the inhibitory effect of CsA on single-cycle HIV-1/GFP infection of normal Jurkat cells was partly relieved by the A92E and G94D mutations, which have previously been shown to confer CsA resistance in a spreading infection assay (Fig. 2A). As expected, these mutations only affected infectivity in Jurkat cells that expressed CypA and did not affect the lack of response of HIV-1/GFP to CsA when CypA-null Jurkat cells were used as targets (Fig. 2A, lower panels).
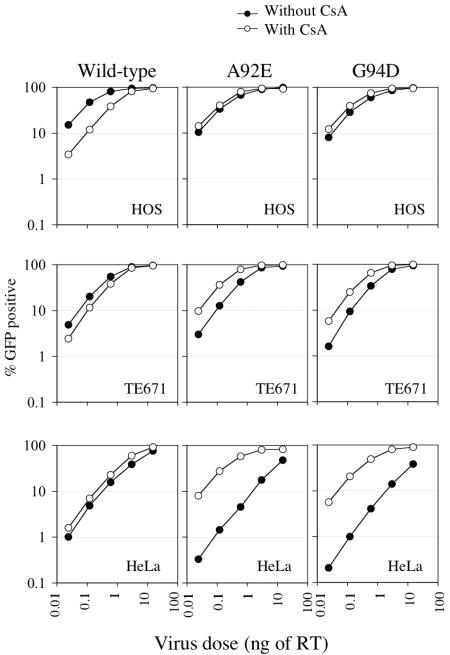
FIG. 3.
Effects of CsA and CypA on the early steps of wild-type and mutant HIV-1 infection in adherent cell lines. Titration of VSV-G-pseudotyped HIV-1/GFP in HOS, TE671, and HeLa cells, as indicated, in the presence (open symbols) or absence (filled symbols) of 5 μM CsA. The percentage of infected GFP-positive cells is plotted as a function of input virus dose, in nanograms of reverse transcriptase (RT).
The same two mutations have previously been shown to confer CsA dependence during spreading HIV-1 infection in another human T-cell line, namely, H9 (45). In fact, as is shown in Fig. 2B, single-cycle infection of H9 cells by HIV-1/GFP bearing A92E or G94D mutations was markedly enhanced (10- to 20-fold) when target H9 cells were treated with CsA. Consistent with previous data obtained in spreading infection assays (45), single-cycle wild-type HIV-1 infection of H9 cells was unaffected by CsA. Clearly, therefore, human cell lines differ in terms of the effect that CsA has on incoming HIV-1 infection. It is noteworthy that infection of H9 cells by wild-type virus is noticeably less efficient than that of either CsA-dependent mutant in the presence of CsA, implying that wild-type infection of H9 cells is intrinsically suboptimal. Also of note is that the enhancing effect of CsA on infection by A92E and G94D mutant HIV-1 is reminiscent of the effect that CsA has on wild-type HIV-1 infection of owl monkey cells (40), where infection is restricted by TRIM-Cyp (36).
Similar single-cycle infection experiments were done in a number of adherent human cells. In HOS cells, CsA inhibited wild-type HIV-1/GFP infection by four- to fivefold, and this effect was completely eliminated by the A92E and G94D mutations. Conversely, in HeLa cells CsA did not inhibit wild-type HIV-1 infection and the A92E and G94D mutations induced marked CsA dependence, with CsA enhancing infection by 20- to 25-fold. As was the case in H9 cells, wild-type virus infection of HeLa cells was less efficient than that of the CsA-dependent mutants in the presence of CsA. TE671 cells exhibited a phenotype that was apparently intermediate between that of HOS and HeLa cells; CsA inhibited infection by wild-type HIV-1 GFP only modestly (about twofold) but enhanced infection by A92E and G94D mutant HIV-1 by about threefold. Overall, inhibition of cyclophilin-CA interactions had both positive and negative effects on HIV-1 infection, and the distinct effects of CA mutations appeared independent of the tissue of origin of the cell line. Importantly, because only the early steps of the viral life cycle were measured in these assays, these effects must be due to cyclophilin-CA interactions involving the incoming viral capsid. Moreover, the effects of these interactions on the incoming capsid are dependent on the sequence of CA proximal to the CypA binding site and the particular target cell context.
CsA dependence is not a consequence of CypA-CA interactions in HIV-1-producing cells.
The finding that single-cycle infection of H9 and HeLa cells by HIV-1 bearing A92E or G94D mutations is dramatically enhanced by CsA is reminiscent of previous reports that documented CsA dependence for spreading infection of these two cell lines by these HIV-1 mutants (1, 45). It was not previously documented, however, whether the effects of mutations conferring CsA dependence were manifested as a consequence of cyclophilin-Gag interactions in the virus-producing cell or in the target cell. The data in Fig. 2 and 3 strongly suggested that CypA inhibits replication of these mutants as a consequence of its recognition of incoming viral capsids. However, it was possible that additional inhibitory effects of cyclophilin-CA interactions on CsA-dependent mutant HIV-1 might occur in virus-producing cells, particularly since previous studies have strongly implicated the CypA-CA interaction that occurs during virus production as the functional target of CsA for inhibition of HIV-1 replication.
Therefore, we next determined whether CsA treatment of HeLa or H9 cells producing wild-type or G94D mutant HIV-1 particles affected HIV-1 infectivity. Because CsA only affects HIV-1 infectivity during production when the natural HIV-1 envelope is used, we used an experimental strategy similar to that described in Fig. 1A. However, in this case we infected HeLa and H9 cells with high-titer VSV-G-pseudotyped HIV-1/HXB/GFP and treated half of the cells with CsA during subsequent virus production. We reasoned that if the G94D mutation conferred a CsA-dependent phenotype during the virus production phase, then we would expect that CsA treatment should enhance infectivity or the yield of particles bearing this mutation from HeLa and H9 cells. However, this proved not to be the case. As can be seen in Fig. 4, CsA modestly inhibited the infectivity of wild-type HIV-1 particles derived from HeLa cells, but not from H9 cells. More importantly, the G94D mutation did not induce CsA-dependent enhancement of HIV-1 infectivity (Fig. 4) or yield (data not shown). Thus, the CsA-dependent HIV-1 replication phenotype induced by the G94D CA mutation is governed entirely by the cyclophilin-CA interactions that occur in the target cell and not in virus-producing cells.
CsA resistance versus dependence and relationship to CypA expression levels.
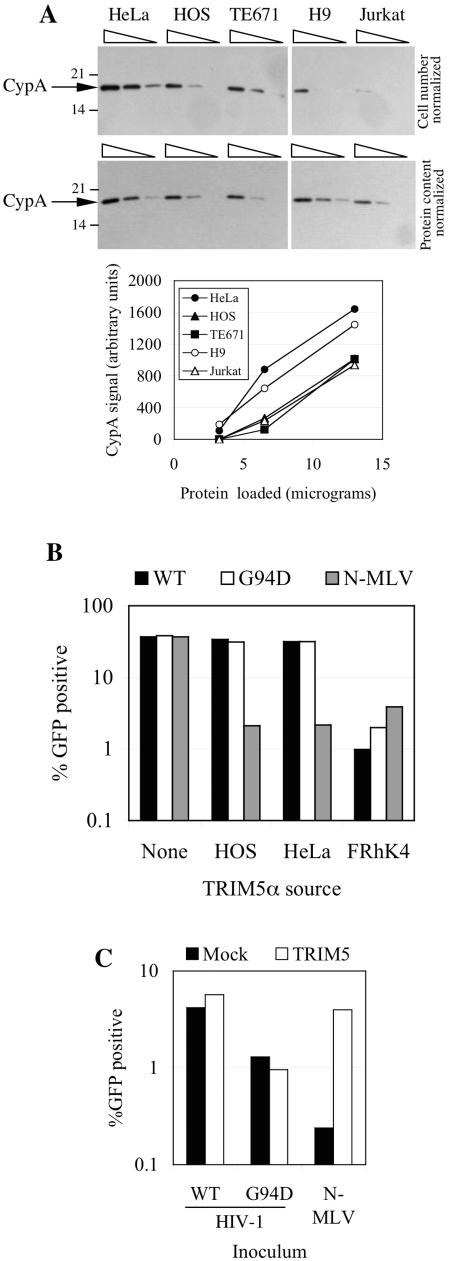
The apparently distinct effects of CypA on HIV-1 infection of different cells could be due to variation in CypA expression level or to other host factors that affect HIV-1 infection and whose activity is modified by CA mutations and the cyclophilin-CA interaction. Previously it was suggested that the differential effects of mutations within and proximal to the CypA binding site on HIV-1 replication in different cell types might be due to variations in CypA expression levels (2, 45). Therefore, we analyzed CypA expression levels in the cell lines used in this study by Western blotting. As can be seen in Fig. 5A, there was variation in the level of CypA in the various cell lines that was particularly evident when the CypA content of equal numbers of cells were analyzed. This variation in CypA expression did not correlate with the occurrence of the CypA-resistant versus CypA-dependent phenotype when the corresponding cell lines were infected with A92E or G94D mutant HIV-1. In fact, each of the adherent cell lines expressed higher levels of CypA than did each of the two T-cell lines, on a per-cell basis, with HeLa cells expressing the highest level, Jurkat the lowest level, and HOS, TE671, and H9 cells expressing intermediate levels. When equal amounts of total protein from each cell line rather than equal numbers of cells were analyzed, the degree of variation in CypA levels was significantly reduced. However, in this case the two cell lines, HeLa and H9, that exhibited the strong CsA-dependent A92E and G94D mutant HIV-1 infection phenotype expressed almost twofold-higher levels of CypA than did HOS, TE671, and Jurkat cells. Thus, there is a correlation between the CsA-dependent phenotype and CypA expression levels but, given the very modest difference, its significance is unclear.
FIG. 5.
Cell type-dependent differences in the effects of CsA and CA mutation and correlations with CypA expression levels or TRIM5α polymorphism. (A) Western blot analysis of CypA expression in various human cell lines. In the upper panels, lysates from 3.3 ×104, 1.65 ×104, and 0.8 ×104 cells (from left to right) for each cell line wereloaded. In the lower panels, 13, 6.5, and 3.25 μg of protein (from left to right) from each cell line was loaded. The migration positions of molecular weight markers and CypA are shown. Exposure times were minimized to avoid saturation of the film, and signals quantitated using NIH Image, for each level of total protein loading, are plotted in the chart below the blots. (B) MDTF cells expressing no TRIM5α (none) or TRIM5α variants from HOS, HeLa, and FRhK4 cells were inoculated with a fixed and equivalent dose of wild-type (WT) or G94D mutant HIV-1/GFP. As a control, cells were also inoculated with an equivalent infectious dose of N-MLV. The percentage of infected (GFP-positive) cells was plotted. (C) HeLa cells that were either mock transfected (mock) or transfected with a TRIM5-specific siRNA duplex (TRIM5) were infected with wild-type or G94D mutant GFP-expressing HIV-1 vectors or a GFP-expressing N-MLV vector, as indicated. The percentage of infected (GFP-positive) cells was plotted.
TRIM5α is not responsible for restriction of HIV-1 bearing CsA-resistant or -dependent mutations in human cells.
Another possibility that could potentially explain why A92E and G94D mutations have different effects in different human cell lines might be polymorphisms in host restriction factors that recognize the incoming HIV-1 capsids. Such factors are known to inhibit HIV-1 infection of nonhuman primate species, and recognition can be markedly affected by mutations in and around the CypA binding site (19, 24, 29, 40). Moreover, in one particular species, namely owl monkeys, wild-type HIV-1 infection is highly CsA dependent (40) due to the presence of a variant form of TRIM5 containing a cyclophilin domain (36). Thus, the phenotype of wild-type HIV-1 in owl monkey cells is similar to that of A92E and G94D mutant HIV-1 in H9 and HeLa cells. We were unable to detect the expression of a similar TRIM5-CypA fusion protein in human cells (data not shown); it was possible therefore that the CsA-dependent phenotype exhibited by H9 and HeLa cells might be due to the presence of a variant form of human TRIM5α that can restrict A92E and G94D mutant, but not wild-type, HIV-1 in a cyclophilin-dependent manner.
We therefore cloned and sequenced several alleles of TRIM5α from HeLa, HOS, H9, and TE671 cells. Among these TRIM5α clones, we found one single amino acid polymorphism in TRIM5α from HeLa cells (R136Q). However, this polymorphism was not detected in H9 cells, whose TRIM5α clones were identical in sequence to those from HOS and TE671 cells and to the database consensus sequence. Nevertheless, we generated murine MDTF cell lines expressing the different TRIM5α variants from HOS cells and HeLa cells and asked whether either variant could inhibit the infectivity of HIV-1 bearing the G94D CA mutation. As a control, we also generated a cell line expressing a TRIM5α from the rhesus monkey cell line FRhK4. In addition, we infected each cell line with an N-tropic MLV vector, because human and nonhuman primate TRIM5α is able to restrict N-MLV infection (20, 22, 30, 44). As can be seen in Fig. 5B, MDTF cells expressing either form of human TRIM5α were as susceptible to infection by wild-type and G94D mutant HIV-1 vectors as were control MDTF cells lacking TRIM5α. Human TRIM5α was functionally expressed in these cells, because N-MLV vector infection was inhibited by about 20-fold. Moreover, MDTF cells can restrict HIV-1 vector infection when an active TRIM5α allele of the appropriate specificity is expressed, because both wild-type and G94D HIV-1 infection was decreased by 20- to 30-fold in MDTF cells expressing rhesus monkey TRIM5α.
In addition, we also tested whether TRIM5α depletion from HeLa cells could enhance G94D mutant HIV-1 infection. As can be seen in Fig. 5C, TRIM5α was effectively depleted from a sufficient number of HeLa cells to induce a 16-fold increase in N-MLV infection. However, infection by both wild-type and G94D mutant HIV-1 was unaffected. We conclude from these experiments that TRIM5α is unlikely to be responsible for the CypA-dependent restriction of the G94D mutant HIV-1 in HeLa or H9 cells.
DISCUSSION
In this study, we show that the CypA that is present in the human target cells, rather than that present in virus-producing cells or the virion particle, is largely responsible for effects on HIV-1 replication. In particular, virions derived from Jurkat cells that are deficient in CypA are equally infectious to those derived from cells that express CypA. Conversely, CypA-deficient Jurkat cells are less sensitive to HIV-1 infection than CypA-expressing cells, irrespective of whether the incoming virion carries CypA or of the route of entry (Fig. 1). Moreover, viruses that contain CA mutations which confer CsA resistance in some cells or CsA dependence in others manifest these phenotypes during the early stages of the virus replication cycle (Fig. 2 and 3) (see also references 24 and 40), not during virus assembly or production (Fig. 4). Thus, while it is possible for CsA to reduce the infectivity of virions if applied during virus production, the predominant effects of CsA and cyclophilin-CA interactions on HIV-1 replication occur during the postentry steps of the HIV-1 life cycle. Notably, the effects of CsA and CA mutations proximal to the CypA binding site mutations were completely absent when CypA-null Jurkat cells were used as targets (Fig. 3).
How could combinations of CA mutations and the choice of different target cells cause CsA to have very different effects on HIV-1 replication? Clearly this variation is not a tissue-specific phenomenon, because the CsA-dependent infection phenotype for A92E and G94D mutants was observed in cells of epithelial (HeLa) and T-cell (H9) origin, while the CsA-resistant phenotype was observed in both fibroblast (HOS) and T-cell (Jurkat) lines. Curiously, the CsA-dependent phenotype for CA mutant HIV-1 infection in H9 and HeLa cells coincided with CsA resistance for wild-type HIV-1 infection (Fig. 2 and 3). One possible explanation for these findings is that different effects of CsA might be governed by different levels of CypA expression. Indeed, differential effects of CsA and CypA binding site mutations on spreading HIV-1 infection in different cell lines have previously been correlated with CypA expression levels (2, 45). Although the two cell lines (HeLa and H9) that exhibited the CsA-dependent infection phenotype for A92E and G94D mutant HIV-1 both had relatively high levels of CypA expression, they were only modestly different (about twofold) from the CypA expression levels in other the cell lines used in this study, and the significance of this finding is unclear.
A possibility that might help to explain the occurrence of CsA-dependent HIV-1 infection in human cells is suggested by studies in nonhuman primate cells. In owl monkey cells, infection by wild-type HIV-1 is inhibited by a restriction factor, TRIM-Cyp, that is a fusion protein containing the amino-terminal portion of TRIM5 fused to CypA as a result of a CypA pseudogene retrotransposition event (36). We wondered whether G94D and A92E mutant HIV-1 had acquired the ability to be restricted by an inhibitor that is specific to HeLa and H9 cells. TRIM5α, an inhibitor that targets incoming HIV-1 capsids, has been identified from rhesus monkey cells, and the human version of this protein exists in at least two variant forms and is capable of inhibiting infection by other retroviruses (20, 22, 30, 44). However, variation in human TRIM5α sequence did not correlate with the CsA-dependent versus CsA-resistant phenotype. Moreover, human or rhesus monkey TRIM5α variants did not discriminate between wild-type and G94D HIV-1, and silencing of TRIM5α expression in HeLa cells increased sensitivity to N-MLV infection but not G94D mutant HIV-1 infection (Fig. 5). Thus, it seems unlikely that TRIM5α plays a role in inhibiting infection by CsA-dependent viruses. We are currently investigating the possibility that some as-yet-unidentified restriction factor, perhaps a different TRIM protein (of which there are many [32]), CypA itself, or a CypA-related protein, might be responsible for preventing infection of CsA-dependent viruses in certain human cells.
Acknowledgments
We thank Greg Towers and Jeremy Luban for reagents.
This work was supported in part by a grant from the National Institutes of Health (RO1AI50111). T.H. is the recipient of a Fellowship award from the American Foundation for AIDS Research. P.D.B. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Aberham, C., S. Weber, and W. Phares. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 70:3536-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerson, B., O. Rey, J. Canon, and P. Krogstad. 1998. Cells with high cyclophilin A content support replication of human immunodeficiency virus type 1 Gag mutants with decreased ability to incorporate cyclophilin A. J. Virol. 72:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bainbridge, J. W., C. Stephens, K. Parsley, C. Demaison, A. Halfyard, A. J. Thrasher, and R. R. Ali. 2001. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 8:1665-1668. [DOI] [PubMed] [Google Scholar]
- 5.Bartz, S. R., E. Hohenwalter, M. K. Hu, D. H. Rich, and M. Malkovsky. 1995. Inhibition of human immunodeficiency virus replication by nonimmunosuppressive analogs of cyclosporin A. Proc. Natl. Acad. Sci. USA 92:5381-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billich, A., F. Hammerschmid, P. Peichl, R. Wenger, G. Zenke, V. Quesniaux, and B. Rosenwirth. 1995. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J. Virol. 69:2451-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIVCPZ GAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 70:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bukovsky, A. A., A. Weimann, M. A. Accola, and H. G. Gottlinger. 1997. Transfer of the HIV-1 cyclophilin-binding site to simian immunodeficiency virus from Macaca mulatta can confer both cyclosporin sensitivity and cyclosporin dependence. Proc. Natl. Acad. Sci. USA 94:10943-10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke, E. K., and J. Luban. 1996. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology 222:279-282. [DOI] [PubMed] [Google Scholar]
- 15.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 16.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 17.Grattinger, M., H. Hohenberg, D. Thomas, T. Wilk, B. Muller, and H. G. Krausslich. 1999. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology 257:247-260. [DOI] [PubMed] [Google Scholar]
- 18.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatziioannou, T., S. Cowan, U. K. Von Schwedler, W. I. Sundquist, and P. D. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karpas, A., M. Lowdell, S. K. Jacobson, and F. Hill. 1992. Inhibition of human immunodeficiency virus and growth of infected T cells by the immunosuppressive drugs cyclosporin A and FK 506. Proc. Natl. Acad. Sci. USA 89:8351-8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan, M., M. Garcia-Barrio, and M. D. Powell. 2003. Treatment of human immunodeficiency virus type 1 virions depleted of cyclophilin A by natural endogenous reverse transcription restores infectivity. J. Virol. 77:4431-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luban, J. 1996. Absconding with the chaperone: essential cyclophilin-Gag interaction in HIV-1 virions. Cell 87:1157-1159. [DOI] [PubMed] [Google Scholar]
- 26.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 28.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pushkarsky, T., G. Zybarth, L. Dubrovsky, V. Yurchenko, H. Tang, H. Guo, B. Toole, B. Sherry, and M. Bukrinsky. 2001. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 98:6360-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenwirth, B., A. Billich, R. Datema, P. Donatsch, F. Hammerschmid, R. Harrison, P. Hiestand, H. Jaksche, P. Mayer, P. Peichl, et al. 1994. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppressive cyclosporine analog. Antimicrob. Agents Chemother. 38:1763-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 2002. Cyclophilin A plays distinct roles in human immunodeficiency virus type 1 entry and postentry events, as revealed by spinoculation. J. Virol. 76:4671-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 37.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 38.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 39.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 41.Wainberg, M. A., A. Dascal, N. Blain, L. Fitz-Gibbon, F. Boulerice, K. Numazaki, and M. Tremblay. 1988. The effect of cyclosporine A on infection of susceptible cells by human immunodeficiency virus type 1. Blood 72:1904-1910. [PubMed] [Google Scholar]
- 42.Wiegers, K., and H. G. Krausslich. 2002. Differential dependence of the infectivity of HIV-1 group O isolates on the cellular protein cyclophilin A. Virology 294:289-295. [DOI] [PubMed] [Google Scholar]
- 43.Wiegers, K., G. Rutter, U. Schubert, M. Grattinger, and H. G. Krausslich. 1999. Cyclophilin A incorporation is not required for human immunodeficiency virus type 1 particle maturation and does not destabilize the mature capsid. Virology 257:261-274. [DOI] [PubMed] [Google Scholar]
- 44.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin, L., D. Braaten, and J. Luban. 1998. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J. Virol. 72:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y. J., T. Hatziioannou, T. Zang, D. Braaten, J. Luban, S. P. Goff, and P. D. Bieniasz. 2002. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J. Virol. 76:6332-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]