Abstract
The hepatitis C virus (HCV) genotype 2a subgenomic replicon can replicate in two human non-hepatocyte-derived cell lines, HeLa and 293, with in vitro-transcribed replicon RNA. Sequencing analysis revealed that mutations in HCV-derived regions were not essential for replication in these cells, as some clones displayed no mutations.
Hepatitis C virus (HCV) was first identified as a causative agent of posttransfusion hepatitis in 1989 (4). The virus is considered hepatotropic and is known to cause liver diseases such as acute or chronic hepatitis, cirrhosis, and hepatocellular carcinoma (11, 16, 17, 21). HCV has been detected not only in liver, but also in peripheral blood mononuclear cells and dendritic cells (7, 12, 19). However, other tissue tropisms and their regulatory factors have yet to be fully elucidated. This lack of progress in the investigations regarding the virus is primarily attributable to a lack of efficient cell culture systems and small animal models of infection. As an important step toward overcoming this disadvantage, a subgenomic HCV RNA replicon system has been developed (18). This replicon system contains the HCV internal ribosome entry site (IRES), which directs expression of the G418 selectable marker, neor, and encephalomyocarditis virus (EMCV) IRES directs the expression of HCV nonstructural (NS) proteins NS3 to NS5B. This enabled assessment of HCV replication in cultured cells. Functional replicons have previously been reported only for genotype 1, and efficient replications of these replicons have been accomplished only in limited human hepatocyte-derived cell lines (2, 3, 9). Attempts to evaluate replication of the HCV replicon in non-hepatocyte-derived cell lines have been made previously (1, 23). Some of these attempts seemed to have succeeded, but efficient replicon replication in nonhepatic cells has not been achieved by synthetic RNA transfection, and other experimental procedures have been required (1, 23). We developed a new HCV replicon system using an HCV genotype 2a clone from a patient with fulminant hepatitis (15). This replicon system provided higher colony formation efficiency and robust replication, not only in Huh7 cells but also in HepG2 and IMY-N9 cells (5, 15), which were established by fusing human primary cultured hepatocytes and HepG2 cells (13). The present study examined whether this replicon, the JFH-1 replicon, can replicate in two non-hepatocyte-derived cell lines: HeLa cells established from human cervical carcinoma (6) and 293 cells established from human embryonic kidney (8).
The HCV genotype 2a clone, JFH-1, was isolated from a patient with fulminant hepatitis, and genotype 2a HCV replicon constructs were built with this clone as reported previously (14, 15). After transfection of RNAs transcribed from the linearized pSGR-JFH1 (DDBJ/EMBL/GenBank accession no. AB114136) into HeLa and 293 cells, transfected cells were cultured for 3 weeks with G418 (Nacalai Tesque, Kyoto, Japan) at working concentrations of 0.8 mg/ml for HeLa and 293 cells and 1.0 mg/ml for Huh7 cells. Visible colonies were observed 3 weeks later in all three transfected cell lines (Fig. 1). Based on three independent assays, colony formation efficiency was lower in HeLa cells, at (5.83 ± 3.18) ×103 CFU/μg of RNA, than in Huh7 cells, at (5.32 ± 5.02) × 104 CFU/μg of RNA (15). Colony formation efficiency was substantially lower in 293 cells than in Huh7 cells, at (1.36 ± 1.11) × 102 CFU/μg of RNA. A total of nine colonies for each line were cloned from pSGR-JFH1 RNA-transfected HeLa and 293 cells and expanded for further analysis.
FIG. 1.
Colony formation of JFH-1 HCV subgenomic RNA replicon in Huh7, HeLa, and 293 cell lines. Transcribed RNAs from pSGR-JFH1 were transfected into each cell line, and cells were cultured with G418 for 3 weeks before staining with crystal violet, as described in Materials and Methods. Representative staining examples are shown for 0.1 μg of synthetic RNA transfected onto Huh7 cells and 3 μg transfected for both HeLa and 293 cells.
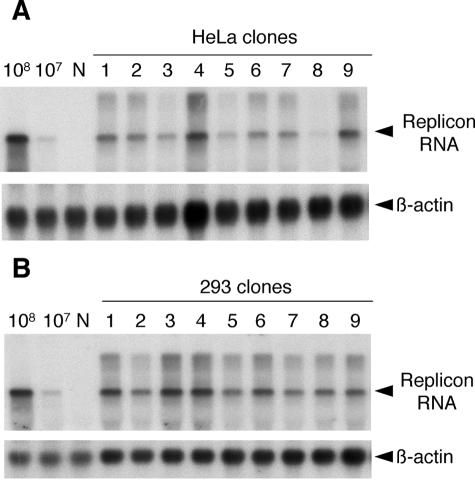
To estimate the correct size of replicating replicon RNA, Northern blot analysis was performed with the nine clones from each of the HeLa and 293 cell lines. DNA probes were synthesized from neor and EMCV-IRES genes using the Megaprime DNA labeling system (Amersham Pharmacia). In all clones, expected sizes of replicon RNAs were detected using neor and EMCV-IRES probes (Fig. 2). The amount of replicon RNA in clones of each cell line varied among clones, particularly in HeLa clones.
FIG. 2.
Detection of replicon RNA in cloned HeLa (A) and 293 (B) cells. Total RNA from cloned cells in each cell line was analyzed by Northern blotting with DNA probes of the neor EMCV IRES and β-actin genes. In vitro synthesis of 108 and 107 copies of transcribed positive-strand RNAs was performed, and RNA was loaded (lanes 108 and 107) as positive controls, as indicated. Arrowheads indicate target positions of replicon RNA and β-actin. N, cellular RNA of HeLa or 293 cells as negative controls.
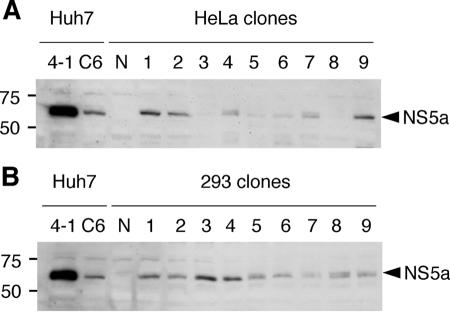
HCV NS protein expression in replicon RNA-transfected cells was tested by Western blotting with HCV NS5A-specific polyclonal antibody obtained by DNA immunization with JFH-1 NS5A-expressing construct (5, 22). In JFH-1 replicon RNA-transfected clones, HCV-specific polyclonal antibody detected NS5A protein as major bands of various intensities at about 56 kDa (Fig. 3). A very faint band above the major 56-kDa band was also detected in some lanes at around 58 kDa. As a positive control, cell lysate from Huh7 replicon cells was also loaded on the left lane of the gel (Fig. 3, lanes 4-1 and C6). Huh7 replicon cell 4-1 displayed strong expression (5; unpublished data), while C6 displayed weak expression (5, 15). The intensity of protein expression in each clone displayed similar trends with regard to the amount of replicating replicon RNA in Northern blotting.
FIG. 3.
Detection of HCV NS5A antigens in cloned cells of HeLa (A) and 293 (B) cells by Western blot analysis. Cell lysates were prepared from pSGR-JFH1 RNA-transfected Huh7 cell clones (lanes 4-1 and C6) as positive controls or untransfected parental HeLa and 293 (lane N) as negative controls. Anti-NS5A polyclonal antibodies were used to detect HCV antigens. Target sizes of NS5A proteins are indicated by arrowheads.
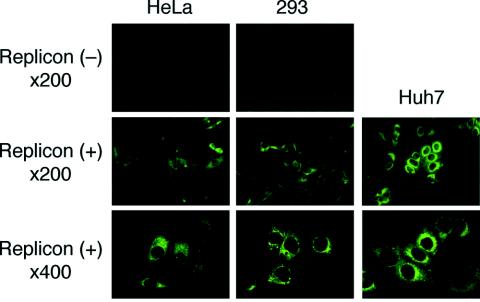
Using recovered phase serum from a patient with acute hepatitis C, HCV antigens in JFH-1 replicon RNA-transfected HeLa and 293 clones were also detected by immunofluorescence assay (Fig. 4). Distributions of HCV-related antigens in both replicon RNA-transfected cell lines resembled those in Huh7, and diffuse and fine reticular patterns with some granular cytoplasmic staining were observed. Signal intensities of HeLa and 293 replicon cells were slightly faint compared to Huh7 replicon cells, but signals were similarly localized within all replicon cells (Fig. 4). No signals were detected in untransfected parental cells. Microscopic morphologies of replicon-containing HeLa and 293 cells were normal and similar to the respective parental cells.
FIG. 4.
Subcellular localization of HCV antigens determined by immunofluorescence. Replicon RNA-untransfected [Replicon(−)] or cloned HeLa, 293, and Huh7 [Replicon(+)] cells were cultured on coverslips, fixed in acetone-methanol, and incubated with patient serum, as described in the Materials and Methods. Representative clones of HeLa, 293, and Huh7 cells are indicated.
To estimate adaptive mutations in HeLa and 293 cells, replicon RNA isolated from each clone was amplified by reverse transcription-PCR (RT-PCR) and sequenced directly (5, 15). Copy numbers of replicating RNA in clones were also determined by real-time detection RT-PCR adjusting for intracellular glyceraldehyde-3-phosphate dehydrogenase concentrations (20). Of the nine HeLa clones, clone 1 displayed no mutation and clone 7 had only one synonymous mutation in the open reading frame (ORF) of HCV for replicating RNA (Table 1). Clone 3 displayed a 1-nucleotide mutation in the 3′ untranslated region (UTR), and clone 8 displayed three synonymous mutations, one located in the core region upstream of the neor gene and two in the HCV ORF. These four clones thus did not contain any mutations resulting in changes to amino acid sequences. The remaining five clones had one to three nonsynonymous mutations in the HCV ORF, and mutations in the NS5a region were prevalent. The mean number of replicon RNA copies in HeLa clones was (5.01 ± 2.87) ×106 copies/μg of RNA (range, 8.31 × 105 to 1.07 × 107 copies/μg of RNA). These data were basically concordant with trends for signal intensities in Northern blot analysis. Mean replicon titers in these clones were slightly lower than those of Huh7 cells, at (2.71 ± 2.11) × 107 copies/μg of RNA (M. Miyamoto, T. Kato, T. Date, and T. Wakita, unpublished data). The mean copy number for replicon RNA in clones with nonsynonymous mutations in the HCV ORF (clones 2, 4, 5, 6, and 9) did not differ significantly from that of clones without nonsynonymous mutations (clones 1, 3, 7, and 8) [(6.03 ± 3.05) × 106 versus (3.74 ± 2.38) × 106 copies/μg of RNA; P = 0.261].
TABLE 1.
Mutations and titers of JFH-1 replicon in HeLa clones
| Clone | Nucleotidea | Amino acidb | Region | Replicon titer (copies/μg of RNA) |
|---|---|---|---|---|
| 1 | None | 6.43 × 106 | ||
| 2 | 5500T→C | 2272S→P | NS5a | 5.69 × 106 |
| 7182T→C | Synonymousc | NS5b | ||
| 7217A→G | 2844H→R | NS5b | ||
| 3 | 7820A→G | NAd | 3′-UTR | 3.05 × 106 |
| 4 | 5681C→A | 2332T→K | NS5a | 1.07 × 107 |
| 6672T→C | Synonymous | NS5b | ||
| 5 | 3643A→G | 1653M→V | NS3 | 2.56 × 106 |
| 5851G→A | 2389A→T | NS5a | ||
| 5914G→A | 2410E→K | NS5a | ||
| 6 | 2474C→G | 1263A→G | NS3 | 4.38 × 106 |
| 7 | 2454A→G | Synonymous | NS3 | 4.66 × 106 |
| 8 | 361T→C | Synonymous | Coree | 8.31 × 105 |
| 5673A→G | Synonymous | NS5a | ||
| 6648G→A | Synonymous | NS5b | ||
| 9 | 7097T→C | 2804L→P | NS5b | 6.81 × 106 |
Position of mutated nucleotide within subgenomic replicon.
Position of mutated amino acid within complete ORF of full-length JFH-1.
Synonymous mutation does not change amino acid sequence.
NA, not applicable.
Sequential region from 5′-UTR upstream of neor gene.
For 293 cell clones, surprisingly, eight of the nine clones displayed no mutation or only one synonymous mutation (Table 2). The remaining clone (clone 6) had one nonsynonymous mutation in the NS5a region and one synonymous mutation. The mean number of replicon RNA copies in 293 clones was (5.30 ± 0.16) × 106 copies/μg of RNA (range, 3.47 × 106 to 8.33 × 106 copies/μg of RNA). The mutation-containing clone, clone 6, showed a replicon titer close to the mean (4.38 × 106 copies/μg of RNA). This mutation was thus not considered to affect replication efficiency. Mutations previously observed in Huh7 and other hepatocyte-derived cell lines were not detected in HeLa and 293 clones (5, 15).
TABLE 2.
Mutations and titers of JFH-1 replicon in 293 clones
| Clone | Nucleotidea | Amino acidb | Region | Replicon titer (copies/μg of RNA) |
|---|---|---|---|---|
| 1 | None | 5.11 × 106 | ||
| 2 | None | 3.47 × 106 | ||
| 3 | None | 6.82 × 106 | ||
| 4 | None | 8.33 × 106 | ||
| 5 | None | 5.93 × 106 | ||
| 6 | 5897T→C | 2404 L→P | NS5a | 5.57 × 106 |
| 6420A→C | Synonymousc | NS5b | ||
| 7 | None | 3.65 × 106 | ||
| 8 | 3195T→C | Synonymous | NS3 | 5.10 × 106 |
| 9 | None | 3.73 × 106 |
Position of mutated nucleotide within subgenomic replicon.
Position of mutated amino acid within complete ORF of full-length JFH-1.
Synonymous mutation results in no change to amino acid sequence.
Our results show that HCV replicon can replicate efficiently in two non-hepatocyte-derived cell lines. Colony formation efficiencies in cell lines HeLa and 293 were lower than in Huh7, but higher than in hepatocyte-derived cell lines HepG2 and IMY-N9 (5, 15). The amounts of replicating replicon RNA in HeLa and 293 cells were comparable to those in HepG2 cells. These results indicate that the JFH-1 replicon can replicate equally well in non-hepatocyte- and hepatocyte-derived cell lines. Sequencing of HCV-derived region in replicons replicating in these cells indicated that no common mutations were observed in these cells. In HeLa clones, five clones displayed nonsynonymous mutations and the other four clones contained no nonsynonymous mutations in the HCV-derived region (Table 1). Amounts of replicating replicon RNAs did not differ between clones with or without nonsynonymous mutations. In 293 cells, surprisingly, most replicon clones had no or only one synonymous mutation (Table 2). As a whole, these results indicate that the JFH-1 replicon can replicate in these cells efficiently without cell-specific mutations and adaptive mutations in these cells might be unnecessary. Furthermore, distributions of HCV antigens in these cells resemble those in Huh7 cells (Fig. 4). Taken together, cell tropism of HCV does not appear to be regulated by cellular factors preventing replication or requiring cell-specific mutation.
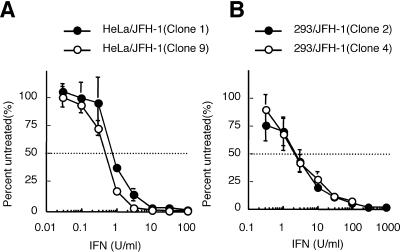
To further characterize JFH-1 replicon-containing HeLa and 293 cells, modifications of cell growth rate were investigated. Temporal evolution of viable cell count was estimated by resazurin reduction assay using the Promega cell titer-blue cell viability assay (Promega, Madison, Wis.). Cell growth rates did not differ significantly between HeLa cells with and without replicon (Fig. 5A). In contrast, in 293 cells, the cell growth rate was slower in replicon-containing cells than in parental cells (Fig. 5B). Therefore, expression of HCV proteins or replication of the JFH-1 replicon seems to suppress cell growth in 293 cells, but this tendency was not clearly observed in HeLa cells. Interferon sensitivities of the two replicon-containing cell lines were also assessed. Administration of interferon suppressed replication of JFH-1 replicon in a dose-dependent manner in both cell lines (Fig. 6). The 50% inhibitory concentrations ranged from 0.6 to 0.8 U/ml in HeLa cells and from 2.2 to 2.4 U/ml in 293 cells. These data are consistent with previously reported interferon sensitivities of genotype 1 HeLa and 293 replicon cells (1, 10).
FIG. 5.
Effect of replicon RNA replication on cell growth rate in HeLa (A) and 293 (B) cell lines. Cell growth rates were compared between parental cells (closed circles) and replicon-containing cell lines (open triangles and diamonds) with a resazurin reduction assay. OD570/600, optical density at 570/600 nm. Names of replicon clones (in parentheses) were consistent with Tables 1 and 2. Experiments were performed with triplicate wells and repeated twice. Mean data and standard deviation bars are shown.
FIG. 6.
Dose-dependent inhibition of replicon RNA replication on interferon administration in the HeLa (A) and 293 (B) cell lines. Amounts of replicating replicon RNA were measured by RT-PCR for serial doses of interferon (IFN) administration and are represented as a percentage of the amount of replicon RNA in cells without interferon administration. Names of replicon clones (in parentheses) are consistent with Tables 1 and 2. Experiments were performed with duplicate wells for each interferon dose and repeated twice. Mean data and standard deviation bars are shown.
Differing results between previous studies and the present study are probably attributable to the HCV clone used in replicon construction (1, 23). Our HCV clone, JFH-1, was isolated from a patient with fulminant hepatitis (14). This replicon displays potent replication ability above that reported for clones with adaptive mutations in their genomes (15). This robust replication ability enabled assessment of replication not only in Huh7 cells but also in other cell lines. Whether this replication ability is specifically associated with fulminant hepatitis and which HCV region or amino acid residues are responsible for this ability remain unclear. Further investigations are thus needed to investigate these issues.
Our results show that nonhepatic cell lines support HCV replication. This suggests that once HCV enters a cell, replication can occur. Hepatotropism of HCV may thus be determined at the step of viral entry into the cell. Specific receptors for HCV may be expressed on the hepatocyte surface. Development of models for HCV infection appears indispensable for further clarification of this hypothesis, although no such system has yet been reported. Establishment of a full-length replicon system using the JFH-1 clone and various cell lines is thus important. When such a model system has been achieved, these two non-hepatocyte-derived cell lines may well prove instrumental in identifying HCV receptors, as these cells can support HCV replication but will not express the receptors necessary for viral hepatotropism.
In summary, the HCV genotype 2a replicon can replicate not only in hepatocytes, but also in the HeLa and 293 non-hepatocyte-derived cell lines. These results provide useful information about HCV replication and cell tropisms.
Acknowledgments
We thank Satoshi Koike for helpful discussion and Kotaro Yasui for support.
This work was partially supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science; grants from Toray Industries, Inc.; the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan; the Research on Health Sciences focusing on Drug Innovation from the Japan Health Sciences Foundation; and The Japanese Society of Gastroenterology.
REFERENCES
- 1.Ali, S., C. Pellerin, D. Lamarre, and G. Kukolj. 2004. Hepatitis C virus subgenomic replicons in the human embryonic kidney 293 cell line. J. Virol. 78:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 5.Date, T., T. Kato, M. Miyamoto, Z. Zhao, K. Yasui, M. Mizokami, and T. Wakita. 2004. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J. Biol. Chem. 279:22371-22376. [DOI] [PubMed] [Google Scholar]
- 6.Gey, G. O., W. D. Coffman, and M. T. Kubicek. 1952. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 12:264-265. [Google Scholar]
- 7.Goutagny, N., A. Fatmi, V. De Ledinghen, F. Penin, P. Couzigou, G. Inchauspe, and C. Bain. 2003. Evidence of viral replication in circulating dendritic cells during hepatitis C virus infection. J. Infect. Dis. 187:1951-1958. [DOI] [PubMed] [Google Scholar]
- 8.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1997. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 9.Gu, B., A. T. Gates, O. Isken, S.-E. Behrens, and R. T. Sarisky. 2003. Replication studies using genotype 1a subgenomic hepatitis C virus replicons. J. Virol. 77:5352-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, J.-T., Q. Zhu, and C. Seeger. 2003. Cytopathic and noncytopathic interferon responses in cells expressing hepatitis C virus subgenomic replicons. J. Virol. 77:10769-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 12.Hu, Y., A. Shahidi, S. Park, D. Guilfoyle, and I. Hirshfield. 2003. Detection of extrahepatic hepatitis C virus replication by a novel, highly sensitive, single-tube nested polymerase chain reaction. Am. J. Clin. Pathol. 119:95-100. [DOI] [PubMed] [Google Scholar]
- 13.Ito, T., K. Yasui, J. Mukaigawa, A. Katsume, M. Kohara, and K. Mitamura. 2001. Acquisition of susceptibility to hepatitis C virus replication in HepG2 cells by fusion with primary human hepatocytes: establishment of a quantitative assay for hepatitis C virus infectivity in a cell culture system. Hepatology 34:566-572. [DOI] [PubMed] [Google Scholar]
- 14.Kato, T., A. Furusaka, M. Miyamoto, T. Date, K. Yasui, J. Hiramoto, K. Nagayama, T. Tanaka, and T. Wakita. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64:334-339. [DOI] [PubMed] [Google Scholar]
- 15.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 16.Kuo, G., Q. L. Choo, H. J. Alter, G. L. Gitnick, A. G. Redeker, R. H. Purcell, T. Miyamura, J. L. Dienstag, M. J. Alter, C. E. Stevens, G. E. Tegtmeier, F. Bonino, M. Colombo, W. S. Lee, C. Kuo, K. Berger, J. R. Shuster, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362-364. [DOI] [PubMed] [Google Scholar]
- 17.Liang, T. J., L. J. Jeffers, K. R. Reddy, M. De Medina, I. T. Parker, H. Cheinquer, V. Idrovo, A. Rabassa, and E. R. Schiff. 1993. Viral pathogenesis of hepatocellular carcinoma in the United States. Hepatology 18:1326-1333. [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 19.Nishiguchi, S., K. Fukuda, S. Shiomi, T. Takeda, T. Kuroki, M. Ogami, H. Morimoto, S. Otani, M. Sakurai, and A. Matsuhisa. 2003. Peripheral blood mononuclear cells are possible extrahepatic replication sites for hepatitis C virus. Hepatogastroenterology 50:1301-1304. [PubMed] [Google Scholar]
- 20.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 21.Tong, M. J., N. S. el-Farra, A. R. Reikes, and R. L. Co. 1995. Clinical outcomes after transfusion-associated hepatitis C. N. Engl. J. Med. 332:1463-1466. [DOI] [PubMed] [Google Scholar]
- 22.Zhao, Z., T. Wakita, and K. Yasui. 2003. Inoculation of plasmids encoding Japanese encephalitis virus PrM-E proteins with colloidal gold elicits a protective immune response in BALB/c mice. J. Virol. 77:4248-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu, Q., J.-T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]