Abstract
The bHLH (basic helix-loop-helix) transcription factor family is the second largest in plants. It occurs in all three eukaryotic kingdoms, and plays important roles in regulating growth and development. However, family members have not previously been studied in apple. Here, we identified 188 MdbHLH proteins in apple “Golden Delicious” (Malus × domestica Borkh.), which could be classified into 18 groups. We also investigated the gene structures and 12 conserved motifs in these MdbHLHs. Coupled with expression analysis and protein interaction network prediction, we identified several genes that might be responsible for abiotic stress responses. This study provides insight and rich resources for subsequent investigations of such proteins in apple.
Keywords: apple, genome-wide analysis, bHLH transcription factor, expression analysis, drought and salt stress, regulation networks
Introduction
Because they are sessile, plants are inevitably affected by environmental factors from which they cannot escape. To optimize their growth and survival, they must utilize a wide range of physiological and biochemical processes in their responses to diverse abiotic stresses (Feller et al., 2011; Sun et al., 2015). Those responses are coordinated by the regulation of gene expression (Agarwal et al., 2006; Pires and Dolan, 2010; Feller et al., 2011). Transcription factors (TFs) play crucial roles in these processes by activating or repressing related downstream genes (Agarwal et al., 2006; Feller et al., 2011; Rehman and Mahmood, 2015). These TF families (Pérez-Rodríguez et al., 2010) include MYBs (Jin and Martin, 1999), bHLHs (basic helix-loop-helix; Massari and Murre, 2000), DREBs (dehydration responsive element-binding; Agarwal et al., 2006), ERFs (ethylene responsive element binding factor; Rehman and Mahmood, 2015), bZIPs (basic region/leucine zipper motif; Corrêa et al., 2008), and WRKY members (Zhang and Wang, 2005), which represent the six major gene families that control stress responses (Kavas et al., 2016).
The bHLH superfamily is the second largest TF family in plants (Feller et al., 2011). Since the discovery of the bHLH motif during research on murine muscle development (Murre et al., 1989), this superfamily has been found in all three eukaryotic kingdoms (Carretero-Paulet et al., 2010). The family is defined by the bHLH domain, which comprises approximately 60 amino acids and contains two functionally distinct regions: the basic region and the HLH region (Toledo-Ortiz et al., 2003). The basic region, with 13–17 amino acids, is located at the N-terminus of the bHLH domain, where it binds the specific E-box (CANNTG) DNA sequence. In addition, this region has six typical basic residues and a highly conserved HER motif (His 5-Glu 9-Arg 13) that are predicted to bind specific DNA sequences (Feller et al., 2011; Kavas et al., 2016). In contrast, the HLH region is composed of two amphipathic α-helices linked by a loop of variable length (Murre et al., 1989; Ferré-D'Amaré et al., 1994). This region functions as a dimerization domain (Massari and Murre, 2000).
As the sequencing of genomes for more species is completed, numerous plant bHLH proteins are being identified and characterized (Pires and Dolan, 2010; Feller et al., 2011; Song et al., 2014; Xu et al., 2014; Chen et al., 2015; Sun et al., 2015; Zhang et al., 2015; Kavas et al., 2016). However, such studies are relatively limited when compared with research efforts with animal species (Song et al., 2014; Kavas et al., 2016). Based on evolutionary relationships, DNA-binding specificity, and conservation of specific amino acids and domains (in addition to the bHLH domain), the animal bHLH proteins have been organized into six groups, A through F (Atchley and Fitch, 1997). Then these bHLH proteins are further divided into small subfamilies (Ledent and Vervoort, 2001; Simionato et al., 2007). In plants, however, the number of groups has not been determined but is thought to cover 15–26 groups (Buck and Atchley, 2003; Pires and Dolan, 2010). Phylogenetic analyses of some atypical bHLH proteins have even extended that number to 32 (Carretero-Paulet et al., 2010). In addition to this bHLH domain, various additional motifs and amino acids are conserved in different bHLH subgroups (Carretero-Paulet et al., 2010; Pires and Dolan, 2010; Feller et al., 2011).
Most of the bHLH proteins already identified and functionally characterized in plants are AtbHLH proteins (Carretero-Paulet et al., 2010), and only a few bHLH family genes have been examined in apple (Malus × domestica; Xie et al., 2012; Qu et al., 2016). Results from studies of plant bHLH-coding genes have suggested that they are involved in regulating a diverse array of biological and biochemical processes, such as light signaling (Roig-Villanova et al., 2007; Leivar et al., 2008), hormone signaling (Friedrichsen et al., 2002; Lee et al., 2006), shoot branching (Komatsu et al., 2001), stomata, and root development (Ohashi-Ito and Bergmann, 2007; Kanaoka et al., 2008), and abiotic stress responses (Chinnusamy et al., 2003; Kiribuchi et al., 2004).
Under stress conditions, some special bHLH TFs are activated and bind to the promoter of the key genes involved in various signaling pathways, and regulate the stress tolerance of plants by regulating the transcription level of these target genes. For example, in response to low temperature, AtICE1 binds to the MYC recognition site (CANNTG; a kind of E-box) in the promoter sequence of CBF3 gene, and induces the expression of CBF3, which functions to increase the resistance of Arabidopsis to cold stress (Chinnusamy et al., 2003). AtNIG1, which was recognized as the first calcium binding TF involved in salt stress signaling pathway, specifically binds to the E-box element in promoter region of some salt stress related genes, and modulates the tolerance to salt stress by regulating downstream gene expression (Kim and Kim, 2006). AtMYC2 promotes the transcription of the drought responsive gene RD22 by binding to its promoter, and thereby increases the drought resistance of Arabidopsis (Abe et al., 2003). Besides, a recent study indicates a direct interaction between AtMYC2 and the ABA receptor AtPYL6, which is inhanced by the presence of ABA, and makes MYC2 important in ABA-JA crosstalk (Aleman et al., 2016). In apple, studies of bHLH TFs are mainly focused on fruit coloration in recent years (An et al., 2012, 2016; Xie et al., 2012; Hu et al., 2016), and just few findings have been reported on the regulation of stress tolerance of MdbHLH genes. Feng et al. (2012) identified an AtICE1 like protein from apple, named MdCIbHLH1. It could bind to the MYC recognition site in the promoter region of a DREB family gene MdCBF2, and regulates apple plants resistance to low temperature by up-regulating the expression of MdCBF2. In addition, MdbHLH104 was proved to enhance the tolerance to iron deficiency by promoting the transcription of MdAHA8 (Zhao et al., 2016).
Even though so many important functions have been recognized, and sequencing of the apple genome has been completed (Velasco et al., 2010) and now improved (Li et al., 2016), no detailed analysis has previously been reported for bHLH family proteins in apple. In this study, we identified and characterized 188 bHLH family genes in apple. They are located on 17 different chromosomes and can be classified into 18 main groups based on phylogenetic analysis. We also investigated their gene structures and conserved domains and motifs. Expression of Arabidopsis homologous genes and MdbHLHs was also analyzed in plants under drought and salt stress, and we predicted the possible interacting proteins and regulatory networks for genes related to those stress responses. Our goal was to provide a useful resource for subsequent research on the functions and regulatory mechanisms of potentially important MdbHLH proteins that are crucial in modulating abiotic stress responses in apple.
Materials and methods
Sequence retrieval and identification of bHLH proteins in apple
We downloaded the file for genome-wide protein sequences in apple (Malus_x_domestica.v1.0.conensus_peptide.fa.gz) from the Genome Database for Rosaceae (GDR; https://www.rosaceae.org/). The Hidden Markov Model (HMM) file of the HLH domain (PF00010) was downloaded from the Pfam database (version 30.0; http://pfam.xfam.org/; Finn et al., 2016), and was used as a query to scan the proteome file via HMMER software (version 3.1b2; http://hmmer.org/) with a default E-value. The protein sequences for genes shown in those HMMER results were obtained from the proteome file using a Bioperl script, and were submitted to the SMART database (http://smart.embl-heidelberg.de) and the online Batch CD-search tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi; Marchler-Bauer et al., 2015) to verify the existence of the conserved bHLH domain. Redundant sequences were removed with online ElimDupes software (http://hcv.lanl.gov/content/sequence/ELIMDUPES/elimdupes.html), and a few sequences with obvious errors were removed manually.
Chromosomal locations, characterizations, and functional annotation of bHLH genes in apple
The GFF file (Malus_x_domestica.v1.0.consensus.gff.gz), containing location data and annotation information for apple genes, was downloaded from the GDR database. MapInspect software was used to map the apple bHLH genes on different chromosomes, and annotation data in the GFF file were exhibited by the online tool WEGO (http://wego.genomics.org.cn/cgi-bin/wego/index.pl). The lengths, masses, isoelectric point (pI)-values, and charge at pH7.0 for these bHLH protein sequences were determined with DNAstar software, and length distributions and functional annotations were analyzed with Blast2GO software (version 2.8.0).
Phylogenetic analysis of apple bHLH proteins
The protein sequences of putative bHLH genes in apple were aligned with ClustalX software (version 1.83) with default parameters, and phylogenetic trees were constructed using the aligned result with MEGA6 software (version 6.06; Tamura et al., 2013) via the Neighbor-Joining (NJ) method (Parameter setting: Bootstrap method-1,000 replicates, Poisson model, Pairwise deletion). Two versions of that tree were produced: the original tree and a bootstrap consensus tree.
Analysis of gene structure (intron-exon) and identification of conserved motifs
Information about the gene structure (intron-exon) of each putative bHLH gene was obtained from the GFF file, downloaded from the GDR database. The schematic structures of these genes were drawn with the online Gene Structure Display Server (GSDS 2.0; http://gsds.cbi.pku.edu.cn). Local MEME software (4.11.2; Bailey and Elkan, 1994) was used to identify conserved motifs in the protein sequences according to the following parameters: -protein, -oc m12, -mod zoops, -nmotifs 12, -minw 6, and -maxw 70. The results from these analyses of gene structure and conserved motifs were arranged according to the order shown on the phylogenetic tree.
Expression analysis of homologous genes in Arabidopsis under drought and salt stress conditions
The Arabidopsis proteome file (TAIR10_pep_20101214) was downloaded from the TAIR database (http://www.arabidopsis.org). Using each putative MdbHLH protein sequence as a query, we performed a local blast (blastp method) to find the homologous genes for these bHLH proteins in Arabidopsis, using BioEdit software (version 7.0.9.0). We also obtained transcriptome data (Affymetrix microarray data; Kilian et al., 2007) for Arabidopsis treated with drought (GSE5624) or salt (GSE5623) stress from the National Center for Biotechnology information (NCBI) database (http://www.ncbi.nlm.nih.gov/). After ID conversion (GenBank ID and Affymetrix ID) and data extraction, the expression patterns of these orthologs under different conditions were displayed in the form of heatmaps (log2-transformed) that were drawn with HemI software (version 1.0.1; Deng et al., 2014). Cluster analysis was conducted with OmicShare tools, a free online platform (www.omicshare.com/tools). The average value of gene expression at each time point was transformed with log2 method, and parameter settings are as follows: number of clusters 5; p < 0.05; fold change > 2.
Protein association network predictions and functional annotations by string
All of the putative MdbHLH protein sequences were submitted to the online server STRING (version 10.0; http://string-db.org), with the organism specified as “Arabidopsis thaliana.” After the blast step was finished, genes with the highest scores (Bitscore) were used to construct the network. Genes that did not interact with any others were removed. The functional annotation information was copied manually from the blast results.
Plant materials and growth conditions
Tissue-cultured apple seedlings (Gala) were grown on MS medium with 0.5 mM 6-BA and 0.1 mM IAA, and grown in controlled environment cabinets under 16 h light/8 h dark conditions at 22°C unless stated otherwise.
Experiments involving drought or salt stress were conducted in a controlled environment chamber. Tissue-cultured apple seedlings were transferred to rooting medium (MS + 0.1 mM IAA) for rooting. Thirty days later, select the plantlets with consistent growth state and transferred into hydroponic conditions (Hoagland nutrient solution) for 7 days. Then these plantlets were treated for various time periods with PEG6000 (10%; W/V) to simulate drought conditions or NaCl (200 mM) to induce salt stress, under continuous white light conditions (Osram L18W/30 tubes; 20 μmol m−2s−1). Roots of these plantlets were sampled at specified time points, frozen in liquid nitrogen and saved in −80°C for subsequent use.
RNA extraction and real-time quantitative RT-PCR analysis
Plant materials were harvested, frozen in liquid nitrogen, and then ground under RNase-free conditions. The RNA was extracted with TRizol reagent (Invitrogen), following the manufacturer's instructions, and then treated with DNase I at 37°C for 30 min. The RNA (1 μg) was then reverse-transcribed using a PrimeScript First-strand cDNA Synthesis Kit (Takara) according to the manufacturer's instructions. A 10-μL aliquot of cDNA was diluted to 100 μL with water, and 2 μL of that diluted cDNA was used for the analyses.
For real-time quantitative RT-PCR (qRT-PCR), gene-specific primers for our selected MdbHLHs were designed and synthesized by Sangon Biotech (Shanghai) Company (product size 110–130 bp; Tm 59–61°C; details are shown in Supplementary Table S1), and Md18S was used as an internal control. All reactions were performed on an Icycler iQ5 system (Bio-Rad), using the SYBR Green Supermix Kit (Bio-Rad) according to the manufacturer's instruction. Expression levels of these genes were calculated as 2−ΔCT values (expression level and standard deviation data were shown in Supplementary Table S2), and heatmaps (Supplementary Figure S6) were drawn with HemI software (log2-transformed). Besides, relative expression levels for each gene along the time series were also calculated as 2−ΔΔCT values (Figure 5). At least three biological replicates were used for the fluorescence-quantitative PCR reactions, with each biological repeat having at least three technical replicates. Each biological repeat contains at least 6 plantlets for mixing. Spearman correlation analysis between the expression patterns of MdbHLHs in apple and MdbHLH orthologs in Arabidopsis were performed using R softwares.
Results
Identification, chromosomal locations, and functional annotation of apple bHLH genes
For genome-wide identification of MdbHLH genes, we used the HMM file as a query to search the apple proteome. With default parameters, 253 putative MdbHLH protein sequences were obtained. After the existence of the conserved bHLH domain was confirmed by SMART and CD-Search, and redundant sequences were removed, we were able to identify 188 sequences as genes in the apple bHLH family. These were named MdbHLH1 to MdbHLH188, based on their chromosomal locations (Figure 1A; Supplementary Table S3). Sequence analysis revealed that these MdbHLH proteins vary widely in length, from 128 to 963aa, and have predicted molecular weights of 14,666–105,632 Da. Their predicted pI-values range from 4.51 to 10.10. Gene IDs, genomic positions, charge at pH 7.0 values, and information about Gene Ontology (GO) annotations were also summarized for these MdbHLH proteins (Supplementary Table S3).
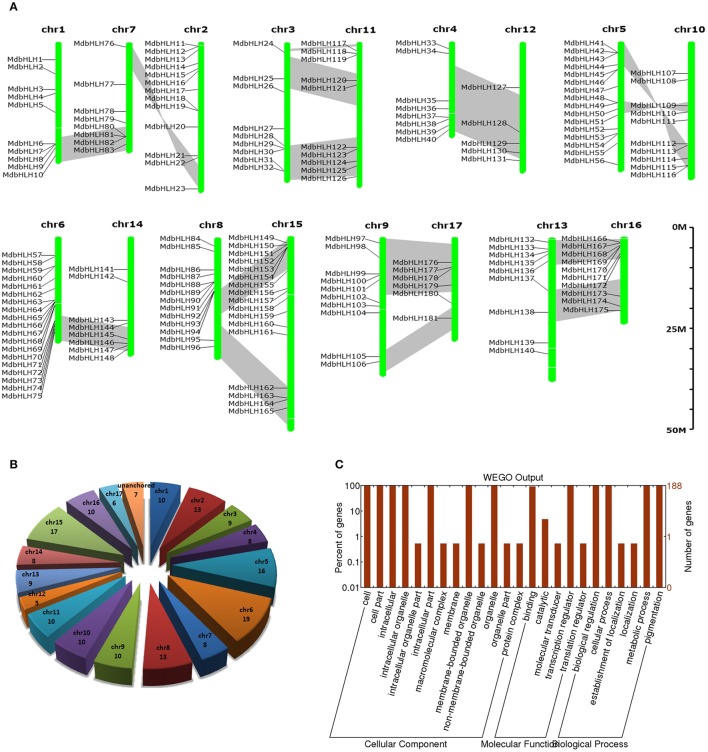
Figure 1.
Summary information for the 188 MdbHLH genes. (A) Locations of 188 MdbHLHs on 17 chromosomes. (B) Distribution of 188 MdbHLHs on chromosomes. (C) Functional annotation (Gene Ontology) of MdbHLH proteins. Segmented duplicate homologous blocks are indicated with gray shadow.
Based on the genomic position information obtained from the GDR database, we showed that the MdbHLH genes were found across all the 17 apple chromosomes, ranging from 5 to 19 per chromosome. However, the other seven bHLHs (MdbHLH182-MdbHLH188) were localized to unassembled genomic scaffolds, and can not be mapped to any particular chromosome according to what we currently know about this genome (Figures 1A,B; Supplementary Table S3). Chromosome 6 has the most MdbHLHs (19 total), followed by chr 15 (17 genes) and chr 5 (16 genes). Furthermore, although chr 12 is longer than either chr 5 or chr 6, it contains only five MdbHLHs, making it the least populated with genes in that family (Figures 1A,B).
The GDR database provided GO annotation information about these MdbHLH proteins (Supplementary Table S3), which we were able to depict by using WEGO software (Figure 1C). There are three aspects of function classifications: CC (cellular component), MF (molecular function), and BP (biological process). The MF and BP aspects mainly described the molecular activities of multiple gene products, and included associated pathways and processes that are enriched by these genes, i.e., binding, transcription regulation, biological regulation, cellular process, metabolic process, and pigmentation. All of these functions and processes are closely related to TFs. In addition, the enrichment results for CC aspects (where gene products are active) were consistent with the primary roles that TFs have. Because the annotation information obtained from the GDR database was simple, we sought more details by running an annotation analysis with Blast2GO software. Those results indicated that individual MdbHLH proteins are 200–600 aa long (average 404 aa; Supplementary Figure S1). These GO annotation results were largely similar to what we obtained from the GDR database. For example, in the MF aspect, the MdbHLH proteins were mainly enriched in binding activities, especially protein-binding and sequence-specific DNA-binding (Supplementary Figure S1; Supplementary Table S4), both of which are the main mechanisms by which bHLH family TFs regulate the expression of downstream genes (Leivar et al., 2008; Feller et al., 2011; Xie et al., 2012).
Phylogenetic analysis, display of gene structure, and prediction of conserved motifs
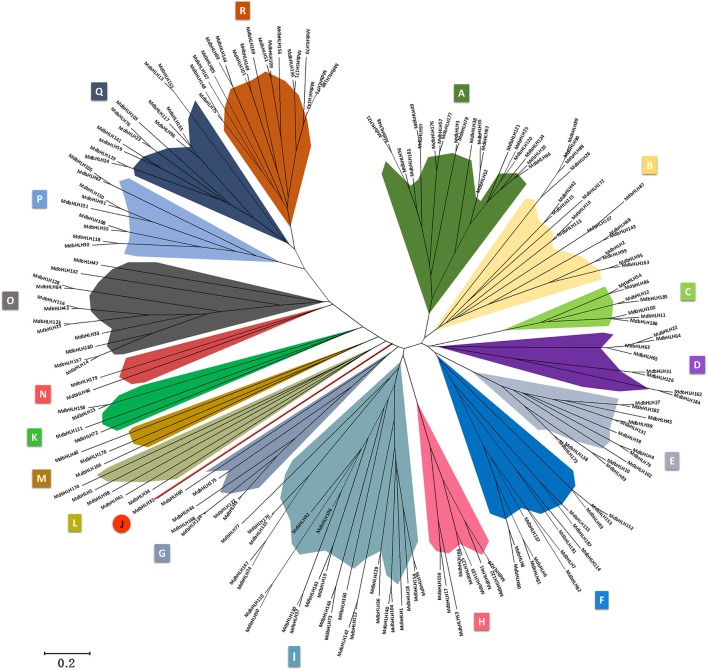
The exact number of subgroup classifications for plant bHLH proteins is unknown, but is thought to be 15–32 (Buck and Atchley, 2003; Carretero-Paulet et al., 2010; Pires and Dolan, 2010). To evaluate the evolutionary relationships of our 188 proteins, we conducted a phylogenetic analysis based on full-length protein sequences. Applying the NJ method, we assigned the proteins to18 main groups: A through R (Figure 2; Supplementary Figure S2), and then subdivided each of the three largest groups (A, I, and R) into two subgroups (Supplementary Figure S2). This resulted in 21 MdbHLH subgroups, which is the same number as those found in tomato (Sun et al., 2015) and Phaseolus vulgaris (Kavas et al., 2016). Unlike the other groups in this phylogenetic tree, Subgroup J contained a single bHLH protein, meaning that MdbHLH32 is also unique (Figure 2). The five smallest groups were J through N, each having one to five MdbHLHs (Figure 2; Supplementary Figure S2).
Figure 2.
Phylogenetic analysis (original tree) and subgroup classifications of MdbHLH proteins.
The evolutionary relationships among these MdbHLH proteins were also determined by analyzing their gene structures and conserved motifs. These composition patterns tended to be consistent with the results from our phylogenetic tree, being nearly identical among genes within the same group, but varying greatly between groups. Except for Motifs 1 and 2, which were conserved in all MdbHLH proteins (Supplementary Figures S3, S4), some of the other 10 motifs were present only in certain groups, which might explain why functions for MdbHLH proteins tend to be specific to a particular group. These included Motifs 4 and 8, only present in Groups A and D; Motifs 7 and 9, in Group A; Motif 6, Groups D and E; Motif 11, Group E; Motif 10, Group R; Motif 3, mainly present in Groups P, Q, and R, except for MdbHLH40, which is in Group M; and Motif 12, mainly present in Groups O and Q but with two exceptions: MdbHLH5 in Group L and MdbHLH160 in Group I (Supplementary Figure S3). Overall, Group A had the most motifs, and the genes in other groups had more complex gene structures.
Although most MdbHLHs within a group showed similar composition patterns, obvious differences were noted for three genes: MdbHLH151 in Group P, MdbHLH13 in Group Q, and MdbHLH70 in Group R. Their genomic sequences were much longer than those of others within the same group. For example, MdbHLH151 had eight introns while most of the others had only four. That gene also lacked the first specific exon that was contained in all of the remaining genes in Group P, and which was also conserved in some other groups (Supplementary Figure S3). For MdbHLH13, its long genomic sequence and protein sequence were very different from others in Group Q. The MdbHLH13 protein also had an additional motif (Motif 12) that was mainly conserved in Group O, and was present only in MdbHLH13 and MdbHLH155 in that group (Supplementary Figure S3). The MdbHLH70 protein lacked Motif 2, which was conserved in 181 of the 188 MdbHLH proteins (Supplementary Figures S3, S4). In addition to these three, MdbHLH176 lacked both Motifs 1 and 2, making it an atypical bHLH protein. Meanwhile, even though MdbHLH152 did have those two motifs, their locations were reversed, with Motif 2 being in the front (Supplementary Figure S3).
Expression profiling and regulatory network predictions for MdbHLH orthologs in Arabidopsis
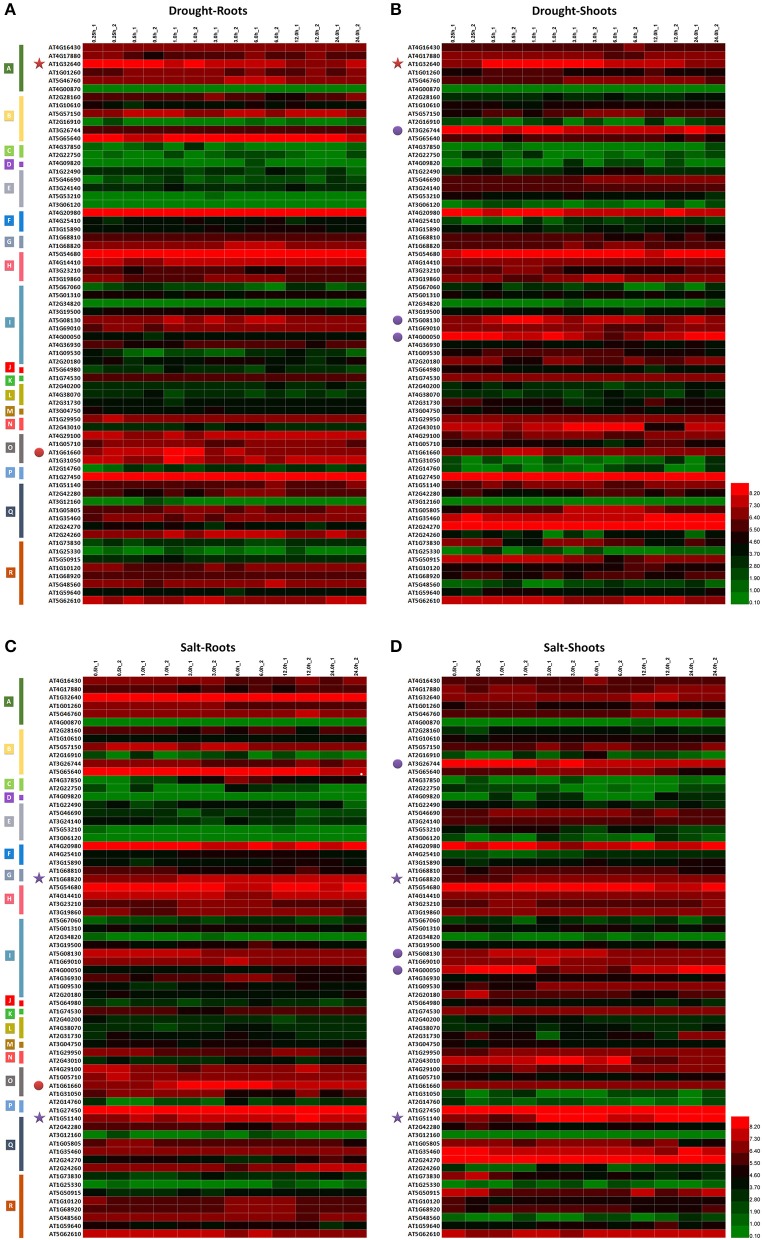
For our preliminary investigation of the functions of MdbHLH genes in regulating plant responses to abiotic stress, we downloaded the transcriptome data of homologous genes in Arabidopsis (Supplementary Table S5), and evaluated their expression patterns (Figure 3; Supplementary Figure S5). Overall, expression of many AtbHLHs was significantly different among tissue types, regardless of which stress conditions were tested (Figure 3). This suggested that their expression was tissue-specific. In contrast, some genes showed different (or even opposite) expression patterns (Figure 3; Supplementary Figure S5), indicating that their responses varied according to the stress conditions applied.
Figure 3.
Expression analysis of MdbHLH orthologs in Arabidopsis. Expression patterns of MdbHLH orthologs under drought (A,B) and salt (C,D) stress. Different colors indicate genes with expression patterns that are similar between tissue types (stars) or treatments (dots).
Several TF genes demonstrate coordinated regulation of responses to drought and salt stress (Jin and Martin, 1999; Agarwal et al., 2006; Feller et al., 2011; Kavas et al., 2016). We also observed this here in responses to drought or salt, as well as in having similar expression patterns in different tissues and/or under different treatment conditions (Supplementary Figure S5). This was all evidence for how essential those genes are in regulating plant responses. For example, under drought conditions, AT1G32640 was down-regulated in both roots and shoots whereas AT1G51140 and AT1G68820 were up-regulated significantly under salt stress (Figure 3). Expression of both AT1G61660 and AT5G08130 under drought or salt stress increased initially in the roots and shoots, respectively, before being down-regulated, whereas AT4G00050 was first down-regulated before expression increased under those same test conditions (Figure 3; Supplementary Figure S5; Supplementary Table S5). These results also suggested that some genes in bHLH family may utilize opposing regulatory mechanisms under abiotic stresses.
Our STRING approach allowed us to predict a protein interaction network using the 188 MdbHLH protein sequences as queries. This produced a complex regulatory network constructed with orthologs in Arabidopsis. Many bHLH proteins interacted with more than one bHLH (Figure 4), which is in accord with previous reports that binding activity with specific DNA sequences depends upon the formation of homodimers or heterodimers of different bHLH proteins (Pires and Dolan, 2010; Feller et al., 2011). Overall, we found 11 proteins that could interact with more than four other bHLH proteins, making them important players in regulating plant growth and stress responses (Supplementary Table S6). Furthermore, our blast results strongly supported our previous classifications of the MdbHLH proteins (Supplementary Table S6). Detailed information about these orthologs is also summarized in Supplementary Table S6.
Figure 4.
Protein interaction network for MdbHLHs based on MdbHLH orthologs in Arabidopsis. This network was predicted by online software STRING. Red lines indicate proteins that are predicted to interact with more than four other bHLH proteins. MdbHLH proteins are shown in brackets with Arabidopsis orthologs.
Expression analysis of MdbHLH responses to salt and drought stress
To identify which of these bHLH genes are most important in the response to abiotic stresses, we studied changes in expression for 30 MdbHLHs (Supplementary Table S7). These were selected because they demonstrated unique composition patterns, and because of their roles in stress responses or in protein interaction networks, as deduced from the expression analysis and predicted regulatory network for homologous genes in Arabidopsis. Besides, correlation analysis (spearman) between the expression patterns of selected MdbHLH genes in apple and MdbHLH orthologs in Arabidopsis were also performed. Because of lacking the expression data for four hmomolgue genes (MdbHLH4-AT2G46810, MdbHLH15-AT5G56960, MdbHLH64-AT2G31220, and MdbHLH152-AT1G49770) in the downloaded file (Supplementary Table S8), we performed a correlation analysis of the remaining 26 genes.
Expression for most of these genes was significantly altered in the early stage of drought or salt treatment (Figure 5; Supplementary Figure S6). Among them, some showed changes in expression that were similar or opposite between drought and salt treatments. For example, under both of those stresses, MdbHLH74 and MdbHLH119 were initially down-regulated significantly before being up-regulated. Their responses tended to be more rapid under drought conditions, usually changing within the first hour (Figure 5; Supplementary Figure S6). The opposite was noted for MdbHLH24 and MdbHLH151. Under drought stress, both were up-regulated at first and then down-regulated, whereas, under salt stress, their expression initially decreased before increasing (Figure 5; Supplementary Figure S6). Some other genes showed changes in expression that were specific to the type of treatment applied. They included MdbHLH4, MdbHLH168, and MdbHLH177, which were down-regulated under salt stress. Under drought conditions, however, their expression was also down-regulated at first, but then increased (Figure 5; Supplementary Figure S6). Other genes were significantly down-regulated by salt treatment but responded in different ways to drought, including MdbHLH2 MdbHLH15, MdbHLH32, MdbHLH43, MdbHLH48, MdbHLH64, MdbHLH77, MdbHLH134, MdbHLH143, MdbHLH147, MdbHLH148, MdbHLH152, MdbHLH160, and MdbHLH172 (Figure 5; Supplementary Figure S6). Among them, MdbHLH143 is exactly the cold-induced bHLH TF gene named MdCIbHLH1 (Feng et al., 2012; Supplementary Table S3), which we have mentioned in introduction. In addition to expression analysis, correlation analysis between MdbHLH genes and their orthologs in Arabidopsis found that several showed a high correlation, such as MdbHLH24, MdbHLH52, MdbHLH74, MdbHLH119, MdbHLH134, and MdbHLH143, with p < 0.05 (Supplementary Table S8; Supplementary Figure S7). Moreover, the expression patterns of MdbHLH134 and its ortholog (AtbHLH30, AT1G68810) showed a significant correlation (p < 0.01), no matter under drought or salt conditions (Supplementary Table S8).
Figure 5.
Relative expression analysis of select genes along the time series. Analysis (2−ΔΔCT method) of 30 selected MdbHLHs expressed in response to induced drought (PEG6000; blue line) or salt stress (NaCl; red line).
Development of protein interaction networks for MdbHLHs that are crucial to the abiotic stress response
Proteins in the bHLH family function primarily by forming homodimers or heterodimers with other proteins, actions that are vital for their binding to related downstream genes (Toledo-Ortiz et al., 2003; Carretero-Paulet et al., 2010; Pires and Dolan, 2010). As part of our preliminary study of the regulatory mechanism(s) of important MdbHLH genes under abiotic stress, we predicted the protein interaction networks for MdbHLH24 and MdbHLH74, which demonstrated either similar or opposite expression patterns under different treatment conditions (Figure 5). We also examined MdbHLH4, which showed a significant response to drought and salt treatments (Figure 5). Besides, its homologous gene, AT2G46810, was located in the center of the predicted gene association network (Figure 4).
Although the predicted network for MdbHLH24 (FBH3 ortholog) was mainly obtained based on text-mining and co-expression analysis (Figure 6A), most of the other proteins were involved in ABA signaling pathways, such as ABI2, HAB1, and 2, PP2CA, and CYP707A3 (Supplementary Table S9). The ABA signal has an important role in regulating the plant response to various abiotic stresses, such as drought, salt, and cold (Agarwal et al., 2006; Rehman and Mahmood, 2015). Therefore, this result was consistent with our previous deduction that MdbHLH24 is a crucial regulator of stress responses in apple, and that these interacting proteins provide useful resources for subsequent research. Moreover, our investigation of MdbHLH74 (ortholog of BIM1) showed that it plays central roles in regulating various proteins with different functions, several of which have been experimentally determined (Figure 6B; Supplementary Table S9). This suggested that, in addition to controlling the abiotic stress response, MdbHLH74 regulates many other aspects of plant growth and development, such as modulating shade avoidance in apple when coupled with BEE1 (Cifuentes-Esquivel et al., 2013). Our findings were again in accord with the various functions identified already for bHLH family genes, especially when combined with genes in other families (Corrêa et al., 2008; Feller et al., 2011; Rehman and Mahmood, 2015). Finally, the results of two predicted networks (Figures 4, 6C) indicated that MdbHLH4 (ortholog of AT2G46810) has crucial roles in the bHLH family and that most of the proteins that possibly interact with MdbHLH4 are also members of that family (Figure 6C; Supplementary Table S9). This implied that MdbHLH4 interacts with more bHLH proteins than do other bHLHs, such as MdbHLH24 and MdbHLH74. Although our network suggested that MdbHLH4 has a wider range of regulatory functions, most genes in this network (Figure 6C) have not been functionally characterized in Arabidopsis, including At2G46810 (AtbHLH70), the MdbHLH4 ortholog (Supplementary Table S9). However, we now recognize that future examinations can begin with MdbHLH4 as part of an extended focus on the potential functions and mechanisms of bHLH family genes in apple.
Figure 6.
Protein interaction networks prediction for crucial MdbHLH proteins. Protein interaction network predictions of MdbHLH24 (A), MdbHLH74 (B), and MdbHLH4 (C), based on MdbHLH orthologs in Arabidopsis. MdbHLH proteins are shown in brackets with Arabidopsis orthologs.
Discussion
Completion of apple genome sequencing (Velasco et al., 2010; Li et al., 2016) has enabled resarchers to identify and characterize many TF genes at the whole-genome level, including MYB (Cao et al., 2013), ERF (Girardi et al., 2013), DREB (Zhao et al., 2012), bZIP (Wang et al., 2015), and WRKY (Meng et al., 2016). However, no such detailed studies have been done with the bHLH family, and only a few examinations have been made of MdbHLHs (An et al., 2012, 2016; Xie et al., 2012). Here, we identified 188 MdbHLH genes in apple. Expression analysis and predictions of interaction networks allowed us to identify genes that have crucial roles in the apple response to salt and drought stresses.
Classification of MdbHLH proteins based on polygenetic relationship, gene structure, and conserved motifs
Based on phylogenetic analysis, intron-exon organizations, DNA-binding motifs, predicted conserved protein motifs, and amino acids, we classified our 188 MdbHLH proteins into 18 groups and then further divided Groups A, I, and R into two subgroups each, such that the total number was 21. This is the same number reported for tomato (Sun et al., 2015), even though that organism has fewer SlbHLHs (159) than the 188 MdbHLHs in apple. Our phylogenetic tree (Figure 2) indicated that Group A has two main branches. The MdbHLH genes in subgroup A-2 have two prominent and similar exons, making them different from A-1. Besides, the patterns of motif composition showed that the MdbHLH proteins in A-2 do not contain Motif 9. Therefore, MdbHLH38 more likely belongs to the A-1 subgroup, which is different from A-2 on the phylogenetic tree (Supplementary Figure S3). If one considers that a significant distance exists between MdbHLH38 and other A-2 proteins (Figure 2), and that this protein has a gene structure and motifs similar to those of other A-1 proteins, then we might propose that MdbHLH38 represents a transitional form in the evolution of those two subgroups. Likewise, if one takes into account the more complex but consistent motif composition patterns of the A-2 subgroup, we can conclude that it might evolved from the A-1 subgroup, thereby acquiring some specific regulatory functions that are common to many large TF families (Zhang and Wang, 2005; Agarwal et al., 2006; Feller et al., 2011; Rehman and Mahmood, 2015). Based on these findings and suppositions, we suggest that MdbHLH38 is a useful gene for studying evolutionary processes within Group A, and it may even provide a model for investigations of other groups within the bHLH family. With regard to Groups I and R, their very obvious differences in gene structure between Subgroups I-1 and I-2, as well as the fact that R-2 contains the specific Motif 10 that R-1 is lacking, means that those particular subgroup classifications are much easier to make.
Identification of important MdbHLH genes that may regulate responses to drought and salt stress in apple
To determine which of the 188 MdbHLHs are the most important regulators of stress responses in apple, we studied how the expression of their orthologs in Arabidopsis was changed when plants were exposed to drought or saline conditions (Figure 3; Supplementary Figure S5). Regulating the functions of bHLH proteins depends upon the formation of homodimers or heterodimers between bHLH proteins, or between bHLH and non-bHLH proteins (Herold et al., 2002; Hernandez et al., 2007). Thus, for genes that show similar expression patterns (Supplementary Figure S5), we might conclude that they likely interact with each other at the protein level (Leivar et al., 2008), and also coordinately control the expression of downstream genes (Feller et al., 2011). Those regulatory mechanisms also depend upon the co-expression of these interacting genes. Nevertheless, it is also possible that the expression of one bHLH gene can be affected, or even regulated, by another one, such that the two appear to have similar changes in expression. Further research is needed to explore the interrelationships between such genes.
We also found here that some MdbHLH genes showed contrasting changes in expression in response to the same stress treatment. Feller et al. (2011) have reported that a single bHLH protein can interact with more than one bHLH protein and some are even able to interact with several bHLHs or non-bHLH proteins. For example, the bHLH protein PIF3 can homodimerize or heterodimerize with PIF4, and also can heterodimerize with an atypical bHLH factor HFR1 (Castillon et al., 2007). And the MYB-bHLH-WDR (MBW) complex (Feller et al., 2011) may be the best proof for the type of interaction between the bHLH family and other families. In Arabidopsis, studies of the bHLH TF gene AtTT8 showed that there is a positive feedback mechanism in regulation of self expression, which is depends on the MBW complex (Baudry et al., 2006). The in vivo binding of TT2 and PAP1 to the TT8 promoter is dependent on the simultaneous expression of TT8, which is crucial for further induction of TT8 expression (Baudry et al., 2006). For PIFs, PIF4, and PIF5 can form homodimers and bind to the promoter of target genes in dark conditions (Xu et al., 2015). Under light conditions, however, another bHLH TF factor HFR1 will directly inhibits these PIFs by forming non-DNA-binding heterodimers with PIF4 and PIF5 (Hornitschek et al., 2009). If we consider the opposite expression patterns that we detected here between different bHLH genes, as well as their complex mechanisms for protein interactions, then we might conclude that the feedback mechanism should be responsible. In other words, some bHLH genes may specifically respond to stress conditions and form heterodimers with specific bHLH proteins, leading to the differentiation in the expression between different bHLH genes. Further research is needed to verify this hypothesis.
Because various bHLH proteins respond differently to environmental factors, we can hypothesize that genes located in the center of regulatory networks are more important, such as MdbHLH4 (AT2G46810) and MdbHLH134 (AT1G68810; Figure 4). In Arabidopsis, the bHLH TF ABS5/T5L1/bHLH30 (ortholog of MdbHLH134; Supplementary Table S6) is essential for leaf morphogenesis, and related to auxin signaling (An et al., 2014). Besides, expression analysis of AtbHLH30 (260034_at) in roots showed that, although did not respond to drought, it was significantly down-regulated by salt stress treatment (Figure 3; Supplementary Table S8). These diverse functions are consistent with its central location in the regulatory network (Figure 4). In this study, the expression pattern of MdbHLH134 and AtbHLH30 showed a significant correlation in spearman correlation analysis (p < 0.01) under drought and salt conditions (Supplementary Table S8; Supplementary Figure S7). Based on this extremely similar expression pattern, and the hypothesis that orthologous proteins are likely to perform similar or identical functions in other organisms (Feller et al., 2011), we can infer that MdbHLH134 should play important roles in leaf development and salt stress response in apple, and may be realted to auxin.
Although MdbHLH143 (MdCIbHLH1) is not located in the center of regulatory networks, its expression is down-regulated by salt treatment obviously (Supplementary Figure S6), and its expression pattern exhibited a significant negative correlation with its ortholog AtICE1 under drought conditions (Supplementary Figure S7; Supplementary Table S8). In Arabidopsis, AtICE1 encodes a MYC-like bHLH TF protein that binds to Myc recognition sequences in the CBF3 promoter under cold stress conditions (Chinnusamy et al., 2003). Following studies in other organisms suggest that, except for cold, ICE1-like genes may also play impotant roles in regulating other plant stress resistance (Chen et al., 2012; Feng et al., 2013), such as drought and salt, by regulating the expression of DREB1 (CBF) genes (Wang et al., 2008; Yang et al., 2011). These results suggest that MdbHLH143 (MdCIbHLH1) should not only regulates plant response to cold (Feng et al., 2012), but also to drought and salt stresses. Besides, the AtMYC2 orthologous gene MdbHLH177 exhibited a significant response to drought and salt conditions (Supplementary Figure S6), indicating a impotrant role of this gene in regulating stress resistance in apple, just like AtMYC2 in Arabidopsis (Abe et al., 2003; Aleman et al., 2016).
Regulatory network predictions of crucial MdbHLH genes that show significant responses to drought and salt stress
Based on our analyses, we selected some MdbHLHs for further expression analysis under drought and salt stress. As we had expected, most of them exhibited significant responses and showed different patterns of expression depending on the type of treatment applied, such as MdbHLH4, MdbHLH24, and MdbHLH74 (Figure 5). Actually, except lacking the expression data of MdbHLH4 orthologous gene, expression of both MdbHLH24 and MdbHLH74 showed significant correlation with their orthologous genes, respectively (Supplementary Figure S7; Supplementary Table S8). These results suggest that these genes play important roles in apple stress response. We also used Arabidopsis orthologs to predict the regulatory networks for these three representative genes, MdbHLH24, MdbHLH74, and MdbHLH4. For the first two, their functions and regulatory mechanisms matched those of their orthologs (Supplementary Table S9), and conincide with the assumption that they are crucial for regulating apple tolerance to drought and salt stresses. For example, most proteins that are predicated to interact with MdbHLH24 are involved in ABA signaling pathway (Supplementary Table S9), which is crucial for abiotic stress response in plants (Agarwal et al., 2006). This result also suggests that MdbHLH24 regulates plant stress tolerance by regulating the expression of key genes in the ABA signaling pathway.
However, most of the proteins that interact with MdbHLH4 have not yet been characterized, even in Arabidopsis. Nonetheless, because the expression of MdbHLH4 is induced by drought and salt treatments significantly, and its ortholog is located in the center of the network and can interact with many bHLH proteins that belong to different signaling pathways, we believe that this protein has a wide range of regulatory roles in apple, and may be a critical component of competitive regulatory mechanisms. The functional annotation of the three known proteins (RGE1, NAI1, and ILR3) also supports this hypothesis (Supplementary Table S9). These three genes are involved in regulating different aspects of plant growth and development, suggesting the extensive functions for proteins that interact with MdbHLH4. The functions and mechanisms of MdbHLH4 and its interacting proteins are largely unknown, but provide a good opportunity for researchers to reveal the potential operations of such mechanisms for bHLH proteins in general, and the unique functions of apple MdbHLH proteins in particular.
In conclusion, this report is the first to describe the identification of 188 bHLH family genes in apple. In addition to investigating their functions and structures, we also performed expression analyses and developed a regulatory network to determine which genes are most active for stress responses in this crop species. Improving our understanding of the competitive regulatory mechanisms of bHLH proteins, and the genes that are central to this proposed mechanism, means that we now have rich resources for subsequent studies of gene cloning and functional characterization of members in this MdbHLH family.
Author contributions
The study was conceived by FM. KM and QD collected the public datasets for Apple and the other researched species. KM, QD, CLi, and CLiu contributed to data analysis, bioinformatics analysis and manuscript preparation. KM, QD, and CLi participated in the qRT-PCR experiment. FM, KM, and CLiu participated in the planning of experiments and revising the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the State Key Program of the National Natural Science Foundation of China (31330068) and the earmarked fund for the China Agriculture Research System (CARS-28). The authors are grateful to Priscilla Licht for help in revising our English composition and Mr. Zhengwei Ma for management of the apple trees.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00480/full#supplementary-material
Annotations of 188 MdbHLH proteins performed by Blast2GO. (A) Length distribution of sequences. (B–D) Annotations for CC, MF, and BP components, respectively.
Phylogenetic analysis (circle tree) and subgroup classifications (21 subgroups) of MdbHLH proteins.
Analysis of gene structure and conserved motifs for MdbHLH proteins. Phylogenetic tree (A), organization of gene structure (B), and distribution of conserved motifs (C) for 188 MdbHLH proteins.
Motif logos for 12 conserved motifs. The motif logos were predicted by local MEME software. Red stars indicate conserved HER motif (His 5- Glu 9-Arg 13).
Cluster analysis of gene expression data for MdbHLH orthologs (fold change >2) in Arabidopsis under drought or salt treatment.
Expression analysis of selected MdbHLH genes. Analysis (2−ΔCT method) of 30 MdbHLHs expressed in response to induced drought (PEG6000) (A) or salt stress (NaCl) (B).
Correlation analysis between the expression patterns of selected MdbHLH genes and MdbHLH orthologs in Arabidopsis, under drought (A) or salt (B) conditions. The value of spearman correlation coefficent in the heatmap was multiplied by one hundred. Positive or negative correlation relationships between the gene expression patterns are indicated with positive or negative values. Color depth (blue or red) of the grids corresponds to the value.
qRT-PCR primers used in this study.
The expression level and standard deviation data for selected MdbHLH family genes.
Chromosomal location and functional annotation of 188 apple bHLH genes.
GO annotation for MdbHLH proteins.
Expression level for MdbHLH orthologs in Arabidopsis under different stress conditions.
Summary information for 188 MdbHLH proteins in STRING database. The predicted important bHLH TFs involved in regulating plant growth and stress responses were marked in red and listed in this table (sheet3).
Name list of the selected 30 MdbHLH genes for subsequent expression analysis.
Correlation analysis between the expression patterns of selected MdbHLH genes and MdbHLH orthologs in Arabidopsis. Genes with significant correlation are marked with asterisks (* for p < 0.05; ** for p < 0.01).
Summary information for proteins that may interact with MdbHLH24, MdbHLH74, or MdbHLH4.
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P. K., Agarwal P., Reddy M. K., Sopory S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25, 1263–1274. 10.1007/s00299-006-0204-8 [DOI] [PubMed] [Google Scholar]
- Aleman F., Yazaki J., Lee M., Takahashi Y., Kim A. Y., Li Z. X., et al. (2016). An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling. Sci. Rep. 6:28941. 10.1038/srep28941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. P., Li H. H., Song L. Q., Su L., Liu X., You C. X., et al. (2016). The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol. Biochem. 108, 24–31. 10.1016/j.plaphy.2016.06.032 [DOI] [PubMed] [Google Scholar]
- An R., Liu X. Y., Wang R., Wu H. C., Liang S., Shao J. X., et al. (2014). The over-expression of two transcription factors, ABS5/bHLH30 and ABS7/MYB101, leads to upwardly curly leaves. PLoS ONE 9:e107637. 10.1371/journal.pone.0107637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An X. H., Tian Y., Chen K. Q., Wang X. F., Hao Y. J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169, 710–717. 10.1016/j.jplph.2012.01.015 [DOI] [PubMed] [Google Scholar]
- Atchley W. R., Fitch W. M. (1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 94, 5172–5176. 10.1073/pnas.94.10.5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. [PubMed] [Google Scholar]
- Baudry A., Caboche M., Lepiniec L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, 768–779. 10.1111/j.1365-313X.2006.02733.x [DOI] [PubMed] [Google Scholar]
- Buck M. J., Atchley W. R. (2003). Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 56, 742–750. 10.1007/s00239-002-2449-3 [DOI] [PubMed] [Google Scholar]
- Cao Z. H., Zhang S. Z., Wang R. K., Zhang R. F., Hao Y. J. (2013). Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 8:e69955. 10.1371/journal.pone.0069955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J. F., Bilbao-Castro J. R., Robertson D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. 10.1104/pp.110.153593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12, 514–521. 10.1016/j.tplants.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Chen L., Chen Y., Jiang J. F., Chen S. M., Chen F. D., Guan Z. J., et al. (2012). The constitutive expression of Chrysanthemum dichrum ICE1 in Chrysanthemum grandiflorum improves the level of low temperature, salinity and drought tolerance. Plant Cell Rep. 31, 1747–1758. 10.1007/s00299-012-1288-y [DOI] [PubMed] [Google Scholar]
- Chen Y.-Y., Li M.-Y., Wu X.-J., Huang Y., Ma J., Xiong A.-S. (2015). Genome-wide analysis of basic helix-loop-helix family transcription factors and their role in responses to abiotic stress in carrot. Mol. Breed. 35:125 10.1007/s11032-015-0319-0 [DOI] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., Agarwal M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. 10.1101/gad.1077503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Esquivel N., Bou-Torrent J., Galstyan A., Gallemí M., Sessa G., Martret M. S., et al. (2013). The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 75, 989–1002. 10.1111/tpj.12264 [DOI] [PubMed] [Google Scholar]
- Corrêa L. G., Riaño-Pachón D. M., Schrago C. G., dos Santos R. V., Mueller-Roeber B., Vincentz M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE 3:e2944. 10.1371/journal.pone.0002944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Wang Y., Liu Z., Cheng H., Xue Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS ONE 9:e111988. 10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E. L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- Feng H. L., Ma N. N., Meng X., Zhang S., Wang J. R., Chai S., et al. (2013). A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Biochem. 73, 309–320. 10.1016/j.plaphy.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Feng X. M., Zhao Q., Zhao L. L., Qiao Y., Xie X. B., Li H. F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 12:22. 10.1186/1471-2229-12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., Burley S. K. (1994). Structure and function of the b/HLH/Z domain of USF. EMBO J. 13, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D. M., Nemhauser J., Muramitsu T., Maloof J. N., Alonso J., Ecker J. R., et al. (2002). Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi C. L., Rombaldi C. V., Dal Cero J., Nobile P. M., Laurens F., Bouzayen M., et al. (2013). Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis. Sci. Hortic. 151, 112–121. 10.1016/j.scienta.2012.12.017 [DOI] [Google Scholar]
- Hernandez J. M., Feller A., Morohashi K., Frame K., Grotewold E. (2007). The basic helix loop helix domain of maize R links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc. Natl. Acad. Sci. U.S.A. 104, 17222–17227. 10.1073/pnas.0705629104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S., Wanzel M., Beuger V., Frohme C., Beul D., Hillukkala T., et al. (2002). Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell 10, 509–521. 10.1016/S1097-2765(02)00633-0 [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902. 10.1038/emboj.2009.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D. G., Sun C. H., Zhang Q. Y., An J. P., You C. X., Hao Y. J. (2016). Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple. PLoS Genet. 12:e1006273. 10.1371/journal.pgen.1006273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H. L., Martin C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. 10.1023/A:1006319732410 [DOI] [PubMed] [Google Scholar]
- Kanaoka M. M., Pillitteri L. J., Fujii H., Yoshida Y., Bogenschutz N. L., Takabayashi J., et al. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20, 1775–1785. 10.1105/tpc.108.060848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavas M., Baloglu M. C., Atabay E. S., Ziplar U. T., Dasgan H. Y., Ünver T. (2016). Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol. Genet. Genomics 291, 129–143. 10.1007/s00438-015-1095-6 [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. 10.1111/j.1365-313X.2007.03052.x [DOI] [PubMed] [Google Scholar]
- Kim J., Kim H. Y. (2006). Functional analysis of a calcium-binding transcription factor involved in plant salt stress signaling. FEBS Lett. 580, 5251–5256. 10.1016/j.febslet.2006.08.050 [DOI] [PubMed] [Google Scholar]
- Kiribuchi K., Sugimori M., Takeda M., Otani T., Okada K., Onodera H., et al. (2004). RERJ1, a jasmonic acid-responsive gene from rice, encodes a basic helix-loop-helix protein. Biochem. Biophys. Res. Commun. 325, 857–863. 10.1016/j.bbrc.2004.10.126 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Maekawa M., Shimamoto K., Kyozuka J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231, 364–373. 10.1006/dbio.2000.9988 [DOI] [PubMed] [Google Scholar]
- Ledent V., Vervoort M. (2001). The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11, 754–770. 10.1101/gr.177001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee S., Yang K. Y., Kim Y. M., Park S. Y., Kim S. Y., et al. (2006). Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 47, 591–600. 10.1093/pcp/pcj026 [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J. M., et al. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337–352. 10.1105/tpc.107.052142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Kui L., Zhang J., Xie Y., Wang L., Yan Y., et al. (2016). Improved hybrid de novo genome assembly of domesticated apple (Malus x domestica). Gigascience 5, 35. 10.1186/s13742-016-0139-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 4, D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M. E., Murre C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429–440. 10.1128/MCB.20.2.429-440.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D., Li Y., Bai Y., Li M., Cheng L. (2016). Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 103, 71–83. 10.1016/j.plaphy.2016.02.006 [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. (1989). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783. 10.1016/0092-8674(89)90682-X [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D. C. (2007). Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 134, 2959–2968. 10.1242/dev.006296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rodríguez P., Riaño-Pachón D. M., Corrêa L. G., Rensing S. A., Kersten B., Mueller-Roeber B. (2010). PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38, D822–D827. 10.1093/nar/gkp805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. 10.1093/molbev/msp288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu D., Yan F., Meng R., Jiang X. B., Yang H. J., Gao Z. Y., et al. (2016). Identification of MicroRNAs and their targets associated with fruit-bagging and subsequent sunlight re-exposure in the “granny smith” apple exocarp using high-throughput sequencing. Front. Plant Sci. 7:27 10.3389/fpls.2016.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman S., Mahmood T. (2015). Functional role of DREB and ERF transcription factors: regulating stress-responsive network in plants. Acta Physiol. Plant. 37:178 10.1007/s11738-015-1929-1 [DOI] [Google Scholar]
- Roig-Villanova I., Bou-Torrent J., Galstyan A., Carretero-Paulet L., Portoles S., Rodriguez-Concepcion M., et al. (2007). Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 26, 4756–4767. 10.1038/sj.emboj.7601890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E., Ledent V., Richards G., Thomas-Chollier M., Kerner P., Coornaert D., et al. (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7:33. 10.1186/1471-2148-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X. M., Huang Z. N., Duan W. K., Ren J., Liu T. K., Li Y., et al. (2014). Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genomics 289, 77–91. 10.1007/s00438-013-0791-3 [DOI] [PubMed] [Google Scholar]
- Sun H., Fan H. J., Ling H. Q. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16:9. 10.1186/s12864-014-1209-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. 10.1105/tpc.013839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42, 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Wang Q. Y., Guang Y. C., Wu Y. R., Chen H. L., Chen F., Chu C. C. (2008). Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 67, 589–602. 10.1007/s11103-008-9340-6 [DOI] [PubMed] [Google Scholar]
- Wang X. L., Zhong Y., Cheng Z. M., Xiong J. S. (2015). Divergence of the bZIP gene family in strawberry, peach, and apple suggests multiple modes of gene evolution after duplication. Int. J. Genomics 2015:536943. 10.1155/2015/536943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. B., Li S., Zhang R. F., Zhao J., Chen Y. C., Zhao Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897. 10.1111/j.1365-3040.2012.02523.x [DOI] [PubMed] [Google Scholar]
- Xu W., Zhang N., Jiao Y., Li R., Xiao D., Wang Z. (2014). The grapevine basic helix-loop-helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis. Mol. Biol. Rep. 41, 5329–5342. 10.1007/s11033-014-3404-2 [DOI] [PubMed] [Google Scholar]
- Xu X., Paik L., Zhu L., Huq E. (2015). Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20, 641–650. 10.1016/j.tplants.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Yang W., Liu X. D., Chi X. J., Wu C. A., Li Y. Z., Song L. L., et al. (2011). Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 233, 219–229. 10.1007/s00425-010-1279-6 [DOI] [PubMed] [Google Scholar]
- Zhang X., Luo H., Xu Z., Zhu Y., Ji A., Song J., et al. (2015). Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci. Rep. 5:11244. 10.1038/srep11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. 10.1186/1471-2148-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Ren Y. R., Wang Q. J., Yao Y. X., You C. X., Hao Y. J. (2016). Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol. J. 14, 1633–1645. 10.1111/pbi.12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Liang D., Wang P., Liu J., Ma F. (2012). Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol. Genet. Genomics 287, 423–436. 10.1007/s00438-012-0687-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotations of 188 MdbHLH proteins performed by Blast2GO. (A) Length distribution of sequences. (B–D) Annotations for CC, MF, and BP components, respectively.
Phylogenetic analysis (circle tree) and subgroup classifications (21 subgroups) of MdbHLH proteins.
Analysis of gene structure and conserved motifs for MdbHLH proteins. Phylogenetic tree (A), organization of gene structure (B), and distribution of conserved motifs (C) for 188 MdbHLH proteins.
Motif logos for 12 conserved motifs. The motif logos were predicted by local MEME software. Red stars indicate conserved HER motif (His 5- Glu 9-Arg 13).
Cluster analysis of gene expression data for MdbHLH orthologs (fold change >2) in Arabidopsis under drought or salt treatment.
Expression analysis of selected MdbHLH genes. Analysis (2−ΔCT method) of 30 MdbHLHs expressed in response to induced drought (PEG6000) (A) or salt stress (NaCl) (B).
Correlation analysis between the expression patterns of selected MdbHLH genes and MdbHLH orthologs in Arabidopsis, under drought (A) or salt (B) conditions. The value of spearman correlation coefficent in the heatmap was multiplied by one hundred. Positive or negative correlation relationships between the gene expression patterns are indicated with positive or negative values. Color depth (blue or red) of the grids corresponds to the value.
qRT-PCR primers used in this study.
The expression level and standard deviation data for selected MdbHLH family genes.
Chromosomal location and functional annotation of 188 apple bHLH genes.
GO annotation for MdbHLH proteins.
Expression level for MdbHLH orthologs in Arabidopsis under different stress conditions.
Summary information for 188 MdbHLH proteins in STRING database. The predicted important bHLH TFs involved in regulating plant growth and stress responses were marked in red and listed in this table (sheet3).
Name list of the selected 30 MdbHLH genes for subsequent expression analysis.
Correlation analysis between the expression patterns of selected MdbHLH genes and MdbHLH orthologs in Arabidopsis. Genes with significant correlation are marked with asterisks (* for p < 0.05; ** for p < 0.01).
Summary information for proteins that may interact with MdbHLH24, MdbHLH74, or MdbHLH4.