Abstract
Gardenia jasminoides is used in traditional Chinese medicine and has drawn attention as a rich source of crocin, a compound with reported activity against various cancers, depression and cardiovascular disease. However, genetic information on the crocin biosynthetic pathway of G. jasminoides is scarce. In this study, we performed a transcriptome analysis of the leaves, green fruits, and red fruits of G. jasminoides to identify and predict the genes that encode key enzymes responsible for crocin production, compared with Crocus sativus. Twenty-seven putative pathway genes were specifically expressed in the fruits, consistent with the distribution of crocin in G. jasminoides. Twenty-four of these genes were reported for the first time, and a novel CCD4a gene was predicted that encodes carotenoid cleavage dioxygenase leading to crocin synthesis, in contrast to CCD2 of C. sativus. In addition, 6 other candidate genes (ALDH12, ALDH14, UGT94U1, UGT86D1, UGT71H4, and UGT85K18) were predicted to be involved in crocin biosynthesis following phylogenetic analysis and different gene expression profiles. Identifying the genes that encode key enzymes should help elucidate the crocin biosynthesis pathway.
Keywords: transcriptome analysis, crocin biosynthesis, Gardenia jasminoides, CCD4a, fruit-specific expression
Introduction
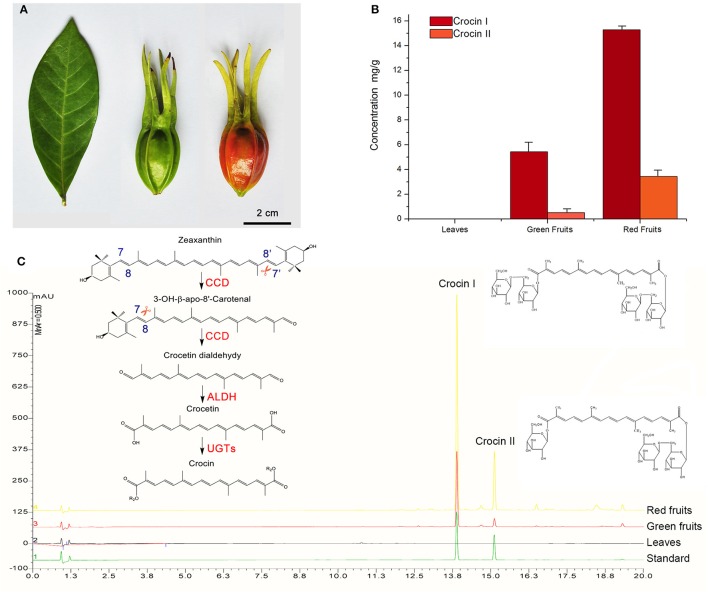
Crocin has received considerable attention in Asia and Europe as a therapeutic agent for the treatment of various diseases. Studies have demonstrated that the growth of cancer cells, such as breast, colorectal, and prostate cancer cells, are altered by crocin via the inhibition of replication (Bhandari, 2015). Crocin has also been shown to significantly improve memory, to have an antidepressant effect in mice (Wang et al., 2010; Hosseinzadeh et al., 2012), and to elicit protective effects against cardiovascular disease (Razavi et al., 2013). Crocin has been found in the stigmas of Crocus sativus L. and the fruit of Gardenia jasminoides (Sheu and Hsin, 1998; Frusciante et al., 2014). Because of the pharmacological benefits and complex harvesting process of C. sativus, the retail price of the red stigmas of C. sativus, which is called “red gold,” ranges between 2,000 and 7,000 £/kg (Frusciante et al., 2014). However, the fruit of G. jasminoides used in Chinese medicine have a low price because of their abundance and ease of cultivation (Figure 1A).
Figure 1.
Quantitative analysis of crocin in various G. jasmonoides tissues. (A) The organs (leaves, green fruits and red fruits) used in this study. (B) Corresponding histograms indicate the difference in concentration of crocin among the different organs. (C) UPLC chromatograms of the crocin content in the different organs and the proposed crocin biosynthesis pathway. Each sample was analyzed with three replicates. Photodiode array detection at a wavelength of 440 nm was used.
Recently, determining the pathway of crocin biosynthesis in plants has received increasing attention because of its high medicinal and commercial value (Figure 1C). The crocin biosynthesis pathway in C. sativus includes the upstream methylerythritol phosphate (MEP) pathway from pyruvate/glyceraldehyde 3-phosphate to geranylgeranyl pyrophosphate (GGPP), the midstream carotenoid pathway from GGPP to zeaxanthin, and the downstream crocin pathway from zeaxanthin to crocin. In the crocin pathway, crocin is generated from the oxidative cleavage of zeaxanthin at the 7,8/7′,8′ positions by carotenoid cleavage dioxygenase (CCD). The cleavage product crocetin dialdehyde is further dehydrogenated and glycosylated to crocin by an aldehyde dehydrogenase (ALDH) and glucosyltransferases (UGTs) (Frusciante et al., 2014). Although CCD, ALDH, and UGT enzymes that may be responsible for crocin biosynthesis are known, the exact genes in C. sativus that encode these enzymes have seldom been subjected to functional analyses. The genes of the MEP and carotenoid pathways have been identified in many plants Nisar et al., 2015; Xu et al., 2015; Xu H. et al., 2016; Xu Z.-C. et al., 2016; however, few of these genes have been functionally analyzed in C. sativus (Ahrazem et al., 2009), and most researchers have explored the functions of the CCD, ALDH and UGT enzymes, such as CsCCD1a, CsCCD1b, CsCCD2, CaCCD2, CsCCD4a, CsCCD4b, CsCCD4c, CsUGT2, UGT707B1, and CsGT45 (Moraga et al., 2004, 2009; Rubio et al., 2008; Nagatoshi et al., 2012; Trapero et al., 2012; Frusciante et al., 2014; Ahrazem et al., 2016). However, only 3 of these enzymes are associated with crocin biosynthesis. One carotenoid cleavage dioxygenase (CsCCD2) of C. sativus was identified through 454 sequencing of transcriptomes, and the expression of the CsCCD2 gene was consistent with the accumulation of crocin (Frusciante et al., 2014). After performing a functional characterization, CsCCD2 was found to catalyze the conversion of zeaxanthin into crocetin dialdehyde via cleavage at the 7,8 and 7′,8′ positions. In Crocus ancyrensis, the expression of CaCCD2 was also correlated with the accumulation of crocin, and CaCCD2 was found to catalyze crocetin formation in bacteria and rice (Ahrazem et al., 2016). Moreover, UGTCs2 was identified in C. sativus, and an in vitro experiment showed that crocetin can be glycosylated by UGTCs2 (Moraga et al., 2004).
In Gardenia jasminoides, two UGT enzymes (UGT75L6 and UGT94E5) were shown to glycosylate crocetin in the last step of the crocin pathway (Nagatoshi et al., 2012). Previous studies revealed that carotenoid accumulation in a specific organ is correlated with the up-regulation of genes that encode key enzymes (Botella-Pavía and Rodríguez-Concepción, 2006). The expression levels of CsCCD2 and CaCCD2 were both correlated with the accumulation of crocin. However, the study by Nagatoshi et al. indicated that the expression of UGT75L6 and UGT94E5 did not differ among various organs, and the authors suggested that these two UGTs were not rate-limiting enzymes for crocin biosynthesis. Considering the correlation between secondary metabolites levels and expression of the putative pathway genes (Ma et al., 2015; Xu et al., 2015; Xu Z. et al., 2016), we speculated that other UGTs may be responsible for crocin biosynthesis in G. jasminoides.
In this study, we propose that G. jasminoides has the potential for use as an alternative plant for research into the crocin biosynthesis pathway. G. jasminoides has been cultivated for at least a thousand years in China for use as an ornamental plant and for traditional medicine. In the mid-eighteenth century, G. jasminoides was introduced to English gardens, and it is now distributed widely around the world (Liu and Yin, 2015). Moreover, studies on its chemicals and pharmacological effects have provided a foundation for further analyses of the crocin biosynthesis pathway (Sheng et al., 2006; Chen et al., 2010). Most importantly, compared with C. sativus, which specifically accumulates crocin in the stigma, obtaining the fruit material of G. jasminoides for research on crocin biosynthesis is much easier. G. jasminoides reportedly accumulates crocin specifically in its fruit, and the content increases during fruit development. The contents of crocin I and crocin II in the pulps are up to 28 mg g−1 (He et al., 2006). Here, a comparative transcriptome analysis was performed for the leaves, green fruit and red fruit of G. jasminoides and candidate crocin biosynthetic genes were screened based on the expression of different genes. Furthermore, quantitative real-time PCR (qRT-PCR) analysis, phylogenetic analysis and quantitative analysis of crocin supported the role of the candidate genes in crocin biosynthesis.
Materials and methods
Plant material and RNA extraction
In this study, G. jasminoides Ellis, which was cultivated at the medicinal garden at the Chongqing Institute of Medicinal Plant Cultivation (China), was used. The leaves, green fruits, and red fruits were collected from three trees, respectively; and immediately frozen in liquid nitrogen for RNA isolation. Total RNA was isolated from each sample using an RNAprep Pure Plant Kit (Tiangen Biotech., Beijing, China) according to the manufacturer's instructions. The quality and quantity of the RNA were analyzed by electrophoresis and a NanoDrop 2000C spectrophotometer. The RNA Integrity Number (RIN) was determined using the BioAnalyzer 2100 (Agilent Technologies) to ensure that the RIN values were >6.5.
RNA library construction and sequencing
The cDNA libraries were created according to the manufacturer's protocol (NEBNext® Ultra™ RNA Library Prep Kit for Illumina®). First strand cDNA was synthesized using ProtoScript II Reverse Transcriptase. After the synthesis of the second-strand cDNA using the Second Strand Synthesis Enzyme Mix, the end repairing, 5′ phosphorylation and dA-tailing were performed in one reaction. Approximately 250 bp cDNA fragments were selected using AxyPrep Mag PCR Clean-up (Axygen) for the library construction. The libraries with different indexes were paired-end sequenced using the Illumina HiSeq 2000 instrument.
Transcriptome data processing and assembly
The raw reads with adaptor and poly-N sequences were removed, and low quality reads (Q-value < 10 in more than 50% of the bases) were also removed. Clean data with high sequence quality were used in all of the downstream analyses. Transcriptome assembly was conducted using Trinity software with the minimum K-mer value set to 1. After removing the redundancy with cd-hit-est software (Li and Godzik, 2006), unigenes were obtained. For the unigene annotation, the unigenes were searched against the NCBI protein sequence database using BLASTp and a cutoff E-value of 10−5. The unigenes were also annotated with the following databases: Pfam (Protein family), KO (KEGG Ortholog database), and GO (Gene Ontology). Benchmarking Universal Single-Copy Orthologs (BUSCO) software was used to assess transcriptome assembly and annotation completeness.
GO and KEGG pathway enrichment analysis
The Gene Ontology annotations were assigned based on their similarity to the A. thaliana proteomic sequences (TAIR10). The transcripts were classified into 45 GO categories under the major categories of Cellular Component, Molecular Function and Biological Process. The transcriptome sequences were BLAST against KEGG GENES databases, and the KEGG classification was analyzed according to the KO number in the annotation results.
Differential expression analysis
The gene expression levels were estimated using RSEM software. The DEGs (differentially expressed genes) analysis among the different organs was performed using DESeq Bioconductor package (1.14.0). The genes with absolute values of log2 fold changes >= 1 (FDR value < 0.001) were identified as DEGs.
qRT-PCR analysis
Total RNA was extracted from the red fruits, green fruits and leaves using an RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions. After analyzing the quality and integrity of the RNA, first-strand cDNA was synthesized using a PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Beijing, China). The specific primers (130–180 bp) were designed using Primer Premier 6.0 (Supplemental Table 1). The qRT-PCR experiment was performed on an ABI 7500 instrument and the reagents and methods were identical to that of our previous study (Luo et al., 2014). Actin and GAPDH were used as internal controls. qRT-PCR analysis was performed with three biological and technical replicates. To detect expression differences of the candidate CCD, ALDH, and UGT genes among various organs, one-way ANOVA test was performed by IBM SPSS 20 software. P < 0.01 was considered highly significant.
Phylogenetic and conserved motif analyses
The CCD, ALDH, and UGT protein sequences of G. jasminoides and other plants were aligned by MEGA 5.0 software. A neighbor-joining (NJ) tree was constructed using a Poisson model with 1,000 bootstrap replicates. The protein sequences included CCD1, CCD4a, NCED1, NCED2, ALDH12, ALDH14, UGT60, UGT67, UGT86, and UGT89 from this study; AtCCD1, AtCCD4, AtNCED3, AtNCED5, AtNCED6, AtCCD7, AtCCD8, AtALDH2C4, AtALDH2B7 UGT73C6, and UGT73B2 from A. thaliana (Skibbe et al., 2002; Jones et al., 2003; Nair et al., 2004; Auldridge et al., 2006; Kim et al., 2006; Alder et al., 2012; Frey et al., 2012; Gonzalez-Jorge et al., 2013); CsCCD2, CsGT45, and UGTCs2 from C. sativus (Moraga et al., 2004; Frusciante et al., 2014); MtCCD1 and UGT73K1 from Medicago truncatula (Achnine et al., 2005; Floss et al., 2008; Moraga et al., 2009); CmCCD1 from Cucumis melo (Ibdah et al., 2006); CmCCD4a from C. morifolium (Ohmiya et al., 2006); CitCCD4 from C. unshiu (Ma et al., 2013; Rodrigo et al., 2013); CaALDH1 from C. annuum (Kim and Hwang, 2015); AaALDH1 from A. annua (Teoh et al., 2009); BoBADH from B. orellana (Bouvier et al., 2003); REF1 from B. napus (Mittasch et al., 2013); Zmrf2 (ALDH2B2) from Z. mays (Cui et al., 1996); BALDH from A. majus (Long et al., 2009); UGT75L6 and UGT94E5 from G. jasminoides (Nagatoshi et al., 2012); VLOGT1, VLOGT2, and VLOGT3 from Vitis labrusca (Hall et al., 2011); Gt5GT7 from Gentiana triflora (Nakatsuka et al., 2008); UGT1, UGTPg29, and UGTPg45 from P. ginseng (Yan et al., 2014; Wang et al., 2015); CrUGT8 from Catharanthus roseus (Asada et al., 2013); AdGT4 from Actinidia deliciosa (Yauk et al., 2014); GAME2 from Solanum lycopersicum (Itkin et al., 2013); ZOG1 from Phaseolus lunatus (Martin et al., 1999b); and ZOX1 from Phaseolus vulgaris (Martin et al., 1999a). Conserved motifs in CCDs, ALDHs and UGTs were detected using motif based sequence analysis tool MEME (Suite version 4.11.2). The Mw and pI of the proteins were predicted using online bioinformatics tool ExPASy proteomics (http://web.expasy.org/protparam/).
Crocin identification by UPLC
Crocin I and crocin II were purchased from the National Institutes for Food and Drug Control (China). Acetonitrile and methanol were of chromatographic purity, and all of the other solvents were analytically pure. The red fruits, green fruits and leaves with three repetitions were freeze dried and then ground to a powder for each sample. The powder of each sample (0.25 g) was accurately weighed and extracted with 50% methanol to a final volume of 25 mL. After treatment with an ultrasonic instrument for 45 min, 50% methanol was added to complement the weight loss and the sample was then filtered. The filtrate was further passed through a 0.22 μm membrane filter to yield the test sample solution. Moreover, the mixed standard solution that included 0.035 mg/ml crocin I and 0.015 mg/ml crocin II was prepared by dissolving the sample in methanol. The chromatographic analysis was performed on a DIONEX Ultimate 3,000 UPLC system with a DAD detector (Thermo Fisher Scientific, USA). The UPLC separation was performed on Waters BEH-C18 column (100 × 2.1 mm, 1.7 μm), operated at 35°C. The mobile phase was acetonitrile (A) and H2O (B) with the flow rate of 0.3 mL/min. The injection volume was 2 μL and the mobile phase gradient was as follows: 0–6 min, 5–12% A; 6–25 min, 12–48% A. The detection wavelength of crocins was 440 nm.
Results
Sequencing, de novo assembly and functional annotation
In this study, 45,224,600, 46,735,458, and 60,458,180 raw Illumina cDNA sequencing reads were obtained from RNA extracted from leaves, green fruits and red fruits, respectively, of G. jasmonoides (Figure 1A). After data filtering, a total of 147,167,330 clean reads of high quality were generated for further analysis (Table 1). The summary of the RNA-Seq data for each sample is illustrated in Supplemental Table 2. The clean reads were assembled into 156,658 contigs using Trinity software. After removing the redundant contigs, 141,665 unigenes were identified, and 58,702 coding sequences (CDSs) longer than 300 bp were detected using a BLAST search against protein databases (Table 1). The size distribution of the contigs, unigenes and CDS is presented in Supplemental Figure 1. In addition, we applied the BUSCO software to assess the assembly and annotation completeness. As 1,440 single copy orthologs for plants were used, our assembly is 84.7% complete, while 6.7% of contigs are fragmented and 8.6% are missing.
Table 1.
Summary of the illumina paired-end sequencing and assembly for Gardenia jasminoides.
| Item | Number | Length (bp) |
|---|---|---|
| Total clean reads | 147,167,330 | 14,370,474,299 |
| Contigs | 156,658 | 141,953,000 |
| Average length of contigs | 906 | |
| Contig size N50 | 1,632 | |
| Unigenes | 141,665 | 122,469,676 |
| Average length of unigenes | 864 | |
| Unigene size N50 | 1,574 | |
| CDS | 58,702 | 51,532,023 |
| Average length of CDS | 878 |
The protein sequences were aligned to the Uniprot, Pfam, KEGG, and GO databases for annotation. A total of 52,816 CDSs (90%) were annotated, including 37,961 (64.7%) in Uniprot, 41,907 (71.4%) in Pfam, 11,375 (19.4%) in KEGG and 45,715 (77.9%) in the Gene Ontology (GO) database (Supplemental Table 3). All of the unigenes were functionally categorized by a GO analysis based on their sequence similarity with Arabidopsis thaliana (Supplemental Figure 2A). A total of 52,296 unigenes were characterized into the following three categories: “Molecular Function” (47,272), “Biological Process” (48,695), and “Cellular Component” (49,743). In addition, a KEGG enrichment analysis was performed to detect the genes in the active compound metabolic pathway (Supplemental Figure 2B). A total of 24,991 unigenes matched to the KEGG database and were assigned to 5 major pathways.
Genes encoding key enzymes in the MEP, carotenoid and crocin biosynthetic pathways
We analyzed the differentially expressed genes (DEGs) among the leaves, green fruits and red fruits. Compared with the leaves, the expression levels of 1,958 and 995 genes were up- and down-regulated in the green fruits, respectively, whereas the expression levels of 3,761 and 967 genes were up- and down-regulated in the red fruits, respectively. In addition, 282 and 421 genes were up- and down-regulated in the red fruits relative to green fruits, respectively (Supplemental Figure 3). All of the DEGs are summarized in Supplemental Table 4.
Twenty-seven genes involved in crocin biosynthesis, including the upstream MEP pathway, the midstream carotenoid pathway, and the downstream crocin pathway, were also identified and selected. Among them, we detected 17 unigenes encoding 9 enzymes in the pathways involved in the synthesis of geranylgeranyl diphosphate (GGPP), a late carotenoid precursor. As shown in Supplemental Figure 4, some enzymes were represented by more than one unigene. Four unigenes were predicted to encode 1-deoxy-D-xylulose-5-phosphate synthase (DXS), two unigenes were predicted to encode isopentenyl-diphosphate delta-isomerase (IDI), and five unigenes were predicted to encode geranylgeranyl diphosphate synthase (GGPPS). Based on the gene expression profiles, five genes (DXS1, DXR, IDI1, GGPPS2, and GGPPS4) were more highly expressed in the red fruit than the green fruit and leaves (Supplemental Figure 4). The expression of these genes is consistent with the distribution of crocin; therefore, these five genes may be responsible for crocin biosynthesis.
In the carotenoid pathway, 13 genes encoding eight enzymes were identified in our transcriptome data (Figure 2A). The expression levels of phytoene synthase 1 (PSY1), phytoene desaturase 2 (PDS2), lycopene β-cyclase 2 (LCYB2), and cytochrome P450-type monooxygenase 97A51 (CYP97A51) were all increased as the fruit ripened, and the fragments per kilobase of transcript per million mapped reads (FPKM) value of PSY1 reached 795.6 in the red fruits (Supplemental Table 5). Moreover, PDS2 presented a nearly 7-fold change in the red fruits compared with that in the leaves. However, ζ-carotene isomerase (Z-ISO), and carotenoid β-hydroxylase (CHY1) were both expressed at higher levels in the green fruits and red fruits compared with the leaves. The gene expression of ζ-carotene desaturase (ZDS) and carotenoid isomerase (CRTISO) did not change in all of the samples. Therefore, PSY1, PDS2, LCYB2, and CYP97A51 are likely to be involved in crocin biosynthesis.
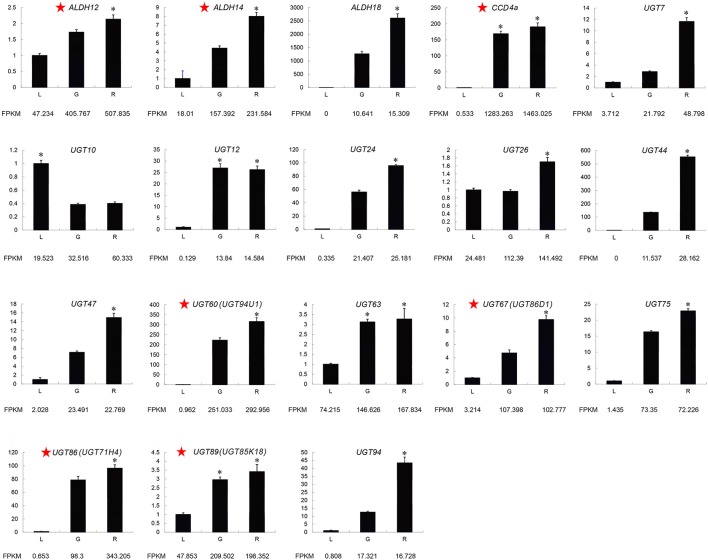
Figure 2.
Expression profile of candidate G. jasmonoides genes for crocin biosynthesis. (A) The proposed carotenoid biosynthetic pathway of G. jasmonoides. (B) The proposed crocin biosynthetic pathway in G. jasmonoides. (C) The expression of candidate CCDs, ALDHs, and UGTs from differential expression analysis.
The crocin biosynthetic pathway includes CCD, ALDH, and UGT enzymes (Figure 2). In this study, a total of 140 unigenes were identified, including 5 CCDs, 34 ALDHs and 101 UGTs (Supplemental Figure 5 and Supplemental Table 5). The plant CCD family is divided into five subfamilies: CCD1, CCD4, CCD7, CCD8, and NCED (nine-cis-epoxy-carotenoid dioxygenase; Tan et al., 2003). Five CCD members from the transcriptome data of G. jasminoides were named CCD1, CCD4a, CCD4b, NCED1, and NCED2 based on their protein sequence similarity with A. thaliana. Compared with the different expression of CCD genes among the various organs, CCD4a demonstrated significant and fruit-specific expression, and the FPKM values in the leaves, green fruit and red fruit were 0.5, 1283.3, and 1463.0, respectively, implying a possible functional link to crocin biosynthesis. However, NCED1 was only slightly expressed in the green fruit, and it was not expressed in the leaves and red fruit. NCED2 was also expressed at low levels in all of the organs, and the expression level in the green fruit was higher than that in the other two organs. In addition, the expression of CCD1 did not change among the various organs, whereas CCD4b had the highest expression in the leaves. This suggests that CCD4a is involved in crocin biosynthesis. ALDHs can catalyze the conversion of crocetin dialdehyde to crocetin (Figure 1). A transcriptome-wide analysis identified 34 ALDHs, and 3 ALDHs (ALDH12, ALDH14, and ALDH18) were highly expressed in the fruits (FPKM > 10). Similarly, the gene expression of 14 UGTs was higher in the fruits than in the leaves. Among them, 6 UGTs have significant and fruit-specific expression, and their FPKM values in the fruit reached over 100 (Supplemental Table 5). CCD4a, 3 ALDHs, and 14 UGTs were selected for further analysis. The summary of the 27 selected genes is illustrated in Supplemental Table 6.
Expression and phylogenetic analysis candidate CCD, ALDH, and UGT genes
The expression patterns of CCD4a, 3 ALDHs, and 14 UGTs were similar based on different internal control genes (actin and GAPDH), indicating the reliability of the RNA-Seq results (Figure 3 and Supplemental Figure 6). We found that ALDH12, ALDH14, ALDH18, CCD4a, UGT7, UGT24, UGT44, UGT47, UGT60 (UGT94U1), UGT67 (UGT86D1), UGT75, UGT86 (UGT71H4), UGT89 (UGT85K18), and UGT94 were more highly expressed in the red fruit than the green fruit and leaves. Moreover, the FPKM values of ALDH12, ALDH14, CCD4a, UGT60 (UGT94U1), UGT67 (UGT86D1), UGT86 (UGT71H4), and UGT89 (UGT85K18) were over 100 in the red fruit. In addition, the UGT proteins related to secondary metabolism were reported to have a PSPG (plant secondary metabolite glycosyltransferase) box. We searched the protein sequences of the 4 UGTs and found that UGT94U1, UGT86D1, UGT71H4, and UGT85K18 all had a PSPG box (Figure 4D). Therefore, we propose that CCD4a, 2 ALDH genes (ALDH12 and ALDH14) and 4 UGT genes (UGT94U1, UGT86D1, UGT71H4, and UGT85K18) may be related to crocin biosynthesis.
Figure 3.
Expression of candidate crocin biosynthesis genes related to crocin biosynthesis determined by qRT-PCR. The characters on the X-axis indicate the leaves (L), green fruits (G), and red fruits (R). The Y-axis represents the fold changes in gene expression. The actin gene was used as an internal reference. The red star next to gene names represents the most likely to be involved in crocin biosynthesis. One-way ANOVA was performed using IBM SPSS 20 software. Asterisks represents significant differences from this comparison. P < 0.01 was considered highly significant.
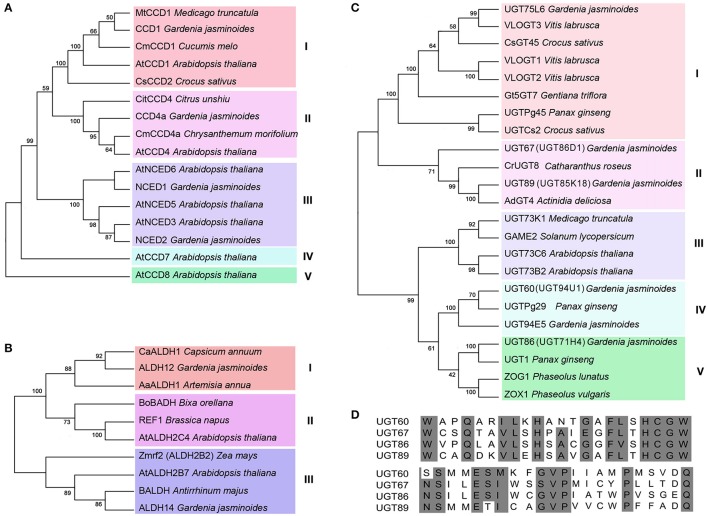
Figure 4.
Phylogenetic and sequence analysis of cadidtate crocin biosynthesis genes. The Phylogenetic trees of the CCDs (A), ALDHs (B), and UGTs (C) between G. jasminoides and other plants, and a PSPG box alignment of the candidate UGT proteins (D) are shown.
Furthermore, the NJ tree of the CCDs was divided into five groups of subfamilies: CCD1, CCD4, NCED, CCD7, and CCD8 (Figure 4A). We found that Gardenia CCD4a was distinct from the CsCCD2 of C. Sativus (Frusciante et al., 2014); however, it was a member of group II and closely related to the CitCCD4 enzyme, which can also cleave zeaxanthin at the 7,8 or 7′,8′ positions (Ma et al., 2013; Rodrigo et al., 2013). Thus, based on the analyses of the expression patterns and phylogenetic relationships, Gardenia CCD4a may be involved in the oxidative cleavage of the 7,8 and 7′,8′ double bonds of zeaxanthin to give crocetin dialdehyde in the crocin biosynthesis pathway. We performed a bioactivity test of CCD4a in E. coli. The plasmid pACCAR25ΔcrtX is capable of producing zeaxanthin. Thus, we co-transformed E. coli BL21 (DE3) with pET28a-CCD4a and pACCAR25ΔcrtX, and we named the strain 25ΔcrtX-CCD4a. After IPTG induction, decoloration was observed in strain 25ΔcrtX-CCD4a compared to the control (CK) (Supplemental Figure 7A). HPLC analysis revealed that zeaxanthin accumulation was decreased significantly in 25ΔcrtX-CCD4a (Supplemental Figure 7B). Therefore, we concluded that CCD4a might cleave zeaxanthin at the 7,8 and 7′,8′ positions, thereby leading to crocin synthesis in G. jasminoides.
The phylogenetic analysis showed that ALDH12 and ALDH14 belong to the ALDH2C and ALDH2B subfamilies, respectively (Figure 4B). ALDH12 was clustered with CaALDH1 of Capsicum annuum, AaALDH1 of Artemisia annua, BoBADH of Bixa orellana, REF1 of Brassica napus and AtALDH2C4 of A. thaliana. These proteins are all ALDH2C members. It is worth noting that AaALDH1 and BoBADH control artemisinin and bixin biosynthesis, respectively, and bixin is specifically derived from the carotenoid pathway as well as the crocin. In addition, ALDH14 was grouped with Zmrf2 of Zea mays, AtALDH2B7 of A. thaliana and BALDH Antirrhinum majus, which are all ALDH2B members. Moreover, BALDH has been reported to be involved in benzoic acid biosynthesis. These results indicate that ALDH12 and ALDH14 may be involved in crocin biosynthesis in G. jasminoides fruit.
The phylogenetic tree of the UGTs was divided into two clades that contained 5 groups (Figure 4C). UGT86D1 and UGT85K18 were members of group II and grouped with AdGT4 of A. deliciosa and CrUGT8 of C. roseus. UGT94U1 was closely related to UGT94E5 of G. jasminoides and UGTPg29 of P. ginseng. Moreover, UGT71H4 was clustered with UGT1 of P. ginseng in group V. The UGT proteins that were grouped with the candidate UGTs in this study were involved in terpenoid biosynthesis. Thus, we suggest that UGT94U1, UGT86D1, UGT71H4, and UGT85K18 may be related to the biosynthesis of crocin.
The multiple sequence alignments were performed using the CCDs, ALDHs, and UGTs sequences of phylogenetic trees, respectively. The results indicated that ALDHs proteins among different species were more conserved than CCDs and UGTs (Supplemental Figure 8). The number of amino acids, Mw and pI of the proteins were also summarized in Supplemental Table 7. Moreover, diverse conserved motifs in CCDs, ALDHs, and UGTs sequences were identified (Supplemental Figure 9). Five conserved motifs were identified in 10 ALDHs, and 9 of them have all 5 motifs indicating their high conservation (Supplemental Figure 9B). The CCDs have 16 conserved motifs and each subfamily has specific motifs. In CCD4 subfamily, motifs 1–8, 10, 11, and 14 corresponded to all members (Supplemental Figure 9A). Thirteen conserved motifs were detected in the UGT proteins (Supplemental Figure 9C). The motifs distribution of UGT86D1 (UGT67) was most similar to CrUGT8, whereas UGT86D1 (UGT67) got 2 more motifs: motif 8 and 13. UGT85K18(UGT89) shared same motifs with AdGT4 except motif 3. UGT94U1(UGT60) got one more motif than UGT94E5. The motifs distribution of UGT71H4(UGT86) was identical with UGT1.
Crocin distribution in G. jasmonoides tissues
To confirm the correlation between the candidate gene expression and crocin distribution, the crocin content in the green fruits, red fruits and leaves was analyzed by ultra-performance liquid chromatography (UPLC). As presented in Figures 1B,C, the distribution of crocin I and crocin II was fruit specific. After the fruit had ripened, the content of crocin I and crocin II increased, and in the red fruits, the content is almost three and seven times higher relative to the content in the green fruit, respectively. These results suggest that crocin biosynthesis in G. jasminoides may be completely conducted in the fruits, and the identification of our 7 candidate genes was confirmed using a co-expression analysis.
Discussion
We initiated the first transcriptome sequencing of the red fruits, green fruits and leaves of G. jasminoides to select genes that encode the key enzymes in the crocin biosynthesis pathway. Twenty-seven pathway genes were expressed at greater levels in the red fruits compared with the green fruits and leaves, and the involvement of 7 of these genes in the crocin pathway was determined by the qRT-PCR and phylogenetic analyses. Moreover, the expression patterns of these 7 genes were consistent with the distribution of crocin in various organs. With the exception of PSY1 (GenBank No. AEF59491.1), LCYB2 (GenBank No. AFI09271.1), and UGT10 (GenBank No. AB555742.1), the 24 candidate genes were described for the first time in this study. Although, the transcriptome analysis of G. jasminoides flowers has been previously studied (Tsanakas et al., 2014), the objective of the previous report was the identification of TFs, which may play a role in petal senescence. In this paper, we report work relevant to the biosynthesis of crocin, a compound with important health benefits. This represents a first major step in the identification of the genes involved in crocin biosynthesis.
A novel CCD4 subfamily member that is different from CCD2 of C. sativus was most likely to be involved in crocin biosynthesis. Five CCDs, including 1 CCD1, 2 CCD4s, and 2 NCEDs, were identified in our study. CsCCD2 of C. sativus and CaCCD2 of C. ancyrensis were the enzymes responsible for the first dedicated step leading to crocin biosynthesis starting with zeaxanthin, and their expression pattern was consistent with crocin production (Frusciante et al., 2014; Ahrazem et al., 2016). CCD2 was not observed in our data. The phylogenetic analysis indicated that CsCCD2 was clustered with the CCD1 subfamily members in group I (Figure 4A). CCD1 of G. jasminoides was also located in group I, although its expression level did not change in the fruit and leaves. Thus, CCD1 may be not associated with crocin accumulation. Remarkably, CCD4a of G. jasminoides was more highly expressed in the red fruit than the green fruit, and it was scarcely expressed in the leaves. In addition, several CCD4s from other plants were also reported to be key determinants of pigmentation. For example, the petal color of C. morifolium changes from white to yellow through RNA interference (RNAi) of a CmCCD4a gene, which is specifically transcribed in flowers (Ohmiya et al., 2006). The carotenoid level of RNAi lines of potato, in which the StCCD4 gene is down-regulated, are 2- to 5-fold higher than that of the control (Campbell et al., 2010; Bruno et al., 2015). Moreover, in A. thaliana, the expression level of AtCCD4 is inversely correlated with pigmentation, and the loss of AtCCD4 function in A. thaliana results in increased β-carotene content (Gonzalez-Jorge et al., 2013). In addition, the CmCCD4a, StCCD4, and AtCCD4 enzymes all cleave their substrates at the 9,10 and 9′,10′ positions (Huang et al., 2009; Bruno et al., 2015). CCD4 appears to act as a negative regulator of carotenoid content and has cleavage position specificity. However, recent studies on a CCD4 from Citrus unshiu demonstrated a consistent expression pattern with the accumulation of β-citraurin, and CitCCD4 was found to cleave the 7,8 or 7′,8′ bond of β-cryptoxanthin and zeaxanthin and contributed to the formation of β-citraurin (Ma et al., 2013; Rodrigo et al., 2013). We also characterized the function of CCD4a in vitro and the results indicated zeaxanthin can be cleaved by CCD4a. Therefore, we conclude that CCD4a might cleave zeaxanthin at the 7,8 and 7′,8′ positions, thereby leading to crocin synthesis in G. jasminoides.
The last step of crocin biosynthesis involves UDP-glucosyltransferases (UGTs), which convert crocetin to crocin. In this study, 101 UGT genes were detected in our RNA-Seq data, and four genes (UGT94U1, UGT86D1, UGT71H4, and UGT85K18) were proposed as candidate genes in crocin biosynthesis. First, the expression of these genes increased during fruit development, and the lowest expression levels were observed in the leaves. UGT94U1 and UGT71H4 presented fruit-specific expression levels, implying their potential functions. Second, these UGTs all had a conserved motif called the PSPG box, which is involved in secondary metabolism. Finally, the phylogenetic analysis indicated that although these genes were clustered in different groups, they were all grouped with the UGTs that were functionally characterized as involved in terpenoid biosynthesis. UGT86D1 and UGT85K18 were in group II with members of AdGT4 from A. deliciosa and CrUGT8 from C. roseus (Asada et al., 2013; Yauk et al., 2014). In A. deliciosa, a range of terpenes are glycosylated by AdGT4; and in C. roseus, CrUGT8 was identified to be responsible for the formation of secologanin. In addition, UGT94U1 was closely related to UGTPg29 from Panax ginseng and UGT94E5 from G. jasminoides, which were involved in ginsenoside and crocin biosynthesis, respectively (Nagatoshi et al., 2012; Wang et al., 2015). Moreover, UGT71H4 was also clustered with a UGT from P. ginseng called UGTPg1, which was the key to the biosynthesis of compound K, a ginsenoside (Yan et al., 2014). We suggest that UGT94U1, UGT86D1, UGT71H4, and UGT85K18 may function as key enzymes in the biosynthesis of crocin. UGT75L6 and UGT94E5 from G. jasminoides were functionally analyzed in a previous study (Nagatoshi et al., 2012). Although they were not included as candidate genes in our study, our transcriptome data identified these genes. The protein sequence of UGT1 has a 92.4% similar identity to UGT75L6, and it was more highly expressed in the leaves than the fruits. In addition, UGT52, which has only one amino acid variation from UGT94E5 (99.8% identity), was highly expressed in the fruits, although the expression level in the fruits was not more than twice that of the leaves. The UGT genes that showed significant or specific expression in the fruits are most likely responsible for crocin biosynthesis. Therefore, a comparative transcriptome analysis of the organs or tissues that show significantly different content provides a more effective method of selecting target genes, especially for elucidating secondary metabolism pathways. In addition, geniposide, which is an iridoid glucoside, is also a major active compound present in G. jasminoides fruits, and UGT proteins are also key enzymes in the biosynthetic pathway of geniposide. The amount of geniposide in green fruits was comparable to that in red fruits (Gao and Zhu, 2013). However, the crocin content increased during fruit development. Following the consistency of chemical contents and biosynthetic gene expression, the candidate UGTs whose expression in red fruits was 1.5 times higher than that in green fruits and whose FPKM value >100 may be involved in crocin biosynthesis. The UGT genes whose expression did not change significantly may be involved in geniposide biosynthesis.
In conclusion, the candidate genes that may encode key enzymes of the MEP, carotenoid and crocin biosynthesis pathways were identified by de novo transcriptome analysis of the green fruits, red fruits and leaves of G. jasminoides. In addition, the comprehensive and detailed relationships between candidate gene expression and crocin distribution were established. This study provides a molecular foundation for determining the crocin pathway. However, future functional characterizations of candidate genes in vivo and in vitro should be performed.
Accession numbers
Transcriptome data from this study has been submitted to Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under the accession number PRJNA321361.
Author contributions
JS, KH, and AJ designed this research. AJ and JJ performed the experiment, data analysis and wrote this paper. ZX and YL contributed to transcriptome data analysis and discussion. JS revised this paper. WB and CH carried out the chemicals detection. KH, FR, and JL collected the materials.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by CAMS Innovation Fund for Medical Sciences (CIFMS, 2016-I2M-3-016) and the Application development project of Chongqing, China (cstc2014yykfA80013). We greatly appreciate Prof. Patrick Covello (National Research Council of Canada, Saskatoon) who improved the writing. We are grateful to Prof. Michael Court (Washington State University College of Veterinary Medicine, Pullman) and Prof. David Nelson (University of Tennessee Health Science Center, Memphis) for deriving the systematic names of candidate UGT and CYP450 sequences, respectively.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00518/full#supplementary-material
Length distribution of the contigs, unigenes and CDSs.
GO (A) and KEGG (B) classifications of the unigenes.
Number of DEGs in comparisons of different G. jasmonoides organs. Red bars represent the number of up-regulated genes in each group. Blue bars represent the number of down-regulated genes in each group. L, leaves; GF, green fruits; RF, red fruits.
Expression profile of the genes in the MEP pathway.
Expression profile of the ALDH and UGT genes in G. jasminoides.
Validation of the expression of the genes related to crocin biosynthesis using qRT-PCR. The characters on the x-axis indicate the leaves (L), green fruits (G), and red fruits (R). The y-axis represents the fold changes in gene expression. The GAPDH gene was used as an internal reference. The red star next to gene names represents the most likely genes involved in crocin biosynthesis. One-way ANOVA was performed using IBM SPSS 20 software. Asterisks represents significant differences from this comparison. P < 0.01 was considered highly significant.
Pigments produced in E. coli during functional analysis of CCD4a. Decoloration of zeaxanthin was observed in CCD4a-expressing cells (A). Results of HPLC analysis of zeaxanthin obtained from E. coli cells (B). CCD4a, the pigments extracted from pET28a-CCD4a and pACCAR25ΔcrtX-expressing cells. CK, the pigments extracted from pACCAR25ΔcrtX-expressing cells.
Comparison of the CCDs (A), ALDHs (B), and UGTs (C) proteins among different species. The 100% conservation of amino acid residues was indicated by gray background. The 75% conservation of amino acid residues was indicated by pink background. The 50% conservation of amino acid residues was indicated by blue background.
The distribution of the conserved motifs of CCDs (A), ALDHs (B), and UGTs (C).
Primers for the qRT-PCR analysis.
Summary of the RNA-Seq data for each G. jasmonoides organ.
Annotation percentage of G. jasminoides.
Overview of the differentially expressed genes of green fruits and leaves (A); red fruits and leaves (B); green fruits and red fruits (C).
FPKM values of the pathway genes.
Summary of the 27 selected genes by DEGs analysis.
The physicochemical property of CCDs, ALDHs, and UGTs proteins, which used in phylogenetic analysis.
References
- Achnine L., Huhman D. V., Farag M. A., Sumner L. W., Blount J. W., Dixon R. A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 41, 875–887. 10.1111/j.1365-313X.2005.02344.x [DOI] [PubMed] [Google Scholar]
- Ahrazem O., Rubio-Moraga A., Berman J., Capell T., Christou P., Zhu C., et al. (2016). The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 209, 650–663. 10.1111/nph.13609 [DOI] [PubMed] [Google Scholar]
- Ahrazem O., Rubio-Moraga A., López R. C., Gómez-Gómez L. (2009). The expression of a chromoplast-specific lycopene beta cyclase gene is involved in the high production of saffron's apocarotenoid precursors. J. Exp. Bot. 61, 105–119. 10.1093/jxb/erp283 [DOI] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., et al. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. 10.1126/science.1218094 [DOI] [PubMed] [Google Scholar]
- Asada K., Salim V., Masada-Atsumi S., Edmunds E., Nagatoshi M., Terasaka K., et al. (2013). A 7-deoxyloganetic acid glucosyltransferase contributes a key step in secologanin biosynthesis in Madagascar periwinkle. Plant Cell 25, 4123–4134. 10.1105/tpc.113.115154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auldridge M. E., Block A., Vogel J. T., Dabney-Smith C., Mila I., Bouzayen M., et al. (2006). Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 45, 982–993. 10.1111/j.1365-313X.2006.02666.x [DOI] [PubMed] [Google Scholar]
- Bhandari P. R. (2015). Crocus sativus L.(saffron) for cancer chemoprevention: a mini review. J. Tradit. Complement. Med. 5, 81–87. 10.1016/j.jtcme.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella-Pavía P., Rodríguez-Concepción M. (2006). Carotenoid biotechnology in plants for nutritionally improved foods. Physiol. Plant. 126, 369–381. 10.1111/j.1399-3054.2006.00632.x [DOI] [Google Scholar]
- Bouvier F., Dogbo O., Camara B. (2003). Biosynthesis of the food and cosmetic plant pigment bixin (annatto). Science 300, 2089–2091. 10.1126/science.1085162 [DOI] [PubMed] [Google Scholar]
- Bruno M., Beyer P., Al-Babili S. (2015). The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch. Biochem. Biophys. 572, 126–133. 10.1016/j.abb.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Campbell R., Ducreux L. J., Morris W. L., Morris J. A., Suttle J. C., Ramsay G., et al. (2010). The metabolic and developmental roles of carotenoid cleavage dioxygenase4 from potato. Plant Physiol. 154, 656–664. 10.1104/pp.110.158733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang H., Li Y. X., Cai L., Huang J., Zhao C., et al. (2010). Crocin and geniposide profiles and radical scavenging activity of gardenia fruits (Gardenia jasminoides Ellis) from different cultivars and at the various stages of maturation. Fitoterapia 81, 269–273. 10.1016/j.fitote.2009.09.011 [DOI] [PubMed] [Google Scholar]
- Cui X., Wise R. P., Schnable P. S. (1996). The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272, 1334. 10.1126/science.272.5266.1334 [DOI] [PubMed] [Google Scholar]
- Floss D. S., Schliemann W., Schmidt J., Strack D., Walter M. H. (2008). RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiol. 148, 1267–1282. 10.1104/pp.108.125062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A., Effroy D., Lefebvre V., Seo M., Perreau F., Berger A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70, 501–512. 10.1111/j.1365-313X.2011.04887.x [DOI] [PubMed] [Google Scholar]
- Frusciante S., Diretto G., Bruno M., Ferrante P., Pietrella M., Prado-Cabrero A., et al. (2014). Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, 12246–12251. 10.1073/pnas.1404629111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Zhu B. (2013). The accumulation of crocin and geniposide and transcripts of phytoene synthase during maturation of Gardenia jasminoides fruit. Evid. Based Complement. Alternat. Med. 2013:686351. 10.1155/2013/686351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge S., Ha S.-H., Magallanes-Lundback M., Gilliland L. U., Zhou A., Lipka A. E., et al. (2013). Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25, 4812–4826. 10.1105/tpc.113.119677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D., Kim K. H., De Luca V. (2011). Molecular cloning and biochemical characterization of three Concord grape (Vitis labrusca) flavonol 7-O-glucosyltransferases. Planta 234, 1201–1214. 10.1007/s00425-011-1474-0 [DOI] [PubMed] [Google Scholar]
- He M.-L., Cheng X.-W., Chen J.-K., Zhou T.-S. (2006). Simultaneous determination of five major biologically active ingredients in different parts of Gardenia jasminoides fruits by HPLC with diode-array detection. Chromatographia 64, 713–717. 10.1365/s10337-006-0099-0 [DOI] [Google Scholar]
- Hosseinzadeh H., Sadeghnia H. R., Ghaeni F. A., Motamedshariaty V. S., Mohajeri S. A. (2012). Effects of saffron (Crocus sativus L.) and its active constituent, crocin, on recognition and spatial memory after chronic cerebral hypoperfusion in rats. Phytother. Res. 26, 381–386. 10.1002/ptr.3566 [DOI] [PubMed] [Google Scholar]
- Huang F. C., Molnár P., Schwab W. (2009). Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 60, 3011–3022. 10.1093/jxb/erp137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibdah M., Azulay Y., Portnoy V., Wasserman B., Bar E., Meir A., et al. (2006). Functional characterization of CmCCD1, a carotenoid cleavage dioxygenase from melon. Phytochemistry 67, 1579–1589. 10.1016/j.phytochem.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Itkin M., Heinig U., Tzfadia O., Bhide A. J., Shinde B., Cardenas P. D., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179. 10.1126/science.1240230 [DOI] [PubMed] [Google Scholar]
- Jones P., Messner B., Nakajima J. I., Schäffner A. R., Saito K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278, 43910–43918. 10.1074/jbc.M303523200 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Kim B. G., Ko J. H., Lee Y., Hur H.-G., Lim Y., et al. (2006). Molecular cloning, expression, and characterization of a flavonoid glycosyltransferase from Arabidopsis thaliana. Plant Sci. 170, 897–903. 10.1016/j.plantsci.2005.12.013 [DOI] [Google Scholar]
- Kim N. H., Hwang B. K. (2015). Pepper aldehyde dehydrogenase CaALDH1 interacts with Xanthomonas effector AvrBsT and promotes effector-triggered cell death and defence responses. J. Exp. Bot. 66, 3367–3380. 10.1093/jxb/erv147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Liu J., Yin F. (2015). Gardenia jasminoides Ellis 栀子 (Zhizi, Capejasmine), in Dietary Chinese Herbs, eds Liu Y., Wang Z., Zhang J.(New York, NY: Springer; ), 379–390. [Google Scholar]
- Long M. C., Nagegowda D. A., Kaminaga Y., Ho K. K., Kish C. M., Schnepp J., et al. (2009). Involvement of snapdragon benzaldehyde dehydrogenase in benzoic acid biosynthesis. Plant J. 59, 256–265. 10.1111/j.1365-313X.2009.03864.x [DOI] [PubMed] [Google Scholar]
- Luo H., Zhu Y., Song J., Xu L., Sun C., Zhang X., et al. (2014). Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol. Plant. 152, 241–255. 10.1111/ppl.12193 [DOI] [PubMed] [Google Scholar]
- Ma D. M., Wang Z., Wang L., Alejos-Gonzales F., Sun M. A., Xie D. Y. (2015). A genome-wide scenario of terpene pathways in self-pollinated Artemisia annua. Mol. Plant 8, 1580–1598. 10.1016/j.molp.2015.07.004 [DOI] [PubMed] [Google Scholar]
- Ma G., Zhang L., Matsuta A., Matsutani K., Yamawaki K., Yahata M., et al. (2013). Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase4 in the flavedo of Citrus fruit. Plant Physiol. 163, 682–695. 10.1104/pp.113.223297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. C., Mok M. C., Mok D. W. (1999a). A gene encoding the cytokinin enzyme zeatinO-xylosyltransferase of Phaseolus vulgaris. Plant Physiol. 120, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. C., Mok M. C., Mok D. W. (1999b). Isolation of a cytokinin gene, ZOG1, encoding zeatin O-glucosyltransferase from Phaseolus lunatus. Proc. Natl. Acad. Sci. U.S.A. 96, 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittasch J., Böttcher C., Frolov A., Strack D., Milkowski C. (2013). Reprogramming the phenylpropanoid metabolism in seeds of oilseed rape by suppressing the orthologs of reduced epidermal fluorescence1. Plant Physiol. 161, 1656–1669. 10.1104/pp.113.215491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga Á., Mozos A., Ahrazem O., Gómez-Gómez L. (2009). Cloning and characterization of a glucosyltransferase from Crocus sativus stigmas involved in flavonoid glucosylation. BMC Plant Biol. 9:109. 10.1186/1471-2229-9-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga A. R., Nohales P. F., Pérez J. A., Gómez-Gómez L. (2004). Glucosylation of the saffron apocarotenoid crocetin by a glucosyltransferase isolated from Crocus sativus stigmas. Planta 219, 955–966. 10.1007/s00425-004-1299-1 [DOI] [PubMed] [Google Scholar]
- Nagatoshi M., Terasaka K., Owaki M., Sota M., Inukai T., Nagatsu A., et al. (2012). UGT75L6 and UGT94E5 mediate sequential glucosylation of crocetin to crocin in Gardenia jasminoides. FEBS Lett. 586, 1055–1061. 10.1016/j.febslet.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Nair R. B., Bastress K. L., Ruegger M. O., Denault J. W., Chapple C. (2004). The Arabidopsis thaliana reduced epidermal fluorescence1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16, 544–554. 10.1105/tpc.017509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka T., Sato K., Takahashi H., Yamamura S., Nishihara M. (2008). Cloning and characterization of the UDP-glucose: anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. J. Exp. Bot. 59, 1241–1252. 10.1093/jxb/ern031 [DOI] [PubMed] [Google Scholar]
- Nisar N., Li L., Lu S., Khin N. C., Pogson B. J. (2015). Carotenoid metabolism in plants. Mol. Plant 8, 68–82. 10.1016/j.molp.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Ohmiya A., Kishimoto S., Aida R., Yoshioka S., Sumitomo K. (2006). Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 142, 1193–1201. 10.1104/pp.106.087130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y., et al. (2014). Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 24, 770–773. 10.1038/cr.2014.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi B. M., Hosseinzadeh H., Movassaghi A. R., Imenshahidi M., Abnous K. (2013). Protective effect of crocin on diazinon induced cardiotoxicity in rats in subchronic exposure. Chem. Biol. Interact. 203, 547–555. 10.1016/j.cbi.2013.03.010 [DOI] [PubMed] [Google Scholar]
- Rodrigo M. J., Alquézar B., Alós E., Medina V., Carmona L., Bruno M., et al. (2013). A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 64, 4461–4478. 10.1093/jxb/ert260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A., Rambla J. L., Santaella M., Gómez M. D., Orzaez D., Granell A., et al. (2008). Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in β-ionone release. J. Biol. Chem. 283, 24816–24825. 10.1074/jbc.M804000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L., Qian Z., Zheng S., Xi L. (2006). Mechanism of hypolipidemic effect of crocin in rats: crocin inhibits pancreatic lipase. Eur. J. Pharmacol. 543, 116–122. 10.1016/j.ejphar.2006.05.038 [DOI] [PubMed] [Google Scholar]
- Sheu S. J., Hsin W. C. (1998). HPLC separation of the major constituents of Gardeniae fructus. J. High Resolut. Chromatogr. 21, 523–526. [DOI] [Google Scholar]
- Skibbe D. S., Liu F., Wen T. J., Yandeau M. D., Cui X., Cao J., et al. (2002). Characterization of the aldehyde dehydrogenase gene families of Zea mays and Arabidopsis. Plant Mol. Biol. 48, 751–764. 10.1023/A:1014870429630 [DOI] [PubMed] [Google Scholar]
- Tan B. C., Joseph L. M., Deng W. T., Liu L., Li Q. B., Cline K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35, 44–56. 10.1046/j.1365-313X.2003.01786.x [DOI] [PubMed] [Google Scholar]
- Teoh K. H., Polichuk D. R., Reed D. W., Covello P. S. (2009). Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany 87, 635–642. 10.1139/B09-032 [DOI] [Google Scholar]
- Trapero A., Ahrazem O., Rubio-Moraga A., Jimeno M. L., Gómez M. D., Gómez-Gómez L. (2012). Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in Crocus sativus. Plant Physiol. 159, 1335–1354. 10.1104/pp.112.198069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanakas G. F., Manioudaki M. E., Economou A. S., Kalaitzis P. (2014). De novo transcriptome analysis of petal senescence in Gardenia jasminoides Ellis. BMC genomics 15:554. 10.1186/1471-2164-15-554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Wei Y., Fan Y., Liu Q., Wei W., Yang C., et al. (2015). Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 29, 97–105. 10.1016/j.ymben.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Wang Y., Han T., Zhu Y., Zheng C.-J., Ming Q.-L., Rahman K., et al. (2010). Antidepressant properties of bioactive fractions from the extract of Crocus sativus L. J. Nat. Med. 64, 24–30. 10.1007/s11418-009-0360-6 [DOI] [PubMed] [Google Scholar]
- Xu H., Song J., Luo H., Zhang Y., Li Q., Zhu Y., et al. (2016). Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. plant 9, 949–952. 10.1016/j.molp.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.-C., Ji A.-J., Zhang X., Song J.-Y., Chen S.-L. (2016). Biosynthesis and regulation of active compounds in medicinal model plant Salvia miltiorrhiza. Chin. Herb. Med. 8, 3–11. 10.1016/S1674-6384(16)60002-3 [DOI] [Google Scholar]
- Xu Z., Luo H., Ji A., Zhang X., Song J., Chen S. (2016). Global identification of the full-length transcripts and alternative splicing related to phenolic acid biosynthetic genes in Salvia miltiorrhiza. Front. Plant Sci. 7:100. 10.3389/fpls.2016.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Peters R. J., Weirather J., Luo H., Liao B., Zhang X., et al. (2015). Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 82, 951–961. 10.1111/tpj.12865 [DOI] [PubMed] [Google Scholar]
- Yauk Y. K., Ged C., Wang M. Y., Matich A. J., Tessarotto L., Cooney J. M., et al. (2014). Manipulation of flavour and aroma compound sequestration and release using a glycosyltransferase with specificity for terpene alcohols. Plant J. 80, 317–330. 10.1111/tpj.12634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Length distribution of the contigs, unigenes and CDSs.
GO (A) and KEGG (B) classifications of the unigenes.
Number of DEGs in comparisons of different G. jasmonoides organs. Red bars represent the number of up-regulated genes in each group. Blue bars represent the number of down-regulated genes in each group. L, leaves; GF, green fruits; RF, red fruits.
Expression profile of the genes in the MEP pathway.
Expression profile of the ALDH and UGT genes in G. jasminoides.
Validation of the expression of the genes related to crocin biosynthesis using qRT-PCR. The characters on the x-axis indicate the leaves (L), green fruits (G), and red fruits (R). The y-axis represents the fold changes in gene expression. The GAPDH gene was used as an internal reference. The red star next to gene names represents the most likely genes involved in crocin biosynthesis. One-way ANOVA was performed using IBM SPSS 20 software. Asterisks represents significant differences from this comparison. P < 0.01 was considered highly significant.
Pigments produced in E. coli during functional analysis of CCD4a. Decoloration of zeaxanthin was observed in CCD4a-expressing cells (A). Results of HPLC analysis of zeaxanthin obtained from E. coli cells (B). CCD4a, the pigments extracted from pET28a-CCD4a and pACCAR25ΔcrtX-expressing cells. CK, the pigments extracted from pACCAR25ΔcrtX-expressing cells.
Comparison of the CCDs (A), ALDHs (B), and UGTs (C) proteins among different species. The 100% conservation of amino acid residues was indicated by gray background. The 75% conservation of amino acid residues was indicated by pink background. The 50% conservation of amino acid residues was indicated by blue background.
The distribution of the conserved motifs of CCDs (A), ALDHs (B), and UGTs (C).
Primers for the qRT-PCR analysis.
Summary of the RNA-Seq data for each G. jasmonoides organ.
Annotation percentage of G. jasminoides.
Overview of the differentially expressed genes of green fruits and leaves (A); red fruits and leaves (B); green fruits and red fruits (C).
FPKM values of the pathway genes.
Summary of the 27 selected genes by DEGs analysis.
The physicochemical property of CCDs, ALDHs, and UGTs proteins, which used in phylogenetic analysis.