Abstract
Small RNAs are a class of short non-coding endogenous RNAs that play essential roles in many biological processes. Recent studies have reported that microRNAs (miRNAs) are also involved in ethylene signaling in plants. LeERF1 is one of the ethylene response factors (ERFs) in tomato that locates in the downstream of ethylene signal transduction pathway. To elucidate the intricate regulatory roles of small RNAs in ethylene signaling pathway in tomato, the deep sequencing and bioinformatics methods were combined to decipher the small RNAs landscape in wild and sense-/antisense-LeERF1 transgenic tomato fruits. Except for the known miRNAs, 36 putative novel miRNAs, 6 trans-acting short interfering RNAs (ta-siRNAs), and 958 natural antisense small interfering RNAs (nat-siRNAs) were also found in our results, which enriched the tomato small RNAs repository. Among these small RNAs, 9 miRNAs, and 12 nat-siRNAs were differentially expressed between the wild and transgenic tomato fruits significantly. A large amount of target genes of the small RNAs were identified and some of them were involved in ethylene pathway, including AP2 TFs, auxin response factors, F-box proteins, ERF TFs, APETALA2-like protein, and MADS-box TFs. Degradome sequencing further confirmed the targets of miRNAs and six novel targets were also discovered. Furthermore, a regulatory model which reveals the regulation relationships between the small RNAs and their targets involved in ethylene signaling was set up. This work provides basic information for further investigation of the function of small RNAs in ethylene pathway and fruit ripening.
Keywords: ethylene, microRNAs, target, high-throughput sequencing, regulatory network
Introduction
Small RNAs are a class of non-coding endogenous RNAs ranged from 20 to 24 nucleotides (nt) that play essential roles in plant growth and development, signal transduction, response to biotic and abiotic stresses and other biological processes (Rhoades et al., 2002; Jones-Rhoades et al., 2006; Tomato Genome Consortium, 2012). MicroRNAs (miRNAs) and small-interfering RNAs (siRNAs) are two mainly classes of small RNAs divided on the difference of their precursor structures and biosynthetic pathways (Carthew and Sontheimer, 2009). Mature miRNAs are evolved from miRNA genes with the action of Dicer-like 1 (DCL1), Hua Enhancer 1 (HEN1), and HASTY proteins (Jones-Rhoades et al., 2006; Xie et al., 2015). SiRNAs are derived from long double-stranded RNAs (dsRNAs) and could be classed to heterochromatic siRNAs (hc-siRNAs), trans-acting short interfering RNAs (ta-siRNAs) and natural antisense siRNAs (nat-siRNAs; Chen, 2009). Recent studies showed that small RNAs can negatively regulate gene expression at the post-transcriptional level based on two possible mechanisms: transcript cleavage and translational repression (Sunkar et al., 2007; Couzigou and Combier, 2016).
As a climacteric fruit model, tomato has been widely used to study the molecular mechanisms of fruit ripening and senescence as well as ethylene biosynthesis and signal transduction. Recently, increasing studies showed that small RNAs are also involved in regulating ethylene signal transduction (Pilcher et al., 2007; Moxon et al., 2008; Zhang et al., 2011; Zuo et al., 2012). For example, Moxon et al. (2008) found that one of the target genes of miR156 was CNR, which belongs to SBP-box family transcription factors (TFs), and the target gene of miR172 was AP2. It has been reported that the expression of genes that encode miRNAs is regulated at the transcriptional level by various transcriptional factors (Yant et al., 2010; Baek et al., 2013). For example, EIN3, a key transcription factor in ethylene signaling, directly binds to the promoter region of miR164 and represses its transcription (Li et al., 2013).
ERFs were a class of TFs located in the downstream of ethylene signal transduction pathways that function in diverse plant growth and metabolism processes as well as in the biotic and abiotic stress response, such as ethylene (Wu et al., 2002; Pirrello et al., 2006), high salt (Park et al., 2001; Wang et al., 2004), drought and low temperature, and so on (Qin et al., 2004; Zhang et al., 2007). Given that the miRNAs were also involved in the ethylene signaling pathways, there may be some relationships between miRNAs and ERFs. The high-throughput sequencing technology has been widely used to explore the functions of miRNA and siRNAs due to its high throughputs and accuracy (An et al., 2011; Cao et al., 2014; Thiebaut et al., 2014). In this study, High-throughput sequencing of small RNAs and degradome sequencing were used to gain a better understanding of the relationship between ethylene and small RNAs using wild type and LeERF1 transgenic tomato fruits. MiRNAs expression patterns were profiled and their targets were conferred; the regulatory network model between the small RNAs and ethylene was set up. This research provides more evidences for understanding the regulatory pathways of miRNAs in the network of fruit ripening.
Materials and methods
Sample collection and preparation
Wild type (Solanum lycopersicum cv. zhongshu4) and sense-/antisense-LeERF1 transgenic tomato plants (Li et al., 2007) were grown in the greenhouse at standard conditions. The Fruits at breaker stage were used in the experiment (Supplementary Figure S1). Pooled mesocarp tissues from three groups were flash frozen in liquid nitrogen and stored at −80°C until further analysis.
Small RNA (sRNA) quantification and qualification
The RNA samples were extracted using Trizol. Nanodrop, Qubit 2.0, and Agilent 2100 bioanalyzer were used to detect the purity, concentration and integrity of RNA samples, respectively, to ensure the use of qualified samples for sequencing. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured using Qubit® RNA Assay Kit in Qubit®2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Library preparation for small RNA sequencing
A total amount of 1.5 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext®Ultra™ small RNA Sample Library Prep Kit for Illumina®(NEB, USA) following manufacturer's recommendations and index codes were added to attribute sequences to each sample. Briefly, first of all, ligated the 3′ SR Adaptor, mixed 3′ SR Adaptor for Illumina, RNA and Nuclease-Free Water, mixture system after incubationa for 2 min at 70 degrees in a preheated thermal cycler, Tube was transferred to ice. Then, add 3′ Ligation Reaction Buffer (2X) and 3′ Ligation Enzyme Mix ligate the 3′ SR Adaptor, incubated for 1 h at 25°C in a thermal cycler. To prevent adaptor-dimer formation, the SR RT Primer hybridizes to the excess of 3′ SR Adaptor (that remains free after the 3′ ligation reaction) and transforms the single stranded DNA adaptor into a double-stranded DNA molecule. sRNAs (18–30 nucleotides in length) were separated from the total RNAs by polyacrylamide gel electrophoresis (PAGE). The small RNA molecules were then ligated with 5′ and 3′ adaptor and used for reverse transcription and subsequent PCR. The final PCR product was purified and sequenced by Illumina Cluster Station and Illumina Genome Analyzer (SanDiego, CA, USA).
Clustering and sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumina) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform and pair-end reads were generated. The sequencing results were deposited in the Sequence Read Archive (SRA) at the NCBI database (accession number: SRP094091).
Quality control
The quality control of raw data (raw reads) in fastq format has been performed by using in-house scripts written in Perl. Reads containing adapter and poly-N sequences and reads with low quality from raw data were removed. Then reads were cleaned by removing the sequences smaller than 18 nt or longer than 30 nt. At the same time, Q20, Q30, GC-content, and sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality.
Bioinformatic analysis of sequencing data
The Clean Reads were aligned with Silva database, GtRNAdb database, Rfam database, and Repbase database respectively to filter ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), repeat sequences, and other ncRNA using Bowtie tools. The remaining reads were used to detect known miRNAs and new miRNAs predicted by comparing with known miRNAs from miRBase. RNAFold tools were used to predict the secondary structure of all new miRNAs.
Identification of siRNA and putative novel miRNA
The adapter reads of the Solexa sequencing results were removed (Supplementary Figure S2). And reads larger than 30 nt and smaller than 18 nt were discarded. All high quality reads were considered as significant and further analyzed. Small RNA reads were mapped to tomato genome with mapping tool bowtie, all tomato genome annotation information is downloaded from ITAG2.3 include repeats and protein-coding regions (http://solgenomics.net/organism/Solanum_lycopersicum/genome). Six libraries are pooled together for miRNA prediction. The potential miRNA loci were analyzed using MIREAP software (version 0.2) with default parameters followed by additional manual check criteria that included: the miRNA sequence length should be between 18 and 26 nt; the maximal free energy allowed for a miRNA precursor (−18 kcal/mol); flank sequence length of miRNA precursor (100 nt); the predicted mature miRNA reads count should be large than 10 and reading counts ratio for miRNA*/miRNA should be small than 0.1. The unique reads left were aligned with known miRNAs from miRBase 21.0 (http://www.mirbase.org/). Phased small RNAs and nat-siRNAs were predicted as described in the previous studies (Chen et al., 2007; Zhou et al., 2009). All the reading counts were normalized to per million of total mapped reads (TPM).
Target gene functional annotation
Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences); Nt (NCBI non-redundant nucleotide sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KO (KEGG Ortholog database); GO (GeneOntology).
Quantification of small RNAs expression levels and differential expression analysis
Small RNA expression levels were estimated by TPM for each sample: sRNA were mapped back onto the reference genome, and read count for each small RNA was obtained from the mapping results. For the samples with biological replicates, differential expression analysis of two conditions/groups was performed using the DESeq R package (1.10.1). DESeq provide statistical routines for determining differential expression in digital miRNA expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. MiRNA with an adjusted p < 0.05 and |log2(fold change)| ≥ 1 were assigned as differentially expressed (Anders and Huber, 2010).
GO enrichment analysis
Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq R packages based on Wallenius non-central hyper-geometric distribution (Young et al., 2010), which can adjust to gene length bias in DEGs.
KEGG pathway enrichment analysis
KEGG (Kanehisa et al., 2008) is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism, and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). We used KOBAS (Mao et al., 2005) software to test the statistical enrichment of differential expression genes in KEGG pathways.
Results
Overview of the small RNA libraries from tomato fruit
To identify small RNAs and analyze their functions in ethylene pathway, the deep sequencing technology with Illumina Hiseq 2500 platform (Biomarker Technologies, China) was performed in wild and sense-/antisense-LeERF1 transgenic tomato fruits at breaker stage. A total of 19.31, 19.80, 17.28, 15.24, 14.61, 14.60 million raw reads in CK1 (wild 1), CK2 (wild 2), F1 (sense-LeERF1 transgenic tomato 1), F2 (sense-LeERF1 transgenic tomato 2), R1 (antisense-LeERF1 transgenic tomato 1), and R2 (antisense-LeERF1 transgenic tomato 2) were generated, respectively. After removing low quality and contaminated reads, poly A-containing sequences, sequences outside of 18–30 nt, 3′ and 5′ adaptors sequences, 15.75 (CK1), 16.88 (CK2), 13.79 (F1), 13.05 (F2), 12.63 (R1), 12.94 (R2) million clean reads were remained for further analysis. Then the small RNAs were categorized into miRNAs, ribosomal (r) RNAs, transfer (t) RNAs, small nuclear (sn) RNAs, small nucleolar (sno) RNAs, repeat regions, exon and intron RNA based on genomic location and function analysis (Table 1).
Table 1.
Small RNAs profiling and classification in six tomato fruit groups.
| Types | CK1 | CK2 | F1 | F2 | R1 | R2 |
|---|---|---|---|---|---|---|
| Total | 15,753,031 | 16,876,616 | 13,793,259 | 13,050,621 | 12,633,453 | 12,938,234 |
| miRNA | 606,491 | 678,439 | 529,661 | 514,194 | 506,601 | 500,709 |
| rRNA | 363,895 | 334,157 | 348,969 | 268,843 | 294,359 | 318,280 |
| tRNA | 73,283 | 69,194 | 68,966 | 58,727 | 42,953 | 60,809 |
| snRNA | 4,726 | 5,906 | 5,517 | 5,220 | 4,802 | 5,752 |
| snoRNA | 7,876 | 8,100 | 8,275 | 7,264 | 5,938 | 6,951 |
| Repeat | 2,451,172 | 2,683,382 | 2,008,299 | 1,958,898 | 1,867,224 | 1,935,560 |
| NAT | 1,228,736 | 1,400,759 | 1,213,807 | 1,070,151 | 1,174,911 | 1,125,626 |
| TAS | 40,958 | 37,129 | 37,242 | 40,457 | 41,690 | 37,521 |
| exon:+ | 677,380 | 573,805 | 537,937 | 587,278 | 517,972 | 491,653 |
| exon:− | 247,323 | 275,089 | 219,313 | 216,640 | 228,665 | 238,064 |
| intron:+ | 849,088 | 916,400 | 760,009 | 726,920 | 720,107 | 720,660 |
| intron:− | 521,425 | 567,054 | 464,833 | 445,026 | 423,221 | 437,312 |
| Other | 8,680,677 | 9,327,202 | 7,590,433 | 7,151,003 | 6,805,010 | 7,059,338 |
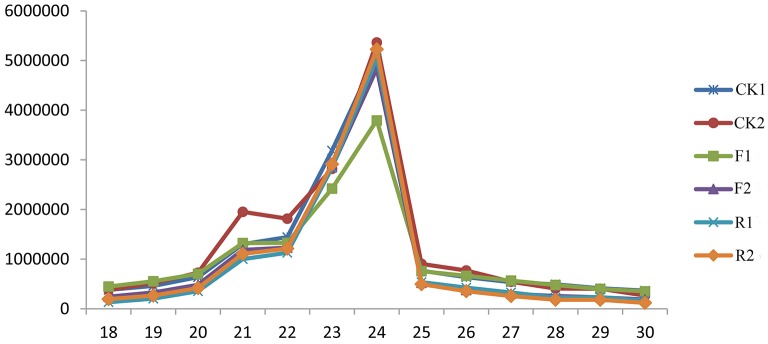
The size distribution is one of the distinct features of the small RNAs libraries from different plants. In our experiments, the length of small RNAs ranges from 18 to 30 nt and the most abundant group of small RNAs have length 21–24 nt in all the six libraries (Figure 1). There is no obvious difference of the length distribution between wild and LeERF1 transgenic tomato. Among the 21–24 nt size small RNAs, 24-nt size class has the highest abundance, accompanied with the 23-nt sRNAs as the second largest groups, which is in accordance with that of rice (Morin et al., 2008), Arabidopsis (Rajagopalan et al., 2006), and our previous results (Zuo et al., 2012, 2013).
Figure 1.
Length distribution of small RNAs in wild (CK1, CK2), sense-LeERF1 (F1, F2), and antisense-LeERF1 (R1, R2) tomato fruit.
Identification of miRNAs and siRNAs in tomato
To identify miRNAs and siRNAs in tomato, the clean sequences were aligned with the tomato small RNAs database (http://ted.bti.cornell.edu/cgi-bin/TFGD/sRNA/home.cgi) and the latest miRNA database (http://www.mirbase.org/, Release 21). In total, 178 known miRNAs belonging to 108 families were obtained in our libraries. Among the 108 families, 46 miRNA families (Supplementary Table S1) were registered in miRBase as belonging to S. lycopersicum and other 62 families (Supplementary Table S2) were less conserved and first identified in tomato (Supplementary Figure S3). Most of the miRNA families belonging to S. lycopersicum in miRBase are composed of more than one member. For instance, miR156 and miR482 were the largest ones with seven members in the families in this study. MiR171 and miR319 were the second largest family with six members. On the other hand, except for the miR548 family, other less conserved miRNA families had only one member detected in this study. Sequence length statistical results showed that the 21-nt miRNAs were the main type of the 178 known miRNAs.
In addition, 36 putative novel miRNAs with hairpin structures renamed as miRZ101 to miRZ136 were predicted and all of them were found to have star sequences (Table 2 and Supplementary Table S3). The length of the putative novel miRNAs were 18–24 and 24 nt miRNAs accounted for the predominance. Most of the first nucleotide of the putative novel miRNAs were A, which was in accordance with previous study that 24 nt miRNAs used to had an A as the first nucleotide (Jain et al., 2014). The minimum folding free energies varied from −149.8 to −28.3 kcal/mol (Supplementary Table S3).
Table 2.
Putative novel miRNAs found in tomato.
| MiRNA | Length | Sequence | Chromosome | Star | MEF (kcal/mol) |
|---|---|---|---|---|---|
| miRZ101 | 22 | uaacuucgucuagcucgccuuc | 10 | + | −70 |
| miRZ102 | 24 | guagagaacucuagaaccuucuag | 10 | + | −84.1 |
| miRZ103 | 24 | aaaggacuccuagauuucucuagu | 11 | + | −93.9 |
| miRZ104 | 24 | aaagacuguucaauuacugcuuga | 11 | + | −28.3 |
| miRZ105 | 24 | uauguccuuuaacuuugagugugc | 12 | + | −110.4 |
| miRZ106 | 24 | uuaguauaguauaagugugucucu | 12 | + | −57.1 |
| miRZ107 | 24 | acacacucugcauucaauuaaauu | 12 | + | −63.4 |
| miRZ108 | 24 | acguugcucagacucuucaaaaau | 12 | + | −60.3 |
| miRZ109 | 22 | auuuauggcuaugaauuugagu | 12 | + | −62.9 |
| miRZ110 | 24 | uuaguauaguauaagugugucucu | 1 | + | −41.7 |
| miRZ111 | 24 | uuaguuuaauuaagaugugucucu | 1 | + | −105.2 |
| miRZ112 | 21 | gcacggcagauaguuauuggc | 1 | + | −114.6 |
| miRZ113 | 24 | guagagaacucuagaaccuucuag | 1 | + | −71.4 |
| miRZ114 | 24 | aagcgaugacuuuagugaccuagu | 1 | + | −39.9 |
| miRZ115 | 22 | cacggucguaccuugacaaggc | 2 | + | −77.8 |
| miRZ116 | 22 | uuguuucuguuuuuguuugagu | 2 | + | −149.8 |
| miRZ117 | 23 | guugcucggacucuucaaaaaug | 2 | + | −69.1 |
| miRZ118 | 20 | auaacacaaaucugagccuc | 2 | + | −56.5 |
| miRZ119 | 22 | agugacucgcucgaucuuucuu | 3 | + | −64 |
| miRZ120 | 24 | uuucgucuugaaaguuugcccaug | 4 | + | −58.3 |
| miRZ121 | 24 | auuuccgaucaaacuuaaacuguu | 4 | + | −40.8 |
| miRZ122 | 23 | guugcucgaacucuucaaaaaug | 5 | + | −62.7 |
| miRZ123 | 24 | augugaucgcuguaaugaccuuac | 5 | + | −132.9 |
| miRZ124 | 24 | ucgagggucuaucagaaacaacau | 6 | + | −50.8 |
| miRZ125 | 18 | accugguugauccugcga | 6 | + | −73.3 |
| miRZ126 | 24 | guugcucgaacucuucaaaaaugu | 6 | + | −78.9 |
| miRZ127 | 24 | uuuucuaucggaacuaucaugugu | 6 | + | −69.3 |
| miRZ128 | 21 | ucaacgcugcacucaaucaug | 7 | + | −75.2 |
| miRZ129 | 24 | aagacguuugaaucugaaaaagau | 8 | + | −57.7 |
| miRZ130 | 23 | uuauacuauacuaagguccuauu | 8 | + | −117 |
| miRZ131 | 24 | cgagugcucauuccacagauaagu | 8 | + | −64.2 |
| miRZ132 | 24 | auacaucgguuacuugauagacgu | 8 | + | −109.4 |
| miRZ133 | 24 | uuaguauaguauaagugugucucu | 8 | + | −103.1 |
| miRZ134 | 24 | ugaaaucgagaugugauguagagg | 9 | + | −59.9 |
| miRZ135 | 23 | uucuucugacucauuuacuuuag | 9 | + | −54.2 |
| miRZ136 | 24 | augcucuagucuuugaacgacagg | 9 | + | −58.7 |
Moreover, several conserved and species-specific endogenous siRNAs were also characterized in our libraries. Ta-siRNAs are a special class of siRNAs that generated from TAS gene transcripts and mediated by miRNA (Xie et al., 2005; Yoshikawa et al., 2005; Li et al., 2012). On the basis of the conservation of the TAS genes in plants, three TAS5 gene family members: TAS5, TAS5b, and TAS5d (TAS5b and TAS5d were found in our previous study; Zuo et al., 2016), all miR482 targets, were identified (Table 3). Surprisingly, one more TAS5 family member (TAS5e) and two more TAS genes (TAS11a and TAS11b), triggered by sly-miR6024, are reported in our results (Table 4). In addition, 19 potential phased small RNAs and 958 nat-siRNAs were also found in this study (Supplementary Tables S4,S5).
Table 3.
The conserved TAS5 family in tomato fruit.
| Name | Chromosome | Length | Start | End | Phased abundance | related miRNA |
|---|---|---|---|---|---|---|
| sly-TAS5 | 6 | 539 | 423,570 | 424,108 | 3,954 | sly-miR482d-3p |
| sly-TAS5b | 2 | 644 | 21,186,658 | 21,187,301 | 530 | sly-miR482d-3p |
| sly-TAS5d | 8 | 917 | 58,262,775 | 58,263,691 | 4,729 | sly-miR482e-3p |
Three TAS5 family members were found and were located in Chromosome 6, 2, and 8 separately. “Phased abundance” means the abundance of phased sequence and the “related miRNA” related the miRNAs that mediated TAS.
Table 4.
The novel TAS families in tomato fruit.
| Name | Chromosome | Start | End | Length | Phased abundance | related miRNA |
|---|---|---|---|---|---|---|
| sly-TAS5e | 11 | 48,467,984 | 48,468,816 | 833 | 22,190 | sly-miR482b |
| sly-TAS11a | 5 | 2,500,975 | 2,501,555 | 581 | 389 | sly-miR6024 |
| sly-TAS11b | 11 | 51,986,458 | 51,986,681 | 224 | 137 | sly-miR6024 |
Three members belong to two TAS families (TAS5, TAS11) were found and located in Chromosome 11, 5, and 11 separately. “Phased abundance” means the abundance of phased sequence and the “related miRNA” related the miRNAs that mediated TAS.
The effect of overexpression sense-/antisense-LeERF1 on small RNA profiles
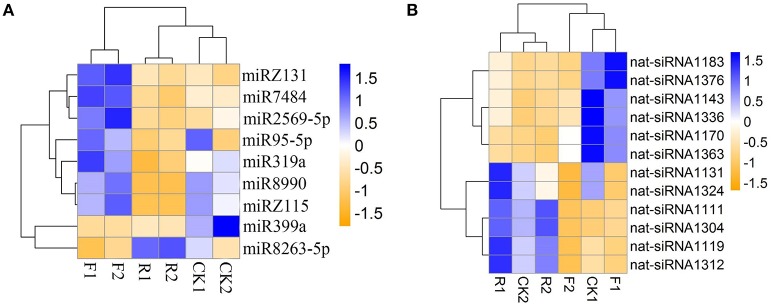
To evaluate the regulatory roles of LeERF1on miRNA expression, differential expression of miRNAs among the wild and sense-/antisense-LeERF1 transgenic tomato were analyzed. After normalization using a RPM method, the miR399a was found to have significant different accumulation between wild type and sense-LeERF1 transgenic tomato fruits. The expression of the miR399a was down-regulated in sense-LeERF1 transgenic fruit (Figure 4A). MiR8990 and the novel miRZ118 were the two miRNAs significant differently expressed between wild type and antisense-LeERF1 transgenic tomato fruits, and their accumulations decreased in the transgenic fruit. Totally, there were nine miRNAs having significant differential expression between sense-LeERF1 and antisense-LeERF1 transgenic fruit. Among them, miR399a and miR8263-5p were up-regulated in antisense-LeERF1 transgenic fruits. Meanwhile, other seven miRNAs including miR7484, miR319a, miR95-5p, miR8990, miR2569-5p, and two putative novel miRNAs (miRZ118 and miRZ131) were down-regulated.
Besides, 12 nat-siRNAs were found to show differential expression patterns. Compared with wild type fruits, most of the nat-siRNAs showed lower expression in sense-LeERF1 tomato fruits, only two of them increased (Figure 4B). However, among the differentially expressed nat-siRNAs, more than half of them had higher expression levels in antisense-LeERF1 transgenic fruits.
Target gene identification of the miRNAs
MiRNAs regulate gene expression mainly through cleaving mRNA or inhibiting the translation process of the targets gene, so identification and analysis of the target genes were the basis to study the function of miRNAs. Bioinformatics prediction and high-throughput degradome sequencing were the two main methods to find the targets gene. Using bioinformatic prediction method, a total of 103 target genes that involved in biological process, cellular component, and molecular function were found and most of them were identified to participate in biological process (Figure 2). Previous studies indicated that the targets of conserved miRNAs were also conservative and most of the miRNA families had not only one target site (Jin et al., 2008; Lu et al., 2008), which was also found in our study. For example, the targets of miR166a are homeobox-leucine zipper protein Revoluta, homeobox-leucine zipper protein ATHB-14 and pentatricopeptide repeat-containing protein At5g25630. Meanwhile, one target gene was often cleaved by two or more miRNAs. For instance, AP2 is the target of miR172 and miR8737, miR319 and miR159 share the same target GAMYB. Among the identified target genes, 16 targets were found to be involved in ethylene (Supplementary Table S6) and most of them were AP2 TFs. Another class of targets was auxin response factors including ARF10, ARF16, ARF17 and ARF18. Two F-box proteins (F-box protein 6, F-box protein At3g07870-like), two ethylene-responsive factors (RAP2-7-like) and an APETALA2-like protein were also predicted.
Figure 2.
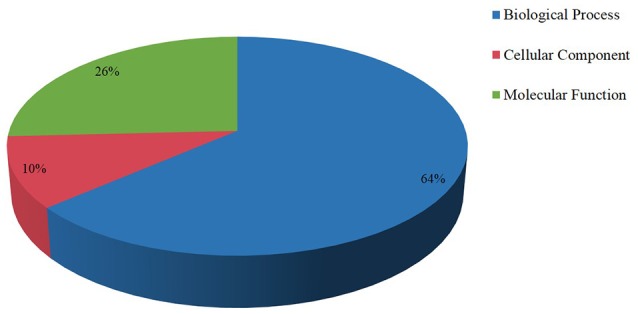
Targets of miRNAs in tomato fruit. Most targets of the miRNAs were involved biological process.
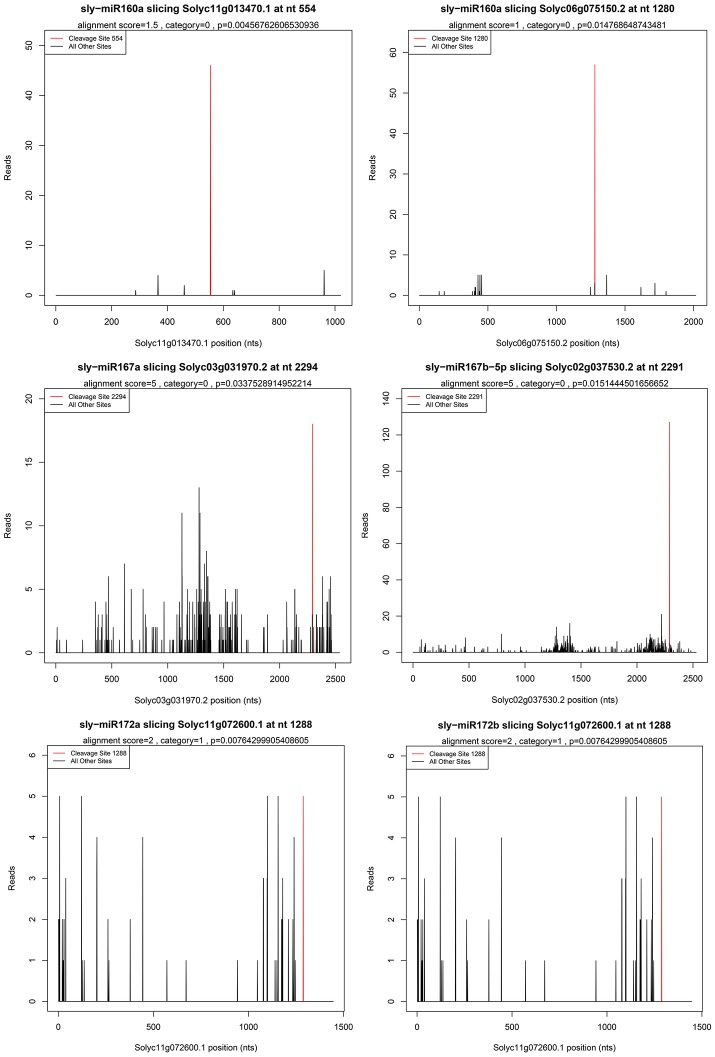
High-throughput degradome sequencing is a new technology to identify miRNAs targets and is successfully applied in Arabidopsis, rice (Addo-Quaye et al., 2008; Li et al., 2010). In this study, a total of 55 cleavage sites associated with 41 miRNAs were detected and seven target genes cleaved by five miRNAs were identified to be related to ethylene synthesis and signal transduction, including five auxin response factors (ARFs), one AP2 TF, and one ERF TF (Supplementary Table S7). Except for the known targets, six new targets were identified. The representative target plots of new targets were shown in Figure 3.
Figure 3.
Target plots of miRNA targets confirmed by degradome sequencing. Six new genes were found to be targets of five miRNAs.
In addition, 389 genes were predicted to be the targets of nat-siRNAs, and 22 of them were found to participate in ethylene pathway (Supplementary Table S8). Ethylene-responsive TFs, MADS-box TFs, F-box proteins were the main targets involved in fruit ripening. Moreover, 55 targets of the ta-siRNAs were also predicted and two of them (Solyc02g092020.1 and Solyc02g082320.1) were related to ethylene pathway.
Target parsing and small RNA regulatory network analysis in tomato
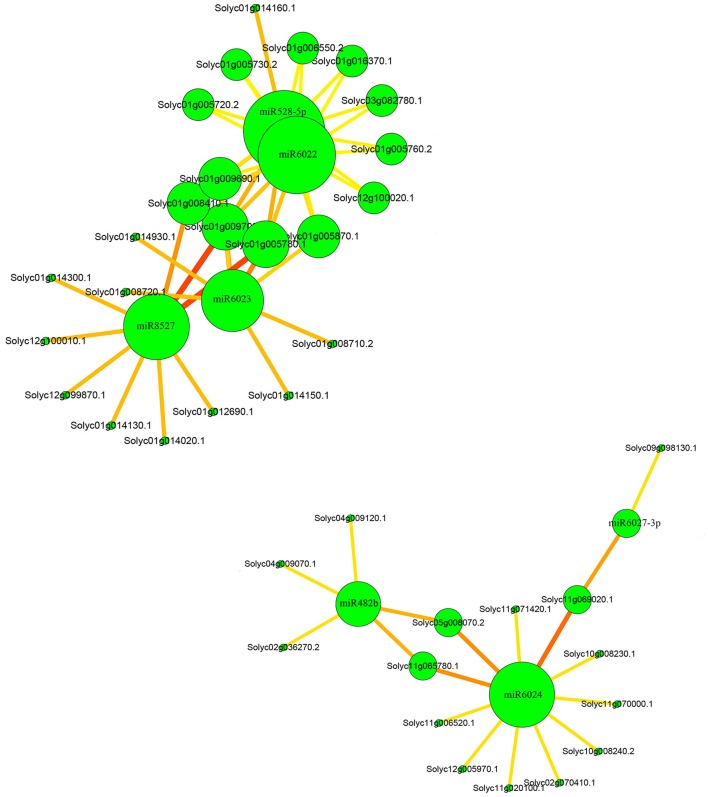
To investigate the network between the miRNAs and their targets, cytoscape platform was employed. In the network, it could be clearly seen that miR6024 had 11 targets and among the targets two of them were also the targets of miR482 (Figure 5). In addition, miR6024 and miR6027-3p shared one common targets. Moreover, miR6022 and miR528 shared most of their targets and they also had common targets with miR6023 and miR8527.
Figure 5.
Relationships between miRNAs and their targets.
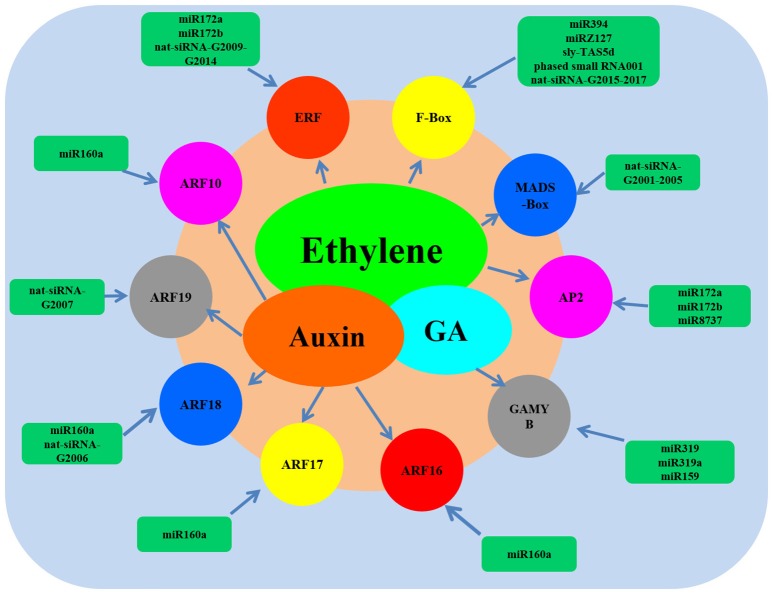
To comprehensively understand the functions of the miRNAs, ta-siRNAs, and nat-siRNAs involved in ethylene synthesis and signal transduction, all the predicted target genes of small RNAs were screened carefully and a regulatory model including small RNAs, the targets and their main functions was set up (Figure 6). As shown in Figure 6, it could clearly been seen that the AP2 TFs that involved in ethylene signaling were the targets of miR172a, miR172b, and miR8737. Ethylene-responsive transcription factors were the target of miR172a, miR172b and the nat-siRNAs renamed as nat-siRNA-G2009, nat-siRNA-G2010, nat-siRNA-G2011, nat-siRNA-G2012, nat-siRNA-G2013, and nat-siRNA-G2014. Meanwhile, the F-Box proteins and MADS-Box TFs that participated in ethylene signaling were the targets of miR394, miRZ131, nat-siRNA-G2015 to nat-siRNA-G2017, sly-TAS5d, phased small RNA001 and nat-siRNA-G2001 to nat-siRNA-G2005, respectively. Moreover, the auxin response factors that indirectly control ethylene signaling were the targets of miR160a and the nat-siRNA-G2006 and nat-siRNA-G2007.
Discussion
Small RNAs are a class of non-coding RNAs that play vital roles in growth and development, signal transduction, biotic and abiotic stresses (Jones-Rhoades et al., 2006; Dalmay, 2010; Mohorianu et al., 2011; Zuo et al., 2012; Pashkovskiy and Ryazansky, 2013). Numerous studies have demonstrated that miRNAs were involved in the regulation of diverse physiological processes by repressing the expression of their target genes. Ethylene is an important endogenous hormone and plays important roles in fruit development and ripening. As a model plant, tomato has been widely used to study the molecular mechanisms of ethylene biosynthesis and signal transduction (Giovannoni, 2004; Osorio et al., 2011), and through the study on ripening-related mutants or transgenic plant, many advances have been achieved. ERFs were a class of TFs located in the downstream of ethylene signal transduction pathways, and as one of the members of ERF class, LeERF1 had been showed to mediate fruit maturation and softening, enhance resistance to osmotic stress and improve plant tolerance to fungal invasion (Li et al., 2007; Lu et al., 2011; Pan et al., 2013). To better understand the relationship between LeERF1 and small RNAs in ethylene pathway, high-throughput sequencing was employed in the sense-/antisense-LeERF1 transgenic tomato fruits and many ethylene-related small RNAs as well as their target genes were found.
High-throughput sequencing of tomato fruit
In the past decades, miRNAs identification and their biological roles analysis were the mainly focused research fields. In tomato, 46 miRNA families were identified and registered in the miRBase database (http://www.mirbase.org/). It is well-known that many small RNAs have temporal expression patterns (Chen, 2009; Rubio-Somoza et al., 2009) and many studies had not detected all the 46 miRNA families (Candar-Cakir et al., 2016; Wu et al., 2016). In this study, the 46 families were all identified though some miRNAs did not found in all libraries, such as miR169 that only detected in wild tomatoes. This result indicated that the high-throughput sequencing had superiority in the identification of small RNA. Meanwhile, we identified 62 less conservative miRNAs that had not previous been found in tomato but documented in the miRBase for other species. For instance, miR861 was found in Arabidopsis (Fahlgren et al., 2007) and miR8010 were registered for potato (Zhang et al., 2013). MiR440, miR528, miR2922 and miR1049, miR1222, miR1063 were detected in rice and moss, respectively (Liu et al., 2005; Sunkar et al., 2005; Talmor-Neiman et al., 2006; Axtell et al., 2007; Sanan-Mishra et al., 2009).
In addition, 41 putative novel miRNAs not identified in other reports were also predicted in this study. The hairpin structures were found and the minimal folding free energies (MFEs) were −149.8 to −28.3 kcal/mol, indicated that the hairpin structures were stable. MiRNAs with detected stars were more likely to predict to be bona fide novel miRNAs (Wu et al., 2016). The renamed putative novel miRNAs in our libraries all had stars, suggested the accuracy of the novel miRNAs. Most of the putative novel miRNAs were 24 nt in length. The 24 nt small RNAs were reported to mainly match to the promoter regions of ripening-associated genes (Tomato Genome Consortium, 2012) and its high percentage in the putative novel miRNAs may imply regulatory roles of the novel miRNAs in ethylene pathway.
Overall, the discovery of less conserved and putative novel miRNAs in tomato provided enriched insight into the plant miRNA dataset. In addition, one previously reported TAS5 gene (Li et al., 2012) and two other TAS5 genes (TAS5b, TAS5d, found in our other study) were detected in our libraries (Table 3). Moreover, another TAS5 (TAS5e) with the target sly-miR482b and two more TAS genes (Named TAS11a and TAS11b) triggered by sly-miR6024 were also found in our libraries (Table 4).
Differential expression profiles of the small RNAs
It is well-known that many small RNAs have temporal expression patterns (Chen, 2009; Rubio-Somoza et al., 2009). Differential expression patterns of small RNAs can be regarded as an index for estimating the regulation contributions. We analyzed small RNAs expression in the wild and sense-/antisense-LeERF1 transgenic tomato. It is worth to noting that 14 miRNAs had significant difference expression between the wild and transgenic tomato. Among them, four miRNA families had a significant different accumulation between wild and sense-LeERF1 transgenic tomato fruits and 10 miRNAs differentially expressed in the response to antisense-LeERF1, indicating their specific roles in fruit ripening (Figure 4A).
Figure 4.
Expression profiles of the significantly differentially expressed miRNAs (A) and nat-siRNAs (B) in wild (CK1, CK2), overexpression sense-LeERF1 (F1, F2), and antisense-LeERF1 (R1, R2) tomato fruit.
It has been reported that miR399 was involved in plant response to phosphate starvation (Fujii et al., 2005; Chiou et al., 2006) and its accumulation increased during fruit development in tomato (Gao et al., 2015). In this study, the miR399a is down-regulated in sense-LeERF1 transgenic fruit, which indicated that miR399 may play an important role in ethylene signal transduction pathway. Totally, there were nine miRNAs having significant different expression between sense-LeERF1 and antisense-LeERF1 transgenic fruit. Among them, miR399a and miR8263-5p were up-regulated in antisense-LeERF1 transgenic fruits, meanwhile, other nine miRNAs, including miR7484, miR319a, miR95-5p, miR8990, miR2569-5p, and two putative novel miRNAs (miRZ118 and miRZ131) were down-regulated. miR319 has been reported to control leaf development and morphogenesis through regulating transmission control protocol (TCP) transcription factors (Palatnik et al., 2003). In this study, the target of miR319 was predicted to be GAMYB, which was also related to ethylene pathway. According to our results, miR319 may also participate in ethylene signaling pathway.
Besides, 12 nat-siRNAs were found to show differential expression patterns. Compared with wild fruits, most of the nat-siRNAs showed lower expression levels in sense-LeERF1 tomatoes, and only two of them increased. However, among the nat-siRNAs, more than half of them had higher expression levels in antisense-LeERF1 transgenic fruits (Figure 4B).
Small RNAs participated in ethylene pathway
To study the function of small RNAs in ethylene pathway, bioinformatic prediction, and degradome sequencing were also used in wild and sense-/antisense-LeERF1 transgenic tomato. Results showed that most of the targets were identified to participate in various biological processes (Figure 2). AP2 transcription factors, AP2-like ethylene-responsive transcription factors, ethylene-responsive transcription factor were TFs belong to AP2/EREBP transcription factors family involved in ethylene signaling pathway and they were the main targets of miR172 family, which was also reported in Arabidopsis and tomato (Wu et al., 2009; Cheng et al., 2016). The AP2/EREBP transcription factors were also the target of miR5658 (Cheng et al., 2016). However, in this study, the miR5658 was not detected and miR8737 were predicted to target AP2/ERF TFs. F-Box proteins were reported to regulate ethylene signaling in Arabidopsis (Wang et al., 2009). It was also been reported that the F-Box proteins were the targets of miR393 in Arabidopsis (Liu et al., 2008). However, in this study, miR393 was not found and F-Box proteins were predicted to be the target of miR394. In addition, auxin response factors genes cleaved by miR160 were also found in our results. In Arabidopsis, ARF6 and ARF8, ARF16, and ARF17 were reported to be the targets of miR167 and miR160, respectively. In this study, ARF8 cleaved by miR167 was found via degradome sequencing. However, ARF6 was not identified for miR167, perhaps because the abundance of cleaved products was too low to be detected. Unexpectedly, ARF10 and ARF18 were identified to be the targets of miR160. Surprisingly, six new targets were found (Figure 3).
Compared with the miRNAs, most targets of the ta-siRNAs and nat-siRNAs in tomato were not verified yet. The distribution of the targets of ta-siRNAs and nat-siRNAs was different from that of the miRNAs, and a great part of the targets were predicted to be involve in all kinds of metabolic processes which were consistent with the previous studies (Zhai et al., 2011; Li et al., 2012; Shivaprasad et al., 2012; Zuo et al., 2013). In this study, several important target genes participating in fruit ripening and senescence were found including Ethylene-responsive transcription factors, F-box proteins, MADS-box TFs, and MADS-box proteins.
Network construction revealed the relationship of small RNAs and ethylene in tomato
To illuminating the network between small RNAs and their target genes involved in ethylene, all the predicted target genes of miRNAs, ta-siRNAs, and nat-siRNAs were screened carefully and a regulatory model was set up (Figure 6). From the network model, it could clearly been seen that miR394, miRZ131, miR172, miR8737, miR319, miR159, miR160, and nat-siRNA-G2001 to nat-siRNA-G2017 as well as their target genes such as auxin response factors, ethylene-responsive transcription factors and GAMYB were involved in ethylene signal pathway. These results indicate that the network of miRNAs are quite complicated, and elucidation of the molecular mechanisms underlying the interplay between miRNA and their target genes involved in ethylene pathway requires further study.
Figure 6.
Network model of the small RNAs and their target genes involved in ethylene.
Conclusion
In summary, ethylene biosynthesis and signal transduction related miRNAs and siRNAs were identified in tomato fruit. These informations broaden the knowledge of the relationship between small RNAs and ethylene regulation. Additionally, many target genes of miRNAs were identified by bioinformatic prediction and degradome sequencing. The result showed that the target genes were involved in various functions and ethylene related targets were also discovered. In addition, a large amount of the target genes of nat-siRNAs were found and some of them were found to participate in ethylene regulation. A regulatory model which reveals the regulation relationship between the small RNAs and their targets was set up. Moreover, 41 putative novel miRNAs were identified and many of them were also involved in ethylene pathway. These findings lay the foundation for exploring the role of small RNAs in ethylene signaling pathway in the plant.
Author contributions
JZ and LG designed the research; YW and JZ carried out the experiments; JZ, YW, QW, and ZJ analyzed the results; YW wrote the manuscript; JZ, BZ, and YL modified the manuscript; all authors have read and approved the manuscript for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31401536), the National Key Research and Development Program of China (No. 2016YFD0400901), the China Agriculture Research System Project (No. CARS-25), the Special Fund for Agro-scientific Research in the Public Interest (No. 201203095), the Young Investigator Fund of Beijing Academy of Agricultural and Forestry Sciences (No. 201404).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00527/full#supplementary-material
The expressions of LeERF1 wild (CK1, CK2), sense-LeERF1 (F1, F2), and antisense-LeERF1 (R1, R2) tomato fruit.
The pipeline for the systematic identification of small RNAs in tomato.
Expression profiles of the known and putative novel miRNAs in wild (CK1, CK2), overexpression sense-LeERF1 (F1, F2), and antisense-LeERF1 (R1, R2) tomato fruit.
Known miRNAs identified in tomato.
Less conserved miRNAs identified in tomato.
Putative novel miRNAs identified in tomato.
Phased small RNAs found in tomato.
Nat-siRNAs found in tomato.
Target genes of the miRNAs involved in ethylene by bioinformatic prediction.
Target genes of the miRNAs involved in ethylene by degradome sequencing.
Target genes of the nat-siRNAs involved in ethylene.
References
- Addo-Quaye C., Eshoo T. W., Bartel D. P., Axtell M. J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762. 10.1016/j.cub.2008.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F. M., Hsiao S. R., Chan M. T. (2011). Sequencing-based approaches reveal low ambient temperature- responsive and tissue-specific microRNAs in phalaenopsis orchid. PLoS ONE 6:e18937. 10.1371/journal.pone.0018937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M. J., Snyder J. A., Bartell D. P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell 19, 1750–1769. 10.1105/tpc.107.051706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Park H. C., Kima M. C., Yun D. J. (2013). The role of Arabidopsis MYB2 in miR399f-mediated phosphate-starvation response. Plant Signal. Behav. 8:e23488. 10.4161/psb.23488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candar-Cakir B., Arican E., Zhang B. H. (2016). Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 14, 1727–1746. 10.1111/pbi.12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Wu Z., Jiang F., Zhou R., Yang Z. (2014). Identification of chilling stress-responsive tomato microRNAs and their target genes by high-throughput sequencing and degradome analysis. BMC Genomics 15:1130. 10.1186/1471-2164-15-1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Sontheimer E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. M., Li Y. H., Wu S. H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 3318–3323. 10.1073/pnas.0611119104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25, 21–44. 10.1146/annurev.cellbio.042308.113417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. Y., Wang Y., Tao X., Fan Y. F., Dai Y., Yang H., et al. (2016). Genomic profiling of exogenous abscisic acid-responsive microRNAs in tomato (Solanum lycopersicum). BMC Genomics 17:423. 10.1186/s12864-016-2591-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T. J., Aung K., Lin S. I., Wu C. C., Chiang S. F., Su C. L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18, 412–421. 10.1105/tpc.105.038943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou J. M., Combier J. P. (2016). Plant microRNAs: key regulators of root architecture and biotic interactions. New Phytol. 212, 22–35. 10.1111/nph.14058 [DOI] [PubMed] [Google Scholar]
- Dalmay T. (2010). Short RNAs in tomato. J. Integr. Plant Biol. 52, 388–392. 10.1111/j.1744-7909.2009.00871.x [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Howell M. D., Kasschau K. D., Chapman E. J., Sullivan C. M., Cumbie J. S., et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of miRNA genes. PLoS ONE 2:e219. 10.1371/journal.pone.0000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chiou T. J., Lin S. I., Aung K., Zhu J. K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15, 2038–2043. 10.1016/j.cub.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Gao C., Ju Z., Cao D., Zhai B., Qin G., Zhu H., et al. (2015). MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol. J. 13, 370–382. 10.1111/pbi.12297 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. J. (2004). Genetic regulation of fruit development and ripening. Plant Cell 16, S170–S180. 10.1105/tpc.019158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Chevala V. V., Garg R. (2014). Genome-wide discovery and differential regulation of conserved and novel microRNAs in chickpea via deep sequencing. J. Exp. Bot. 65, 5945–5958. 10.1093/jxb/eru333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Li N., Zhang B., Wu F., Li W., Guo A., et al. (2008). Identification and verification of microRNA in wheat (Triticum aestivum). J. Plant Res. 121, 351–355. 10.1007/s10265-007-0139-3 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P., Bartel B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Orban R., Baker B. (2012). SoMART, a web server for plant miRNA, tasiRNA and target gene analysis. Plant J. 70, 891–901. 10.1111/j.1365-313X.2012.04922.x [DOI] [PubMed] [Google Scholar]
- Li Y., Zhu B., Xu W., Zhu H., Chen A., Xie Y., et al. (2007). LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep. 26, 1999–2008. 10.1007/s00299-007-0394-8 [DOI] [PubMed] [Google Scholar]
- Li Y. F., Zheng Y., Addo-Quaye C., Zhang L., Saini A., Jagadeeswaran G., et al. (2010). Transcriptome-wide identification of microRNA targets in rice. Plant J. 62, 742–759. 10.1111/j.1365-313X.2010.04187.x [DOI] [PubMed] [Google Scholar]
- Li Z. H., Peng J. Y., Wen X., Guo H. W. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25, 3311–3328. 10.1105/tpc.113.113340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li P., Li X., Liu C., Cao S., Chu C., et al. (2005). Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139, 296–305. 10.1104/pp.105.063420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. H., Tian X., Li Y. J., Wu C. A., Zheng C. C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843. 10.1261/rna.895308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. W., Shao Y., Li L., Chen A. J., Xu W. Q., Wu K. J., et al. (2011). Overexpression of SlERF1 tomato gene encoding an ERF-type transcription activator enhances salt tolerance. Rus. J. Plant Physiol. 58, 118–125. 10.1134/S1021443711010092 [DOI] [Google Scholar]
- Lu S., Sun Y. H., Chiang V. L. (2008). Stress-responsive microRNAs in Populus. Plant J. 55, 131–151. 10.1111/j.1365-313X.2008.03497.x [DOI] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J., Wei L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- Mohorianu I., Schwach F., Jing R., Lopez-Gomollon S., Moxon S., Szittya G., et al. (2011). Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. Plant J. 67, 232–246. 10.1111/j.1365-313X.2011.04586.x [DOI] [PubMed] [Google Scholar]
- Morin R. D., Aksay G., Dolgosheina E., Ebhardt H. A., Magrini V., Mardis E. R., et al. (2008). Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 18, 571–584. 10.1101/gr.6897308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S., Jing R., Szittya G., Schwach F., Rusholme Pilcher R. L., Moulton V., et al. (2008). Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 18, 1602–1609. 10.1101/gr.080127.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S., Alba R., Damasceno C. M., Lopez-Casado G., Lohse M., Zanorc M. I., et al. (2011). Systems biology of tomato fruit development: combined transcript, protein and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157, 405–425. 10.1104/pp.111.175463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Pan X. Q., Fu D. Q., Zhu B. Z., Lu C. W., Luo Y. B. (2013). Overexpression of the ethylene response factor SlERF1 gene enhances resistance of tomato fruit to Rhizopus nigricans. Postharvest Biol. Technol. 75, 28–36. 10.1016/j.postharvbio.2012.07.008 [DOI] [Google Scholar]
- Park J. M., Park C. J., Lee S. B., Ham B. K., Shin R., Paek K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. 10.1105/tpc.13.5.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkovskiy P. P., Ryazansky S. S. (2013). Biogenesis, evolution, and functions of plant microRNAs. Biochemistry 78, 627–637. 10.1134/s0006297913060084 [DOI] [PubMed] [Google Scholar]
- Pirrello J., Jaimes-Miranda F., Sanchez-Ballesta M. T., Tournier B., Khalil-Ahmad Q., Regad F., et al. (2006). Sl-ERF2 a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol. 47, 1195–1205. 10.1093/pcp/pcj084 [DOI] [PubMed] [Google Scholar]
- Qin J., Zhao J. Y., Zuo K. J., Cao Y. F., Ling H., Sun X. F., et al. (2004). Isolation and characterization of an ERF-like gene from Gossypium barbadense. Plant Sci. 167, 1383–1389. 10.1016/j.plantsci.2004.07.012 [DOI] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425. 10.1101/gad.1476406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. W., Reinhart B. J., Lim L. P., Burge C. B., Bartel B., Bartel D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. 10.1016/s0092-8674(02)00863-2 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Cuperus J. T., Weigel D., Carrington J. C. (2009). Regulation and functional specialization of small RNA-target nodes during plant development. Curr. Opin. Plant Biol. 12, 622–627. 10.1016/j.pbi.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Pilcher R. L., Moxon S., Pakseresht N., Moulton V., Manning K., Seymour G., et al. (2007). Identification of novel small RNAs in tomato (Solanum lycopersicum). Planta 226, 709–717. 10.1007/s00425-007-0518-y [DOI] [PubMed] [Google Scholar]
- Sanan-Mishra N., Kumar V., Sopory S. K., Mukherjee S. K. (2009). Cloning and validation of novel miRNA from basmati rice indicates cross talk between abiotic and biotic stresses. Mol. Genet. Genomics 282, 463–474. 10.1007/s00438-009-0478-y [DOI] [PubMed] [Google Scholar]
- Shivaprasad P. V., Chen H. M., Patel K., Bond D. M., Santos B. A., Baulcombe D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. 10.1105/tpc.111.095380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Chinnusamy V., Zhu J., Zhu J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309. 10.1016/j.tplants.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Girke T., Jain P. K., Zhu J. K. (2005). Cloning and characterization of microRNAs from rice(W). Plant Cell 17, 1397–1411. 10.1105/tpc.105.031682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmor-Neiman M., Stav R., Frank W., Voss B., Arazi T. (2006). Novel micro-RNAs and intermediates of micro-RNA biogenesis from moss. Plant J. 47, 25–37. 10.1111/j.1365-313X.2006.02768.x [DOI] [PubMed] [Google Scholar]
- Thiebaut F., Rojas C. A., Grativol C., Motta M. R., Vieira T., Regulski M., et al. (2014). Genome-wide identification of microRNA and siRNA responsive to endophytic beneficial diazotrophic bacteria in maize. BMC Genomics 15:766. 10.1186/1471-2164-15-766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Huang Z., Chen Q., Zhang Z., Zhang H., Wu Y. M., et al. (2004). Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol. Biol. 55, 183–192. 10.1007/s11103-004-0113-6 [DOI] [PubMed] [Google Scholar]
- Wang X., Kong H., Ma H. (2009). F-box proteins regulate ethylene signaling and more. Genes Dev. 23, 391–396. 10.1101/gad.1781609 [DOI] [PubMed] [Google Scholar]
- Wu G., Park M. Y., Conway S. R., Wang J. W., Weigel D., Scott Poethig R. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K. Q., Tian L. N., Hollingworth J., Brown D. C. W., Miki B. (2002). Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 128, 30–37. 10.1104/pp.010696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Wu Y., Liu C. C., Liu L. W., Ma F. F., Wu X. Y., et al. (2016). Identification of Arbuscular Mycorrhiza (AM)-Responsive microRNAs in Tomato. Front. Plant Sci. 7:429. 10.3389/fpls.2016.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Jones D. C., Wang Q., Sun R., Zhang B. (2015). Small RNA sequencing identifies miRNA roles in ovule and fibre development. Plant Biotechnol. J. 13, 355–369. 10.1111/pbi.12296 [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S. A., Carrington J. C. (2005). Expression of Arabidopsis miRNA genes. Plant Physiol. 138, 2145–2154. 10.1104/pp.105.062943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T. T., Ott F., Lanz C., Wollmann H., et al. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170. 10.1105/tpc.110.075606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M. Y., Poethig R. S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175. 10.1101/gad.1352605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:r14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J. X., Jeong D. H., De Paoli E., Park S., Rosen B. D., Li Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. 10.1101/gad.177527.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Y., Broeckling C. D., Sumner L. W., Wang Z. Y. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64, 265–278. 10.1007/s11103-007-9150-2 [DOI] [PubMed] [Google Scholar]
- Zhang R. X., Marshall D., Bryan G. J., Hornyik C. (2013). Identification and characterization of miRNA transcriptome in potato by high-throughput sequencing. PLoS ONE 8:e57233. 10.1371/journal.pone.0057233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zou Z., Zhang J., Zhang Y., Han Q., Hu T., et al. (2011). Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 585, 435–439. 10.1016/j.febslet.2010.12.036 [DOI] [PubMed] [Google Scholar]
- Zhou X. F., Sunkar R., Jin H., Zhu J. K., Zhang W. X. (2009). Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res. 19, 70–78. 10.1101/gr.084806.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Fu D., Zhu Y., Qu G., Tian H., Zhai B., et al. (2013). SRNAome parsing yields insights into tomato fruit ripening control. Physiol. Plant. 149, 540–553. 10.1111/ppl.12055 [DOI] [PubMed] [Google Scholar]
- Zuo J., Zhu B., Fu D., Zhu Y., Ma Y., Chi L., et al. (2012). Sculpting the maturation, softening and ethylene pathway: the influences of microRNAs on tomato fruits. BMC Genomics 13:7. 10.1186/1471-2164-13-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J. H., Wang Q., Han C., Ju Z., Cao D., Zhu B., et al. (2016). SRNAome and degradome sequencing analysis reveals specific regulation of sRNA in response to chilling injury in tomato fruit. Physiol Plant. [Epub ahead of print]. 10.1111/ppl.12509 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expressions of LeERF1 wild (CK1, CK2), sense-LeERF1 (F1, F2), and antisense-LeERF1 (R1, R2) tomato fruit.
The pipeline for the systematic identification of small RNAs in tomato.
Expression profiles of the known and putative novel miRNAs in wild (CK1, CK2), overexpression sense-LeERF1 (F1, F2), and antisense-LeERF1 (R1, R2) tomato fruit.
Known miRNAs identified in tomato.
Less conserved miRNAs identified in tomato.
Putative novel miRNAs identified in tomato.
Phased small RNAs found in tomato.
Nat-siRNAs found in tomato.
Target genes of the miRNAs involved in ethylene by bioinformatic prediction.
Target genes of the miRNAs involved in ethylene by degradome sequencing.
Target genes of the nat-siRNAs involved in ethylene.