Abstract
Defective interfering (DI) RNAs are subviral replicons originating from the viral genome and are associated with many plant RNA viruses and nearly all animal RNA viruses. The presence of DI RNAs in tombusvirus-infected plants reduces the accumulation of helper virus RNA and results in the development of attenuated symptoms similar to those caused by tombusviruses defective in p19, the posttranscriptional gene silencing (PTGS) suppressor. In situ analysis of infected plants containing DI RNAs revealed that the extent of virus infection was spatially restricted as was found for p19-defective tombusvirus. Previously, p19 was shown to suppress PTGS by sequestering the small interfering RNAs (siRNAs), which act as the specificity determinant for PTGS. Our results demonstrate that DI RNAs dramatically elevate the level of virus-specific siRNAs in viral infections, resulting in the saturation of p19 and the accumulation of unbound siRNAs. Moreover, we showed that, at low temperature, where PTGS is inhibited, DI RNAs are not able to efficiently interfere with virus accumulation and protect the plants. These data show that the activation of PTGS plays a pivotal role in DI RNA-mediated interference. Our data also support a role for 21-nucleotide siRNAs in PTGS signaling.
Defective interfering (DI) RNAs are deletion mutant RNAs of the parental viral genome generated spontaneously by replicase errors and are associated with many plant RNA viruses and nearly all animal RNA viruses (17). DI RNAs generally have lost essential viral genes for movement, replication, and encapsidation and thus require the presence of a helper virus for providing all the trans-acting proteins necessary for these functions. Interference with the helper virus by the DI RNA frequently results in remarkable symptom modification associated with a substantial decrease in helper virus levels (22).
Some of the most extensively studied plant virus DI RNA systems are those found in association with species of the Tombusvirus genus (18). Tombusviruses have a plus-sense RNA genome of about 4.7 kb that contains five open reading frames (ORF). ORF5 encodes a 19-kDa protein (p19) that is an important symptom determinant (1, 19). In addition, p19 has been identified as a potent posttranscriptional gene silencing (PTGS) suppressor (21, 23, 30). A number of DI RNAs from tombusvirus infections have been described (32), and all possess common structural features, which include noncontiguous elements corresponding to the terminal regions and an internal segment of the parental genome (18). DI RNAs do not code for any proteins and replicate by utilizing helper virus-encoded RNA-dependent RNA-polymerase. The presence of DI RNAs in virus-infected plants dramatically suppresses virus accumulation and attenuates the lethal necrotic symptoms normally associated with infection of the helper virus. A general assumption is that the reduction in helper virus levels by DI RNAs is due to competition for replication components, which in turn leads to development of attenuated symptoms (17, 18). Protoplast transfection experiments demonstrated that DI RNAs interfere with the accumulation of helper virus (2, 10, 14). Analyses of protoplasts cotransfected with tomato bushy stunt virus (TBSV) and DI RNAs revealed that suppression of the viral genomic RNA was mediated by a reduction in the rate at which the viral genomic RNA accumulated (10). Other studies suggested that the presence of TBSV DI RNAs specifically reduces the level of subgenomic (sg) RNA 2, which encodes the movement protein and p19 (20). Recently it was demonstrated that DI RNAs of cymbidium ringspot virus (CymRSV) activate PTGS, which efficiently targets the helper virus genome, while DI RNAs are poor targets for degradation (23). These results strongly suggested the involvement of PTGS in DI RNA-mediated symptom modulation.
PTGS is an adaptive, sequence-specific RNA degradation system that plays a role in the control of transposons, preservation of genome integrity, and defense against viruses (28, 31). This mechanism is activated by double-stranded (ds) RNA (dsRNA), which is cleaved into 21- to 26-nucleotide (nt) ds small interfering RNAs (siRNAs) (4) by an RNase III-like enzyme called DICER (5). The siRNAs generated are associated with an enzyme complex called RNA-induced silencing complex (RISC) and guide the RISC to degrade any RNA with sequence homology to the inducer dsRNA. In plants, in addition to its cell-autonomous defense function, PTGS is associated with a mobile signal that instructs target RNA degradation at a distance (29). The sequence specificity of PTGS implies that the signal must consist of nucleic acid components homologous to the target RNA. Previously it has been reported that longer siRNAs are associated with PTGS long-distance signaling (3); however, a recent study suggests that 21-nt siRNAs play a central role in short-distance and probably long-distance signaling as well (9). Consistent with the antiviral function of PTGS, many viruses, including tombusviruses, developed gene silencing suppressor proteins (13, 27). p19 of CymRSV has been demonstrated to bind ds siRNAs in vitro (21, 26, 33), while in infected cells p19 sequesters the majority of viral (21- to 22-nt) ds siRNAs (12). Consequently, the presence of p19 prevents the programming of silencing effector complexes, including short-distance systemic signaling complexes; thus plants fail to confine the virus. In contrast, in plants infected with suppressor-defective virus, siRNAs are not bound by p19 (12) and plants develop a typical PTGS-associated recovery phenotype (15, 16, 23). Moreover, biochemical analyses of siRNAs not bound by p19 revealed that these siRNAs are free in solution, with no discernible interaction with host proteins (12). In situ analyses of recovering plants revealed that virus accumulation was confined to the veins and surrounding cells (6).
Here we show that the presence of DI RNAs efficiently activates PTGS, confining the virus infection to veins and surrounding tissues. In addition, our data demonstrate that the presence of DI RNAs increases the generation of virus-specific siRNAs beyond the amount that can be sequestered by p19, leading to the accumulation of free siRNAs. We suggest that, due to the saturation of p19 by enhanced levels of siRNAs, DI RNAs induce development of a recovery-like phenotype. Moreover, we also demonstrate that, at low temperatures, where PTGS is not effective, DI RNAs are not able to efficiently restrict the spread of the helper virus and induce attenuated symptoms. In addition, our data suggest a central role for 21-nt siRNAs in the signaling of systemic PTGS.
MATERIALS AND METHODS
Plant materials and inoculation.
Four-week-old Nicotiana benthamiana plants grown in soil under normal growth conditions were used for virus inoculation with in vitro RNA transcripts of TBSV-P (24), p19-defective TBSV-P (T19stop), and mixed RNA transcripts of TBSV-P and DI-5 (24), as described previously (24). The T19stop mutant was constructed as described previously (23) with oligonucleotide AACCATGGAATGAGCTATATAAGGAAACG. Virus-infected plants were grown in environmental test chambers (MLR-350; Sanyo, Tokyo, Japan) under a 16-h light and 8-h dark regime at different temperatures (15, 21, and 24°C). The inoculations were repeated in several independent experiments at every investigated temperature with 5 to 10 plants.
RNA isolation and Northern blotting.
Total RNA extraction and Northern blot analyses of high-molecular-weight RNAs were performed as described previously (23). Randomly priming probes of TBSV-P genomic sequences and DI-5 sequences were used for detecting high-molecular-weight viral RNAs. Detection of virus specific siRNAs was carried out as described previously (23) with a radioactively labeled in vitro RNA transcript of plus-sense TBSV-P genomic RNA.
Protein separation and Western analysis.
Proteins were separated in a sodium dodecyl sulfate-12% polyacrylamide gel and then transferred onto a Hybond C Extra filter (Pharmacia-Biotech). Western blot analysis of protein samples was performed as described previously (8).
Immunoprecipitation and siRNA distribution assayed by gel filtration.
For immunoprecipitations, 2 g of infected N. benthamiana leaves showing systemic symptoms was used to prepare extracts (12). For gel filtration, extracts were prepared from 0.5 g of systemic leaves of infected N. benthamiana plants. Sixty 200-μl fractions were collected and used for Western and Northern blot analyses as described previously (12).
In situ hybridization and immunochemistry.
In situ hybridization of 12-μm paraffin-embedded leaf cross sections was carried out with digoxigenin-11-UTP-labeled virus-specific RNA probes as described previously (6). Two systemically infected leaves per plant were taken at 7 and 14 days postinoculation (dpi). The leaf samples were collected and processed for paraffin embedding. A digoxigenin-11-UTP-labeled minus-sense probe corresponding to the TBSV-P coat protein (CP)-encoding region was hybridized to tissue sections and detected with an alkaline phosphatase-conjugated antidigoxigenin antibody as described previously (7).
In situ immunohistochemistry of paraffin sections from systemically infected leaves at 7 dpi was conducted as described previously with a diluted (1:200) anti-p19 antibody (6).
Protoplast transfection and band quantification.
Mesophyll protoplasts of N. benthamiana were prepared as described previously (11). Quantification of RNA and protein bands was carried out by using Analysis, version 2.0 (Soft-Imaging Software GmbH) as described previously (8).
RESULTS
The presence of DI RNAs in infected tissue limits the extent of virus infection.
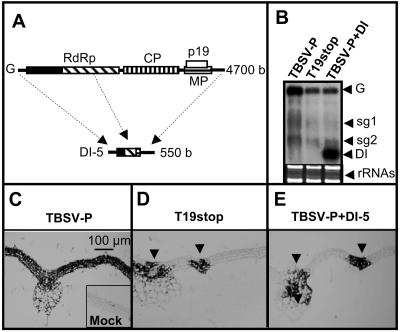
A p19-defective TBSV pepper isolate (TBSV-P; (24)) mutant was generated as described previously (23) and designated T19stop. Infection of N. benthamiana with T19stop resulted in reduced accumulation of viral genomic RNA and the development of a recovery phenotype at constant 21°C, similar to the phenotype caused by p19-defective CymRSV (21). N. benthamiana plants were infected with in vitro-synthesized RNA transcripts of TBSV-P (24) and T19stop and mixed RNA transcripts of TBSV-P and DI-5 (TBSV-P+DI) (24) as described previously (1). The infected plants showed intensive symptoms on the first systemically infected leaf after 6 to 7 dpi (data not shown). As described previously, TBSV-P infection leads to apical necrosis, which eventually culminates in the death of the plant (24). Although T19stop- and TBSV-P+DI-inoculated plants also displayed severe symptoms on the first systemically infected leaf, no apical necrosis was observed. By 14 dpi, TBSV-P+DI-infected plants began recovery from virus infection, similar to T19stop-infected plants (data not shown). In line with previous observations (6, 23), Northern blot analyses of RNA samples from the first systemically infected leaf at 7 dpi showed significant reduction in the accumulation of genomic RNA in both TBSV-P+DI- and T19stop-infected plants compared with that in TBSV-P-infected plants (Fig. 1B).
FIG. 1.
Accumulation of virus and DI RNAs in systemically infected leaves of N. benthamiana at 7 dpi. (A) Schematic representation of the organization of the TBSV-P genome and DI RNAs. RdRp, RNA-dependent RNA polymerase; MP, movement protein. (B) Northern blot analysis of TBSV-P, T19stop, and TBSV-P+DI accumulation in systemically infected leaves. Relative gel loadings are shown by ethidium bromide staining of rRNAs (bottom). G, genomic RNA; sg, sg RNA; DI, DI RNA. (C to E) In situ hybridization of leaf cross sections at 7 dpi. (C) TBSV-P-infected tissue. (Inset) Control mock-inoculated tissue. (D) T19stop-infected tissue. (E) TBSV-P+DI-infected tissue. The bar in panel C applies also to panels D and E. Black triangles, sites of virus accumulation.
To understand the development of DI RNA-mediated symptom attenuation, in situ hybridization was performed on the first systemic leaf of infected plants at 7 dpi with a CP-specific probe (6). Since DI RNAs do not contain sequence elements homologous to the CP-coding region, this probe detects only the accumulation of genomic RNA. The majority of the investigated TBSV-P-infected leaves showed uniform high-level accumulation of virus RNA throughout the whole tissue, indicating the establishment of a successful systemic infection (Fig. 1C). In contrast, all the investigated TBSV-P+DI-infected leaves displayed marked reduction in the extent of virus infection. Typical representatives of these samples showed viral RNA accumulation confined to the veins and neighboring mesophyll cells (Fig. 1E). This phenomenon is surprisingly similar to the previously described virus accumulation pattern in plants infected with p19-defective CymRSV (Cym19stop) (6) and T19stop (Fig. 1D).
Accumulation of virus-derived products at the cellular level in the presence of DI RNAs.
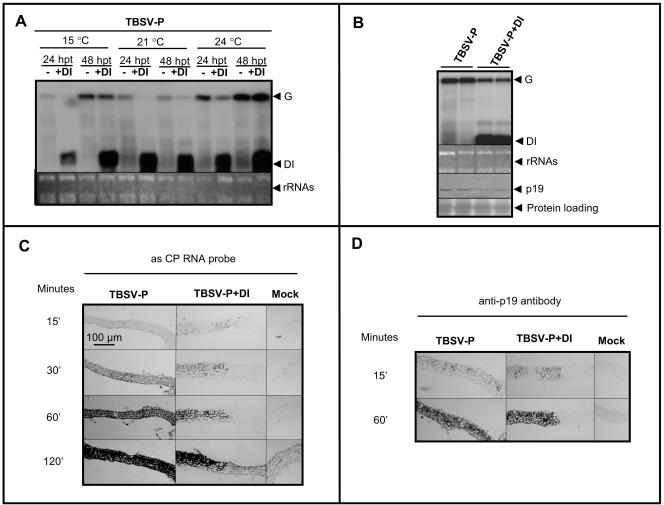
It has been previously demonstrated that the presence of DI RNAs interferes with the accumulation of genomic RNA at the single-cell level (2, 10, 14). To assess the effect of DI RNAs on helper virus accumulation in our system, transfection of mesophyll protoplasts was carried out with TBSV-P and DI-5 RNA transcripts as described previously (11). Since PTGS has been demonstrated to be temperature sensitive (25), the transfected protoplasts were incubated at different temperatures (15, 21, and 24°C). Results demonstrated that the presence of equimolar amounts of DI and genomic RNA in the inoculum resulted in reduced accumulation of genomic RNA at 24 h posttransfection (hpt). However, by 48 hpt, this suppression was less pronounced, allowing increased accumulation of genomic RNA (Fig. 2A). This phenomenon was observed at every investigated temperature. Our findings support previous observations that demonstrated that the presence of DI RNAs reduced the rate of TBSV genomic RNA synthesis (10). Moreover, we also showed that, in infected protoplasts at 24°C, p19 protein accumulated approximately proportionally to the level of genomic RNA at 24 hpt (Fig. 2B).
FIG. 2.
Accumulation of viral RNA and p19 in TBSV-P- and TBSV-P+DI-transfected protoplasts and infected tissues. (A) Northern analyses of helper virus accumulation in transfected protoplasts at different temperatures at 24 and 48 hpt. (B) Western blot analyses of p19 accumulation in transfected protoplasts in two independent samples at 24°C. (C) In situ hybridization of leaf cross sections detecting the accumulation of plus-sense viral RNA at 7 dpi. An antisense (as) CP RNA probe detects the genomic RNAs but not DI RNAs. The color reactions of nearly consecutive sections of TBSV-P-, TBSV-P+DI-, and mock-inoculated tissues were halted at different time points before reaching the saturation level. The bar applies to all images in panels C and D. (D) Immunohistochemistry applying an anti-p19 antibody to nearly consecutive sections of TBSV-P-, TBSV-P+DI-, and mock-inoculated tissues. The color reactions were stopped at the times indicated.
To reveal the level of helper virus accumulation in TBSV-P+DI-infected tissue relative to TBSV-P infection, systemically infected leaves were analyzed by in situ hybridization at 7 dpi. Nearly consecutive sections of TBSV-P- and TBSV-P+DI-infected leaves were applied to microscopic slides and hybridized with a probe detecting the plus strand of the CP-coding region. To visualize the accumulation of virus-derived products in TBSV-P+DI-infected plants, a tissue section where the mutant virus infected a high number of cells was chosen. To avoid the misinterpretation of the intensity of hybridization signals, the color reaction was stopped at different time points (15, 30, 60, and 120 min) before reaching the saturation level. TBSV-P- and TBSV-P+DI-infected cells displayed similar signal intensities at each time point, indicating that these cells accommodate comparable amounts of virus RNA (Fig. 2C).
Our finding that the presence of DI RNAs induces a phenotype characteristic of p19-defective virus infection prompted us to also investigate the accumulation of p19. Immunohistochemical staining of nearly consecutive paraffin sections was carried out to detect the accumulation of p19 in systemically infected leaves at 7 dpi. To assess the accumulation of p19 in infected cells, the color reaction was stopped before reaching the saturation point. Detection of p19 by immunohistochemistry revealed that, at the level of sensitivity of this technology, p19 accumulated at similar levels in TBSV-P+DI- and TBSV-P-infected cells (Fig. 2D).
Data from protoplast transfection studies indicate that at early stages of infection the presence of DI RNAs slows down the rate of genomic RNA synthesis, probably due to competition for trans-acting factors necessary for replication. However, at later time points, the infected cells have the capacity to accommodate a high level of virus-derived products. Altogether, our results demonstrate that DI RNA-mediated interference at the cellular level cannot satisfactorily explain the spatial restriction of the helper virus in infected plants and development of the recovery phenotype.
The presence of DI RNAs enhances the generation of virus-specific siRNAs, leading to accumulation of p19-unbound siRNAs.
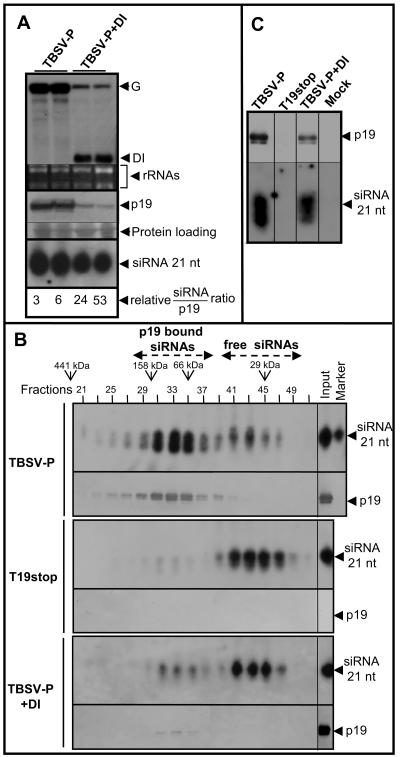
The presence of DI RNAs in TBSV-P-infected plants results in the development of a recovery-like phenotype characteristic of p19-defective virus (T19stop) infection even though the p19 silencing suppressor was present in the infected cells. Since the recovery phenotype induced by p19-defective tombusvirus was shown to be the consequence of activated PTGS (23), the connection between DI RNA-mediated interference and PTGS was further investigated. To clarify the mechanism of DI RNA-mediated symptom attenuation, the accumulation of virus-specific siRNAs, which are the hallmark of PTGS, was analyzed along with the accumulation of p19. Since p19-mediated silencing suppression is based on siRNA sequestering and was shown to be dose dependent (12, 21), the siRNA/p19 ratio was also investigated. To avoid sampling mistakes and to keep samples comparable, all systemically infected leaves from TBSV-P- and TBSV-P+DI-infected plants at 7 dpi were collected, homogenized, and then divided into two parts for either RNA or protein analyses (8). Northern blot analyses revealed that DI RNAs accumulated to a high level and dramatically suppressed helper virus accumulation relative to the DI RNA-free TBSV-P accumulation (Fig. 3A). We also assessed the accumulation of all virus-specific siRNAs in the same RNA samples by using a genomic-RNA-specific probe. This experiment revealed that, in DI RNA-containing samples, the level of virus-specific siRNAs, including DI RNA-derived siRNAs, increases to a level similar to that for TBSV-P infection (Fig. 3A). p19, however, accumulated approximately proportionally with the viral genome in these samples. The relative siRNA/p19 ratios in several independent experiments increased at least sevenfold when TBSV-P infections included DI RNA. These data demonstrate that the presence of DI RNAs significantly enriches the virus-specific siRNA content relative to the accumulation of helper virus genomic RNA and the corresponding accumulation of p19. This observation predicts that a portion of virus-specific siRNAs exist in free form, similar to what is found for Cym19stop-infected plants (12).
FIG. 3.
Accumulation of virus-specific siRNAs and p19 in the systemically infected leaves at 7 dpi. (A, top) Northern blot analysis of the accumulation of genomic and DI RNAs in TBSV-P- and TBSV-P+DI-infected plants. (Middle) Accumulation of p19 in the corresponding samples. (Bottom) siRNA accumulation in the corresponding samples, as shown with a genomic-RNA-specific probe. The calculated siRNA/p19 ratios are indicated at the bottom. (B) Fractionation of crude extracts prepared from TBSV-P-, T19stop-, and TBSV-P+DI-infected plants with a Superdex-200 gel filtration column. Collected fractions were tested for the presence of virus-specific siRNAs and p19 by Northern and Western blot analyses. A γ-ATP-labeled 21-nt synthetic RNA oligonucleotide was used as a size marker. (C) Extracts prepared from leaves of TBSV-P-, T19stop-, TBSV-P+DI-, and mock-inoculated plants were immunoprecipitated with an anti-p19 antibody. Immunoprecipitates were analyzed for the accumulation of p19 and virus-specific siRNAs by Western and Northern blot analyses.
To test whether the presence of elevated levels of siRNAs results in the accumulation of free siRNAs, crude extracts were prepared from TBSV-P-, TBSV-P+DI-, and T19stop-infected plants. These extracts were loaded onto a gel filtration column, and the fractions were analyzed for the presence of either siRNAs or p19, as described previously (12). As expected, the majority of siRNAs and p19 cofractionated in samples originating from TBSV-P-infected plants, indicating that siRNAs were bound by p19 (Fig. 3B). Only very low levels of free siRNAs could be detected in samples representing low-molecular-weight fractions. In contrast, the lack of p19 in T19stop infections resulted in the accumulation of free siRNAs. In the presence of DI RNA, we detected p19-bound siRNAs; however, the majority of siRNAs accumulated in free form in samples of low-molecular-weight fractions. The low level of p19 accumulating in the presence of DI RNAs is the result of the reduced accumulation of genomic RNA due to the spatially restricted spread of helper virus.
We next investigated whether DI RNAs are able to interfere with the ability of p19 to bind siRNAs. The p19 suppressor was immunoprecipitated from extracts of TBSV-P-, T19stop-, and TBSV-P+DI-infected plants. The immunoprecipitated samples were divided into two parts and used for the analyses of either siRNA or p19 accumulation. The results demonstrated that the presence of DI RNAs did not interfere significantly with the binding ability of p19 because the relative amounts of siRNAs versus p19 in samples obtained from TBSV-P+DI- and TBSV-P-infected plants were similar (Fig. 3C). As expected no immunoprecipitated siRNAs and p19 were detected in samples from T19stop-infected plants.
In conclusion, we demonstrated that the increased accumulation of siRNAs relative to the level of p19 in TBSV-P+DI infections saturates p19 with siRNAs, which leads to the accumulation of high levels of virus-specific free siRNAs (Fig. 3B).
DI RNAs are not able to modulate virus infection at low temperature, where RNA silencing is impaired.
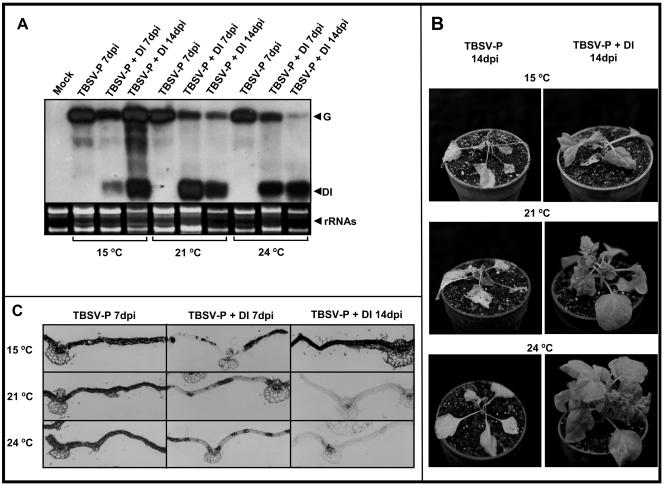
A previous study reported that low temperature (15°C) strongly inhibits PTGS compared to the standard temperature (24°C) (25). Our hypothesis predicts that, at 15°C, where PTGS is very weak, the presence of DI RNAs should not interfere with the virus infection process significantly. To test this hypothesis, TBSV-P- and TBSV-P+DI-infected plants were grown at 15, 21, and 24°C. Samples were taken from systemically infected leaves showing symptoms at 7 and 14 dpi and used for Northern blot analyses and in situ hybridization. Northern blot analyses revealed that samples from TBSV-P+DI infections at 7 dpi contained reduced levels of helper genomic RNA at all temperatures, although some variation was observed (Fig. 4A). However, by 14 dpi, the differences in accumulation of helper virus increased dramatically. Helper virus accumulation was close to DI RNA-free-infection levels at 15°C, was strongly reduced at 21°C, and was barely detectable at 24°C. DI RNAs accumulated to high levels at every investigated time point and temperature (Fig. 4A). The TBSV-P-infected plants developed general necrosis at every investigated temperature, as expected (Fig. 4B). However, TBSV-P+DI-infected plants displayed dramatically different symptoms depending on the applied temperature. At 21 and 24°C the TBSV-P+DI-infected plants developed recovery-like symptoms. The development of a recovery phenotype was most pronounced at 24°C. In contrast, TBSV-P+DI-infected plants incubated at 15°C developed severe necrotic symptoms, which in most cases culminated in the death of the plants (8 out of 10), indicating that the DI RNA-mediated defense system was not operating. In situ hybridization revealed that genomic RNA accumulation levels correlated with the extent of virus infections (Fig. 4C). Most importantly, at 15°C the virus was able to colonize the entire tissue even in the presence of DI RNA, similar to helper virus alone. This phenomenon is very similar to that described for heat sensitivity of p19-defective-CymRSV infection (25). The above results further confirm the pivotal role of PTGS in DI RNA-mediated symptom modulation.
FIG. 4.
Effect of temperature on DI RNA-containing-TBSV-P infection. (A) Northern blot analysis of TBSV-P- and TBSV-P+DI-infected plants. Samples were taken at 7 and 14 dpi at 15, 21, and 24°C as indicated. Mock, mock-inoculated tissue. (B) Symptoms induced by TBSV-P and TBSV-P+DI infections at 15, 21, and 24°C at 14 dpi. (C) TBSV-P genomic RNA accumulation in systemically infected leaves at 7 and 14 dpi. Sections were hybridized with a CP ORF-specific RNA probe. Samples and the applied temperatures are indicated.
DISCUSSION
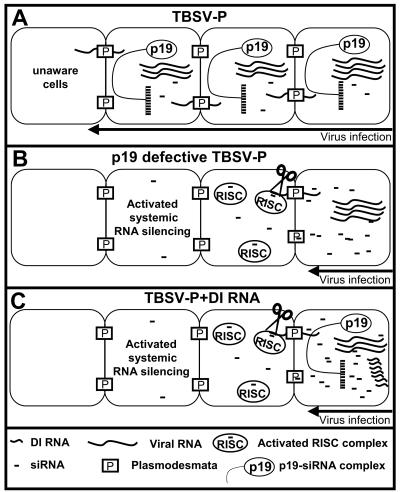
PTGS was recently integrated into the model for the evolution and symptom attenuation effects of tombusvirus DI RNAs (23). According to this current model, DI RNAs are generated from the viral genome by replicase errors and are selected for by the presence of cis-acting elements required for replication and by the absence of sequences that are targeted by PTGS. In this paper we refine and extend our knowledge about the interaction between the plant and virus during infection. We suggest a model in which the presence of DI RNAs slows down the replication of the helper virus genome and generation of an excess of siRNAs, which saturates the binding capacity of the available p19 suppressor proteins. This in turn leads to accumulation of siRNAs not bound by p19, allowing the activation of RISC ahead of the virus infection front (Fig. 5). The activated RISC prevents the further spread of the virus by destroying the entering viral RNA before the establishment of virus replication. DI RNAs are poorly targeted by PTGS (23), which ensures an additional advantage in the competition between helper virus and DI RNAs for the replicase. The race between the DI RNA-generated mobile siRNAs and viral genomic RNA gradually leads to a block of virus spread in the infected tissue and eventually results in the development of a PTGS-associated recovery phenotype similar to that associated with p19-defective-virus infection.
FIG. 5.
Proposed model for the PTGS-based mechanism of DI RNA-mediated interference. (A) In wild-type virus infections, the viral genomic RNA accumulates to high levels in infected mesophyll cells and the PTGS machinery of the host produces virus-specific siRNAs. However, at the same time, the virus translates p19, which physically binds the generated siRNAs, inhibiting their spread out of the infected cell. Consequently the cells ahead the infection front remained unprotected against the spreading virus. (B) Cells infected with p19-defective virus accommodate the same level of virus genomic RNA as cells infected with wild-type virus because RNA silencing has no capacity to cope with the invasive accumulation of virus-derived products. However, the absence of p19 results in the accumulation of free virus-specific siRNAs, which are able to traffic through plasmodesmata to cells beyond the infection front. These virus-specific mobile siRNAs are incorporated into the RISC of cells ahead of the infection front. Thus the already-activated RISCs destroy the entering viral RNA before the establishment of virus replication. (C) In the presence of DI RNAs, the infected cells accumulate significant amounts of genomic RNA and p19. However, because DI RNAs are poor targets of PTGS, they accumulate to extremely high levels and provide a source for the generation of extra amounts of virus-specific siRNAs, saturating the p19-binding capacity. The remaining free siRNAs are able to induce a similar process, which has been described for p19-defective-virus infection. Other previously described DI RNA-associated factors, such as suppressed rate of accumulation of genomic RNA and reduced p19 accumulation in early stages of virus infection, may further enhance the efficiency of PTGS-based defense by increasing the amount of free siRNA. The synergistic mode of action of different factors results in restricted spread of the virus and development of attenuated symptoms.
Our results provide the experimental support for the model described above. In situ analysis revealed that, while DI RNA-free TBSV-P invades throughout systemically infected leaves, the presence of DI RNAs in the virus infection results in the spatially restricted accumulation of helper virus in and around the veins. This phenomenon is surprisingly similar to that described for p19-defective-virus infection (6) (Fig. 1, compare panels D and E), suggesting that PTGS is involved in DI RNA-mediated host defense. In addition we showed that, at low temperature, where PTGS is disabled (25), the presence of DI RNAs could neither substantially inhibit the accumulation of helper virus nor protect the infected plants from systemic necrosis. In contrast, at higher temperatures, where PTGS is more active, the presence of DI RNAs induced a dramatic recovery phenotype, with little virus accumulation in the recovered tissue (data not shown). These phenomena are similar to that described for the temperature dependency of Cym19stop infection (25) and also to those associated with T19stop infection (data not shown). These data demonstrate that the presence of active PTGS machinery is indispensable for DI RNA-mediated interference.
Several studies demonstrated that DI RNAs reduce the accumulation of helper virus by competing for trans-acting factors necessary for replication. However, our observation showed that the presence of DI RNAs slows down helper virus accumulation and spread only when PTGS is disabled by low temperature (Fig. 4C, compare infections with DI RNA-containing virus at 7 and 14 dpi at 15°C). Therefore, competition for the replication apparatus cannot be directly responsible for the dramatic restriction in helper virus accumulation observed at higher temperatures. Our results demonstrate that DI RNAs exert their control over virus infection by blocking the spread of the helper virus utilizing the power of PTGS-based host defense.
The similarity between infections with DI RNA-containing TBSV-P and T19stop suggests that DI RNAs can interfere with the accumulation or activity of p19. Since it was previously demonstrated that p19 specifically binds the majority of PTGS-generated 21-nt siRNAs (21, 26, 33) in virus-infected plants (12), we investigated the siRNA/p19 ratio in infections with DI RNA-containing virus. We showed that the presence of DI RNAs dramatically elevates the siRNA accumulation relative to the amount of p19. More importantly, enhanced accumulation of siRNAs results in the saturation of p19 and the appearance of free siRNAs, similar to Cym19stop infection. Recent findings showed that the presence of 21-nt siRNAs correlated with the establishment of PTGS signaling, and it has been proposed that 21-nt siRNAs act as PTGS signaling molecules for short distances (9). Moreover, it has also been suggested that infection with p19-defective tombusvirus results in the spread of free 21-nt siRNAs beyond the infection front, providing sequence specificity for the RISC (6, 12).
A previous study reported that DI RNAs selectively reduce the accumulation of p19 via suppression of sg RNAs (20). We were not able to detect obvious selective inhibition of p19 accumulation in our system (8). However, early time points of virus infection were not investigated. The ability of DI RNAs to reduce the relative accumulation of p19 at early stages of viral replication would support our model since it could further increase the accumulation of free siRNAs, resulting in more-effective activation of PTGS. Moreover, the ability of DI RNAs to outcompete helper virus during replication generates high-level siRNA accumulation and decreased p19 accumulation at early stages of virus replication, which can create a window of opportunity for the host to outcompete the virus by utilizing the generated free siRNAs. The DI RNA-mediated host defense is greatly influenced by the synergistic dialog between PTGS and replication.
The presence of another subviral replicon, satellite RNA of turnip crinkle virus (TCV), enhanced viral pathogenesis, which was correlated with reduced virion accumulation and abundance of free CP (34). Since the CP of TCV is a PTGS suppressor, it was suggested that the accumulation of free CP may augment PTGS suppression. These results suggest that interference with PTGS could be a common mechanism utilized by different subviral replicons.
Acknowledgments
We thank Anne Simon and Alan Herr for critical reading and helpful comments on the manuscript. We thank László Szabó for quantitation of RNA and protein accumulation levels by Analysis, version 2.0 (Soft-Imaging Software GmbH).
This research was supported by grants from the Hungarian Scientific Research Fund (OTKA; T038313) and the RIBOREG EU project (LSHG-CT-2003503022). Z.H. is a recipient of a Bolyai Janos Fellowship.
REFERENCES
- 1.Burgyan, J., C. Hornyik, G. Szittya, D. Silhavy, and G. Bisztray. 2000. The ORF1 products of tombusviruses play a crucial role in lethal necrosis of virus-infected plants. J. Virol. 74:10873-10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, Y. C., M. Borja, H. B. Scholthof, A. O. Jackson, and T. J. Morris. 1995. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology 210:41-53. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 5.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 6.Havelda, Z., C. Hornyik, A. Crescenzi, and J. Burgyan. 2003. In situ characterization of Cymbidium Ringspot Tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 77:6082-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havelda, Z., and A. J. Maule. 2000. Complex spatial responses to cucumber mosaic virus infection in susceptible Cucurbita pepo cotyledons. Plant Cell 12:1975-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havelda, Z., G. Szittya, and J. Burgyan. 1998. Characterization of the molecular mechanism of defective interfering RNA-mediated symptom attenuation in tombusvirus-infected plants. J. Virol. 72:6251-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Himber, C., P. Dunoyer, G. Moissiard, C. Ritzenthaler, and O. Voinnet. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, R. W., A. O. Jackson, and T. J. Morris. 1990. Defective-interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts. Virology 176:539-545. [DOI] [PubMed] [Google Scholar]
- 11.Kollar, A., and J. Burgyan. 1994. Evidence that ORF 1 and 2 are the only virus-encoded replicase genes of cymbidium ringspot tombusvirus. Virology 201:169-172. [DOI] [PubMed] [Google Scholar]
- 12.Lakatos, L., G. Szittya, D. Silhavy, and J. Burgyan. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 14.Qiu, W., J. W. Park, A. O. Jackson, and H. B. Scholthof. 2001. Retention of a small replicase gene segment in tomato bushy stunt virus defective RNAs inhibits their helper-mediated trans-accumulation. Virology 281:51-60. [DOI] [PubMed] [Google Scholar]
- 15.Qiu, W., J. W. Park, and H. B. Scholthof. 2002. Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol. Plant-Microbe Interact. 15:269-280. [DOI] [PubMed] [Google Scholar]
- 16.Qu, F., and T. J. Morris. 2002. Efficient infection of Nicotiana benthamiana by tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol. Plant-Microbe Interact. 15:193-202. [DOI] [PubMed] [Google Scholar]
- 17.Roux, L., A. E. Simon, and J. J. Holland. 1991. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv. Virus Res. 40:181-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of Tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 19.Scholthof, H. B., K. B. Scholthof, and A. O. Jackson. 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7:1157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholthof, K. B., H. B. Scholthof, and A. O. Jackson. 1995. The effect of defective interfering RNAs on the accumulation of tomato bushy stunt virus proteins and implications for disease attenuation. Virology 211:324-328. [DOI] [PubMed] [Google Scholar]
- 21.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon, A. E., M. J. Roossinck, and Z. Havelda. 2004. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu. Rev. Phytopathol. 42:415-437. [DOI] [PubMed] [Google Scholar]
- 23.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szittya, G., P. Salamon, and J. Burgyan. 2000. The complete nucleotide sequence and synthesis of infectious RNA of genomic and defective interfering RNAs of TBSV-P. Virus Res. 69 :131-136. [DOI] [PubMed] [Google Scholar]
- 25.Szittya, G., D. Silhavy, A. Molnar, Z. Havelda, A. Lovas, L. Lakatos, Z. Banfalvi, and J. Burgyan. 2003. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. T. Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 27.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 28.Voinnet, O. 2002. RNA silencing: small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol. 5:444-451. [DOI] [PubMed] [Google Scholar]
- 29.Voinnet, O., and D. C. Baulcombe. 1997. Systemic signalling in gene silencing. Nature 389:553. [DOI] [PubMed] [Google Scholar]
- 30.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]
- 32.White, K. A. 1997. Formation and evolution of Tombusvirus defective interfering RNAs. Semin. Virol. 7:409-416. [Google Scholar]
- 33.Ye, K., L. Malinina, and D. J. Patel. 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, F., and A. E. Simon. 2003. Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology 312:8-13. [DOI] [PubMed] [Google Scholar]