Abstract
The morphogenesis of rotaviruses follows a unique pathway in which immature double-layered particles (DLPs) assembled in the cytoplasm bud across the membrane of the endoplasmic reticulum (ER), acquiring during this process a transient lipid membrane which is modified with the ER resident viral glycoproteins NSP4 and VP7; these enveloped particles also contain VP4. As the particles move towards the interior of the ER cisternae, the transient lipid membrane and the nonstructural protein NSP4 are lost, while the virus surface proteins VP4 and VP7 rearrange to form the outermost virus protein layer, yielding mature infectious triple-layered particles (TLPs). In this work, we have characterized the role of NSP4 and VP7 in rotavirus morphogenesis by silencing the expression of both glycoproteins through RNA interference. Silencing the expression of either NSP4 or VP7 reduced the yield of viral progeny by 75 to 80%, although the underlying mechanism of this reduction was different in each case. Blocking the synthesis of NSP4 affected the intracellular accumulation and the cellular distribution of several viral proteins, and little or no virus particles (neither DLPs nor TLPs) were assembled. VP7 silencing, in contrast, did not affect the expression or distribution of other viral proteins, but in its absence, enveloped particles accumulated within the lumen of the ER, and no mature infectious virus was produced. Altogether, these results indicate that during a viral infection, NSP4 serves as a receptor for DLPs on the ER membrane and drives the budding of these particles into the ER lumen, while VP7 is required for removing the lipid envelope during the final step of virus morphogenesis.
Rotaviruses are nonenveloped icosahedral viruses whose capsid is formed by three concentric layers of protein. The innermost layer is formed by 60 dimers of VP2 that surrounds the viral genome composed of 11 segments of double-stranded RNA and 12 copies of each VP1, the virus polymerase, and VP3, the virus capping enzyme. The second layer of protein is formed by 280 trimers of VP6, which sits on top of VP2 to form double-layered particles (DLPs). Finally, the addition of 280 trimers of glycoprotein VP7 which constitute the outermost layer of the virus and 60 dimeric spikes of the VP4 protein to DLPs form triple-layered particles (TLPs) that represent the mature infectious virus (13).
Rotavirus morphogenesis occurs by an unusual process where DLPs, which are thought to assemble in cytoplasmic inclusions termed viroplasms, bud across the membrane of the endoplasmic reticulum (ER). During this process, the DLPs acquire a transient lipid envelope which is subsequently lost to yield the mature infectious TLPs (33). The ER membrane through which DLPs bud is modified by two viral proteins, the virion surface protein VP7 and the nonstructural polypeptide NSP4 (5, 7, 21). NSP4 has a large cytosolic domain that interacts with DLPs, and it has been proposed that this interaction drives the translocation of the double-layered particles into the lumen of the ER (3, 35). NSP4 is also known to associate with the lumen-oriented VP7 and with VP4, forming a heterotrimeric complex that is thought to participate in the budding of DLPs into the ER (22).
During the last step of rotavirus morphogenesis, the transient lipid envelope is lost and the viral proteins are rearranged so that VP4 and VP7 assemble to form the outer layer of the virus, while NSP4 is excluded from the virus particle. The mechanisms underlying the loss of the lipid envelope and the viral protein NSP4 as well as the correct assembly of the outer-layer proteins are not understood, although it has been suggested that the membrane-destabilizing activities of NSP4 and/or VP4 and the relatively high calcium concentration present in the lumen of the ER could be implicated in the removal of the lipid envelope (10, 30, 36). The produced mature virus is then released either by cell lysis in MA104 cells (13) or after transport along an atypical trafficking pathway from the ER to the plasma membrane in polarized epithelial Caco-2 cells (18). It has recently been shown that the expression of rotavirus genes can be efficiently silenced by RNA interference using small interfering RNAs (siRNAs) that have a sequence complementary to the viral gene to be silenced (1, 9, 32). siRNA has proven to be a very useful tool to dissect the function of the viral genes in the context of a natural infection (1, 20). Silencing the expression of the rhesus rotavirus VP4 gene showed that in the absence of VP4, the DLPs were still able to bud into the ER and to lose the lipid envelope, yielding TLPs that lack VP4 spikes. These results indicated that VP4 is required neither for the translocation of DLPs into the ER nor for the removal of the lipid membrane from the transiently enveloped particles. In this work, we evaluated the role that the viral glycoproteins VP7 and NSP4 play in the morphogenesis of rotavirus. We found that silencing the NSP4 gene affected the accumulation and the cellular distribution of several viral proteins and that little or no virus was assembled in its absence. VP7 silencing, in contrast, did not affect the expression or the distribution of other viral proteins, but when it was not present, enveloped particles accumulated within the lumen of the ER, and very little, if any, mature infectious virus was produced.
MATERIALS AND METHODS
Cells and viruses.
The rhesus monkey epithelial cell line MA104 was grown in Eagle's minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and was used for all experiments carried out in this work. Rhesus rotavirus (RRV) was obtained from H. B. Greenberg (Stanford University, Stanford, Calif.), simian rotavirus SA11 was obtained from H. H. Malherbe, and porcine rotavirus YM was isolated in our laboratory. All rotavirus strains were propagated in MA104 cells as described previously (26).
Antibodies.
Monoclonal antibodies (MAbs) to VP2 (3A8), VP6 (255/60), VP7 (60), and NSP4 (B4) were kindly provided by H. B. Greenberg. MAb HS2 directed to VP4 (25), the rabbit anti-rotavirus polyclonal serum raised against purified RRV TLPs, and the rabbit anti-vimentin sera were produced in our laboratory. Rabbit polyclonal sera to NSP2, NSP3, and NSP5 have been described previously (16). Alexa-488- and Alexa-568-conjugated secondary antibodies and phalloidin-Alexa-546 were purchased from Molecular Probes (Eugene, Oreg.). Horseradish peroxidase-conjugated goat anti-rabbit polyclonal antibody was from Perkin-Elmer Life Sciences (Boston, Mass.), and horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin G and rabbit anti-tubulin antibody were from Zymed (San Francisco, Calif.).
siRNA transfection.
The siRNAs were obtained from Dharmacon Research (Lafayette, Colo.) as annealed duplexes. The sequence and relative positions of the siRNAs (shown in Table 1) were chosen based on the rules proposed by Elbashir et al. (12). As irrelevant controls, siRNAs with sequences from the caveolin-1, lamin A/C, or green fluorescent protein gene were used during this work. None of these siRNAs affected the viral protein synthesis or viral infectivity, and no differences were found between them; thus, they are generically referred to as control siRNAs (Table 1). Transfection of siRNAs was carried out in nearly confluent cell monolayers by using 2 μl of Lipofectamine (Invitrogen) per 100 μl of siRNAs at 600 pmol/ml in MEM without antibiotics. The transfection mixture was added to cells previously washed with MEM and incubated for 8 h at 37°C. After this time, the transfection mixture was removed and the cells were washed with MEM and kept in this medium for 48 h at 37°C before virus infection.
TABLE 1.
Sequences and nucleotide positions of the siRNAs for RRV VP7 and NSP4
| siRNA | Nucleotide positions | Sense sequence | Relative inhibitionb |
|---|---|---|---|
| VP7-1 | 317-337 | AAU UCG UGG AAG GAU ACA CUC | + |
| VP7-2 | 390-410 | AAU ACA CGG AUA UUG CUU CCU | + |
| VP7-3c | 681-701 | AAG UCG CUA CAG CUG AAA AAC | +++ |
| VP7-4 | 751-771 | AAC UGC UAC UUG CAC UAU CAG | − |
| VP7-5 | 217-237 | AAU GGA CAC UGC AUA CGC UAA | ++ |
| NSP4-1c | 183-203 | AAG GCC UCG GUU CCA ACC AUG | ++ |
| NSP4-2 | 352-372 | AAG AAA UGA GAC GUC AGC UGG | + |
| Controla | |||
| Lamin A/C | AAC UGG ACU UCC AGA AGA ACA | − | |
| Caveolin-1 | AAG CCC AAC AAC AAG GCC AUG | − | |
| GFP | AAC UUA CCC UGA AGU UCA UCU | − |
The sequences of the irrelevant controls used througouht this work are listed (they were purchased from Dharmacon). GFP, green fluorescent protein.
Relative inhibition of the production of viral progeny compared to cells transfected with an irrelevant siRNA. +++, >70%; ++, 50%; +, <25%; −, no effect.
The most effective siRNAs for silencing the expression of these genes were named siRNANSP4 and siRNAVP7 throughout the text.
Infection of cells and titration of viral progeny.
Transfected cell monolayers in 24- or 48-well plates were infected with 3 virus focus-forming units per cell and then incubated for 24 h at 37°C. At this time, the cells were then lysed by two freeze-thaw cycles, and the lysates were treated with 10 μg of trypsin/ml for 30 min at 37°C. The infectious titer of the viral preparations was obtained by an immunoperoxidase focus assay (26). Briefly, confluent MA104 cells in 96-well plates were washed twice with phosphate-buffered saline (PBS), and serial dilutions (n-fold) of the above-mentioned viral lysate were adsorbed to the cells for 60 min at 37°C. After the adsorption period, the virus inoculum was removed, the cells were washed once with PBS, MEM was added, and the infection was left to proceed for 14 h at 37°C for RRV. RRV-infected cells were detected by an immunoperoxidase focus detection assay using a rabbit hyperimmune serum to rotavirus as described previously (26). The focus-forming units were counted with the help of a Visiolab 1000 station (Biocom, France), as has been previously reported (17).
Immunoblots.
Cells were transfected with siRNAs and infected with rotavirus RRV as described above. Twelve hours postinfection (hpi), the cells were lysed with Laemmli sample buffer, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes (Millipore, Bedford, Mass.). Membranes were blocked with 5% nonfat dried milk in PBS and incubated at 4°C with the indicated primary antibodies in PBS containing 0.1% milk followed by an incubation with secondary, species-specific, horseradish peroxidase-conjugated antibodies. The peroxidase activity was developed by the Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences) according to the manufacturer's instructions.
Immunofluorescence.
MA104 cells grown on glass coverslips were transfected as mentioned above, and 48 h posttransfection, the cells were infected with rotavirus RRV at a multiplicity of infection (MOI) of 3. Eight hours postinfection, the cells were fixed with 2% paraformaldehyde in PBS for 20 min at 37°C. After this time, the cells were washed twice with PBS containing 50 mM NH4Cl and were permeabilized by incubation with PBS-0.5% Triton X-100-50 mM NH4Cl for 15 min at room temperature and washed twice with PBS with gentle swirling. The coverslips were then incubated for 1 h at room temperature with primary antibodies diluted in blocking buffer (50 mM NH4Cl and 1% bovine serum albumin in PBS) and then rinsed four times with PBS. The coverslips were then incubated with the appropriate Alexa-labeled secondary antibodies in blocking buffer for 1 h at room temperature. The cells were washed four times with PBS and mounted onto glass slides with Fluoprep (BioMérieux) and the antifading agent DABCO (100 mg/ml; Sigma, St. Louis, Mo.). The slides were analyzed with a Bio-Rad MRC-600 confocal microscope and CoMOS MPL-1000 software or with a Nikon E600 epifluorescence microscope coupled to a DXM1200 digital still camera (Nikon). The images were then digitally captured and prepared with Adobe Photoshop software version 7.0.
Radiolabeling, isolation, and analysis of viral particles.
Cells grown in 48-well plates were transfected with siRNAs and infected with rotavirus RRV as described above. At 4 hpi, the medium was replaced by MEM without methionine, supplemented with 50 μCi of Easy Tag EXPRESS-35S labeling mix (Dupont, NEN)/ml, and incubated for 8 h; after this period, the cells were washed and lysed with Laemmli sample buffer. To prepare purified viral particles, cells grown in 75-cm2 flasks were transfected and infected as described previously and were harvested until complete cytopathic effect was attained; the virus in the cell lysates was pelleted by centrifugation for 1 h at 25,000 rpm at 4°C with an SW28 rotor (Beckman). The virus pellet was resuspended in TNC buffer (10 mM Tris-HCl [pH 7.5], 140 mM NaCl, 10 mM CaCl2), sonicated once for 20 s, and extracted with Genetron (trichloro-monofluoro-ethane). CsCl was then added to obtain a density of 1.36 g/cm3, the mixture was centrifuged for 18 h at 36,000 rpm with an SW50.1 rotor, and the virus bands were collected by puncture. The CsCl in the bands was removed by desalting-centrifugation on Sephadex G-25, and the protein composition of each band was analyzed by SDS-PAGE and silver staining.
Electron microscopy.
Cells grown in 75-cm2 flasks were transfected with siRNAs and infected with rotavirus RRV at an MOI of 3 as described above. Eight hours postinfection, the cells were fixed in 2.5% glutaraldehyde-0.1 M cacodylate (pH 7.2), postfixed with 1% osmium tetroxide, and embedded in Epon 812 resin. The ultrathin sections obtained were stained with 2% uranyl acetate-1% lead citrate (Reynolds mix) (31). The grids were examined with a Zeiss EM-900 electron microscope at 80 kV.
RESULTS
siRNAs directed to rotavirus NSP4 and VP7 genes inhibit the synthesis of the encoded glycoproteins.
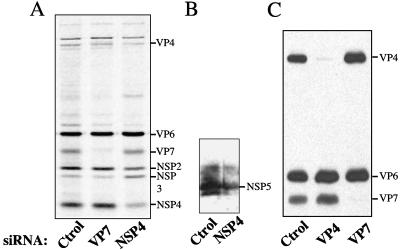
To evaluate the effect of siRNAs on the synthesis of viral glycoproteins NSP4 and VP7, siRNAs corresponding to the sequences of the NSP4 and VP7 genes of RRV were transfected into MA104 cells. Forty-eight hours after transfection, the cells were infected with rotavirus RRV, and 4 hpi, the proteins were metabolically labeled with EXPRESS-35S labeling mix for 8 h and analyzed by gel electrophoresis to evaluate the synthesis of viral proteins. Initially, five and two different siRNAs were tested for VP7 and NSP4, respectively. Their sequences and their relative effect on the specific inhibition of protein synthesis are shown in Table 1. The most effective siRNAs for silencing the expression of these genes were named siRNANSP4 and siRNAVP7 (Table 1) and were used throughout this work. When cells were infected with rotavirus RRV in the presence of siRNANSP4, the synthesis of NSP4 was clearly diminished to about 20% of that of the control (Fig. 1A). Unexpectedly, in the presence of this siRNA, the accumulation of other viral proteins was altered. Scanning analysis from three different experiments indicated that the level of some proteins consistently decreased to 80% (VP2), 75% (NSP2), 70% (VP7), 60% (VP4), and 50% (NSP5) compared to the levels found in control cells (Fig. 1A and B, lanes NSP4), while the production of NSP3 was consistently enhanced about twofold (170%) (Fig. 1A, lane NSP4). The level of VP1 and VP6 was not significantly altered.
FIG. 1.
Effect of siRNAVP7 and siRNANSP4 on the synthesis of viral proteins. MA104 cells in 48-well plates were transfected with the indicated siRNA, and after 8 h, the Lipofectamine-siRNA mixture was removed and replaced by MEM. Forty-eight hours posttransfection, the cells were infected with rotavirus RRV at an MOI of 3, and 12 h postinfection, the cells were harvested and processed for SDS-PAGE. (A) Fluorography of cells transfected with the indicated siRNAs in which the cells were labeled with 50 μCi of Easy Tag EXPRESS-35S/ml at 4 hpi and harvested 12 hpi. (B) A lysate of cells transfected with the indicated siRNAs was harvested at 12 hpi and subjected to SDS-PAGE. The proteins were transferred to nitrocellulose membrane and immunostained with a rabbit polyclonal antibody to rotavirus NSP5. (C) Immunoblot analysis of lysates of cells transfected with the indicated siRNAs and harvested at 12 hpi. The proteins were incubated with a rabbit anti-rotavirus polyclonal antiserum. The relative positions of the viral proteins are indicated in each gel. Ctrol, control.
In contrast with the previous findings, in the presence of siRNAVP7, the synthesis of VP7 was barely detectable, whereas the production of the other viral proteins was not modified (Fig. 1A, lane VP7). This observation was confirmed when the viral proteins synthesized in the presence of siRNAVP7 were analyzed by immunoblot using a polyclonal serum to the structural proteins of the virus (Fig. 1C). As an additional control in this assay, an siRNA to VP4, which has been previously shown to efficiently inhibit the synthesis of this protein (9), was included.
Silencing the expression of NSP4 and VP7 reduces the yield of rotavirus progeny.
The effect of inhibiting the synthesis of either NSP4 or VP7 on the generation of infectious rotavirus progeny was evaluated. Either siRNANSP4 or siRNAVP7 was transfected into MA104 cells; 48 h after transfection, the cells were infected with rotavirus RRV, and 24 h later, the progeny virus produced under these conditions was recovered and its titer was determined. Transfection with either siRNANSP4 or siRNAVP7 decreased the yield of viral progeny by about 75%, while an irrelevant siRNA used as a control did not affect the production of virus (Table 2). The inhibition of RRV progeny by the siRNA to NSP4 was shown to be specific, since siRNANSP4 did not affect the replication of the porcine rotavirus strain YM, whose NSP4 gene differs in the siRNA target site by 4 nucleotides with respect to the RRV sequence. Likewise, siRNAVP7 did not block the production of simian rotavirus SA11 infectious particles, whose target sequence in the VP7 gene differs by 3 positions from that of RRV (Table 2).
TABLE 2.
Viral progeny recovered from cells transfected with the indicated siRNA
| siRNA/virus | % Infectivitya |
|---|---|
| siRNActrolb | 97.2 (±6.2) |
| siRNANSP4/RRV | 17.8 (±5.1) |
| siRNANSP4/YM | 92.7 (±4.9) |
| siRNAVP7/RRV | 20.1 (±5.5) |
| siRNAVP7/SA11 | 108 (±19.0) |
Data are expressed as the percentage of the infectivity obtained when the cells were mock transfected. The standard deviations of at least three independent experiments are shown in parentheses.
siRNActrol, control.
The reduced synthesis of NSP4 alters the distribution of other viral proteins.
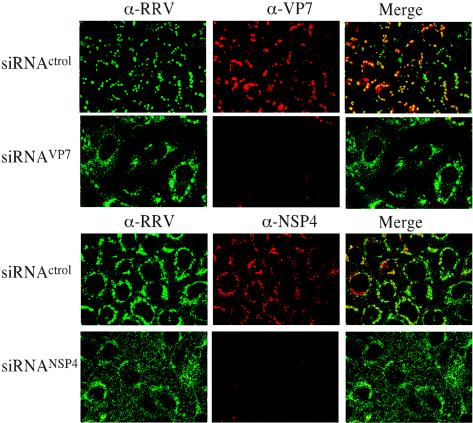
To evaluate the synthesis of NSP4 and VP7 in individual cells, monolayers of MA104 cells were lipofected with either siRNANSP4 or siRNAVP7 or with an irrelevant siRNA as a negative control. The cells were then infected with RRV, and 8 hpi, the cells were fixed and immunostained with a mixture of MAb B4 (to NSP4) and a rabbit hyperimmune serum to purified RRV TLPs when the cells were transfected with siRNANSP4 or with MAb 60 (to VP7) and the rabbit hyperimmune serum to RRV particles when transfected with siRNAVP7. The hyperimmune serum was used to detect the infected cells. In the cell monolayers transfected with siRNAVP7, about 80% of the infected cells (as judged by their reactivity with the anti-RRV serum) had either very low or undetectable levels of VP7. Similarly, transfection with siRNANSP4 caused a large number of infected cells to have barely detectable levels of NSP4 (Fig. 2).
FIG. 2.
Effect of siRNAs to NSP4 and VP7 on the expression of the corresponding protein in infected cells. MA104 cells transfected with the indicated siRNAs were infected 48 h posttransfection, and at 8 hpi, the cells were fixed and processed for immunofluorescence. The expression levels of VP7 and NSP4 were monitored by using MAbs 60 (α-VP7) and B4 (α-NSP4), respectively, and the cell infection by RRV was monitored by using a rabbit polyclonal antiserum to rotavirus (α-RRV) followed by anti-mouse Alexa-568 and anti-rabbit Alexa-488 antibodies, respectively.
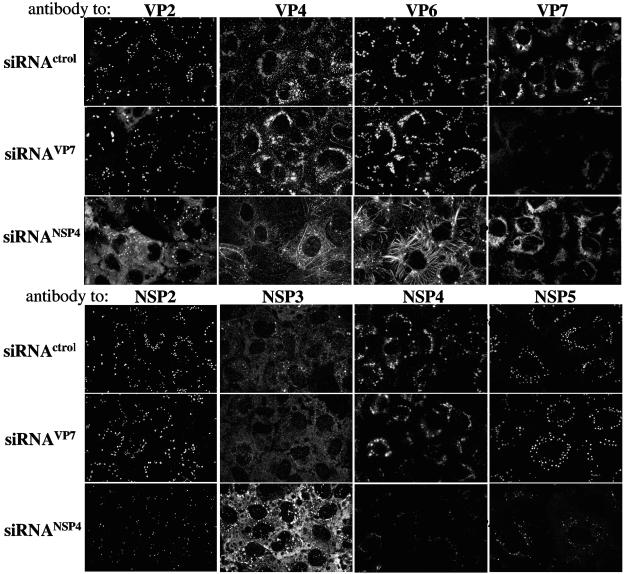
Of interest, when the polyclonal serum to RRV was used to stain the virus-infected, siRNANSP4-transfected cells, a striking filamentous pattern which was not present in cells transfected with either siRNAVP7 or the irrelevant siRNA was observed (Fig. 2). To find out which viral protein(s) was responsible for this filamentous signal, a detailed analysis of the distribution of the individual proteins was carried out by using MAbs to VP2, VP4, VP6, VP7, and NSP4 and monospecific polyclonal sera to NSP2, NSP3, and NSP5. This analysis showed that the intracellular distribution of several viral proteins was altered in siRNANSP4-transfected cells. VP6 was found to extensively redistribute, forming filaments that appear to extend to the periphery of the cell instead of being localized to viroplasms, as was observed in cells transfected with siRNAVP7 or with the siRNA control (Fig. 3). These filaments are not the result of an association of VP6 with tubulin, actin, or vimentin, since the signal of none of these proteins, as detected with specific antibodies or with phalloidin in the case of actin, colocalized with VP6 (data not shown).
FIG. 3.
Cellular distribution of viral proteins in cells transfected with siRNAVP7 or siRNANSP4. MA104 cells grown on coverslips were transfected with the indicated siRNAs and infected 48 h posttransfection. At 8 hpi, the cells were fixed and processed for immunofluorescence. The distribution of VP2, VP4, VP6, VP7, and NSP4 was monitored by using MAbs 3A8, HS2, 255/60, 60, and B4, respectively. NSP2, NSP3, and NSP5 were stained by using rabbit monospecific antibodies. Anti-mouse Alexa-568 and anti-rabbit Alexa-488 were used as secondary antibodies. siRNActrol, control.
VP2 was found to distribute homogeneously in the cytoplasm rather than in viroplasms (Fig. 3). The mixed pattern of VP4 that has been previously reported (15, 24), which consists of perinuclear ring-like or semicircular structures closely associated to viroplasms (15) and a filamentous array due to its association with microtubules (24), was observed in cells transfected with the control siRNA and siRNAVP7; however, in the presence of siRNANSP4, most of the perinuclear structures disappeared while the filamentous signal of VP4 remained and was even more evident than that of control cells (Fig. 3). In cells transfected with siRNANSP4, VP7 was no longer detected in semicircular structures around viroplasms (similar to VP4) (15); rather, it showed a more diffuse although still perinuclear pattern of distribution, probably due to its homogeneous distribution along the ER membrane.
As far as the nonstructural proteins are concerned, in NSP4-silenced cells, NSP5 and NSP2 were still restricted to viroplasms, but these structures were significantly smaller and probably fewer than those found in control cells. On the other hand, the distribution pattern of NSP3 did not change in siRNANSP4-transfected cells compared to control cells or cells transfected with siRNAVP7, but the signal of this protein was significantly more intense, in agreement with the increased level of NSP3 observed in cells with reduced amounts of NSP4 (Fig. 1A).
In contrast to the findings described above, in the presence of siRNAVP7, the level and intracellular distribution of the viral proteins (other than VP7), as detected by immunofluorescence, were not evidently affected (Fig. 3), in agreement with the analysis of viral proteins by gel electrophoresis and immunoblot (Fig. 1A and C).
Reduced levels of NSP4 decrease the formation of both DLPs and TLPs, whereas reduced levels of VP7 prevent the formation of TLPs.
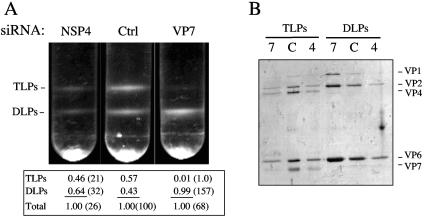
To evaluate the effect of silencing NSP4 on virus particle assembly, cells lipofected with either a control siRNA or siRNANSP4 were infected with RRV, and the virus particles produced were purified by CsCl density gradients 24 hpi. The viral particles obtained from siRNA control-transfected cells yielded, as regularly observed, two bands (Fig. 4A). By using gel electrophoresis, these two bands were shown to correspond to DLPs and TLPs (Fig. 4B), since the heavier band was formed by particles containing VP1, VP2, and VP6, while the lighter band contained, in addition to the DLP proteins, the virus surface proteins VP4 and VP7. Both TLPs and DLPs were also obtained from cells infected in the presence of siRNANSP4; however, the yield of viral particles (both DLPs and TLPs) was consistently about four- to fivefold lower than that obtained in control cells (Fig. 4).
FIG. 4.
Virus particles synthesized in the presence of siRNAVP7 or siRNANSP4. (A) Isopycnic CsCl gradients of viral particles assembled in the presence of the indicated siRNAs. The density of each band in the CsCl gradients was determined by scanning digital photographs of the gradients. The numbers below the gradients represent the relative amount of each band with respect to the total amount of viral particles detected in each gradient. The numbers in parentheses indicate the percentage of each band with respect to the amount of the corresponding band found in the control gradient (Ctrl). (B) Gel electrophoresis analysis of the viral particles present in the bands detected in the isopycnic gradients shown in panel A. The same proportion of each collected band was loaded onto the gel, which was silver stained. 4, C, and 7 correspond to the gradients labeled NSP4, Ctrl, and VP7, respectively. The migration of the viral structural proteins in the gels is indicated.
On the other hand, density gradient analysis of cells infected with rotavirus RRV in the presence of siRNAVP7 showed that although the yield of total particles assembled (TLPs plus DLPs) was similar (about 70 to 80%) to that of particles assembled in control cells, DLPs represented more than 99% of these particles, while TLPs were not, or very poorly, assembled (Fig. 4). These results indicate that, as expected, in the absence of or with reduced levels of VP7, TLPs cannot be produced, while the assembly of DLPs was not perturbed.
The reduced synthesis of NSP4 and VP7 blocks rotavirus morphogenesis at different stages.
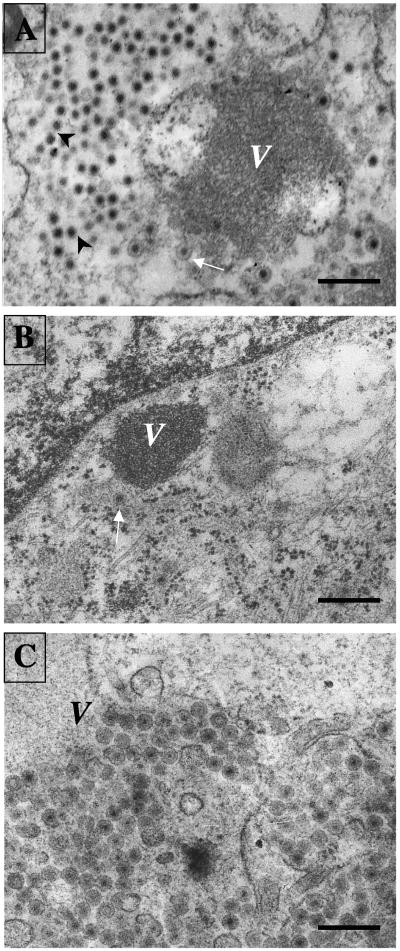
The type of virus particles assembled in the presence of siRNANSP4 or siRNAVP7 was examined by transmission electron microscopy (TEM). In siRNA control-transfected cells, typical electrodense viroplasm structures were evident in the cytoplasm in close apposition to the ER membrane (Fig. 5A). DLPs in the periphery of viroplasms were shown to bud into the lumen of the ER, leading to the formation of membrane-enveloped particles, while many of the viral particles observed within the ER had lost their lipid envelope. In contrast with these findings, in most siRNANSP4-treated cells, only small, compact viroplasms were found, with few or no viral particles around them (Fig. 5B). Analysis of the EM images showed that while only 17% (6 out of 35 cells counted) of the cells transfected with the control siRNA had less than 50 viral particles per cell, 62% (25 out of 40 cells) of the siRNANSP4-transfected cells had less than 50 viral particles; in fact, 35% (14 out of 40) of the cells had no viral particles at all. These observations are in agreement with the small viroplasms detected by immunofluorescence and with the low yield of virions detected by density gradients. Altogether, these data suggest that with reduced levels of NSP4, viral particles are not or are very inefficiently formed.
FIG. 5.
Morphogenesis of RRV rotavirus particles in MA104 cells transfected with siRNAs. MA104 cells transfected with (A) the siRNA control (siRNActrol), (B) siRNANSP4, or (C) siRNAVP7 were infected 48 h posttransfection, and at 8 hpi, the cells were fixed and prepared for electron microscopy as indicated in Materials and Methods. Dense viroplasmic inclusions (V) are present in the cytoplasm of rotavirus-infected cells, adjacent to the ER. From these structures, DLPs bud into the lumen of the ER, resulting in membrane-enveloped particles (arrows) which later lose the membrane to produce mature triple-layered virions (arrowheads). The pictures shown are representative of at least 20 different virus-infected cells. Magnification, ×15,000. Scale bars, 400 nm.
To find out if NSP4 was able to promote the budding of DLPs through the membrane of the ER in the absence of VP7, siRNAVP7-transfected cells were infected with RRV and analyzed by TEM at 8 hpi. Interestingly, in cells that were infected in the presence of siRNAVP7, most of the particles observed within the ER lumen appeared to have a lipid coat (Fig. 5C). This type of particle is very similar to those observed in cells treated with the drugs tunicamycin and thapsigargin, which cause the transiently enveloped rotavirus particles to accumulate into the lumen of the ER, preventing the maturation of the viral particles into infectious virions (2, 23, 28, 30; M. Zayas et al., unpublished data). In the presence of siRNAVP7, the nonenveloped particles that are usually observed in a regular infection (Fig. 5A) were not observed. Given the transfection efficiency of about 75%, at least 20 siRNAVP7-transfected cells were analyzed to confirm the phenotype described. Altogether, these results suggest that in the absence of VP7, DLPs are assembled in viroplasms and bud into the lumen of the ER, most probably driven by NSP4, but that once inside the ER, the lipid envelope is not removed, suggesting that VP7 is critical for this maturation step of the virus.
DISCUSSION
In this work, we have shown that the expression of rotavirus genes that encode NSP4 and VP7 is efficiently silenced by RNA interference. In cells transfected with either siRNANSP4 or siRNAVP7, the yield of viral progeny was reduced by 75 to 80%, a reduction that is in the range of the efficiency of transfection which was estimated to be achieved with siRNAs in MA104 cells (9). These results suggest that the synthesis of the viral glycoproteins is essentially abolished in the cells that incorporate the corresponding siRNA. This conclusion is supported by immunofluorescence microscopy analysis, since about 75% of cells transfected with siRNAs showed very poor or no synthesis of the silenced protein. Even though the silencing of either glycoprotein resulted in a marked decrease in the production of progeny virus, the underlying mechanism for the reduction of viral yield was different in each case.
Silencing NSP4.
Given the role that NSP4 is thought to play as the receptor for DLPs in the ER membrane during the budding of these particles into the lumen of the ER, it was expected that in its absence, DLPs would accumulate in the cytosol while TLPs would not form. Surprisingly, when the NSP4 gene was silenced, little or no viral particles (neither DLPs nor TLPs) were assembled, and the amount and distribution of several viral proteins were altered. Thus, in cells containing reduced amounts of NSP4, the levels of VP2, VP4, VP7, NSP2, and NSP5 were decreased by 20 to 50%, depending on the protein, while NSP3 was increased about twofold. It has been previously reported that NSP4 alters the intracellular concentration of calcium when expressed in insect cells (37) or when added exogenously to cells (4). In addition, it is known that during a virus infection, the cytosolic concentration of this cation increases (27). Although it has not been formally proven that NSP4 is responsible for the observed calcium increase during a viral infection, it is conceivable that the cytosolic calcium concentrations in infected cells are lower in the absence of this protein, and this in turn could alter the accumulation level of some viral proteins by modifying either their synthesis or degradation rates. It would be interesting to test if the translation rate or stability of the viral mRNAs can be differentially regulated by cytoplasmic calcium levels. The modification in the cytosolic accumulation of some viral proteins correlated with the presence of smaller and fewer viroplasms in siRNANSP4-transfected cells compared to control cells, which may in turn cause a decrement of de novo assembled, transcriptionally active, double-layered replicative intermediate particles. This would prevent the second wave of transcription and translation that is thought to occur during the virus infectious cycle, with the consequent reduction in the final assembly of both DLPs and TLPs (1).
An alternative explanation for the low amount of DLPs and TLPs assembled in the absence of NSP4 is based on the interesting observation that no defined viral particles, or at least DLPs, can be distinguished within viroplasms which are usually observed as amorphous, electrodense structures under the electron microscope. Well-defined particles are apparent only at the periphery of viroplasms during the process of budding into the lumen of the ER (Fig. 5) (23, 28, 30). This observation suggests that NSP4 could have a role in facilitating the assembly of the VP6 protein layer on top of core particles. This hypothesis is supported by the observation that DLPs incubated with soluble NSP4 aggregate and change their appearance by TEM (34). These results were suggested to represent a temporary conformational change of DLPs induced by NSP4 that could facilitate the correct assembly of the outer capsid proteins (34). If NSP4 does indeed have a role in promoting the assembly of VP6, the synthesis of DLPs in the absence of NSP4 would be affected, with the consequent reduction of the secondary transcription and translation of viral proteins as described above. However, it is interesting that even though the formation of viral particles in the cells transfected with the siRNA to NSP4 was reduced by up to 75% compared to that of control cells, the cytosolic accumulation of the viral proteins did not decreased accordingly. This observation suggests that either the half-life of the mRNAs synthesized by the entering virus particle is long enough to direct the synthesis of viral proteins up to 12 h postinfection (time at which the cells were harvested for protein analysis) (Fig. 1A) or the viral particle responsible for the second round of mRNA synthesis is not the DLP per se, as has been suggested previously (13), but rather a subviral replication intermediate particle that contains little or no VP6.
In addition to the modification of the level of synthesis of several viral proteins, the silencing of NSP4 also caused a marked intracellular redistribution of some of these proteins. The relocalization of VP7 in the ER membrane from discrete sites around viroplasms (15) to a more diffuse pattern (Fig. 3) is most likely the direct result of the absence of NSP4 since VP7 (an ER lumen-oriented protein) has been shown to interact with NSP4 (22), and this protein, which has domains located at both sides of the ER membrane (33), is the only link of VP7 with the rest of the viral proteins in the cytoplasm. The large decrease in the signal of VP4 around viroplasms, as well as the relocalization of VP2 to a more homogenous distribution in the cytosol, and of VP6 from viroplasms to fibers could also be the direct result of the absence of NSP4, since the cytoplasmic carboxy-terminal domain of this protein is known to interact with VP4 and VP6 (and VP6 in turn interacts with VP2). Alternatively, the reduced size and number of viroplasms in NSP4-silenced cells might be the cause of VP2 and VP6 not being present in these structures. This could also explain the redistribution of VP4, since this protein has been described to be closely associated to viroplasms (15). The filaments formed by VP6 in the absence of NSP4 are not the result of its interaction with tubulin microtubules, actin filaments, or vimentin intermediate filaments, since no colocalization was observed by immunofluorescence microscopy (data not shown). Apparently, the VP6 filamentous structures do not result from the self-polymerization of the protein since they are substantially longer and thicker than the VP6 tubes that have been described previously (14, 19). In addition, these VP6 fibers could not be observed by TEM. Thus, it seems that VP6 probably associates with some type of cellular fibers that have not yet been identified. Since a similar redistribution of viral proteins was observed when gene 11 of RRV, which encodes NSP5 and NSP6 proteins, was silenced (López et al., unpublished), it is not possible to determine whether the altered distribution of the viral proteins is the direct effect of the absence of NSP4 or the indirect effect of the decreased synthesis of NSP5 and NSP2 as a result of the silencing of NSP4.
Silencing VP7.
When the cells were transfected with siRNAVP7, the amount of viral proteins synthesized and their intracellular distribution were not affected, and the viral cycle proceeded apparently undisturbed until the morphogenetic process was arrested within the lumen of the ER, where enveloped particles that failed to produce mature infectious virus accumulated. Similar results were obtained by Silvestri et al., who found that silencing the expression of the VP7 gene of rotavirus strain DXRRV led to the accumulation of DLPs and the inhibition of the production of infectious virus (32). The lack of formation of TLPs in the absence of VP7 was expected since this protein forms the outermost protein layer of the virus. However, it was surprising to find that the lipid envelope was not lost from the usually transient enveloped particles in the absence of VP7. VP4 and NSP4 have both been demonstrated to have membrane-destabilizing activities (6, 10, 36), and they have both been suggested as candidates for removing the lipid envelope acquired during the budding of DLPs into the lumen of the ER. However, the fact that the lipid envelope is retained in the absence of VP7, even when NSP4 is present (Fig. 5C), strongly suggests that VP7 is essential for this maturation step of rotavirus morphogenesis. The central role of VP7 in the morphogenetic process of the virus is supported by the fact that the lipid envelope was removed in the absence of VP4 and spikeless TLPs were assembled in which the structure of the VP7 surface layer was indistinguishable by cryoelectron microscopy from the VP7 layer present in complete infectious virus (B. V. V. Prasad et al., unpublished data).
It has been previously reported that VP7 is arranged on the surface of the virion as trimers (8, 38), and more recently, it was more recently shown that its trimerization is dependent on the concentration of calcium (11). It is tempting to propose that soon after the budding of DLPs into the lumen of the ER, VP7, which is embedded in the envelope surrounding the intermediary particles, assembles in a calcium-dependent process into a tight protein layer that could exclude the lipid envelope from the virion as well as NSP4 and other nonstructural proteins that have been shown to be present in small amounts in these intermediary particles (29), yielding the mature infectious virus. The role of VP7 in removing the lipid membrane from the enveloped particles is supported by the fact that virus morphogenesis is also arrested at the enveloped stage of the particles, when the cells are treated with thapsigargin (23, 28) or the calcium ionophore A23187 (30), which decreases the concentration of calcium in the ER, or with tunicamycin that blocks the glycosylation of VP7 and NSP4 (2, 30). As mentioned above, it cannot be excluded, however, that the glycosylated form of NSP4 or an N-glycosylated cellular protein participates in this step, since an SA11 rotavirus variant (clone 28) that has a nonglycosylated VP7 is also arrested at the enveloped intermediate stage when treated with tunicamycin (28).
It is also important that in the absence of VP7 (this work) as well as in the absence of VP4 (9), viral DLPs are still able to bud into the lumen of the ER, indicating that NSP4, and not the heterotrimer VP4/VP7/NSP4, is sufficient to serve as a receptor for DLPs on the ER membrane and to drive the budding of these particles into the ER lumen during the late steps of virus morphogenesis.
Based on the data described in this work, together with previous observations, we propose the following working model for the final stages of rotavirus morphogenesis. DLPs assemble at the periphery of viroplasms and bud across the membrane of the ER with the help of NSP4. During the process of budding, VP4 (that interacts with the cytosolic domain of NSP4), NSP4, and VP7 are incorporated into the intermediary enveloped particles. Once the particles are inside the ER, the high-calcium environment of the lumen triggers the lateral interaction of VP7 molecules (located on the outside of the lipid envelope) promoting the surface protein layer to tighten and making it interact with the VP6 layer, excluding the lipids and NSP4 during this process. This model does not rule out, however, the possibility that NSP4 or cellular proteins might be needed in this process. The exclusion of lipids and NSP4 from the intermediary particles by an as-yet-undetermined mechanism would leave the VP4 protein in place, yielding the mature infectious virus. This model should serve as a framework to design experiments to elucidate the complex morphogenetic process of rotaviruses.
Acknowledgments
We are grateful to Rafaela Espinosa and Pedro Romero for their excellent technical assistance with the cell cultures and virus purification, respectively, and to Xóchitl Alvarado for her help with the confocal microscope.
This work was partially supported by grants 55000573 and 55000613 from the Howard Hughes Medical Institute and G37621N from Conacyt Mexico.
REFERENCES
- 1.Arias, C. F., M. A. Dector, L. Segovia, T. Lopez, M. Camacho, P. Isa, R. Espinosa, and S. Lopez. 2004. RNA silencing of rotavirus gene expression. Virus Res. 102:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, C. F., S. Lopez, and R. T. Espejo. 1982. Gene protein products of SA11 simian rotavirus genome. J. Virol. 41:42-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Au, K. S., W. K. Chan, J. W. Burns, and M. K. Estes. 1989. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 63:4553-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, J. M., P. Tian, C. Q. Zeng, A. P. Morris, and M. K. Estes. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101-104. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann, C. C., D. Maass, M. S. Poruchynsky, P. H. Atkinson, and A. R. Bellamy. 1989. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. EMBO J. 8:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne, E. P., A. R. Bellamy, and J. A. Taylor. 2000. Membrane-destabilizing activity of rotavirus NSP4 is mediated by a membrane-proximal amphipathic domain. J. Gen. Virol. 81:1955-1959. [DOI] [PubMed] [Google Scholar]
- 7.Chan, W. K., K. S. Au, and M. K. Estes. 1988. Topography of the simian rotavirus nonstructural glycoprotein (NS28) in the endoplasmic reticulum membrane. Virology 164:435-442. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, S. E., S. K. Mukherjee, M. K. Estes, J. A. Lawton, A. L. Shaw, R. F. Ramig, and B. V. V. Prasad. 2001. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 75:6052-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dector, M. A., P. Romero, S. Lopez, and C. F. Arias. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 3:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denisova, E., W. Dowling, R. LaMonica, R. Shaw, S. Scarlata, F. Ruggeri, and E. R. Mackow. 1999. Rotavirus capsid protein VP5* permeabilizes membranes. J. Virol. 73:3147-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2000. Purified recombinant rotavirus VP7 forms soluble, calcium-dependent trimers. Virology 277:420-428. [DOI] [PubMed] [Google Scholar]
- 12.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199-213. [DOI] [PubMed] [Google Scholar]
- 13.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1786. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 14.Estes, M. K., S. E. Crawford, M. E. Penaranda, B. L. Petrie, J. W. Burns, W. K. Chan, B. Ericson, G. E. Smith, and M. D. Summers. 1987. Synthesis and immunogenicity of the rotavirus major capsid antigen using a baculovirus expression system. J. Virol. 61:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez, R. A., R. Espinosa, P. Romero, S. Lopez, and C. F. Arias. 2000. Relative localization of viroplasmic and endoplasmic reticulum-resident rotavirus proteins in infected cells. Arch. Virol. 145:1963-1973. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez, R. A., M. A. Torres-Vega, S. Lopez, and C. F. Arias. 1998. In vivo interactions among rotavirus nonstructural proteins. Arch. Virol. 143:981-996. [DOI] [PubMed] [Google Scholar]
- 17.Guerrero, C. A., S. Zarate, G. Corkidi, S. Lopez, and C. F. Arias. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74:9362-9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jourdan, N., M. Maurice, D. Delautier, A. M. Quero, A. L. Servin, and G. Trugnan. 1997. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J. Virol. 71:8268-8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lepault, J., I. Petitpas, I. Erk, J. Navaza, D. Bigot, M. Dona, P. Vachette, J. Cohen, and F. A. Rey. 2001. Structural polymorphism of the major capsid protein of rotavirus. EMBO J. 20:1498-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez, S., and C. F. Arias. 2004. Preface. Virus gene silencing by RNA interference. Virus Res. 102:1-2. [DOI] [PubMed] [Google Scholar]
- 21.Maass, D. R., and P. H. Atkinson. 1994. Retention by the endoplasmic reticulum of rotavirus VP7 is controlled by three adjacent amino-terminal residues. J. Virol. 68:366-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maass, D. R., and P. H. Atkinson. 1990. Rotavirus proteins VP7, NS28, and VP4 form oligomeric structures. J. Virol. 64:2632-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michelangeli, F., F. Liprandi, M. E. Chemello, M. Ciarlet, and M. C. Ruiz. 1995. Selective depletion of stored calcium by thapsigargin blocks rotavirus maturation but not the cytopathic effect. J. Virol. 69:3838-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nejmeddine, M., G. Trugnan, C. Sapin, E. Kohli, L. Svensson, S. Lopez, and J. Cohen. 2000. Rotavirus spike protein VP4 is present at the plasma membrane and is associated with microtubules in infected cells. J. Virol. 74:3313-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padilla-Noriega, L., R. Werner-Eckert, E. R. Mackow, M. Gorziglia, G. Larralde, K. Taniguchi, and H. B. Greenberg. 1993. Serologic analysis of human rotavirus serotypes P1A and P2 by using monoclonal antibodies. J. Clin. Microbiol. 31:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pando, V., P. Isa, C. F. Arias, and S. Lopez. 2002. Influence of calcium on the early steps of rotavirus infection. Virology 295:190-200. [DOI] [PubMed] [Google Scholar]
- 27.Perez, J. F., M. C. Ruiz, M. E. Chemello, and F. Michelangeli. 1999. Characterization of a membrane calcium pathway induced by rotavirus infection in cultured cells. J. Virol. 73:2481-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrie, B. L., M. K. Estes, and D. Y. Graham. 1983. Effects of tunicamycin on rotavirus morphogenesis and infectivity. J. Virol. 46:270-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poruchynsky, M. S., and P. H. Atkinson. 1991. Rotavirus protein rearrangements in purified membrane-enveloped intermediate particles. J. Virol. 65:4720-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poruchynsky, M. S., D. R. Maass, and P. H. Atkinson. 1991. Calcium depletion blocks the maturation of rotavirus by altering the oligomerization of virus-encoded proteins in the ER. J. Cell Biol. 114:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, J. A., and A. R. Bellamy. 2003. Interaction of the rotavirus nonstructural glycoprotein NSP4 with the viral and cellular components, p. 225-235. In U. Desselberger and J. Gray (ed.), Viral gastroenteritis. Elsevier Science, Amsterdam, The Netherlands.
- 34.Taylor, J. A., J. A. O'Brien, V. J. Lord, J. C. Meyer, and A. R. Bellamy. 1993. The RER-localized rotavirus intracellular receptor: a truncated purified soluble form is multivalent and binds virus particles. Virology 194:807-814. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, J. A., J. A. O'Brien, and M. Yeager. 1996. The cytoplasmic tail of NSP4, the endoplasmic reticulum-localized non-structural glycoprotein of rotavirus, contains distinct virus binding and coiled coil domains. EMBO J. 15:4469-4476. [PMC free article] [PubMed] [Google Scholar]
- 36.Tian, P., J. M. Ball, C. Q. Zeng, and M. K. Estes. 1996. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J. Virol. 70:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian, P., M. K. Estes, Y. Hu, J. M. Ball, C. Q.-Y. Zeng, and W. P. Schilling. 1995. The rotavirus nonstructural glycoprotein NSP4 mobilizes Ca2+ from the endoplasmic reticulum. J. Virol. 69:5763-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeager, M., K. A. Dryden, N. H. Olson, H. B. Greenberg, and T. S. Baker. 1990. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J. Cell Biol. 110:2133-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]