Abstract
We recently demonstrated that the cytomegalovirus (CMV) immediate-early 1 (IE1) protein induces transcription of the gene encoding the RelB NF-κB subunit. The mechanism of this activation has been explored here. We report that the induction of the relB promoter by IE1 protein is mediated via activation of JNK and AP-1. The region controlling relB promoter induction was mapped to the upstream ∼600-bp region between −1694 and −1096 bp. IE1 stimulated AP-1 activity in NIH 3T3 cells. Competition electrophoretic mobility shift assay (EMSA) confirmed the presence of one bona fide AP-1 element centered at −1503 bp. Introduction of a G-to-C mutation in the AP-1 binding site within the distal region of the relB promoter eliminated its activation by IE1 in both NIH 3T3 fibroblasts and vascular smooth muscle cells (SMCs). Supershift EMSA identified c-Jun, Fra-2, and c-Fos in AP-1 binding complexes in IE1 transfected NIH 3T3 cells. IE1 induced c-Jun phosphorylation, and treatment with SP600125, a selective JNK inhibitor, as well as overexpression of JNK-binding domain of JIP1, blocked IE1-mediated induction of AP-1 and relB promoter activity in NIH 3T3 cells and SMCs. Ectopic expression of c-Jun plus Fra-2, but not c-Fos, induced relB promoter activity. The relB promoter has two proximal NF-κB elements, and c-Jun/Fra-2 worked in synergy with p50/p65 NF-κB complexes. Overall, these findings demonstrate for the first time the role of AP-1 in transcriptional regulation of a gene encoding an NF-κB subunit, and its involvement in induction of RelB activity by the CMV IE1 protein.
Cytomegalovirus (CMV) is a betaherpesvirus that latently infects a majority of adults worldwide (24). Although most healthy immunocompetent individuals are asymptomatic (6), enhanced risk of atherosclerosis and restenosis after coronary angioplasty has been observed in patients with prior human CMV (HCMV) infection (23, 40). Infection by CMV has also been shown to stimulate migration and proliferation of smooth muscle cells (SMCs) in culture (38, 58, 60). HCMV has also been implicated in the transformation and pathogenesis of many malignancies (13, 19, 26, 46, 50, 53). Furthermore, increased morbidity and mortality is frequent in immunocompromised and immunosuppressed individuals, e.g., patients with AIDS or after transplantation (6). The CMV immediate-early protein 1 (IE1), a 491-amino-acid nuclear phosphoprotein, is one of two major products of the ie gene and is a predominant protein expressed during the immediate-early phase of CMV infection (22, 40). Although IE1 was first found to regulate viral gene expression by positive feedback regulation of the ie promoter (29), other functions have been reported recently, including the induction of NF-κB activity (14, 30, 47) and posttranscriptional activation of the c-Fos and c-Jun subunits of AP-1 (35).
NF-κB/Rel is a family of dimeric transcription factors whose DNA-binding domains have considerable homology with an approximately 300-amino-acid region of the v-Rel oncoprotein and was thus termed the Rel homology region (28, 64). NF-κB plays a pivotal role in the control of cell proliferation, survival, neoplastic transformation, and immune and inflammation responses (25, 27, 43, 56). Members of the mammalian Rel family include p65 (RelA), RelB, c-Rel, p50, and p52 (28). In most non-B cells, NF-κB factors are inactive and sequestered in the cytoplasm by direct interaction with specific inhibitor proteins, termed IκBs, which need to be phosphorylated and degraded in order for NF-κB to transfer into the nucleus (12, 64). IE1 has been found to induce NF-κB activity, as judged by NF-κB element driven constructs, in fibroblasts and Jurkat T cells (47). Previous work from our laboratory found that IE1 selectively induces nuclear RelB and p50 in SMCs and NIH 3T3 cells (30). The increase in RelB protein mediated by IE1 could be related to an increase in steady-state relB mRNA levels. Although the relB promoter contains two proximal NF-κB elements (11), IE1 was unable to activate an NF-κB element driven construct in relB−/− mouse embryo fibroblasts. Thus, the data suggested that relB was the primary NF-κB target gene of IE1 activity (30), although the mechanism controlling relB induction by IE1 was unclear. Interestingly, RelB/p50 complexes are somewhat unique among the NF-κB family in that IκB-α binds only poorly (39), allowing them to almost freely translocate to the nucleus (20). In the mouse, high levels of RelB expression are restricted to specific regions of the thymus and spleen (65), and RelB complexes represent constitutive NF-κB activity in these tissues (66), suggesting a role for RelB in control of gene expression in these tissues. Importantly, RelB has recently been shown to display specificity of NF-κB regulated gene transcription (30).
AP-1 is a transcription factor composed of members of the Jun and Fos families (2) that binds and activates transcription at TPA response elements (TRE), with the consensus sequence of 5′-TGACTCA-3′ (1). These proteins belong to the bZIP group of DNA-binding proteins, which associate to form a variety of homo- and heterodimers (2). Members of the Jun family include c-Jun, JunB, and JunD, and members of the Fos family include c-Fos, FosB, Fra-1, and Fra-2. The transcriptional activity of c-Jun is regulated by phosphorylation at Ser63 and Ser73 (9, 54). Extracellular signals, including growth factors, transforming oncoproteins, and UV irradiation, stimulate this phosphorylation of c-Jun, thereby activating c-Jun-dependent transcription (17, 35, 36). For example, the mitogen-activated protein kinase homologue JNK/SAPK binds to the N-terminal region of c-Jun and phosphorylates c-Jun at Ser63/73 (17, 36). Furthermore, Kim et al. (35) reported that HCMV IE1 protein induced AP-1 activity via a posttranscriptional, rather than transcriptional, mechanism. Here we have explored the mechanism of RelB induction by IE1. We report that IE1 activates JNK and thereby AP-1 activity, which leads to enhanced relB promoter transcription via binding to an upstream AP-1 element.
MATERIALS AND METHODS
Cell culture conditions.
NIH 3T3 fibroblasts, which were maintained as described previously (30) were passaged prior to reaching confluence. Bovine aortic SMCs were isolated by the explant method and maintained as described previously (7). The cell culture medium was changed every 2 days, and cultures were passaged every 3 to 4 days. SMCs were used by passage 6 from the primary explant. Where indicated, cells were treated with SP600125 (Calbiochem), a selective inhibitor of JNK (8), which was dissolved in dimethyl sulfoxide (DMSO).
Plasmids.
The pON2205 construct containing a full-length CMV IE1 protein cDNA driven by the simian virus 40 (SV40) early promoter was kindly provided by Edward Mocarski (Stanford University, Palo Alto, Calif.) (14). The human p1.7 relB promoter-Luc (−1694 to +1), p1.1 relB promoter-Luc (−1096 to + 1), and p0.6 relB promoter-Luc (−608 to +1) vectors containing the indicated lengths of human relB promoter sequences driving a luciferase reporter in pGL3 basic vector, were kindly provided by Carlos V. Paya (Mayo Clinic, Rochester, Minn.) (11). The AP-1 reporter, 4XTRE-Luc construct containing two copies of the oligonucleotide with two human collagenase TPA-responsive elements (TRE) driving a luciferase reporter, was kindly provided by Marcello Arsura (University of Tennessee College of Medicine, Memphis) (4, 44). The pCMV FJIP1 vector containing the JNK-binding domain of JIP1 tagged with FLAG was kindly provided by Roger Davis (University of Massachusetts Medical School, Worcester) (67). The SV40 β-galactosidase (β-Gal) reporter vector was as reported previously (3). The AP-1 family members constructs containing c-Jun, c-Fos, Fra-1, or Fra-2 in pCI expression vector were kindly provided by Dany Chalbos (INSERM, Montpellier, France) (62).
Transfection analysis.
Cultures of NIH 3T3 cells or SMCs at 50 to 70% confluence were transiently transfected by using FUGENE transfection reagent, according to the manufacturer's instructions (Boehringer Mannheim). After 24 or 48 h, cells were harvested, and cellular extracts were prepared for reporter assays as described previously (37). Cells were harvested 24 h after transfection for immunoblot analysis and sonicated in ice-cold radioimmunoprecipitation assay extraction buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% sodium lauryl sarcosine, 1% NP-40, 0.1% sodium dodecyl sulfate, 1 mM EDTA, 1 mM dithiothreitol plus 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, 10 mM para-nitrophenylphosphate, 4 mM sodium orthovanadate, 10 mM NaF, 10 mM β-glycerol phosphate). After centrifugation to remove cell debris, supernatants were collected and analyzed for protein content by using the Bio-Rad protein quantitation kit for detergent lysis according to the manufacturer's directions.
Immunoblot analysis.
Samples (30 μg) were separated by electrophoresis through a 10% polyacrylamide-sodium dodecyl sulfate gel, and transferred and processed as we have described previously (5). Rainbow markers (Amersham) were used to determine the molecular weight. The c-Jun and phospho-Ser63 c-Jun antibodies were purchased from Cell Signal. Rabbit anti-Fra-2 and rabbit anti-c-Fos antibodies were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-IE1 72 and β-actin (AC-15) antibodies were purchased from Vancouver Biotech, Ltd., and Sigma, respectively. Rabbit anti-Fra-1, rabbit anti-JunB, and rabbit anti-JunD antibodies were kindly provided by Judith A. Foster (Boston University School of Medicine, Boston, Mass.). Quantitation by scanning densitometry was performed by using KDS1D version 2.0 (Kodak).
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts were prepared as described previously (57). The sequences of the oligonucleotides containing the consensus AP-1, putative AP-1 site 1 [puta-AP-1(1)], and putative AP-1 site 2 [puta-AP-1(2)] from the human relB promoter were as follows: consensus AP-1, 5′-GATCCGCTTGATGACTCAGCCGGAAG-3′; puta-AP-1(1), 5′-GATCCTCTCACTCTGTCACCCAGG-3′; puta-AP-1(2), and 5′-GATCCTGCAATAGCGTCATCACAG-3′ (the underlined regions indicate the core binding element). For the binding reaction, 5 μg of nuclear extract was used in binding reactions, and DNA-protein complexes were separated as described previously (57). For competition gel shift assays, 10- or 30-fold excess unlabeled oligonucleotide was incubated with nuclear extracts for 30 min at room temperature prior to the addition of the probe. For supershift EMSA, antibodies for AP-1 family members were incubated with nuclear extracts for 20 h at 4°C prior to addition of the probe.
Site-directed mutagenesis.
The single base mutation in the puta-AP-1 (ATAGCGTCATC → ATAGCCTCATC) element in the AP-1 mutant p1.7 (Mut-p1.7) relB promoter-Luc was generated by site-directed mutagenesis of the p1.7 relB promoter-Luc by using the QuickChange site-directed mutagenesis kit (Stratagene) and verified by sequencing. To construct the κB-Mut or AP-1/κB-Mut-p1.7, the κB-Mut p1.1 construct, kindly provided by Carlos V. Paya, was digested with SacI and BglII, and the 1.1-kb fragment containing two mutant κB sites was isolated and cloned into the wild-type (WT) or AP-1 mutant p1.7, from which the corresponding fragment had been removed. The mutant p1.7 constructs were verified by sequencing.
RNA isolation and analysis.
Total cellular RNA was isolated by the Ultraspec-II RNA isolation kit (Biotecx Lab, Inc.) from transfected NIH 3T3 cultures in P-100 dishes, and samples (20 μg) subjected to Northern blot analysis, as described previously (48). The 2.1-kb EcoRI insert of the RelB expression vector was used as a probe. Quantitation by scanning densitometry was performed as described above.
RESULTS
relB promoter induction by IE1 is mediated by distal elements.
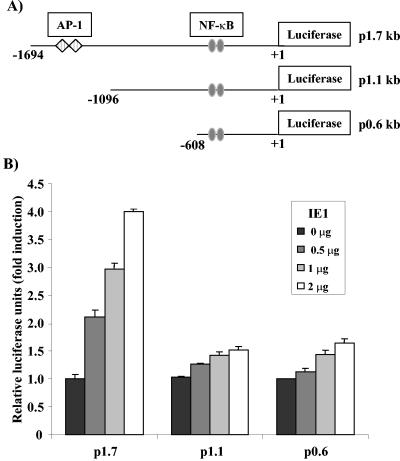
To determine whether IE1 induces the relB promoter and to map the required region(s), three human relB promoter reporter constructs—p1.7, p1.1, and p0.6—were used in cotransfection analysis (Fig. 1A). IE1 expression resulted in a dose-dependent increase in activity of the p1.7 relB promoter, with a 4.0-fold increase at the highest amount of IE1 (Fig. 1B). The shorter relB promoters were not induced to the same extents, with only a slight induction of about 1.5- to 1.7-fold at the highest dose of IE1. The basal level of activity of each of the three promoters was nearly identical (data not shown). In three separate experiments, average increases of 4.2 ± 0.5-, 1.5 ± 0.3-, and 1.8 ± 0.2-fold were observed at the highest dose for the p1.7, p1.1, and p0.6 relB promoter constructs, respectively. All of the vectors contained two NF-κB binding sites (Fig. 1A), suggesting that activation of the relB promoter by IE1 did not involve NF-κB, which is consistent with our previous findings (30). Since only the longest promoter can be induced by IE1, the functional element(s) appears to be within the ∼600-bp region between −1.7 and −1.1 kb.
FIG. 1.
The induction of the relB promoter by IE1 requires the distal region of the promoter. (A) Structures of the p1.7, p1.1, and p0.6 relB promoter luciferase reporter vectors. The positions of the NF-κB elements and putative AP-1 sites are indicated. (B) NIH 3T3 cells were transiently transfected, in triplicate, with the indicated dose of pON2205 vector expressing IE1 protein; 0.5 μg of either p1.7, p1.1, or p0.6 relB promoter-Luc vector; 0.5 μg of SV40 β-Gal expression vector; and pBluescript for a total of 3 μg of DNA. Values of luciferase normalized to β-Gal activity (Relative luciferase units) are presented ± the standard deviation. Untreated p1.7, p1.1, and p0.6 relB promoter-Luc activities were set at 1.
The 600-bp region contains putative AP-1 elements.
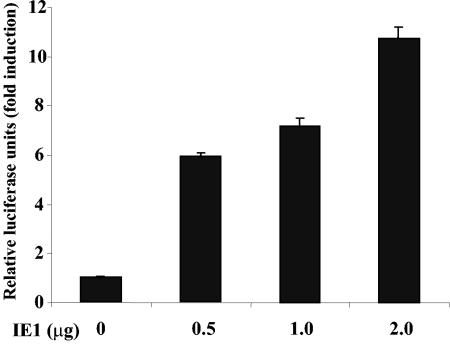
Since it was previously reported that c-Jun and c-Fos induction occurs after HCMV infection (10), we used Transfac software (http://transfac.gbf.de/homepage/databases/transfac/transfac.html) to analyze the 0.6-kb DNA sequence for AP-1 sites. Two putative sites were detected: −1532 bp (5′-ACTCTGTCACC-3′) and −1508 bp (5′-ATAGCGTCATC-3′), which were termed puta-AP-1(1) and puta-AP-1(2), respectively. To first test whether AP-1 was activated by IE1 in NIH 3T3 fibroblasts, cells were cotransfected with a four-copy AP-1 element construct 4XTRE-Luc and increasing amounts of IE1 expression vector. A dose-dependent increase in the luciferase reporter activity was observed with a 10.8-fold increase at the highest amount of transfected IE1 DNA (Fig. 2), with an average value of 11.2 ± 0.8-fold obtained in three separate experiments. Thus, AP-1 activity is induced by IE1.
FIG. 2.
IE1 induces activity of 4XTRE-Luc, four-copy AP-1 element-driven reporter construct, in NIH 3T3 cells. NIH 3T3 cells were transiently transfected, in triplicate, with the indicated dose of pON2205 vector expressing IE1 protein, 0.5 μg of 4XTRE-Luc vector, 0.5 μg of SV40 β-Gal expression vector, and pBluescript for a total of 3 μg of DNA. Relative luciferase units are presented ± the standard deviation. Activity of the control 4XTRE-Luc vector DNA was set at 1.
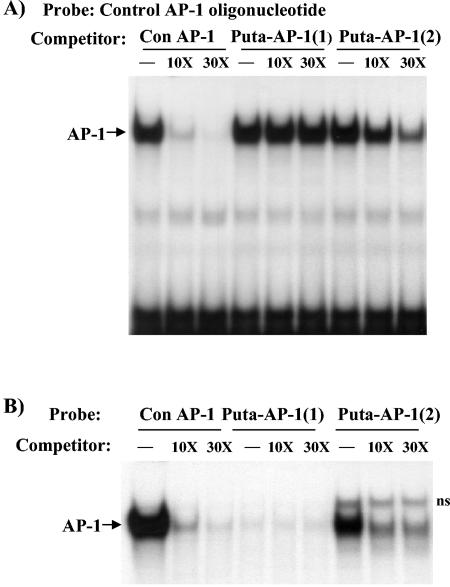
We next sought to determine whether the putative AP-1 sites in the distal 0.6-kb region are legitimate AP-1 sites. Competition EMSA was performed by using a consensus AP-1 site (core sequence TGACTCA) as radiolabeled probe and excess unlabeled oligonucleotides containing the puta-AP-1(1) and puta-AP-1(2) sites. The oligonucleotides containing the puta-AP-1(2) site and the consensus AP-1 element successfully competed for binding, whereas puta-AP-1(1) did not compete for binding (Fig. 3A). We next used the three oligonucleotides as probes and competed with excess unlabeled oligonucleotide containing the consensus AP-1 element. The puta-AP-1(2) element gave a band that comigrated with the AP-1 complex, and the consensus oligonucleotide successfully competed for binding to this element (Fig. 3B). Another slower-migrating band was not competed away by the consensus oligonucleotide, suggesting that the puta-AP-1(2) oligonucleotide can bind a non-AP-1 specific complex as well. In contrast, the puta-AP-1(1) failed to display binding of an AP-1 complex. These data suggest that only puta-AP-1(2), and not puta-AP-1(1), contains a legitimate AP-1 binding site.
FIG. 3.
The distal region of the relB promoter contains an AP-1 binding site. (A) NIH 3T3 cell nuclear extracts (5 μg) were subjected to competition EMSA by using the oligonucleotide containing the consensus AP-1 element as a probe and 10- or 30-fold excess unlabeled oligonucleotides containing either the consensus (Con) AP-1 element or the putative AP-1 element Puta-AP-1(1) or Puta-AP-1(2), as indicated. (B) NIH 3T3 cell nuclear extracts (5 μg) were subjected to competition EMSA by using either the consensus AP-1 element, Puta-AP-1(1), or Puta-AP-1(2) as a probe, and 10- or 30-fold excess unlabeled consensus AP-1 oligonucleotide. ns, Position of non-AP-1 band.
The upstream AP-1 site regulates relB promoter activity.
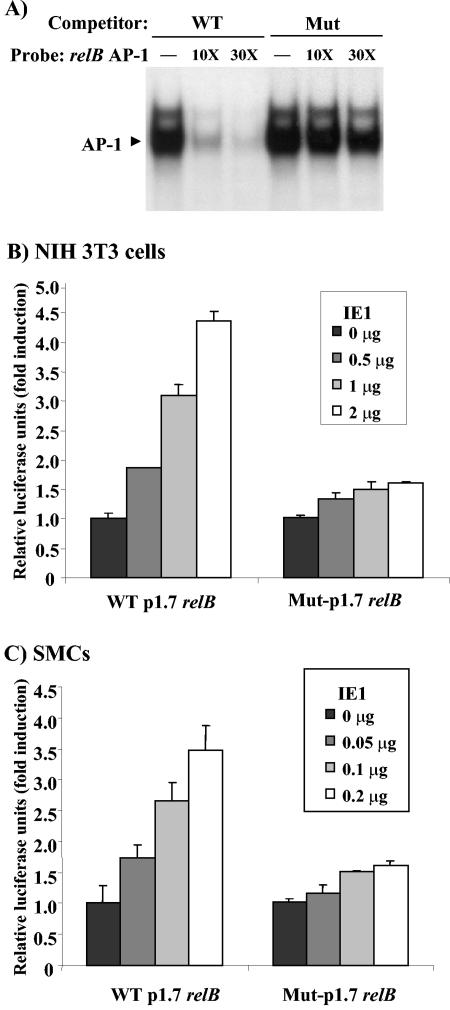
The identified AP-1 site in the distal region of the relB promoter was mutated to test its functional involvement. A G-to-C point mutation was introduced within the core of this element and tested for its affect on binding via competition EMSA. Although WT AP-1 successfully competed against itself for binding, the mutated sequence (Mut AP-1) was only minimally effective even at a 30-fold molar excess (Fig. 4A). Thus, the mutation almost completely eliminates the ability of the element to bind AP-1 protein. This mutation was then introduced within the p1.7 DNA via site-directed mutagenesis, generating the AP-1 Mut-p1.7 relB-Luc construct. NIH 3T3 cells were cotransfected with either the p1.7 relB-Luc or the AP-1 Mut-p1.7 relB-Luc reporter construct and increasing amounts of IE1 expression vector (Fig. 4B). Although the WT p1.7 relB promoter activity was induced ∼4.4-fold at the highest IE1 dose, only a 1.6-fold induction was seen with the AP-1 Mut-p1.7 relB-Luc reporter construct at this dose: 4.2 ± 0.3-fold versus 1.7 ± 0.1-fold in two separate experiments. The basal level of activity of the mutant and WT promoters were similar (data not shown). A similar assay was performed in SMCs (Fig. 4C). Mutation of the AP-1 site again resulted in a substantial reduction in the ability of IE1 to induce relB transcription (3.6 ± 0.1-fold versus 1.6 ± 0.1-fold in two separate experiments). These findings demonstrate that the distal AP-1 site plays a critical role in IE1-mediated induction of the relB promoter in both NIH 3T3 and vascular SMCs.
FIG. 4.
Mutation of the AP-1 element blocks AP-1 binding and reduces RelB induction by IE1. (A) NIH 3T3 cells nuclear extracts (5 μg) were subjected to competition EMSA with labeled relB promoter WT AP-1 element as probe, and excess unlabeled oligonucleotides containing either a WT or mutant (Mut) relB promoter AP-1 site. (B and C) NIH 3T3 cells or SMCs at 50 to 70% confluence were transiently transfected, in triplicate, with the indicated dose of pON2205 vector expressing IE1 protein, 0.5 μg of WT p1.7 or AP-1 Mut-p1.7 relB promoter-Luc, 0.5 μg of SV40 β-Gal expression vector and pBluescript for a total of 3 μg of DNA. Relative luciferase units are presented ± the standard deviation. Untreated WT and AP-1 Mut-p1.7 relB promoter-Luc activities were set at 1 in NIH 3T3 cells (B) or SMCs (C).
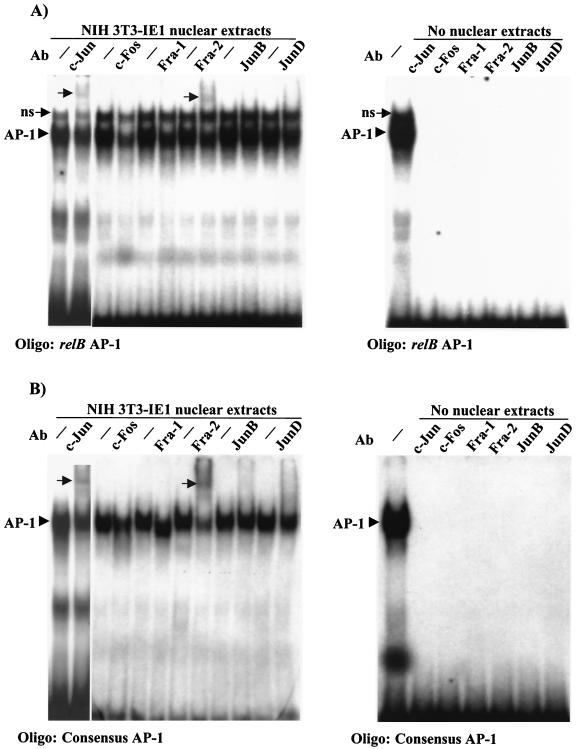
The AP-1 element of the relB promoter binds c-Jun, Fra-2, and c-Fos subunits.
Next, we sought to determine which AP-1 family members are present in nuclear extracts from NIH 3T3 cells transfected with CMV IE1 expression vectors and bind to the relB promoter AP-1 site. As control, the consensus AP-1 site was similarly analyzed. CMV IE1 expression did not induce the level of AP-1 binding (data not shown), in agreement with the findings of Kim et al. (35). Addition of an antibody against c-Jun or Fra-2 resulted in a supershifted complex with the relB AP-1 oligonucleotide and nuclear extracts from IE1 transfected cells (Fig. 5A) or untransfected NIH 3T3 cells (data not shown). Similar data were obtained with the control consensus AP-1 oligonucleotide (Fig. 5B). The addition of an antibody against c-Fos caused a substantial reduction in binding, while antibodies against Fra-1, JunB, or JunD had only modest effects on binding and failed to yield a supershift. As a negative control, incubation of antibodies with either probe alone showed no binding (Fig. 5A and B, right panels). Taken together, these data indicate that c-Jun, Fra-2, and c-Fos are the major AP-1 subunits binding to the AP-1 site upstream of the relB promoter in NIH 3T3 cells.
FIG. 5.
c-Jun, Fra-2, and c-Fos bind to the distal AP-1 element of the relB promoter. Nuclear extracts were isolated from NIH 3T3 cells 48 h after transfection with 10 μg of pON2205 vector expressing IE1 protein, and samples were subjected to EMSA in the absence (−) or presence of the indicated antibody using as a probe oligonucleotides containing either the AP-1 element upstream of the relB promoter (A) or the consensus AP-1 (B). Alternatively, in the right panels, antibodies were added to the oligonucleotides, as indicated, in the absence of extracts to verify the specificity of the supershifted bands. ns, Nonspecific band.
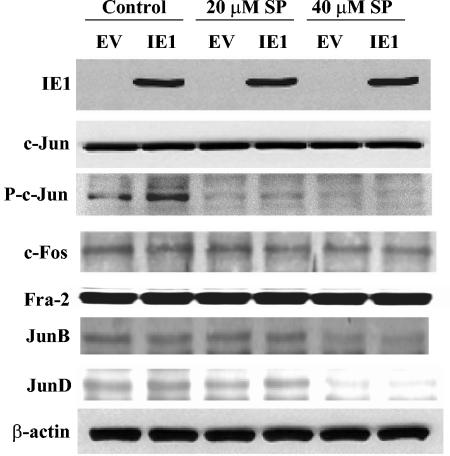
c-Jun phosphorylation is necessary for AP-1 activation and RelB induction by IE1.
We next evaluated the effects of IE1 on expression of AP-1 family members via immunoblot analysis. NIH 3T3 cells were transfected with either pCMV IE1 expression or empty vector DNA, and IE1 expression was confirmed (Fig. 6). IE1 did not affect the overall levels of c-Jun, c-Fos, Fra-2, JunB, or JunD protein (Fig. 6, control lanes). Since AP-1 activity depends mainly on phosphorylation of the subunit factors, the effects of IE1 expression on the level of phosphorylated c-Jun was examined. NIH 3T3 cells transfected with IE1 had a large increase in Ser63 phosphorylated c-Jun compared to empty vector DNA, although the overall level of c-Jun protein did not change (Fig. 6). To evaluate the role of JNK, the selective inhibitor SP600125 (8) was used in immunoblot and cotransfection analyses. NIH 3T3 cells were transfected with empty vector or IE1 expression vectors and, after 4 h, were treated with either 20 or 40 μM SP600125 (Fig. 6). The increase in phosphorylated c-Jun protein was reduced from 2.9-fold to 1.4- and 1.0-fold with 20 and 40 μM SP600125, respectively. (Similar results were obtained by using a phospho-Ser73 c-Jun antibody [data not shown].)
FIG. 6.
CMV IE1 protein activates c-Jun phosphorylation. NIH 3T3 cells were transfected with 10 μg of pON2205 vector expressing IE1 protein (IE1) or pCMV empty vector DNA (EV). After 4 h, cells were treated with DMSO, 20 μM SP600125, or 40 μM SP600125 for 20 h. Total cell lysates were prepared, and samples (30 μg) were subjected to immunoblot analysis for IE1, c-Jun, phospho-Ser63 c-Jun (P-c-Jun), c-Fos, Fra-2, JunB, JunD, and β-actin (as a loading control).
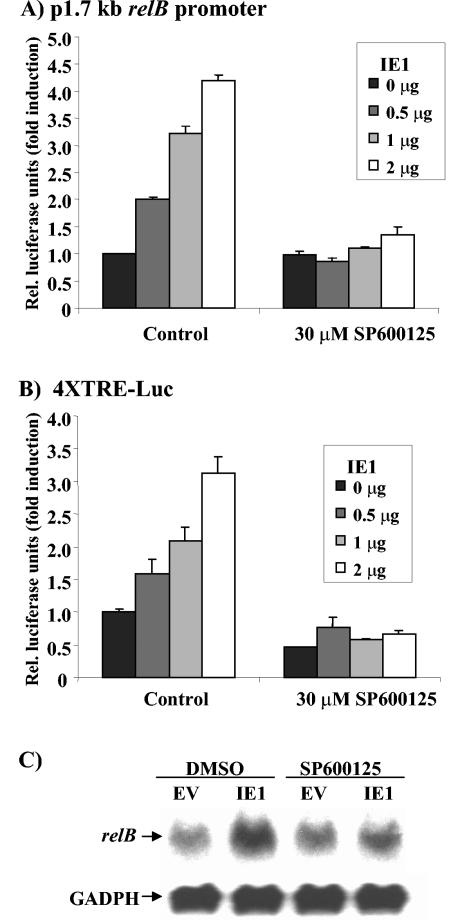
To test whether c-Jun phosphorylation is necessary for transcriptional induction of the relB promoter by IE1, NIH 3T3 cells were cotransfected with either the p1.7 relB promoter or the 4XTRE-Luc reporter and an increasing amount of IE1 expression vector, and after 4 h treated with either 0 or 30 μM SP600125. The normal 4.2-fold induction of the relB promoter mediated by IE1 was almost completely inhibited by the JNK selective inhibitor: 4.0 ± 0.3-fold versus 1.5 ± 0.1-fold in two separate experiments (Fig. 7A). Activation of the AP-1 element reporter was also essentially completely inhibited by SP600125: 3.6 ± 0.5-fold versus 1.6 ± 0.3-fold in two separate experiments (Fig. 7B). These findings suggest that most of the induction of the relB promoter by IE1 can likely be attributed to enhanced c-Jun phosphorylation and increased AP-1 activity.
FIG. 7.
SP600125, the selective inhibitor of the JNK signaling pathway, blocks relB promoter induction by IE1. NIH 3T3 cells were transiently transfected, in triplicate, with the indicated dose of pON2205 vector expressing IE1 protein, and either 0.5 μg of p1.7 relB promoter-Luc (A) or 4XTRE-Luc (B), 0.5 μg of SV40 β-Gal expression vector, and pBluescript for a total of 3 μg of DNA. After 4 h, cells were treated with carrier DMSO (control) or 30 μM SP600125. Cells were harvested after 20 h, the luciferase and β-Gal activities were measured, and normalized values were obtained. Relative (Rel.) luciferase units are presented ± the standard deviation. Control p1.7 relB promoter-Luc activity (A) and untreated 4XTRE-Luc activity (B) were set at 1. (C) NIH 3T3 cells were transiently transfected with 10 μg of pON2205 vector expressing IE1 protein (IE1) or pCMV empty vector DNA (EV). After 4 h, cells were treated with DMSO or 30 μM SP600125 for 20 h, the RNA was isolated, and samples (20 μg) were subjected to Northern blot analysis for relB mRNA (upper panel). Equal loading was confirmed by GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA level (lower panel).
To assess whether c-Jun phosphorylation is necessary for the previously observed increased relB mRNA levels (30), NIH 3T3 cells were treated with 30 μM SP600125 or control DMSO 4 h after transfection with pON2205 IE1 expression vector or pCMV empty vector. RNA was isolated after 20 h and subjected to Northern blot analysis (Fig. 7C). The levels of relB mRNA were substantially increased upon expression of IE1, and the JNK inhibitor potently reduced this induction from 4.2- to 1.4-fold (Fig. 7C).
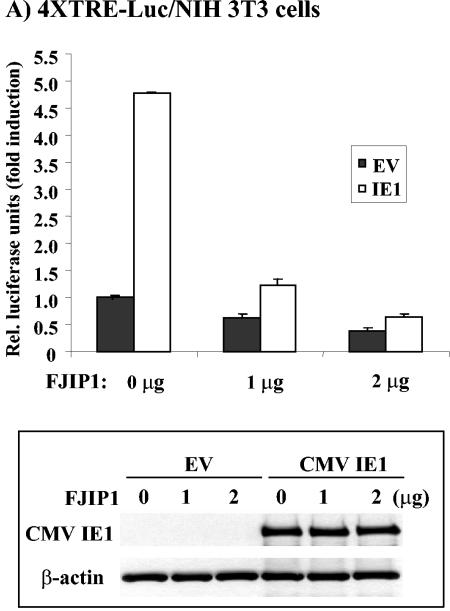
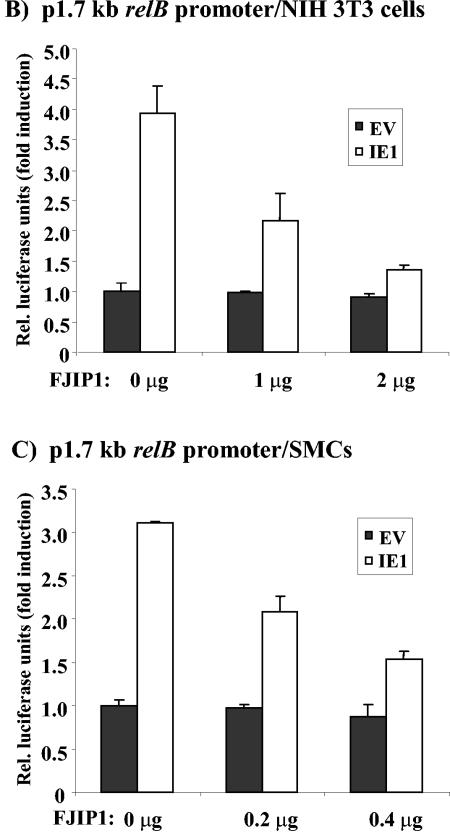
Since the pharmacologic inhibitor SP600125 may also affect other kinases, a vector expressing a dominant-negative inhibitor of JNK, containing the JNK-binding domain of JIP1, pCMV-FJIP1 (68), was used to test directly its role in relB promoter activation. First, we confirmed that overexpression of the JNK-binding domain of JIP1 blocked AP-1 activation by using the 4XTRE-Luc construct and an increasing dose of pCMV-FJIP1 (Fig. 8A). The 4.4 ± 0.6-fold induction seen in NIH 3T3 cells in the absence of FJIP1 was reduced to 1.6 ± 0.1-fold in the presence of 2 μg of pCMV-FJIP1. Immunoblot analysis showed that the cotransfection of pCMV-FJIP1 did not reduce the expression of CMV IE1 (Fig. 8A, lower panel). We next tested the effects of pCMV-FJIP1 on relB promoter activation by IE1. NIH 3T3 cells or SMCs were cotransfected with the p1.7 relB promoter reporter, the parental vector, or the IE1 expression vector, and an increasing amount of pCMV-FJIP1 vector DNA. The normal induction of the relB promoter by IE1 in NIH 3T3 cells was reduced from 4.2 ± 0.4-fold down to 1.7 ± 0.3-fold with the higher dose of pCMV-FJIP1 (Fig. 8B). Similarly, pCMV-FJIP1 caused a similar dose-dependent decrease in the ability of IE1 to induce relB promoter activity in SMCs: 3.2 ± 0.2-fold versus 1.6 ± 0.2-fold at the higher dose (Fig. 8C). Thus, JNK signaling plays a critical role in AP-1-mediated induction of relB promoter activity by IE1 in fibroblasts and SMCs.
FIG. 8.
Inhibition of the JNK pathway blocks IE1-mediated activation of the relB promoter. (A) 4XTRE-Luc. NIH 3T3 cells were transiently transfected, in triplicate, with 1 μg of pCMV empty vector or pON2205 vector expressing IE1 protein; 0, 1, or 2 μg of pCMV FJIP1; 0.25 μg of 4XTRE-Luc; 0.25 μg of SV40 β-Gal expression vector; and pBluescript for a total of 3.5 μg of DNA. After 24 h, cells were harvested, and the luciferase activity and β-Gal activity were measured. Relative luciferase units are presented ± the standard deviation. The control, empty vector DNA sample was set at 1. For the lower part of panel A, samples of cell lysates (20 μg) from reporter transfection analyses were subjected to immunoblot analysis for IE1 and β-actin. (B) p1.7 relB promoter. NIH 3T3 cells were transiently transfected, in triplicate, with the 1 μg of pCMV empty vector or pON2205 vector expressing IE1 protein, the indicated dose of pCMV FJIP1, 0.25 μg of p1.7 relB promoter-Luc, 0.25 μg of SV40 β-Gal expression vector, and pBluescript for a total of 3.5 μg of DNA. After 24 h, samples were processed, and data are presented as described above. (C) SMCs were transiently transfected, in triplicate, with 0 or 0.2 μg of pON2205 vector expressing IE1 protein, the indicated dose of pCMV FJIP1, 0.5 μg of p1.7 relB promoter-Luc, 0.5 μg of SV40 β-Gal expression vector, and pBluescript for a total of 2 μg of DNA. After 24 h, samples were processed, and data are presented as described above.
c-Jun/Fra-2 activate the relB promoter via its AP-1 element.
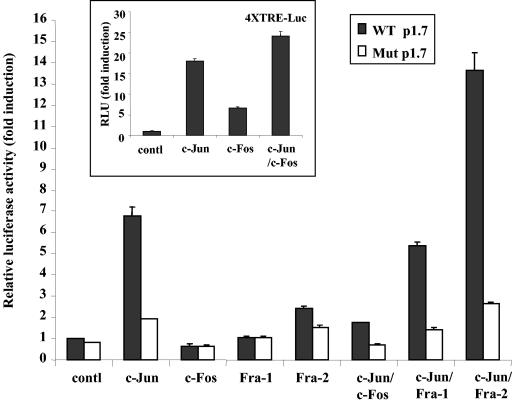
To verify the ability of AP-1 family members to induce relB promoter activity, NIH 3T3 cells were cotransfected with either the p1.7 relB-Luc or AP-1 Mut-p1.7 relB-Luc reporter construct, and vectors expressing c-Jun, c-Fos, Fra-1, or Fra-2 alone or in various combinations. In two separate experiments, the WT p1.7 relB promoter was induced by c-Jun alone, c-Jun/Fra-1, or c-Jun/Fra-2, with an induction of 6.8 ± 0.5-, 5.4 ± 0.6-, or 13.6 ± 1.3-fold, respectively, whereas the AP-1 Mut-p1.7 relB promoter was induced to a much lower extent (Fig. 9). Surprisingly, c-Jun/c-Fos appeared to have little effect on WT p1.7 relB promoter. To confirm the activity of the vectors expressing c-Jun and c-Fos, we tested their effects on the 4XTRE-Luc AP-1 reporter construct. Cotransfection of vectors expressing c-Jun and c-Fos in NIH 3T3 cells increased the activity of the 4XTRE-Luc by 24.8 ± 3.6-fold (Fig. 9, inset). Thus, c-Jun/c-Fos can induce the activity of the 4XTRE-Luc AP-1 element reporter but not the relB promoter. c-Jun/Fra-2 AP-1 complexes, which are activated by IE1, can therefore potently induce the relB promoter activity.
FIG. 9.
Expression of c-Jun and Fra-2 induces the relB promoter. NIH 3T3 cells were transiently transfected, in triplicate, with 0.5 μg of the indicated AP-1 subunit expression vector, 0.5 μg of WT p1.7 or AP-1 Mut-relB promoter-Luc, 0.5 μg of SV40 β-Gal expression vector, and pCI empty vector for a total of 3 μg of DNA. (Inset) NIH 3T3 cells were transiently transfected, in triplicate, with 0.5 μg of the indicated AP-1 subunit expression vector, 0.5 μg of 4XTRE-Luc vector, 0.5 μg of SV40 β-Gal expression vector, and pCI empty vector for a total of 3 μg of DNA. Values of luciferase normalized to β-Gal activity (in relative luciferase units) are presented ± the standard deviation. Control p1.7 relB promoter- and 4XTRE-Luc activities were set at 1.
c-Jun/Fra-2 synergize with p50/p65 classical NF-κB to potently activate the relB promoter.
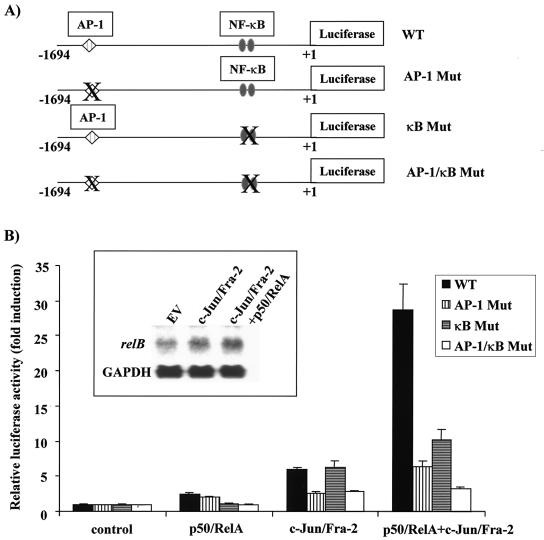
It was previously reported that NF-κB and AP-1 can functionally synergize to transactivate transcription of several specific genes (21, 41, 42). The relB promoter contains two proximal NF-κB elements, in addition to the distal AP-1 element. To check whether these two transcription factor families can function in synergy on the relB promoter, we introduced mutations into the two NF-κB sites in both the WT p1.7 relB-Luc and the AP-1 Mut-p1.7 relB-Luc constructs (Fig. 10A). NIH 3T3 cells were cotransfected with the WT and κB Mut-p1.7 relB-Luc reporter constructs in the presence of p50/RelA or c-Jun/Fra-2 alone or in combination (Fig. 10B). For this analysis, suboptimal doses of NF-κB and AP-1 expression vectors were used. In two separate experiments, the p50/RelA and c-Jun/Fra-2 expression vectors alone induced the WT promoter to 2.5 ± 0.3- and 6.0 ± 0.5-fold, respectively, whereas the combination resulted in a 28.0 ± 2.2-fold induction. Mutation of either the κB or AP-1 sites alone blocked the activation seen upon expression of p50/RelA or c-Jun/Fra-2, respectively. Furthermore, an almost complete inhibition was observed with the AP-1/κB double mutant relB promoter. Thus, c-Jun/Fra-2 synergize with p50/RelA to potently activate the relB promoter. To assess whether AP-1 expression caused an increase in relB mRNA levels, NIH 3T3 cultures were transfected with c-Jun/Fra-2 expression vectors alone or in combination with p50/RelA expression vectors, and after 48 h RNA was isolated (Fig. 10B, inset). Northern blot analysis demonstrated that relB mRNA levels were increased 1.8-fold upon expression of c-Jun/Fra-2 alone and 2.5-fold when cotransfected in combination with p50/RelA. Thus, c-Jun/Fra-2 synergize with p50/RelA to induce relB mRNA levels.
FIG. 10.
Synergy between c-Jun/Fra-2 AP-1 complexes and p50/p65 NF-κB complexes potently activate the relB promoter. (A) Scheme of the structures of the WT and NF-κB/AP-1 mutant p1.7 relB promoter luciferase reporter vectors. Mutant elements are indicated by an X. (B) NIH 3T3 cells were transiently transfected, in triplicate, with 0.25 μg of the indicated AP-1 or NF-κB subunit expression vector, 0.5 μg of WT or the indicated NF-κB/AP-1 mutant p1.7 relB promoter-Luc, 0.5 μg of SV40 β-Gal expression vector, and pCI empty vector for a total of 3 μg of DNA. Values of luciferase normalized to β-Gal activity (in relative luciferase units) are presented ± the standard deviation. Control p1.7 relB promoter-Luc activities were set at 1. (Inset) NIH 3T3 cells were transiently transfected with 5 μg of EV and c-Jun/Fra-2 alone or in combination with 5 μg of p50/RelA expression vectors. After 48 h, the RNA was harvested, and samples (20 μg) were subjected to Northern blot analysis for relB mRNA or GAPDH as a control for equal loading.
DISCUSSION
Here we demonstrate for the first time that induction of relB NF-κB subunit gene transcription by the CMV IE1 protein is mediated via c-Jun/Fra-2 complexes binding to an upstream AP-1 element; furthermore, these AP-1 complexes can synergize with classical NF-κB transcription factors to potently induce relB promoter activity. The sequences mediating activation were mapped to an ∼600-bp region between bp −1694 and −1096 of the relB promoter, which contained a bona fide AP-1 element at bp −1503. IE1 expression induced c-Jun phosphorylation and AP-1 activity as judged by a four-copy element construct. Mutation of the upstream AP-1 element of the relB promoter prevented its induction by IE1 in both NIH 3T3 cells and vascular SMCs. The transcriptional activity of c-Jun is regulated by phosphorylation at Ser63 and Ser73 by JNK (9, 54), and addition of the pharmacological inhibitor SP600125 or overexpression of the JNK-binding domain of JIP1 blocked relB promoter activation. Binding of c-Jun, Fra-2, and c-Fos was demonstrated by using antibody supershift EMSA of IE1 transfected NIH 3T3 cells, but only ectopic expression of c-Jun plus Fra-2 induced relB promoter activity. Although the NF-κB elements, which are located at bp −245 and −178 of the relB promoter (11), were not required for IE1-mediated activation, classical NF-κB complexes synergized with c-Jun and Fra-2 in promoter induction. Overall, our findings demonstrate for the first time the role of AP-1 in the activation of NF-κB RelB subunit expression. Furthermore, given the role of CMV IE1 in control of SMC migration and proliferation, these findings suggest a role for RelB in these processes.
In our previous work, we showed that the NF-κB activity induced by IE1 was predominantly RelB and p50 nuclear proteins, as judged by immunohistochemistry and supershift EMSA (30). Furthermore, IE1 was unable to induce an NF-κB element driven reporter construct in relB−/− cells. In agreement with these findings, we observed that IE1 protein was unable to induce either the p1.1 or p0.6 relB promoter constructs, which contain two functional NF-κB binding sites at −245 and −178 bp (11). Furthermore, although RelA was a very potent inducer, p50/RelB complexes only modestly activated the relB promoter and failed to synergize with c-Jun/Fra-2 complexes (data not shown). Thus, indicating the relB promoter is not activated indirectly via delayed induction RelB binding. Recently, an alternative pathway leading specifically to activation of RelB/p52 complexes has been elucidated (15, 45, 55). This pathway involves IKKα phosphorylation of p100 in RelB/p100 cytoplasmic complexes, leading to proteasome-mediated processing of p100 to p52 and to p52/RelB migration to the nucleus. Interestingly, we observed that expression of CMV IE1 failed to induce either IKKα activation or a change of the ratio of p52 to p100 (data not shown). These results indicate that CMV IE1 does not activate this alternative signal pathway.
AP-1 is an important transcription factor involved in control of cell proliferation, death, survival, and differentiation (2, 31, 51, 52). Its activity can be regulated at the transcriptional or posttranslational level (33, 34). It was reported previously that IE1 does not raise the mRNA levels of c-jun or c-fos (35), indicating posttranslational control of AP-1 activation. Consistent with these observations, no increase in the protein levels of AP-1 family members or AP-1 binding was seen upon IE1 expression in NIH 3T3 cells. The mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 1 (MEKK1) had a highly synergistic effect on AP-1 activation by IE1 (35). The mechanism of the synergy between IE1 and MEKK1 is unclear. MEKK1 is one important upstream protein kinase of JNK and Erk signal pathway (32). In our study, IE1 induces AP-1 activity mostly via c-Jun phosphorylation by JNK. Since Erk signaling is responsible for the expression and activation of Fra-1 and Fra-2 (63), the synergistic effect may depend on the activation of the substrates of both pathways.
The AP-1 and NF-κB family of factors have been found to interact physically and functionally at multiple levels. Stein et al. (59) first reported p65 could physically interact with both c-Jun and c-Fos and functionally synergize to transactivate the human immunodeficiency virus type 1 5′ long terminal repeat. Consistent with our findings, Murayama et al. reported that HCMV induces interleukin-8 gene expression through concurrent AP-1 and NF-κB activation (41). AP-1 and NF-κB factors also synergistically cooperate in the transcriptional regulation of interleukin-6 expression by transforming growth factor β1 in prostate carcinoma cells (42). PPARα has been shown to repress the activities of both AP-1 and NF-κB via physical interaction with c-Jun, p65, and CBP (16). More recently, Gelinas and coworkers (21) demonstrated that binding of the NF-κB c-Rel subunit to the promoter region of bfl-1 induced the assembly and concerted binding of both AP-1 and C/EBPβ to form an enhanceosome-like regulatory complex. Interestingly, interaction of both NF-κB and AP-1 with glucocorticoid receptor has also been reported to release repression mediated by these factors individually, although the mechanism has not been fully elucidated (49). In addition to cooperation between NF-κB and AP-1, several groups have demonstrated that NF-κB can inhibit JNK signaling induced by tumor necrosis factor alpha (18, 61). Consistent with these reports, we showed that the transient activation of NF-κB through TAK1/IKK kinase pathway by transforming growth factor β1 inhibits JNK signaling and inhibits AP-1/SMAD signaling in hepatocytes (4). Our findings here demonstrate for the first time the role of AP-1 in direct regulation of relB gene transcription, providing an additional important linkage between these two transcription factor families critical for cell survival and proliferation.
Acknowledgments
We thank E. Mocarski, R. Davis, C. Paya, M. Arsura, and D. Chalbos for generously providing cloned DNAs. We thank Karine Belguise for helpful comments on the manuscript.
This study was supported by NIH grants HL13262 (G.E.S.) and ES11624 (G.E.S.).
REFERENCES
- 1.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 2.Angel, P., and M. Karin. 1991. The role of Jun, Fos, and the AP-1 complex in cell proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 3.Arsura, M., M. J. FitzGerald, N. Fausto, and G. E. Sonenshein. 1997. Nuclear factor-κB/Rel blocks transforming growth factor β1-induced apoptosis of murine hepatocyte cell lines. Cell Growth Differ. 8:1049-1059. [PubMed] [Google Scholar]
- 4.Arsura, M., G. R. Panta, J. D. Bilyeu, L. G. Cavin, M. A. Sovak, A. A. Oliver, V. Factor, R. Heuchel, F. Mercurio, S. S. Thorgeirsson, and G. E. Sonenshein. 2003. Transient activation of NF-κB through a TAK1/IKK kinase pathway by TGF-β1 inhibits AP-1/SMAD signaling and apoptosis: implications in liver tumor formation. Oncogene 22:412-425. [DOI] [PubMed] [Google Scholar]
- 5.Arsura, M., M. Wu, and G. E. Sonenshein. 1996. TGF-β1 inhibits NF-κB/Rel activity inducing apoptosis of B cells: transcriptional activation of I κB alpha. Immunity 5:31-40. [DOI] [PubMed] [Google Scholar]
- 6.Becker, Y., G. Darai, and E. S. Huang. 1993. Molecular aspects of human cytomegalovirus diseases. Springer-Verlag, New York, N.Y.
- 7.Beldekas, J. C., L. Gerstenfeld, G. E. Sonenshein, and C. Franzblau. 1982. Cell density and estradiol modulation of procollagen type III in cultured calf smooth muscle cells. J. Biol. Chem. 257:12252-12256. [PubMed] [Google Scholar]
- 8.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binetruy, B., T. Smeal, and M. Karin. 1991. Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature 351:122-127. [DOI] [PubMed] [Google Scholar]
- 10.Boldogh, I., S. AbuBakar, C. Z. Deng, and T. Albrecht. 1991. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J. Virol. 65:1568-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bren, G. D., N. J. Solan, H. Miyoshi, K. N. Pennington, L. J. Pobst, and C. V. Paya. 2001. Transcription of the RelB gene is regulated by NF-κB. Oncogene 20:7722-7733. [DOI] [PubMed] [Google Scholar]
- 12.Brown, K., S. Gerstberger, L. Carlson, G. Franzoso, and U. Siebenlist. 1995. Control of IκBα proteolysis by site-specific, signal-induced phosphorylation. Science 267:1485-1488. [DOI] [PubMed] [Google Scholar]
- 13.Castillo, J. P., and T. F. Kowalik. 2004. HCMV infection: modulating the cell cycle and cell death. Int. Rev. Immunol. 23:113-139. [DOI] [PubMed] [Google Scholar]
- 14.Cherrington, J. M., and E. S. Mocarski. 1989. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coope, H. J., P. G. Atkinson, B. Huhse, M. Belich, J. Janzen, M. J. Holman, G. G. Klaus, L. H. Johnston, and S. C. Ley. 2002. CD40 regulates the processing of NF-κB2 p100 to p52. EMBO J. 21:5375-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delerive, P., K. De Bosscher, S. Besnard, W. Vanden Berghe, J. M. Peters, F. J. Gonzalez, J. C. Fruchart, A. Tedgui, G. Haegeman, and B. Staels. 1999. Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-κB and AP-1. J. Biol. Chem. 274:32048-32054. [DOI] [PubMed] [Google Scholar]
- 17.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 18.De Smaele, E., F. Zazzeroni, S. Papa, D. U. Nguyen, R. Jin, J. Jones, R. Cong, and G. Franzoso. 2001. Induction of gadd45beta by NF-κB downregulates proapoptotic JNK signalling. Nature 414:308-313. [DOI] [PubMed] [Google Scholar]
- 19.DeVita, V. T., S. Hellman, and S. A. Rosenberg. 2001. Cancer, principles and practice of oncology. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 20.Do, R. K., E. Hatada, H. Lee, M. R. Tourigny, D. Hilbert, and S. Chen-Kiang. 2000. Attenuation of apoptosis underlies B lymphocyte stimulator enhancement of humoral immune response. J. Exp. Med. 192:953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edelstein, L. C., L. Lagos, M. Simmons, H. Tirumalai, and C. Gelinas. 2003. NF-κB-dependent assembly of an enhanceosome-like complex on the promoter region of apoptosis inhibitor Bfl-1/A1. Mol. Cell. Biol. 23:2749-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett, R. D. 1984. Trans activation of transcription by herpesvirus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 3:3135-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett, R. D., and M. Dunlop. 1984. Trans activation of plasmid-borne promoters by adenovirus and several herpes group viruses. Nucleic Acids Res. 12:5969-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fields, B. N., D. M. Knipe, and P. M. Howley. 1996. Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 25.Foo, S. Y., and G. P. Nolan. 1999. NF-κB to the rescue: RELs, apoptosis, and cellular transformation. Trends Genet. 15:229-235. [DOI] [PubMed] [Google Scholar]
- 26.Fritschy, J. M., S. Brandner, A. Aguzzi, M. Koedood, B. Luscher, and P. J. Mitchell. 1996. Brain cell type specificity and gliosis-induced activation of the human cytomegalovirus immediate-early promoter in transgenic mice. J. Neurosci. 16:2275-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilmore, T. D., M. Koedood, K. A. Piffat, and D. W. White. 1996. Rel/NF-κB/IκB proteins and cancer. Oncogene 13:1367-1378. [PubMed] [Google Scholar]
- 28.Grilli, M., J. J. Chiu, and M. J. Lenardo. 1993. NF-κB and Rel: participants in a multiform transcriptional regulatory system. Int. Rev. Cytol. 143:1-62. [DOI] [PubMed] [Google Scholar]
- 29.Jenkins, D. E., C. L. Martens, and E. S. Mocarski. 1994. Human cytomegalovirus late protein encoded by ie2: a transactivator as well as a repressor of gene expression. J. Gen. Virol. 75:2337-2348. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, H. Y., C. Petrovas, and G. E. Sonenshein. 2002. RelB-p50 NF-κB complexes are selectively induced by cytomegalovirus immediate-early protein 1: differential regulation of Bcl-x(L) promoter activity by NF-κB family members. J. Virol. 76:5737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang, D. C., M. Motwani, and P. B. Fisher. 1998. Role of the transcription factor AP-1 in melanoma differentiation. Int. J. Oncol. 13:1117-1126. [DOI] [PubMed] [Google Scholar]
- 32.Karandikar, M., S. Xu, and M. H. Cobb. 2000. MEKK1 binds raf-1 and the ERK2 cascade components. J. Biol. Chem. 275:40120-40127. [DOI] [PubMed] [Google Scholar]
- 33.Karin, M. 1995. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 270:16483-16486. [DOI] [PubMed] [Google Scholar]
- 34.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 35.Kim, S., S. S. Yu, I. S. Lee, S. Ohno, J. Yim, and H. S. Kang. 1999. Human cytomegalovirus IE1 protein activates AP-1 through a cellular protein kinase(s). J. Gen. Virol. 80:961-969. [DOI] [PubMed] [Google Scholar]
- 36.Kyriakis, J. M., P. Banerjee, E. Nikolakaki, T. Dai, E. A. Rubie, M. F. Ahmad, J. Avruch, and J. R. Woodgett. 1994. The stress-activated protein kinase subfamily of c-Jun kinases. Nature 369:156-160. [DOI] [PubMed] [Google Scholar]
- 37.La Rosa, F. A., J. W. Pierce, and G. E. Sonenshein. 1994. Differential regulation of the c-myc oncogene promoter by the NF-κB rel family of transcription factors. Mol. Cell. Biol. 14:1039-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemstrom, K. B., J. H. Bruning, C. A. Bruggeman, I. T. Lautenschlager, and P. J. Hayry. 1993. Cytomegalovirus infection enhances smooth muscle cell proliferation and intimal thickening of rat aortic allografts. J. Clin. Investig. 92:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lernbecher, T., U. Muller, and T. Wirth. 1993. Distinct NF-κB/Rel transcription factors are responsible for tissue-specific and inducible gene activation. Nature 365:767-770. [DOI] [PubMed] [Google Scholar]
- 40.Mocarski, E. S., M. Bonyhadi, S. Salimi, J. M. McCune, and H. Kaneshima. 1993. Human cytomegalovirus in a SCID-hu mouse: thymic epithelial cells are prominent targets of viral replication. Proc. Natl. Acad. Sci. USA 90:104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murayama, T., Y. Ohara, M. Obuchi, K. S. Khabar, H. Higashi, N. Mukaida, and K. Matsushima. 1997. Human cytomegalovirus induces interleukin-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1- and NF-κB-binding sites of the interleukin-8 gene. J. Virol. 71:5692-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, J. I., M. G. Lee, K. Cho, B. J. Park, K. S. Chae, D. S. Byun, B. K. Ryu, Y. K. Park, and S. G. Chi. 2003. Transforming growth factor-beta1 activates interleukin-6 expression in prostate cancer cells through the synergistic collaboration of the Smad2, p38-NF-κB, JNK, and Ras signaling pathways. Oncogene 22:4314-4332. [DOI] [PubMed] [Google Scholar]
- 43.Rayet, B., and C. Gelinas. 1999. Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18:6938-6947. [DOI] [PubMed] [Google Scholar]
- 44.Rincon, M., and R. A. Flavell. 1994. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 13:4370-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitoh, T., M. Nakayama, H. Nakano, H. Yagita, N. Yamamoto, and S. Yamaoka. 2003. TWEAK induces NF-κB2 p100 processing and long lasting NF-κB activation. J. Biol. Chem. 278:36005-36012. [DOI] [PubMed] [Google Scholar]
- 46.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambucetti, L. C., J. M. Cherrington, G. W. Wilkinson, and E. S. Mocarski. 1989. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T-cell stimulation. EMBO J. 8:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schauer, S. L., Z. Wang, G. E. Sonenshein, and T. L. Rothstein. 1996. Maintenance of nuclear factor-κB/Rel and c-myc expression during CD40 ligand rescue of WEHI 231 early B cells from receptor-mediated apoptosis through modulation of I κB proteins. J. Immunol. 157:81-86. [PubMed] [Google Scholar]
- 49.Scheinman, R. I., A. Gualberto, C. M. Jewell, J. A. Cidlowski, and A. S. Baldwin, Jr. 1995. Characterization of mechanisms involved in transrepression of NF-κB by activated glucocorticoid receptors. Mol. Cell. Biol. 15:943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholz, M., R. A. Blaheta, B. Wittig, J. Cinatl, J. U. Vogel, H. W. Doerr, and J. Cinatl, Jr. 2000. Cytomegalovirus-infected neuroblastoma cells exhibit augmented invasiveness mediated by β1α5 integrin (VLA-5). Tissue Antigens 55:412-421. [DOI] [PubMed] [Google Scholar]
- 51.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 52.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 53.Shen, Y., H. Zhu, and T. Shenk. 1997. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc. Natl. Acad. Sci. USA 94:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smeal, T., B. Binetruy, D. A. Mercola, M. Birrer, and M. Karin. 1991. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature 354:494-496. [DOI] [PubMed] [Google Scholar]
- 55.Solan, N. J., H. Miyoshi, E. M. Carmona, G. D. Bren, and C. V. Paya. 2002. RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 277:1405-1418. [DOI] [PubMed] [Google Scholar]
- 56.Sonenshein, G. E. 1997. Rel/NF-κB transcription factors and the control of apoptosis. Semin. Cancer Biol. 8:113-119. [DOI] [PubMed] [Google Scholar]
- 57.Sovak, M. A., R. E. Bellas, D. W. Kim, G. J. Zanieski, A. E. Rogers, A. M. Traish, and G. E. Sonenshein. 1997. Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J. Clin. Investig. 100:2952-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 59.Stein, B., A. S. Baldwin, Jr., D. W. Ballard, W. C. Greene, P. Angel, and P. Herrlich. 1993. Cross-coupling of the NF-κB p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 12:3879-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Streblow, D. N., C. Soderberg-Naucler, J. Vieira, P. Smith, E. Wakabayashi, F. Ruchti, K. Mattison, Y. Altschuler, and J. A. Nelson. 1999. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell 99:511-520. [DOI] [PubMed] [Google Scholar]
- 61.Tang, G., Y. Minemoto, B. Dibling, N. H. Purcell, Z. Li, M. Karin, and A. Lin. 2001. Inhibition of JNK activation through NF-κB target genes. Nature 414:313-317. [DOI] [PubMed] [Google Scholar]
- 62.Teyssier, C., K. Belguise, F. Galtier, and D. Chalbos. 2001. Characterization of the physical interaction between estrogen receptor alpha and JUN proteins. J. Biol. Chem. 276:36361-36369. [DOI] [PubMed] [Google Scholar]
- 63.Treinies, I., H. F. Paterson, S. Hooper, R. Wilson, and C. J. Marshall. 1999. Activated MEK stimulates expression of AP-1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal to stimulate DNA synthesis. Mol. Cell. Biol. 19:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verma, I. M., J. K. Stevenson, E. M. Schwarz, D. Van Antwerp, and S. Miyamoto. 1995. Rel/NF-κB/I κB family: intimate tales of association and dissociation. Genes Dev. 9:2723-2735. [DOI] [PubMed] [Google Scholar]
- 65.Weih, D. S., Z. B. Yilmaz, and F. Weih. 2001. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J. Immunol. 167:1909-1919. [DOI] [PubMed] [Google Scholar]
- 66.Weih, F., D. Carrasco, and R. Bravo. 1994. Constitutive and inducible Rel/NF-κB activities in mouse thymus and spleen. Oncogene 9:3289-3297. [PubMed] [Google Scholar]
- 67.Whitmarsh, A. J., J. Cavanagh, C. Tournier, J. Yasuda, and R. J. Davis. 1998. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science 281:1671-1674. [DOI] [PubMed] [Google Scholar]
- 68.Yasuda, J., A. J. Whitmarsh, J. Cavanagh, M. Sharma, and R. J. Davis. 1999. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol. Cell. Biol. 19:7245-7254. [DOI] [PMC free article] [PubMed] [Google Scholar]