Abstract
The hepatitis C virus (HCV) is a positive-strand RNA virus belonging to the Flaviviridae. Its genome carries at either end highly conserved nontranslated regions (NTRs) containing cis-acting RNA elements that are crucial for replication. In this study, we identified a novel RNA element within the NS5B coding sequence that is indispensable for replication. By using secondary structure prediction and nuclear magnetic resonance spectroscopy, we found that this RNA element, designated 5BSL3.2 by analogy to a recent report (S. You, D. D. Stump, A. D. Branch, and C. M. Rice, J. Virol. 78:1352-1366, 2004), consists of an 8-bp lower and a 6-bp upper stem, an 8-nucleotide-long bulge, and a 12-nucleotide-long upper loop. Mutational disruption of 5BSL3.2 structure blocked RNA replication, which could be restored when an intact copy of this RNA element was inserted into the 3′ NTR. By using this replicon design, we mapped the elements in 5BSL3.2 that are critical for RNA replication. Most importantly, we discovered a nucleotide sequence complementarity between the upper loop of this RNA element and the loop region of stem-loop 2 in the 3′ NTR. Mismatches introduced into the loops inhibited RNA replication, which could be rescued when complementarity was restored. These data provide strong evidence for a pseudoknot structure at the 3′ end of the HCV genome that is essential for replication.
The hepatitis C virus (HCV) is the only member of the Hepacivirinae within the Flaviviridae family (46). These viruses possess an RNA genome of positive polarity that in the case of HCV has a length of about 9,600 nucleotides (nt). Its genome carries a single long open reading frame (ORF) that is flanked at either end by highly conserved nontranslated regions (NTRs) (2, 37). The 5′ NTR harbors an internal ribosome entry site (IRES) directing the synthesis of the viral polyprotein which is cleaved into 10 different products. RNA replication takes place in the cytoplasm in a distinct compartment (12) and requires viral proteins NS3 to NS5B as well as host cell factors (28, 43). In addition, cis-acting replication elements (CREs) are important for the synthesis of RNA progeny. Such CREs may reside at various positions in an RNA genome but are most often found in the NTRs. In the case of HCV, the minimal signals that are essential for RNA replication have been mapped within the NTRs. The 5′ NTR is composed of four distinct domains, with domains 1 and 2 being sufficient for RNA replication (10, 19). The 3′ NTR has a tripartite structure and is composed of a highly variable region immediately downstream of the stop codon of the polyprotein, a polypyrimidine [poly(U/UC)] tract of variable length, and a highly conserved 98-nt-long RNA element designated the X-tail (22, 41, 42, 52). The latter has the potential to form three stem-loop (SL) structures, of which the most 3′ terminal, designated SL1, has been confirmed experimentally (5, 15). In contrast, the structures of the two other SLs that are located upstream are much less clear (5, 15, 56). Both the X-tail and a minimal poly(U/UC) tract of about 40 nt are essential for RNA replication in cell culture and in vivo (9, 23, 54-56). Removal of part of or the complete variable region from HCV genomes or replicons is tolerated but significantly reduces RNA replication and in vivo infectivity (9, 23, 54-56).
For several positive-strand RNA viruses, in addition to RNA signals located in the NTRs, CREs residing within the coding sequence have been described. One of the best-studied examples with respect to RNA replication is the CRE of picornaviruses, including the human rhinovirus-14 (HRV-14), the poliovirus (PV), the Mengo virus, Theiler's virus, and the foot-and-mouth disease virus (FMDV) (11, 25, 30, 31, 34). In the cases of PV and HRV-14, the CRE is located in the coding region of 2C and in the capsid protein, respectively, whereas in FMDV the CRE resides in the 5′ NTR. These CREs serve as a template for the uridylylation of VPg, which is the protein primer for genome replication (35). The picornaviral CRE acts independently of position and can be translocated to different regions within the viral genome (11).
By using various biocomputational methods, including phylogenetic analyses and thermodynamic calculations, several groups analyzed the HCV genome for the presence of highly conserved RNA secondary structures within the coding region. Potential SL structures were identified primarily in the 5′ terminal core region and the coding sequence of NS5B (13, 38, 39, 45). Taking advantage of the subgenomic replicon system, in this study we analyzed the role of the SL structures in the 3′ coding region of NS5B for RNA replication. We identify a stem-loop that is essential for replication and provide evidence that the loop region of this RNA element interacts with SL2 in the X-tail of the 3′ NTR.
MATERIALS AND METHODS
Cell cultures.
Huh-7 cells were grown in Dulbecco's modified Eagle medium (Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 10% fetal calf serum. Transient replication assays were performed with a Huh-7 cell clone that was generated by treating luc-ubi-neo cells (48) with a selective inhibitor. After several passages in the presence of this drug and the absence of G418, no replicon RNA could be detected. Compared to naive Huh-7 cells, these “cured ” cells supported higher RNA levels and had a more stable permissiveness.
Plasmid constructions.
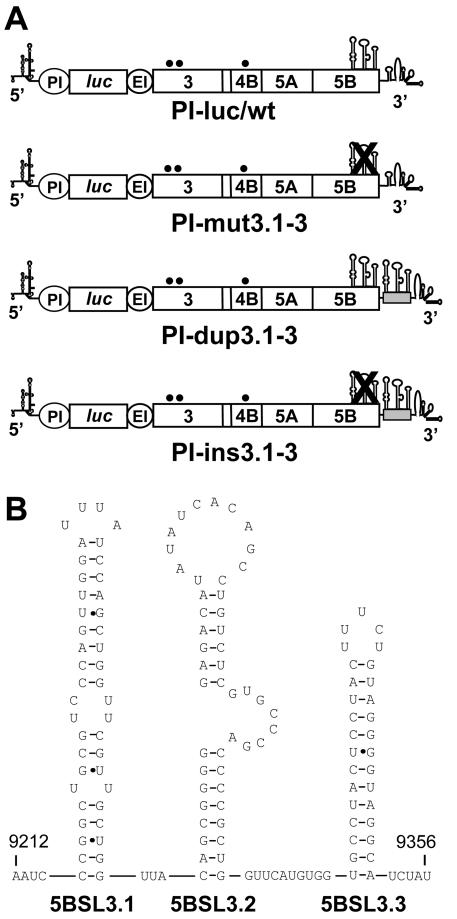
Unless otherwise stated, standard recombinant DNA technologies were used for all cloning procedures (1). The basic constructs pFK-nt341-sp-PI-lucEI3420-9605/ET (abbreviated rep PI-luc/ET) and pBSK8499-9605/Stu have been described recently (9, 10, 26) (Fig. 1A). To generate an appropriate transfer vector, the latter was modified by inactivation of the XbaI restriction site in the multiple cloning site and generation of a novel XbaI restriction site in the variable region of the 3′ NTR. For this purpose, cytidine 9393 was replaced by a uridine, and guanosine 9396 was replaced by an adenosine, resulting in plasmid pBSK8499-9605/Xba-Stu. Mutations affecting 5BSL3.1 to 5BSL3.3 or the 3′ NTR were introduced into this transfer vector, and SfiI-SpeI fragments isolated therefrom were transferred into the replicon construct rep PI-luc/ET-vXS (referred to as PI-luc/wt in this report). This construct corresponds to rep PI-luc/ET but carries the analogous XbaI and StuI restriction sites in the 3′ NTR as the transfer vector. We found that these modifications in the variable region of the 3′ NTR did not affect RNA replication (P. Friebe and R. Bartenschlager, unpublished data).
FIG. 1.
(A) Schematic presentation of the basic replicon constructs used in this study. The 5′ and 3′ NTRs are given with their secondary structures. The PV IRES (PI) directing the translation of the reporter gene firefly luciferase (luc) is depicted as a circle. The EMCV IRES (EI) directs the translation of the NS3 to 5B coding sequence. The three SL structures in NS5B are indicated with their proposed secondary structures. They are crossed out in cases of constructs in which the structures were disturbed by silent nucleotide substitutions. Names of the corresponding replicons are given below each graph. The grey box in replicons PI-dup3.1-3 and PI-ins3.1-3 indicates the insertion of part of the 3′ coding region of NS5B that carries the SL structures. (B) Predicted RNA secondary structure of the three stem-loops, 5BSL3.1, 5BSL3.2, and 5BSL3.3, used throughout this study as a basis for the design of mutants. Numbers refer to the nucleotide positions of the HCV Con1 isolate (21).
Mutations affecting the SL structures in the coding region of NS5B were generated by two consecutive PCRs by using primers in the first PCR that carry the desired mutations and primers S8996NcoI (ATATACCATGGCCTTAGCGC) and A9383Xba/Stu (TATATAGGCCTATTGGTCTAGAGTGTTTAGCTCCCCGTTC) for the second PCR, which was used to combine both fragments. After restriction with NheI and XbaI, fragments were inserted into the transfer vector pBSK8499-9605/Xba-Stu. Nucleotide sequences of the subcloned fragments were verified by using cycle sequencing and capillary electrophoresis on an ABI Prism 310 genetic analyzer. Fragments were inserted into the basic replicon vector PI-luc/ET-vXS, resulting in constructs PI-mut3.1, PI-mut3.2, and PI-mut3.3. Combinations of mutations in the different SLs were achieved by two consecutive PCRs with primers carrying the desired mutations in the first reaction and primers S8996NcoI and A9383Xba/Stu for the fragment combination. In this way, replicon construct PI-mut3.1-3 was obtained (Fig. 1A).
Insertion of SLs 3.1 to 3.3 into the variable region of the 3′ NTR was done by PCR using the primer pair S3.1Xba (ATATTCTAGACCGGCTGCGTCCCAGTTGG) and A3.3Stu (TATATAGGCCTGCCTACCCCTACAG) and rep PI-luc/ET as template. Fragments were restricted with XbaI and StuI and inserted into either pBSK8499-9605/Xba/Stu or pBSK8499-9605/Xba/Stu-mut3.1-3, resulting in the transfer vectors pBSK8499-9605/Xba/Stu-dup3.1-3 or pBSK8499-9605/Xba/Stu-mut3.1-3/dv3.1-3. Upon restriction with SfiI and SpeI, HCV fragments from these plasmids were inserted into PI-luc/ET-vXS, resulting in constructs PI-dup3.1-3 and PI-ins3.1-3 (Fig. 1A). Plasmid PI-ins3.2 was obtained by insertion of two hybridized oligonucleotides (S3.2Xba/Stu, ATATTCTAGACAGCGGGGGAGACATATATCACAGCCTGTCTCGTGCCCGACCCCGCTGAGGCCTATAT; and A3.2Xba/Stu, ATATAGGCCTCAGCGGGGTCGGGCACGAGACAGGCTGTGATATATGTCTCCCCCGCTGTCTAGAATAT) into pBSK8499-9605/Xba/Stu-mut3.1-3 after restriction with XbaI and StuI. All mutations that were introduced into SL3.1 to 3.3 that are present in the variable region of the 3′ NTR were generated by two consecutive PCRs by using primers carrying the desired mutations and PI-luc/ET-vXS as a template for the first reaction. Fragments were combined in a second PCR using primers S3.1Xba and A3.3Stu, and after restriction with XbaI and StuI fragments were inserted into pBSK8499-9605/Xba/Stu-mut3.1-3. This strategy was used to generate the following replicon constructs: PI-SL-up, PI-SL-low, PI-ΔSLup-ACA:UGU, PI-ΔSLup-AG:UC, PI-ΔSLlow-CAG:CUG, PI-Δbulge-UGC, PI-bulgeG→A, PI-bulge-random, PI-Δloop-CACAG, PI-Δloop-UAUAU, PI-5B3.2x, PI-loop-AG, PI-loop-ACG, PI-loop-AGG, PI-5B2+1, PI-5B1+1, PI-5B1+2, and PI-5B-N. Mutations in the X-tail were introduced by overlap PCR using primers carrying the desired mutations for the first PCR and rep PI-luc/ET-vXS as template and primers S8996NcoI and A9605+100 (CACCTGACGTCTAAGAAACC) for combination of the fragments. After restriction with StuI and SpeI, fragments were inserted into pBSK8499-9605/Xba/Stu-mut3.1-3/dv3.1-3 and, after verification of the nucleotide sequence of the transferred fragment, inserted into the replicon vector PI-mut3.1-3. In this way, we obtained replicon constructs PI-XT-x, PI-XT2+1, PI-XT1+1, PI-XT1+2, and PI-XT-N. In order to obtain replicon constructs with restored complementarity in the loop regions of 5BSL3.2 and SL2, SfiI-StuI and StuI-SpeI fragments derived from these plasmids were inserted into PI-mut3.1-3 In this way, we obtained replicon constructs PI-5B3.2x+XTx, PI-5B+XT2+1, PI-5B+XT1+1, PI-5B+XT1+2, and PI-5B+XT-N.
Bicistronic replicons that lack the PV IRES and monocistronic replicons were generated by transferring the SfiI-SpeI fragments from PI-mut3.1-3, PI-ins3.1-3, or PI-5B3.2+XT-x into pFK-I389luc/NS3-3′/ET (26) to generate the bicistronic RNAs or into pFK-I389luc-ubi-NS3-3′/ET (N. Appel and R. Bartenschlager, unpublished data) to generate the monocistronic replicons.
The construct pFK-nt341-sp-PI-lucUbiMyc-shiftEI3420-9605/ET (designated PI-lucubi/dup3.1-3 in this study) carries an insertion of SLs 3.1 to 3.3 upstream of the encephalomyocarditis virus (EMCV) IRES. This insertion comprises the C-terminal region of NS5B (nt 9128 to 9383) with a deletion of a guanosine residue at position 9192, which was necessary to avoid expression of a C-terminal NS5B fragment that we found to be inhibitory for RNA replication (T. Pietschmann and R. Bartenschlager, unpublished data). Replicon construct PI-lucubi/ins3.1-3 was generated by insertion of the SfiI-SpeI fragment that was isolated from PI-mut3.1-3 into PI-lucubi/dup3.1-3.
Insertion of 5BSL3.1-3 or 5BSL3.2 into the NS5A coding sequence was done by using the transfer vector pUCNS3-5B/Δ-ISDR carrying the NS3 to NS5A coding sequence in which nt 6964 to 7104 had been replaced by an in-frame linker sequence containing restriction sites for XbaI and PmeI (N. Appel, P. Friebe, and R. Bartenschlager, unpublished data). This deletion was analogous to a cell culture adaptive mutation described recently (4). Insertion of SLs 3.1 to 3.3 into the NS5A coding sequence was done by PCR using the primer pair S3.1Xba and A3.3Stu (see above) and rep PI-luc/ET as template. Fragments were restricted with XbaI and StuI and inserted in frame into this transfer vector via the XbaI and PmeI restriction site. This resulted in construct pUC NS3-5B/Δ-ISDR+3.1-3. To generate construct pUC NS3-5B/Δ-ISDR+3.2, two complementary oligonucleotides spanning the SL3.2 region were hybridized and inserted in frame into this transfer vector via the XbaI and PmeI restriction site. Plasmids pFK-nt341-sp-PI-lucEI3420-9605/T/Δ-ISDR, pFK-nt341-sp-PI-lucEI3420-9605/T/Δ-ISDR+3.2, and pFK-nt341-sp-PI-lucEI3420-9605/T/Δ-ISDR+3.1-3 (abbreviated in Results to ΔISDR, ΔISDR/dup3.2, and ΔISDR/dup3.1-3, respectively) were generated by combining the EcoRI-XhoI fragment of the corresponding donor plasmid (pUC NS3-5B/Δ-ISDR, pUC NS3-5B/Δ-ISDR+3.2, or pUC NS3-5B/Δ-ISDR+3.1-3) with the EcoRI-NotI and the NotI-XhoI fragments, obtained by restriction of pFK-nt341-sp-PI-lucEI3420-9605/T (26). Plasmids pFK-nt341-sp-PI-lucEI3420-9605/T/Δ-ISDR/mut3.1-3 and pFK-nt341-sp-PI-lucEI3420-9605/T/Δ-ISDR+3.2/mut3.1-3 (abbreviated in Results to ΔISDR/mut3.1-3 and ΔISDR/ins3.2, respectively) were obtained by transferring the SfiI-SpeI fragment of PI-mut3.1-3 into the vectors pFK-nt341-sp-PI-lucEI3420-9605/T/Δ-ISDR or pFK-nt341-sp-PI-lucEI3420-9605/T/Δ-ISDR+3.2, respectively.
In vitro transcription, RNA transfection, and transient replication assays.
Five micrograms of in vitro transcript generated from DNA templates after linearization with ScaI was mixed with 400 μl of a suspension of 107 Huh-7 cells per ml in a cuvette with a gap width of 0.4 cm (Bio-Rad, Munich, Germany). After one pulse at 975 μF and 270 V with a Gene pulser system II (Bio-Rad), cells were immediately transferred into 14 ml of complete Dulbecco's modified Eagle medium. Aliquots of the cell suspension were seeded in 3-cm-diameter culture dishes and harvested at various time points. To determine the luciferase activity, cells were washed three times with phosphate-buffered saline and scraped off the plate into 350 μl of ice-cold lysis buffer (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM dithiothreitol). One hundred microliters of lysate was mixed with 360 μl of assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM dithiothreitol, 2 mM ATP, 15 mM K2PO4, pH 7.8) and, after addition of 200 μl of a 200 μM luciferin stock solution, measured in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany) for 20 s. Values that were obtained with cells harvested 4 h after electroporation were used to correct for the transfection efficiency. All luciferase assays were done in duplicate measurements. Since the absolute number of relative light units (RLU) differed between individual experiments due to fluctuations of transfection efficiency and cell permissiveness, luciferase activities were expressed as the ratio of RLU measured 4 and 48 h after transfection. Moreover, in each experiment the relevant positive and negative control replicons were used.
Preparation of total RNA and quantification of HCV RNA by Northern hybridization.
Total RNA was prepared by a single-step isolation method as described previously (7). Five to 10 μg of total RNA was denatured by treatment with 5.9% glyoxal in 50% dimethyl sulfoxide and 10 mM sodium phosphate buffer (pH 7.0) and analyzed after denaturing agarose gel electrophoresis by Northern hybridization. Positive-strand replicon RNA was detected by hybridization with a 32P-labeled negative-sense riboprobe complementary to nt 5989 to 6699 of the HCV Con1 isolate. Hybridization with a β-actin-specific riboprobe was used to correct for total RNA amounts loaded in each lane of the gel. Northern blots were quantified by phosphoimaging using an 880E imager (Molecular Dynamics, Sunnyvale, Calif.).
Determination of RNA secondary structures by NMR spectroscopy.
The two RNAs, 5′ GGG GAG ACA UAU AUC ACA GCC UGU CUC GUG CCC GAC CC 3′ (corresponding to 5BSL3.2) and 5′ UCA CGG CUA GCU GUG AAA GGU CCG UGA 3′ (corresponding to SL2 of the X-tail), were chemically synthesized and purified by high-performance liquid chromatography. RNAs were dissolved in 20 mM NaCl-5 mM acetic buffer (pH 5.5) in 90% H2O-10% D2O at concentrations of 1.3 mM for 5BSL3.2 and 1.4 mM for SL2. One- and two-dimensional nuclear overhauser effect spectroscopy (NOESY) experiments were performed at 5 and 25°C and repeated after buffer exchange against 20 mM phosphate buffer (pH 7.0) and 20 mM NaCl. All nuclear magnetic resonance (NMR) experiments were recorded on Varian INOVA 600- and 800-MHz spectrometers equipped with a triple-resonance probe and shielded z-gradients. Water suppression was achieved in one- and two-dimensional NOESY experiments by using a Watergate sequence (36). Mixing time was set to 150 ms. Data processing was performed by using the FELIX program version 2000 (Accelrys Inc., San Diego, Calif.) and NMRpipe (8).
RESULTS
Prediction of RNA secondary structures in the 3′ coding sequence of NS5B.
Several groups have recently predicted a number of highly conserved SL structures in the HCV core and NS5B region (13, 38, 39, 45). In order to study the functional importance of these SLs for replication, we used the subgenomic replicon system. Since in this system the core coding sequence is not required for RNA replication, we focused our analysis on the SL elements in the NS5B region. By using the boundaries of the RNA structures identified primarily in the study by Tuplin and coworkers (45), we reinvestigated the 3′ terminal NS5B coding sequence of the HCV Con1 isolate from which the first replicons were derived (28). An analysis with the widely used secondary structure prediction software Mfold (59) revealed three potential SL structures in the HCV Con1 isolate (Fig. 1B). Since during the preparation of this manuscript, You and coworkers described analogous results (57), for reasons of consistency and comparison we will use their nomenclature for the different SLs throughout this report. Stem-loops 5BSL3.1 and 5BSL3.3 are analogous to the ones described by Tuplin and coworkers and were originally designated SL9011 and SL9118, respectively (45). The novel stem-loop 5BSL3.2 is clamped in between these two other RNA structures and spans nt 9263 to 9310. It consists of an 8-bp lower and a 6-bp upper stem, an 8-base bulge, and a 12-base-long upper loop.
Essential role of stem-loop 5BSL3.2 for HCV RNA replication.
In order to analyze the role of these potential SLs for RNA replication, we used a highly efficient replicon that is composed of the HCV 5′ NTR, the IRES of the poliovirus that directs the translation of the firefly luciferase reporter gene, the EMCV IRES directing translation of the HCV replicase proteins NS3 to NS5B, and the HCV 3′ NTR (9) (Fig. 1A). The highest replication levels were achieved by introducing a combination of cell culture adaptive mutations in NS3 and NS4B that were found to enhance RNA replication in a cooperative manner (E1202G, T1280I, and K1846T [26]). We have recently shown that with this replicon design, HCV RNA replication can be measured with high sensitivity and accuracy by using luciferase assays (9, 26). To facilitate genetic manipulation in the variable region of the 3′ NTR (see below), we introduced two convenient restriction sites at positions 9391 and 9402. In agreement with our recent observation that the variable region is not essential for RNA replication, we found that these nucleotide substitutions did not affect RNA amplification (data not shown). Therefore, in most subsequent analyses the replicon referred to as PI-luc/wt (Fig. 1A) was used as a reference and as a recipient for the insertion of the various mutations.
Having developed a highly efficient replicon, we next addressed the question of whether stem-loops 5BSL3.1, 5BSL3.2, and 5BSL3.3 play a role for RNA replication. To this end, we introduced a maximum of silent nucleotide substitutions in order to disturb the base pairing and thereby the RNA structures. Care was taken for the codon usage, because the introduction of rare codons might interfere with proper RNA translation, resulting in premature translation termination and C-terminally truncated NS5B proteins. Since the C-terminal membrane anchor of NS5B that is encoded in this region is essential for RNA replication, the presence of rare codons might interfere with our studies (24, 32). Finally, the mutations were tested for their impact on disturbing the SL structures by using the Mfold algorithm (data not shown).
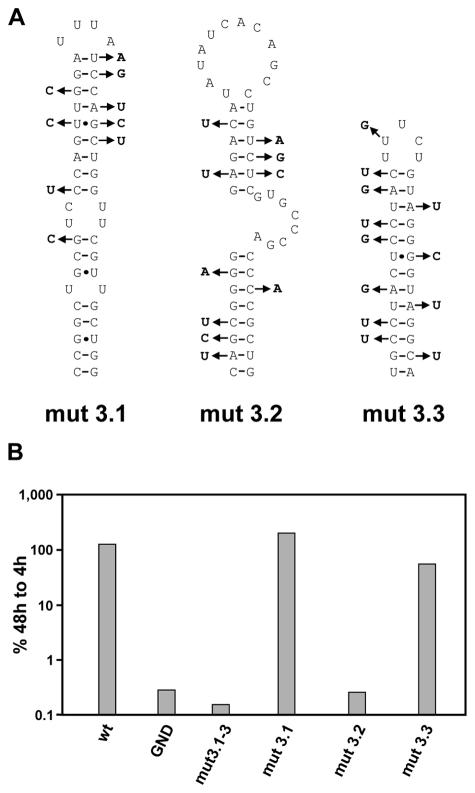
As a result of these precautions, we replaced 9 nt in 5BSL3.1, 10 nt in 5BSL3.2, and 12 nt in 5BSL3.3 (Fig. 2A). Replicon RNAs harboring these nucleotide substitutions were transfected into Huh-7 human hepatoma cells and harvested after 4, 24, 48, and 72 h, and luciferase activities were determined. In this and all subsequent experiments, replication efficiency is expressed as the ratio of RLU measured at 48 and 4 h after transfection. Since the 4-h value, which was set 100%, reflects only the translation of the input RNA, given ratios are corrected for transfection efficiency. As a negative control we used a replicon with an inactivating mutation in the GDD motif of the NS5B RNA-dependent RNA polymerase (RdRp) (PI-GND). Luciferase activities obtained with this replicon were used to determine background values. In the first set of experiments, we introduced all silent nucleotide substitutions simultaneously into the basic replicon and tested the resulting mutant, PI-mut3.1-3, in the transient replication assay. As shown in Fig. 2B, these mutations completely prevented RNA replication. To determine which of the potential SLs contributes to replication, we generated three replicons, each carrying only one disrupted SL structure. While the substitutions in 5BSL3.1 and 5BSL3.3 had no effect, the replicon carrying the mutations affecting 5BSL3.2 did not multiply to a detectable level. In summary, these results indicate that stem-loop 5BSL3.2 in the 3′ coding region of NS5B is a novel RNA element that is essential for HCV replication.
FIG. 2.
Stem-loop 3.2 is essential for HCV RNA replication. (A) Summary of the nucleotide substitutions each introduced simultaneously into the given SL structures. Note that the mutations do not affect the NS5B coding sequence. (B) Results of a transient replication assay using luciferase reporter-gene replicons specified in Fig. 1. The mutations represented in panel A were each introduced into one or all three SLs simultaneously (PI-mut3.1-3). Values represent the ratio of RLU measured at 48 and 4 h after electroporation. The 4-h values, which reflect the transfection efficiency, were set to 100%. The replicons PI-luc/wt and PI-GND served as positive and negative controls, respectively. A representative result from five independent experiments is shown.
Functional rescue of HCV replicons with disturbed SLs in NS5B.
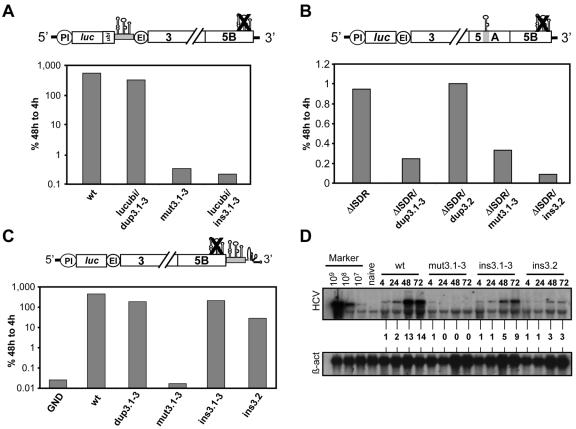
A further analysis of the essential replication elements in 5BSL3.2 was complicated by the fact that this element was part of the NS5B coding sequence. Consequently, mutations affecting the integrity of this element in most cases would alter the amino acid sequence of NS5B and thereby might interfere with its function. To overcome this limitation, we attempted to rescue replication of an RNA with scrambled 5BSLs by insertion of an intact copy of these elements or only 5BSL3.2 at different positions of the RNA. For this purpose, we chose three different positions: the region upstream of the EMCV IRES, a region in the coding sequence on NS5A that was found to be dispensable for RNA replication (4), and the variable region in the 3′ NTR that is nonessential for RNA replication (9, 10, 54-56). As shown in Fig. 3A, the insertion of all three SLs upstream of the EMCV IRES was well tolerated and reduced RNA replication only about fourfold (compare the parental replicon wild type with lucubi/dup3.1-3). In this replicon, the NS5B coding sequence spanning all three SLs was fused in the +1 frame of the luciferase reporter gene via a ubiquitin protease cleavage site. This design allowed the efficient synthesis of enzymatically active luciferase without influence by the C-terminal fusion (T. Pietschmann and R. Bartenschlager, unpublished data). Unfortunately, the insertion of 5BSL3.1 to 3.3 at this position did not rescue replication of an analogous replicon in which these RNA elements at their original position were scrambled by silent nucleotide substitutions (replicon lucubi/ins3.1-3).
FIG. 3.
Rescue of HCV RNA replication by insertion of 5BSL3.2 into the variable region of the 3′ NTR. (A) Attempts to rescue RNA replication by insertion of all three SLs upstream of the EMCV IRES. The corresponding replicon construct (PI-lucubi/dup3.1-3) directs the expression of a luciferase-ubiquitin fusion protein carrying in the +1 NS5B reading frame the 3′ coding region of NS5B in the context of the wild-type replicon. In replicon lucubi/ins3.1-3, the three 5BSLs were inserted into the mut3.1-3 replicon at the same position. The design of the lucubi/ins3.1-3 replicon is depicted in the upper panel. (B) No rescue of HCV RNA replication by insertion of 5BSL3.2 into the middle of NS5A. The replicon carrying this insertion (ΔISDR/ins3.2) is depicted above the plot, which shows representative results from a transient replication assay. In replicon ΔISDR/dup3.2, 5BSL3.2 was inserted into the same position of the parental replicon (ΔISDR), which results in a duplication of this SL. Replicon ΔISDR/dup3.1-3 carries an insertion of all 3 SLs in NS5A of the same parental replicon, which results in a duplication of all three SLs. (C) In the upper panel, the design of the replicon construct in which the three SLs in the NS5B coding region were inactivated and an intact version was inserted into the variable region of the 3′ NTR is depicted (replicon ins3.1-3). In replicon ins3.2, only SL3.2 was inserted into the 3′ NTR. Replicon dup3.1-3 carries the intact SLs in NS5B and an additional insertion of all three SLs in the variable region of the 3′ NTR. Results in panels A to C are representative of at least three independent experiments. (D) Analysis of HCV RNA replication by Northern hybridization. Replicons specified above the lanes were transfected into Huh-7 cells and harvested at given time points. HCV-specific RNA present in total RNA was quantitated by Northern hybridization and densitometry scanning. Known amounts of in vitro transcripts (Marker) spiked with total RNA from naive Huh-7 cells as carrier and total RNA from naive Huh-7 cells were used as controls. Numbers between the panels refer to relative signal intensities of HCV RNAs. Signals determined at 4 h after transfection were set to 1. β-act, β-actin RNA.
Attempts to rescue RNA replication by insertion of 5BSL3.1-3 into the center of NS5A were not possible, because this insertion into the parental replicon abolished RNA replication (ΔISDR/dup3.1-3) (Fig. 3B). In this particular construct that carried an adaptive mutation only in NS3, we generated an in-frame insertion of the NS5B coding sequence that spans SL3.1-3 into a deletion in the center of NS5A that encompasses the interferon sensitivity-determining region (ISDR) and that confers cell culture adaptation (4). When the insertion into NS5A was reduced to only 5BSL3.2, it was well tolerated when tested in the context of the parental replicon ΔISDR (mutant ΔISDR/dup3.2) (Fig. 3B). However, insertion of 5BSL3.2 in the context of the mutant with scrambled SL3.1-3 was unable to rescue replication (ΔISDR/ins3.2) (Fig. 3B).
Owing to the low replication level of RNAs with an adaptive mutation in NS3 and the NS5A deletion (4) and the possibility that rescue of RNA replication is rather inefficient, we might have missed a rescue. Therefore, we analyzed the ΔISDR/ins3.2 mutation in the context of a selectable replicon which is more sensitive than transient assays. However, in several independent experiments no G418-resistant colonies were obtained, supporting the notion that insertion of 5BSL3.2 into the center of NS5A does not restore replication of a RNA in which 5BSL3.1-3 is scrambled.
Insertion of all three SLs into the 3′ NTR of the wild-type replicon was well tolerated and reduced replication only about fivefold, showing again that the presence of two SL3.2 elements does not cause a dramatic interference. More importantly, the same insertion into the replicon in which the authentic SL3.1-3 was scrambled by silent nucleotide substitutions efficiently rescued RNA replication to a level that was only about three- to fivefold lower than that of the wild type (Fig. 3C, replicon ins3.1-3). Moreover, the insertion of only 5BSL3.2 (replicon ins3.2) also restored RNA replication, although in this case replication was about 10- to 15-fold lower than that of the wild type.
The analogous result was found when we measured RNA accumulation in a transient assay by using Northern hybridization (Fig. 3D). High-level RNA replication was found with the parental RNA, whereas only input RNA was detected 4 h after transfection in the case of the mutant in which all three SLs were disturbed (mut3.1-3) as well as the negative control with the inactivated NS5B RdRp (GND) (data not shown). In agreement with the luciferase assays, an unambiguous rescue of RNA replication was observed, with the replicons carrying intact copies of 5BSL3.1 to 3.3 or only 5BSL3.2 in the variable region (ins3.1-3 and ins3.2, respectively). These results identify 5BSL3.2 as a novel RNA replication element that is not strictly position dependent and can be translocated to a different region in the 3′ NTR of the HCV genome.
Mapping of the regions in 5BSL3.2 essential for RNA replication.
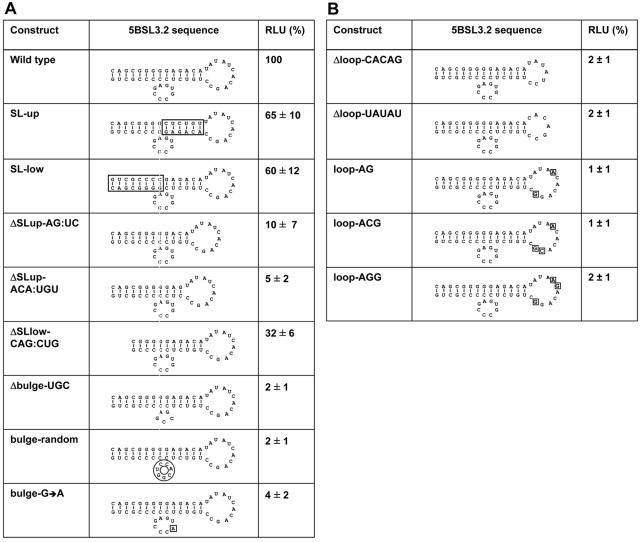
The development of a replicon that allows the manipulation of 5BSL3.2 without affecting the NS5B coding sequence enabled a more detailed mutation analysis of the elements in this SL that are crucial for RNA replication. Since the highest RNA levels were achieved with the replicon in which all three SLs were inserted into the variable region of the 3′ NTR, this replicon was used for the mutation analysis summarized in Fig. 4. When the primary sequence of the upper or the lower part of the stem of 5BSL3.2 was altered by inverting its orientation, RNA replication was only moderately affected (replicons SL-up and SL-low). In contrast, shortening the upper stem by 2 or 3 bp drastically reduced or almost completely blocked replication (ΔSL-up-AG:UC or ΔSL-up-ACA:UGU, respectively). A similar result was found when the lower stem was truncated by 3 bp (ΔSL-low-CAG:CUG). Mutations affecting the bulge were in all cases inhibitory. A deletion of 3 nt of the bulge (Δbulge-UGC) or a randomization of its sequence completely blocked RNA replication, providing evidence that the bulge plays a very important role. In fact, even a single nucleotide exchange reduced RNA replication about 25-fold (bulge-G→A). Of equal importance was the upper loop, where two deletions (Δloop-CACAG and Δloop-UAUAU) and several nucleotide substitutions completely blocked RNA replication (Fig. 4B). These observations show that the lengths but not the primary sequences of the upper and lower stem are important for RNA replication. Moreover, the data suggest that both the bulge and the loop region are the crucial determinants for replication.
FIG. 4.
Effect of mutations targeting the stem or the bulge (A) or the loop (B) region of 5BSL3.2 on RNA replication. All given mutations were introduced into 5BSL3.2 in the variable region in the context of replicon PI-ins3.1-3, which has an unaltered structure (wild type). The names of the replicons are given on the left, the introduced mutations are shown in the middle in the context of 5BSL3.2, and the results of transient replication assays are given in the right. Percent values of RLU represent means and variation ranges from three independent experiments. Note that conservation of the structure of 5BSL3.2 with the mutants serves only for clarity and does not consider the possibility that individual mutations may affect the folding of 5BSL3.2.
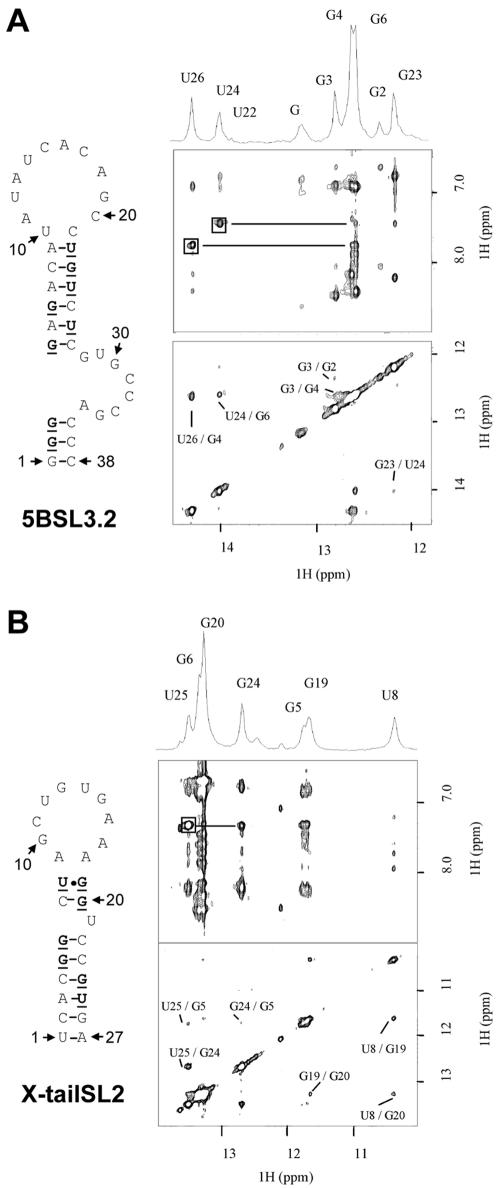
Determination of secondary structures of stem-loops 5BSL3.2 and SL2 in the 3′ NTR by using NMR.
In the course of our genetic study of 5BSL3.2, we realized that the upper loop region was complementary to 7 nt in the loop of SL2 in the X-tail. This observation suggested that a kissing-loop interaction might take place and, given the importance of 5BSL3.2 for RNA replication, is of functional relevance. An essential prerequisite of this assumption is that the complementary region in SL2 of the X-tail is single stranded. For 5BSL3.2, the results of our genetic analysis are in full agreement with the secondary structure prediction depicted in Fig. 1B, but no experimental evidence was yet available to demonstrate the presence and stability of this type of secondary structure in solution. The same is true for SL2 in the X-tail region, and therefore we wanted to determine the secondary structures of these two RNA elements by using homonuclear NMR analysis. This method allows the identification of imino protons involved in different types of base pairing. Under standard conditions, imino protons of unfolded nucleotides exchange too fast with water to be detectable by NMR. In contrast, when hydrogen is bonded to or shielded from the solvent, the imino protons are protected from exchange and give rise to detectable NMR signals. For Watson-Crick base pairing, an imino proton which is hydrogen bonded to a nitrogen gives a resonance between 11.5 and 13.5 ppm and 13 to 15 ppm for A-U base pairs (50). In base pairs where the imino protons are hydrogen bonded to a carboxyl oxygen, such as U-U, G-U, and G-A, the imino protons are observed between 10 and 12 ppm. To completely assign the imino region, two-dimensional NOESY can be used to identify the proximity between imino protons of adjacent base pairs. Furthermore, the NOESY cross-peak pattern permits one to distinguish between G-C and A-U base pairs by using the characteristic cross peak between the uracil imino and the H2 amino proton of the adenosine versus the guanine imino and the H2 amino protons of the cytosine. The expected G-U wobble base pair can be discerned by its characteristic strong imino-imino NOE cross peaks.
One- and two-dimensional spectra recorded on the sequence of 5BSL3.2 are shown in Fig. 5A. Nine imino proton resonances can be observed. Two strong peaks at 13.98 and 14.25 ppm and a weak peak at 13.95 ppm are assigned to U-A base pairs. The other six imino resonances can be assigned to Watson-Crick G-C base pairs. Five of them can be correlated by sequential NOE with the three U-A base pairs. This network of sequential correlation between adjacent base pairs is in very good agreement with the stem shown in Fig. 5A (G2 to A9 and U22 to C37). For the last G imino observed with a small intensity at 13.12 ppm, we found a strong intensity dependence on pH and temperature. A likely assignment for this imino resonance is the G1 at the 5′ terminus exposed to exchange. One-dimensional spectra recorded at different salt concentrations and pHs show no drastic differences in the imino region. The conservation of base pairing under different conditions indicates a relatively high stability of 5BSL3.2 secondary structure.
FIG. 5.
NMR analysis of the secondary structure of 5BSL3.2 (A) and SL2 of the X-tail (B). The predicted secondary structures are each shown in the left panel, and the NMR spectra are shown on the right. Bold characters in the secondary structure indicate nucleotides with a detectable imino proton. One- and two-dimensional NOESY spectra recorded on 5BSL3.2 (A) and SL2 (B) are presented. For the two-dimensional NOESY spectra, only the imino-imino region (bottom) and imino-amino/H2 region (top) are shown. Imino-imino region spectra are labeled with sequential assignments, and complete assign-ments are reported on the one-dimensional spectrum. The imino-imino region contains NOE cross peaks used to connect adjacent base pairs. For the imino-amino/aromatic region, NOE connectivities between U imino and A H2 are squared. Imino-amino NOE expected for G-C base pairs are also observed in this region, as well as sequential imino-amino/H2 NOE. Samples were dissolved at pH 5.5, and experiments were recorded at 25°C.
One- and two-dimensional spectra recorded on the sequence of the X-tail SL2 are shown in Fig. 5B. Imino protons of a G-U base pair can be identified at 11.63 and 10.34 ppm and can correspond to U8 to G19 base pairing shown in the predicted structure. From those iminos, sequential connectivity was made through four successive G-C base pairs (G20, G6, G5, and G24) followed by an A-U base pair with an imino resonance at 13.5 ppm (U25). Three other weak peaks can be observed in the one-dimensional spectrum at 12.07, 12.5, and 13.6 ppm and could be assigned to base pairs positioned at the extremity of a stem (G26 and U1) or in the loop. Exchange with water caused by fraying of the terminal base pair of a stem can explain the weak intensity of those imino resonances. Based on this evidence, the secondary structure presented in Fig. 5B is in good agreement with the NMR data.
Genetic evidence for a kissing-loop interaction between 5BSL3.2 and SL2 in the 3′ NTR.
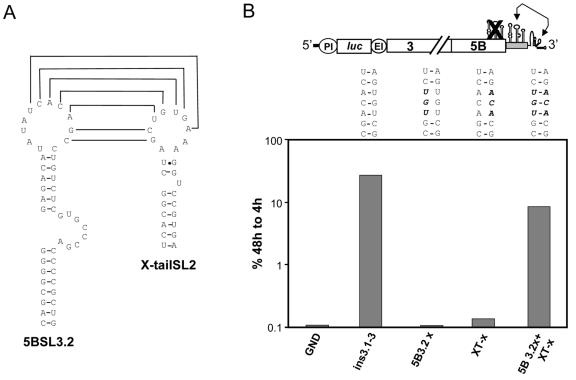
The structure analysis described above confirmed that the complementary sequences in 5BSL3.2 and SL2 reside in a single-stranded loop region, which is a prerequisite for base pairing (Fig. 6A). This result prompted us to test whether the complementarity is required for RNA replication. For this purpose, we created three mismatches by introducing nucleotide substitutions either into 5BSL3.2 or SL2 in the context of the replicon carrying an intact copy of the NS5B SLs in the 3′ variable region (Fig. 1A, replicon ins3.1-3). As shown in Fig. 6B, the reduction of the total complementarity to each 2 nt at the borders of the complementarity region led to a complete block of RNA replication (replicons 5B3.2x and XT-x). However, when full complementarity was restored, RNA replication was rescued even though the primary sequences of both loops had been altered (replicon 5B3.2x+XT-x). This result supported our assumption that a kissing-loop interaction required for RNA replication takes place at the 3′ end of the HCV genome. The fact that replication was not rescued to the level of the wild-type replicon suggests that the primary sequence plays an important role in determining the efficiency of RNA replication.
FIG. 6.
Evidence for a kissing-loop interaction between the loops of 5BSL3.2 and SL2 in the X-tail sequence. (A) Predicted secondary structures of 5BSL3.2 and X-tail SL2, with complementary nucleotides in the loop regions indicated by connecting lines. (B) The nucleotide sequences of the complementary loop regions in 5BSL3.2 and X-tail SL2 are depicted in the middle panel. Noncomplementary nucleotides are indicated in bold letters. A representative result from a transient replication assay is shown in the lower panel. Mutations specified in the upper panel were introduced into the replicon PI-ins3.1-3 and analyzed for transient replication as described in the legend to Fig. 1. The basic replicon used for this analysis is depicted at the top.
Complementarity but not primary sequence of the interacting loop sequences is the major determinant for RNA replication.
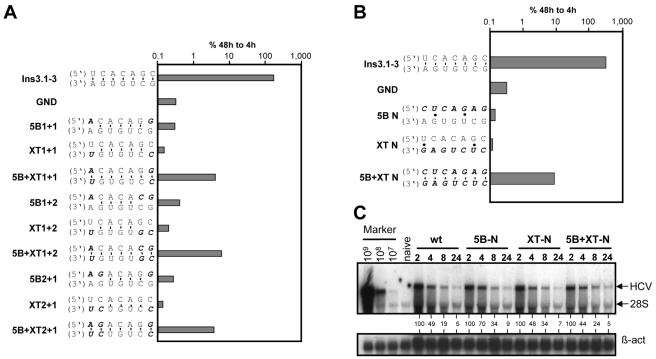
In order to characterize the sequence requirements of the interacting loops in more detail, a series of replicons that were derived from PI-ins3.1-3 (Fig. 1A) was generated, in which various nucleotides in the complementarity region were exchanged in 5BSL3.2, SL2, or both. In the latter case, substitutions were designed to restore the possibility for base pairing. As shown in Fig. 7A, whenever complementarity was affected, RNA replication was reduced to below the detection limit. This result was irrespective of whether nucleotides were exchanged in 5BSL3.2 (replicons 5B1+1, 5B1+2, and 5B2+1) or SL2 in the X-tail (replicons XT1+1, XT1+2, and XT2+1). When base pairing was fully restored, RNA replication was restored, too, albeit to about 50-fold-lower levels than in the wild type (replicons 5B+XT1+1, 5B+XT1+2, and 5B+XT2+1).
FIG. 7.
Primary sequence requirements for the kissing-loop interaction. (A) Analysis of the minimal length required for the kissing-loop interaction. The mutations introduced into PI-ins3.1-3 are specified on the left, and a representative result of a transient replication assay is shown in the graph on the right. (B) Randomization of the loop sequences in either 5BSL3.2 (replicon 5B-N) or SL2 in the X-tail (replicon XT-N) or both loops (replicon 5B+XT-N). Mutations were introduced and tested in the context of replicon PI-ins3.1-3. Representative results from three independent experiments are shown. (C) Determination of the RNA stability of different mutants in Huh-7 cells as determined by Northern hybridization. Ten micrograms of the given replicon RNA was transfected into Huh-7 cells that were harvested at the time points specified above the lanes. Positive-strand RNA was detected by Northern hybridization as described in the legend to Fig. 3. Numbers below the lanes refer to the relative HCV RNA signal intensities after correction for RNA amounts by using β-actin (β-act). The value obtained with the 2-h sample was set at 100%. Given amounts of in vitro transcripts (Marker) and total RNA from naive Huh-7 cells were used as positive and negative controls for this blot, respectively.
Finally, we wanted to know whether the primary sequence of the complementarity region or the possibility for base pairing is the major determinant for RNA replication. Therefore, we generated three replicon constructs in which the seven complementary nucleotides were randomized in 5BSL3.2 or SL2 or both in a way to restore complementarity. The result of a transient assay shown in Fig. 7B demonstrates that RNA replication occurs as long as full complementarity is preserved (replicon 5B+XT-N), even when most of the loop sequences of 5BSL3.2 and SL2 are randomized. However, replication of this RNA was reduced about 50-fold compared to the parental reference replicon (ins3.1-3).
Several possibilities could account for this reduction. First, the mutations may affect the stability of the different replicon RNAs. To address this possibility, we determined the half-lives of the replicons with the randomized loop regions after transfection into Huh-7 cells by Northern hybridization. In order to exclude changes of RNA levels due to replication, in all three replicons the same amino acid substitution was introduced into the GDD motif of NS5B that inhibits RdRp activity. As shown in Fig. 7C, all RNAs decayed with comparable kinetics, demonstrating that the mutations in the loop sequences did not affect RNA half-lives. Second, the transposition of the three SLs into the variable region of the 3′ NTR and thereby the alteration of the spacing between 5BSL3.2 and SL2 might have caused the reduction of RNA replication. To address this possibility, we introduced four silent nucleotide substitutions into the loop of 5BSL3.2 of the parental PI-luc/wt replicon (Fig. 1A) in order to disturb the complementarity to SL2 in the X-tail without affecting the positions of the interacting loops. These mutations completely blocked RNA replication (data not shown). However, when complementarity was restored in this replicon by corresponding substitutions in SL2, RNA replication was restored, too; the efficiency was again about 100- to 200-fold lower than that of the wild type but still at least 10-fold above background (data not shown). This result argues that the altered spacing in replicon ins3.1-3 is not responsible for the reduction of RNA replication (data not shown). Third, a thermodynamically less favorable interaction may occur between the kissing loops. This could be caused by an alteration of the folding of 5BSL3.2 or SL2 in the X-tail. However, it should be pointed out that the number of purines and pyrimidines in the loops was the same for all constructs (Fig. 7B). Fourth, the loop region of 5BSL3.2 may be involved in some protein interaction for which the primary sequence is important (see Discussion). Presently, we cannot distinguish between these possibilities. However, the results clearly show that a kissing-loop interaction between a 7-nt-long complementary sequence in 5BSL3.2 and SL2 in the X-tail is required for RNA replication. This result implies that at least a transient long-range RNA-RNA interaction takes place at the 3′ end of the HCV genome.
Rescue of RNA replication is independent from replicon context.
The analyses described thus far were performed with a bicistronic replicon carrying the PV IRES to direct translation of the reporter gene. To determine whether the RNA-RNA interaction at the 3′ end of this replicon would be affected by this heterologous sequence at the 5′ end, a representative set of mutations affecting the complementarity region of 5BSL3.2 was analyzed in two alternative replicon vectors: first, a bicistronic replicon in which the luciferase reporter gene was translated under control of the HCV IRES, and second, a monocistronic RNA that lacked any heterologous regulatory sequence (Fig. 8). In both replicon settings, disruption of the structures of the three SLs in the NS5B coding region by the silent nucleotide substitutions depicted in Fig. 2A blocked RNA replication (HI/mut3.1-3 and mono/mut3.1-3 [Fig. 8B and C, respectively]). Upon insertion of an intact version of SL3.1-3 into the variable region of the 3′ NTR of these replicons, RNA replication was restored (Fig. 8, ins3.1-3 replicons). RNA replication was also restored in both replicon systems when the complementarity region was altered by 3 nt without affecting base pairing (5B3.2+XT-x replicons) (nucleotide sequences of the altered loops are depicted in Fig. 6B). Thus, the kissing-loop interaction in the 3′ NTR was not affected by the heterologous elements present in the 5′ regions of the replicons.
FIG. 8.
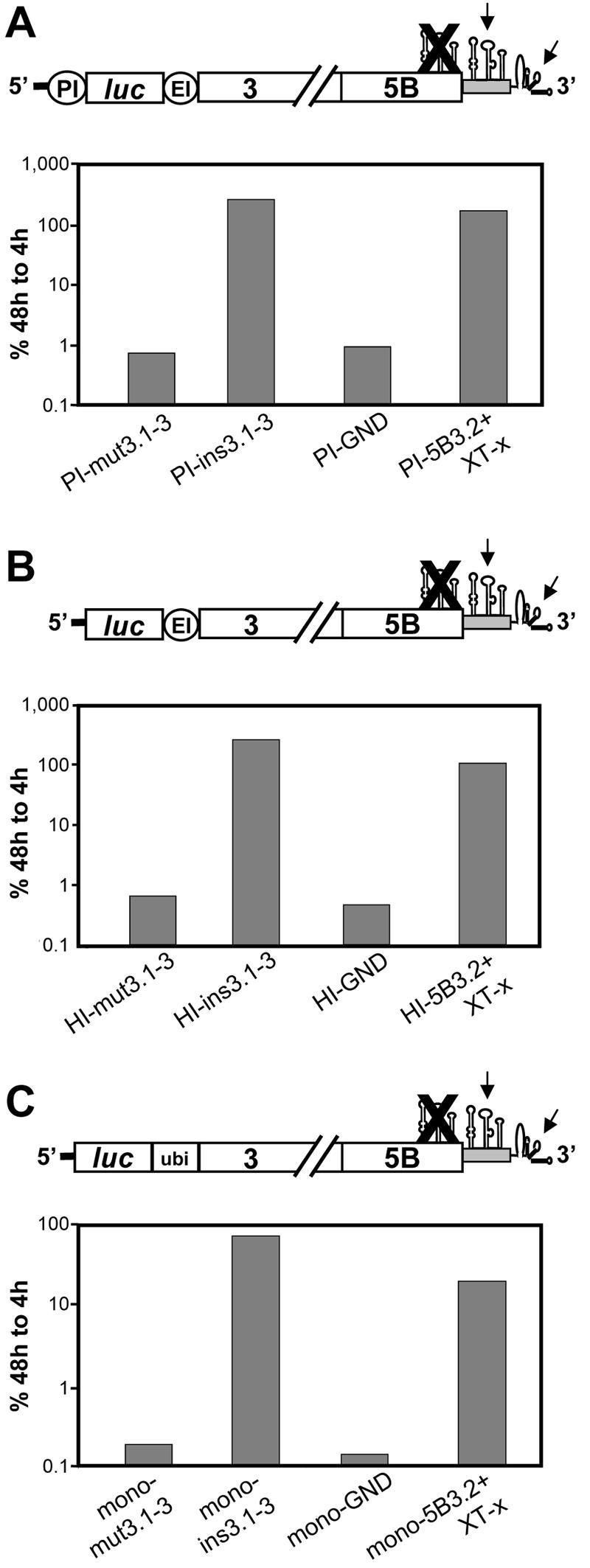
The kissing-loop interaction is not affected by the replicon design. Representative results from transient replication assays using replicons with disturbed 5BSL3.1 to 3.3 structures (mut3.1-3 constructs), insertion of the three SLs into the variable region (ins3.1-3 constructs), an inactive NS5B RdRp (PI-GND, HI-GND, or mono-GND), or altered loop sequences in 5BSL3.2 and SL2 that are fully complementary (5B3.2+XT-x constructs). The replicons shown in panel A carry the PV IRES, replicons in panel B carry only the HCV IRES (solid line), and replicons in panel C are monocistronic and do not contain heterologous IRES elements. In the case of the latter, the luciferase reporter is released from the polyprotein by a ubiquitin-dependent pathway. This result was achieved by in-frame fusion of luciferase with ubiquitin and the NS3 to 5B polyprotein fragment. The ins3.1-3 constructs are each depicted in the top panels.
DISCUSSION
In this report, we identified a novel RNA element in the 3′ terminal coding sequence of NS5B that is essential for RNA replication. This result is in remarkable agreement with the results described recently by You and coworkers (57). By using computer-based secondary structure prediction and enzymatic and chemical probing, they identified the very same RNA element in the NS5B coding region. Moreover, they also found that only 5BSL3.2 out of several other SLs in close proximity, including 5BSL3.1 and 5BSL3.3, is essential for RNA replication. However, in contrast to their study, we were able to efficiently rescue RNA replication by insertion of 5BSL3.1 to 3.3 or only 5BSL3.2 into the variable region of the 3′ NTR. This approach allowed us to analyze the individual elements within 5BSL3.2 in more detail, because we could genetically separate the coding function of this RNA sequence from its role in RNA replication. By using this replicon construct design we were able to demonstrate that the stem structures but not their primary sequences are required for RNA replication. Nucleotide sequences comprising the stems could be inverted, resulting in a complete alteration of their primary sequences without affecting the stem structure of 5BSL3.2 as predicted by Mfold and with little effect on RNA replication. In contrast, truncations of the stem, which resulted in ambiguous predictions of alternative structures, reduced RNA replication severely. Deletions in the bulge or randomization of its sequence were inhibitory, and even a single nucleotide substitution drastically reduced RNA replication. Presently, it is unclear whether the bulge sequence per se or its impact on folding of 5BSL3.2 is important. You and coworkers (57) reported mutations in this region that could influence the overall folding of the SL, suggesting that the bulge has a very important structural role.
Insertion of 5BSL3.2 alone into the variable region of the 3′ NTR was sufficient to rescue RNA replication of a replicon in which all three SLs in the NS5B coding region were disturbed, showing that 5BSL3.2 can act as an autonomous RNA element. However, replication efficiency was about fivefold lower than with the insertion of all three SLs into the variable region. This reduction could be due to the fact that the SLs flanking 5BSL3.2 promote its proper folding, e.g., by preventing unfavorable base pairings with neighboring sequences in the variable region. Alternatively, the alteration of the spacing between SL2 in the X-tail and 5BSL3.2 transposed into the 3′ NTR may be less favorable for the kissing-loop interaction, which is to some extent compensated by the presence of flanking 5BSL3.1 and 5BSL3.3. In its original position in the NS5B coding sequence, these requirements may be more relaxed due to an adequate spacing between 5BSL3.2 and SL2 in the X-tail. Finally, it should be kept in mind that the replicon system used here can only measure effects on RNA replication. We can therefore not exclude other functions for 5BSL3.1 and 5BSL3.3 in the HCV life cycle, like RNA packaging and virion assembly.
It is noteworthy that the novel HCV RNA replication element is positioned within the coding region, whereas, until recently, all other elements known resided within the NTRs. In principle, this positioning may be required for regulating the translation of NS5B, e.g., by slowing down ribosome movement along the RNA template, which in turn would affect RNA replication due to reducing the amounts of RdRp. Alternatively, the location at the 3′ end of the ORF could facilitate the binding of newly synthesized NS5B in order to initiate negative-strand RNA synthesis which would interfere with translation. In this study, we did not analyze in detail the effects of the mutations in 5BSL3.2 on RNA translation. However, even an altered 5BSL3.2 sequence that disturbed the RNA structure still allowed expression of a functional NS5B, because the insertion of the intact version of 5BSL3.2 into the variable region rescued RNA replication. In addition, You and coworkers (57) reported no significant differences in the expression of viral proteins from the wild type and 5BSL3.2 mutants in vitro.
One of the best-studied groups of CREs that reside within the coding region are those of viruses that initiate replication with a protein primer, like adenoviruses, hepadnaviruses, and picornaviruses. For instance, in the case of hepatitis B virus, reverse transcription is primed by the addition of the first 4 nt to the terminal protein in a reaction that uses the bulge of the RNA packaging SL structure close to the 5′ end of the RNA pregenome as a template (17, 49). Upon addition of the first 4 nt to the primer, it is translocated to a complementary sequence close to the 3′ end of the pregenome to allow synthesis of minus-strand DNA. A very similar mechanism is operating with picornaviruses (11, 30, 31). Moreover, the CRE that serves as a template for the nucleotidylation of the protein primer VPg can be translocated to different regions within the genome. For instance, in the case of HRV-14, the 3′ NTR appears to interact with the CRE site some 5,000 nt upstream (31). For the FMDV, it was shown that the CRE can be translocated from the 5′ NTR into the 3′ NTR, although the resulting viruses appear to replicate at significantly lower levels (30). A similar result was found with the PV, where the translocation of CRE into the 5′ NTR also rescued replication of a CRE mutant (11). In contrast to these observations, 5BSL3.2 of HCV can be translocated only within a rather small region of the 3′ NTR, whereas its translocation to distant sites (the middle of NS5A or upstream of the EMCV IRES) does not restore replication of replicons with scrambled 5BSLs.
Only few examples for interactions between a viral CRE in the coding region and a CRE in the 3′ NTR have been described. One is bacteriophage Qβ, where a long-range pseudoknot that is formed by base pairing between 8 nt in the loop of the 3′ terminal hairpin and a single-stranded interdomain sequence located about 1,200 nt upstream in the coding region of the replicase subunit was repeated (20). A similar long-range interaction was found with the arterivirus porcine reproductive and respiratory syndrome virus (47). Within ORF7 of the viral positive-strand RNA genome, a stable hairpin structure composed of a 12-base upper loop and a 7-bp stem was found. RNA replication depends on an interaction of 7 nt of this loop with a complementary sequence in the loop of an SL structure located about 300 nt downstream in the 3′ NTR. Mutational disruption of the complementarity inhibited RNA replication, which was restored upon reestablishment of the possibility to form base pairings (47). It was speculated that this kissing-loop interaction stabilizes an RNA conformation within the 3′ terminal region of the viral genome, onto which the viral replicase complex assembles. By analogy, the complex pseudoknot interaction at the 3′ end of the HCV genome may also be part of the promoter for negative-strand RNA synthesis.
Our results are very similar to those described for porcine reproductive and respiratory syndrome virus replication. We found that a complementarity of at least 6 nt is required for HCV RNA replication, because the introduction of just two mismatches led to a complete inactivation of an HCV replicon. Moreover, You and coworkers (57) described a replication-defective mutant in which only 1 nt in the upper loop of 5BSL3.2 was altered. Based on our data, the most likely explanation is that this single nucleotide substitution also affected the kissing-loop interaction.
Within the upper loop of 5BSL3.2, a CACAGC sequence motif that is involved in the kissing-loop interaction is found. Interestingly, this sequence, which is virtually invariant among HCV genotypes, is also found in CREs of even distantly related flaviviruses, like Kunjin virus, West Nile virus, or Dengue virus (reviewed in reference 29). The 3′-terminal ∼95 nt of these viruses form a conserved SL structure that in the upper loop carries the highly conserved pentanucleotide motif CACAG that at least for Kunjin virus was shown to be crucial for RNA replication (18). Given this high conservation be-tween different flaviviruses, we assume that cellular proteins that are expressed in various tissues and conserved between different species are involved in the formation of a replication complex at the 3′ end of these genomes. In fact, it has been shown that HCV RNA replication is possible in mouse cells, showing that liver cell-specific factors are not required for RNA replication (reference 58 and N. Appel and R. Barten-schlager, unpublished data).
In spite of the genetic evidence for the kissing-loop interaction, attempts to demonstrate it by NMR analysis were not successful. You and coworkers found that in the context of a subgenomic replicon RNA, at least the upper loop of 5BSL3.2 was susceptible to cleavage by lead(II) and RNase T1, indicating that this region was single stranded under their experimental conditions (57). One likely possibility is that the kissing-loop interaction is mediated by viral or host cell proteins. In fact, multiple proteins have been reported to bind to the 3′ NTR, including polypyrimidine tract-binding protein, which binds to SL1 and SL2 in the X-tail, and the poly(U/UC) tract (16, 44), autoantigen La (40), and several ribosomal proteins (51). Thus far, the role of these proteins for HCV RNA replication has not been studied. However, more recently, cellular proteins binding to the 3′ NTR of the closely related pestivirus bovine viral diarrhea virus were identified and shown to be required for RNA replication (14). These proteins belong to a class of nuclear factors that appear to bring about a circular conformation of the bovine viral diarrhea virus genome via interactions with the 3′ and the 5′ NTR. It will be interesting to determine whether a protein(s) of this class is involved in the kissing-loop interaction of the HCV genome.
Apart from cellular proteins, the viral RdRp may be involved in the kissing-loop interaction in the HCV genome, too. On one hand, several groups described rather nonspecific binding of NS5B to homo- and heteropolymeric templates (3, 27). On the other hand, Oh and coworkers reported a high-affinity binding of this protein to the poly(U/UC) tract and, in particular, to SL2 of the X-tail (33). Yamashita and colleagues found that the level of in vitro RNA synthesis with a 98-nt X-tail RNA was relatively low compared to that of a 480-nt RNA containing the 3′ terminal coding region of NS5B (53). In addition, Cheng and coworkers reported a strong binding of NS5B to a ∼300-nt-long RNA spanning the 3′-terminal NS5B coding region and including part of the poly(U/UC) tract (6). Interestingly, this interaction could be completed with the same RNA much more efficiently than with RNAs corresponding to the X-tail or the 5′ NTR. Based on these observations, we speculate that NS5B is involved in the kissing-loop interaction by binding to sequences that include 5BSL3.2. This interaction may be the first step in the initiation of negative-strand RNA synthesis and may facilitate the transfer of the viral polymerase to the very 3′ end of the viral genome.
Acknowledgments
We thank U. Herian for excellent technical assistance; I. L. Hofacker and P. F. Stadler, University of Vienna, Vienna, Austria, and C. J. Pleij and R. Olsthoorn, University of Leiden, Leiden, The Netherlands, for initial advice on RNA structures; and J. Neyts, University of Leuven, Leuven, Belgium, for provision of the NS5B-specific inhibitor. The critical reading of the manuscript by S. Sparacio and T. Pietschmann and help in preparing the figures by M. Frese are gratefully acknowledged.
This work was supported by grants from the Sonderforschungsbereich 638 (Teilprojekt A5) and a grant from the Bristol-Myers Squibb Foundation.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Behrens, S. E., L. Tomei, and R. DeFrancesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J.-U., M.-F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 9.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofacker, I. L., M. Fekete, C. Flamm, M. A. Huynen, S. Rauscher, P. E. Stolorz, and P. F. Stadler. 1998. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 26:3825-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, T., and M. M. C. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, T., and M. C. Lai. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254:288-296. [DOI] [PubMed] [Google Scholar]
- 17.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khromykh, A. A., N. Kondratieva, J. Y. Sgro, A. Palmenberg, and E. G. Westaway. 2003. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 77:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, Y. K., C. S. Kim, S. H. Lee, and S. K. Jang. 2002. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 290:105-112. [DOI] [PubMed] [Google Scholar]
- 20.Klovins, J., and J. van Duin. 1999. A long-range pseudoknot in Qbeta RNA is essential for replication. J. Mol. Biol. 294:875-884. [DOI] [PubMed] [Google Scholar]
- 21.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, K. J., J. Choi, J. H. Ou, and M. M. Lai. 2004. The C-terminal transmembrane domain of hepatitis C virus (HCV) RNA polymerase is essential for HCV replication in vivo. J. Virol. 78:3797-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 29.Markoff, L. 2003. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 59:177-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason, P. W., S. V. Bezborodova, and T. M. Henry. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76:9686-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moradpour, D., V. Brass, E. Bieck, P. Friebe, R. Gosert, H. E. Blum, R. Bartenschlager, F. Penin, and V. Lohmann. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 78:13278-13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh, J. W., T. Ito, and M. C. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 36.Piotto, M., V. Saudek, and V. Sklenar. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2:661-665. [DOI] [PubMed] [Google Scholar]
- 37.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 38.Rijnbrand, R., P. J. Bredenbeek, P. C. Haasnoot, J. S. Kieft, W. J. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, D. B., and P. Simmonds. 1997. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J. Mol. Evol. 45:238-246. [DOI] [PubMed] [Google Scholar]
- 40.Spangberg, K., L. Goobar-Larsson, M. Wahren-Herlenius, and S. Schwartz. 1999. The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3′ untranslated region. J. Hum. Virol. 2:296-307. [PubMed] [Google Scholar]
- 41.Tanaka, T., N. Kato, M. J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419-427. [DOI] [PubMed] [Google Scholar]
- 44.Tsuchihara, K., T. Tanaka, M. Hijikata, S. Kuge, H. Toyoda, A. Nomoto, N. Yamamoto, and K. Shimotohno. 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 71:6720-6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuplin, A., J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8:824-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the VIIth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 47.Verheije, M. H., R. C. Olsthoorn, M. V. Kroese, P. J. Rottier, and J. J. Meulenberg. 2002. Kissing interaction between 3′ noncoding and coding sequences is essential for porcine arterivirus RNA replication. J. Virol. 76:1521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vrolijk, J. M., A. Kaul, B. E. Hansen, V. Lohmann, B. L. Haagmans, S. W. Schalm, and R. Bartenschlager. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201-209. [DOI] [PubMed] [Google Scholar]
- 49.Wang, G. H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wijmenga, S., M. Mooren, and C. W. Hilbers. 1993. NMR macromolecules: a practical approach, p. 217-288. IRL Press, New York, N.Y.
- 51.Wood, J., R. M. Frederickson, S. Fields, and A. H. Patel. 2001. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 75:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada, N., K. Tanihara, A. Takada, T. Yorihuzi, M. Tsutsumi, H. Shimomura, T. Tsuji, and T. Date. 1996. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology 223:255-261. [DOI] [PubMed] [Google Scholar]
- 53.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 54.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]