Abstract
Colony opacity phase variation in Acinetobacter baumannii strain AB5075 is regulated by a reversible high‐frequency switch. Transposon mutagenesis was used to generate mutations that decreased the opaque to translucent switch and a gene encoding a predicted periplasmic membrane fusion component of a resistance–nodulation–cell division (RND)‐type efflux system was isolated. This gene was designated arpA and immediately downstream was a gene designated arpB that encodes a predicted membrane transporter of RND‐type systems. A nonpolar, in‐frame deletion in arpA resulted in a 70‐fold decrease in the opaque to translucent switch. An arpB::Tc mutant exhibited a 769‐fold decrease in the opaque to translucent switch. However, the translucent to opaque switch was largely unchanged in both the arpA and arpB mutants. The arpA and arpB mutants also exhibited increased surface motility in the opaque form and the arpB mutant exhibited increased susceptibility to aminoglycosides. The arpA and arpB mutants were both attenuated in a Galleria mellonella model of virulence. A divergently transcribed TetR‐type regulator ArpR was capable of repressing the arpAB operon when this TetR regulator was overexpressed. The arpR gene was also involved in regulating the opaque to translucent switch as an in‐frame arpR mutation decreased this switch by 1,916‐fold.
Keywords: Acinetobacter, phase variation, RND efflux system
1. Introduction
Acinetobacter baumannii is a Gram‐negative pathogen typically associated with infections in hospital settings (Bergogne‐Berezin & Towner, 1996; Gootz & Marra, 2008; Joly‐Guillou, 2005; Peleg, Seifert, & Paterson, 2008). Although most A. baumannii infections are seen in immunocompromised patients or those with severe injuries, community acquired infections and infections in otherwise healthy patients have increased in recent years (Antunes, Visca, & Towner, 2014; Charnot‐Katsikas et al., 2009; Guerrero et al., 2010; Lowman, Kalk, Menezes, John, & Grobusch, 2008). This development, combined with the increasing frequency of multidrug resistance, has made A. baumannii an extremely problematic pathogen for clinicians to treat and mortality rates for these infections has approached 70% (Lee, Chen, Wu, Huang, & Chiu, 2014). It has become widely recognized that new therapies are needed to help combat these infections (Gootz & Marra, 2008; Hujer et al., 2006; Joly‐Guillou, 2005; Scott et al., 2007).
Members of the resistance–nodulation–cell division (RND) class of efflux systems in Gram‐negative bacteria are composed of three proteins: an inner membrane transporter, an outer membrane protein that serves as a pore, and a periplasmic adapter protein that interacts with both the inner and outer membrane proteins to form a conduit for the extrusion of small molecules. RND‐type systems typically capture toxic compounds or metabolites and remove them from the cell, and because of this function they can be involved in resistance to antibiotics, disinfectants, and heavy metals (Alvarez‐Ortega, Olivares, & Martinez, 2013; Anes, McCusker, Fanning, & Martins, 2015; Delmar, Su, & Yu, 2014; Magnet, Courvalin, & Lambert, 2001; Routh et al., 2011; Venter, Mowla, Ohene‐Agyei, & Ma, 2015). They have also been shown to have roles in virulence, resistance to host antimicrobial peptides, and in cellular homeostasis by removing excess metabolites (Helling et al., 2002; Warner, Folster, Shafer, & Jerse, 2007). Acinetobacter baumannii possesses a number of RND‐type efflux systems that have roles in antibiotic resistance, virulence, and biofilm formation (Damier‐Piolle, Magnet, Bremont, Lambert, & Courvalin, 2008; Magnet et al., 2001; Yoon et al., 2015), and these systems are reviewed in Coyne, Courvalin, and Perichon (2011). The expression of these genes in A. baumannii is often regulated at the level of transcription by activator and/or repressor proteins (Lin, Lin, & Lan, 2015; Marchand, Damier‐Piolle, Courvalin, & Lambert, 2004; Rosenfeld, Bouchier, Courvalin, & Perichon, 2012).
Recently, our laboratory described a high‐frequency switch that results in the interconversion between opaque and translucent colony opacity phenotypes (Tipton, Dimitrova, & Rather, 2015). This switch is mediated by an unknown mechanism, but is stimulated at high cell density. Unique phenotypes are associated with each colony variant. For example, the opaque variants are more motile, highly virulent, and exhibit higher levels of resistance to aminoglycosides (Tipton et al., 2015). In contrast, translucent variants are more adept at forming biofilms on both polystyrene and glass.
To begin understanding the mechanism underlying this high‐frequency colony opacity switch, transposon mutagenesis was used to generate mutations in strain AB5075 that greatly reduced the frequency of phase variation from opaque to translucent. One mutant revealed a role for a previously uncharacterized RND‐type system in this process. Mutations in the genes encoding this RND system significantly decreased phase variation in the opaque to translucent direction, but had little to no effect on phase variation in the translucent to opaque direction. Moreover, mutations inactivating this RND system were pleiotropic and resulted in altered surface motility, aminoglycoside resistance, and virulence in a Galleria mellonella waxworm model.
2. Experimental Procedures
2.1. Bacterial strains, plasmids, and growth conditions
Both A. baumannii and Escherichia coli were grown in modified Luria Broth containing 10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter. For screening opaque and translucent colonies of AB5075, LB was prepared at 0.5× of the normal concentration with 8 g agar per liter. Escherichia coli transformants were selected with chloramphenicol (25 μg/ml), ampicillin (200 μg/ml), or kanamycin (20 μg/ml) when appropriate. Acinetobacter baumannii AB5075 transformants were selected with tetracycline (3 μg/ml). Plamsid pEX18Tc was used for allelic replacement in AB5075 (Hoang, Karkhoff‐Schweizer, Kutchma, & Schweizer, 1998). For all experiments involving opaque and translucent variants, cells were obtained from freezer stocks that were grown to low density and contained <0.5% of the opposite cell type. Strain AB00075 arpB::Tc was obtained from the A. baumannii transposon insertion library maintained by Dr. Colin Manoil's laboratory at the University of Washington (Gallagher et al., 2015).
2.2. Phase variation assays
Freezer stocks of each strain that were verified as 99.99% of a single colony type were grown in LB broth to an OD of 1.8–1.9. Colonies were resuspended in LB broth and serial dilutions were plated on 0.5× LB, 0.8% agar plates. After overnight growth, the total number of viable cells per ml was determined and the number of opaque or translucent colonies present was determined on plates with at least 200 colonies/plate by using oblique light to illuminate the colonies.
2.3. Transposon mutagenesis
A culture of the A. baumannii AB5075 opaque variant was grown in 25 ml of LB broth at 37°C with vigorous shaking. Cells were harvested from cultures at an OD600 ~0.5 by centrifugation at 4°C for 10 min. Pellets were washed twice with 10% glycerol to prepare electrocompetent cells. Transposon (0.1 pmole) and transposase (1 unit) components of an EZ‐Tn5 <TET‐1> kit (Epicentre Biotechnologies) were combined with 10% glycerol and incubated at 37°C for approximately 45 min. Transposome complex mixture was then cooled on ice prior to electroporation. Aliquots of 1.25 μl transposome mixture and 1 μl of TypeOne Restriction Inhibitor (Epicentre Biotechnologies) were electroporated into 60 μl of competent AB5075 O cells at 2.5 kV. Following electroporation, 1 ml of room temperature LB broth was added following electroporation. This cell suspension was transferred to tubes and incubated at 37°C with shaking for 1 hr. Mutagenized cells were plated in 100 μl aliquots on 0.5× LB, 0.8% agar plates supplemented with 3 μg/ml tetracycline to select for insertional mutants and plates were incubated at 37°C. Colonies were evaluated at 24 and 48 hr postplating for altered colony morphology or reduced colony sectoring when viewed under a dissecting scope with oblique illumination from below. Putative mutants were restreaked on 0.5× LB, 0.8% agar with Tet3 to ensure colony morphology was stable and that picked colonies were pure. Identification of insertion sites was accomplished by rescue cloning of the tetracycline resistance gene and DNA sequencing of plasmids generated from rescue cloning.
2.4. Construction of in‐frame deletions in arpA and arpR
An in‐frame deletion in arpA was generated as described previously (Hoang et al., 1998) from AB5075 genomic DNA by PCR amplification of two approximately 1 kb fragments containing small portions of the arpA coding region using the primers arpA Up‐1 (5′‐AAAAAGGATCCATACTACGTTGTACCGCTAC‐3′), arpA Up‐2 (5′‐GTGAAAATTCAGGGAGCCA‐3′), arpA Down‐1 (5′‐TTGTTCGTGAAGTGTGGTT‐3′), and arpA Down‐2 (5′‐AAAAAGGATCCCACCTAATAAATTGCCAAGTAAGC‐3′). Oligonucleotide primers arpA Up‐1 and arpA Down‐2 were engineered to contain BamHI restriction sites at the 5′ end. The up‐ and downstream fragments were ligated together to produce an approximately 2‐kbp fragment containing the ΔarpA allele which was subsequently gel purified. The ΔarpA allele contains an in‐frame deletion in the arpA coding sequence corresponding to amino acids 48–359 of the 366 (85% of coding region). Purified fragment was reamplified with outer primers (arpA Up‐1 and arpA Down‐2) and gel purified. The ΔarpA fragment and pEX18Tc were digested with BamHI and gel purified. Digested fragments were then ligated and transformed into competent E. coli DH5α cells. To transfer the mutant alleles to the chromosome of A. baumannii AB5075, the suicide vector containing the in‐frame arpA deletion was electroporated into competent AB5075 cells which had been grown overnight in LB and washed with 300 mM sucrose as described previously (Choi & Schweizer, 2006). Integrants were selected on LB + tetracycline at 5 μg/ml. Counterselection to select for excision of the integrated plasmid was carried out at room temperature on LB without NaCl supplemented with 10% sucrose. Potential mutants were screened by PCR and confirmed by DNA sequencing.
The arpR in‐frame deletion allele was generated from AB5075 genomic DNA by PCR amplification (Phusion Hot‐Start Polymerase, Thermo Scientific) of two approximately 1 kbp fragments containing small portions of the arpR coding region. Oligonucleotide primers arpR Up‐1 (5′‐AAAAAGGATCCCGTGATAACCACAATACTTC‐3′) and arpR Down‐2 (5′‐AAAAAGGATCCATGACATTAGTTTGAGTCGA‐3′) were engineered to contain BamHI restriction sites at the 5′ end and were paired with arpR Up‐2 (5′‐CTCAAATATCGGCATTAAACC‐3′) and arpR Down‐1 (5′‐ATTAACTGTTTGCACGAAAC‐3′), respectively. The up‐ and downstream fragments were ligated (Fast‐Link DNA Ligation Kit, Epicentre Biotechnologies) together to produce an approximately 2 kbp fragment containing the ΔarpR allele which was subsequently gel purified (UltraClean 15 DNA Purification Kit, MoBio Laboratories). The ΔarpR mutant allele contains an in‐frame deletion corresponding to amino acids 6–202 of the 207 aa protein (94% of protein sequence). Purified deletion fragment was reamplified with outer primers (acrR Up‐1 and Down‐2) and gel purified. The ΔarpR fragment and pEX18Tc were digested with BamHI and gel purified. Digested fragments were ligated and transferred into competent E. coli EC100D Transformax cells (Epicentre Biotechnologies). This ligation produced the suicide vector pΔarpR/EX18Tc. This construct was then used to create an in‐frame deletion in arpR as described above for arpA.
2.5. Complementation of the arpA mutant
The full‐length arpA allele including 91 bp of sequence upstream from the ATG start codon and 42 bp downstream from the stop codon were amplified from A. baumannii AB5075 genomic DNA by PCR with the following oligonucleotide primers: 5′‐CCTTATTGTTCAGTGCCCAT‐3′ and 5′‐GTGCCGTCGGGTATATTAATTA‐3′. Primers were phosphorylated prior to PCR amplification to add 5′ phosphates with T4 polynucleotide kinase (New England Biolabs). The arpA fragment was gel purified and ligated with the shuttle vector pWH1266 (Hunger, Schmucker, Kishan, & Hillen, 1990) which had been digested with ScaI and treated with recombinant shrimp alkaline phosphatase (New England Biolabs). The ligation mixture was introduced into E. coli EC100D Transformax cells (Epicentre) via electroporation and clones were screened for tetracycline resistance and ampicillin sensitivity. The following plasmid pArpA was confirmed by DNA sequencing prior to introduction into A. baumannii. Competent A. baumannii AB5075 opaque and ΔarpA opaque cells were prepared by washing cells from overnight LB plates in dH2O two times. pArpA plasmid was electroporated into competent cells and transformants were selected on LB + tetracycline at 5 μg/ml. For switching frequency assays, overnight starter cultures of LB + tetracycline (3 μg/ml) were inoculated with AB5075 or ΔarpA harboring pWH1266 or pArpA and incubated at room temperature overnight without shaking. Strains were subcultured into LB without antibiotics and incubated at 37°C with vigorous shaking. CFU/ml and number of translucent colonies (switching frequency) were quantified at OD600 0.7 and 1.7.
2.6. Preparation of conditioned media
To prepare conditioned media, cells were grown in 25 ml LB cultures at an optical density of 1.7. At this time, aliquots were restreaked to verify that the cultures remained at least 95% opaque or translucent. Cells were removed by centrifugation and the resulting media was adjusted to pH 7.0 with HCl and filter sterilized by passing through a 0.22‐μm filter. Aliquots were frozen at −80°C and used within 2 weeks. To grow cells in conditioned media, a 2‐ml aliquot was thawed and a 10× concentrate of tryptone and yeast extract (TY) was added back to a final concentration of 0.25×. Cells were grown by shaking at 270 rpm at 37°C to an optical density of 1.1 and dilutions were then plated to determine phase variation frequencies. Growth of cells in the LB control was done at 0.25× LB, a concentration that gave a similar growth rate as the TY‐supplemented conditioned media.
2.7. Galleria mellonella infections
Galleria mellonella larvae between 200 and 250 mg were utilized for infection studies. Acinetobacter baumannii strains were grown in LB broth at 37°C with shaking to an OD600 ~0.5. Cultures were serially diluted in LB broth and plated to determine CFU/ml for each bacterial strain. Strains diluted to 10−2 were chilled on ice prior to injection into larvae. Four μl aliquots of each strain were injected into the hemolymph of G. mellonella larvae (10 larvae per strain in three replicates, ~30 total larvae per strain). The average CFU for the injected wild‐type cells in the three experiments was 8.2 × 103 and 9.5 × 103 for ∆arpA mutant. Infected larvae were incubated at 37°C for up to 5 days in a humidified incubator and mortality was assessed at daily intervals.
2.8. Construction of an arpR expression plasmid
The full‐length arpR gene including 146 bp of sequence upstream from the GTG start codon and 71 bp downstream from the stop codon were amplified from AB5075 genomic DNA by PCR (Phusion Hot‐Start Polymerase, Thermo Scientific). Primers arpR Exp.1 (5′‐CATTTAAATCGCTTATAACAC‐3′) and arpR Exp.2 (5′‐TTATCGCTCTTATTCAAACT‐3′) were phosphorylated prior to PCR amplification to add 5′ phosphates with T4 polynucleotide kinase (New England Biolabs). The arpR fragment was gel purified and ligated (Fast‐Link Ligation Kit, Epicentre Biotechnologies) into pWH1266 that had been digested with ScaI and treated with recombinant shrimp alkaline phosphatase (rSAP, New England Biolabs). The ligation was electroporated into competent E. coli EC100D Transformax cells (Epicentre Biotechnologies). Plasmids that conferred tetracycline resistance but not ampicillin resistance were purified and confirmed by DNA sequencing prior to introduction into A. baumannii. This produced the expression vector parpR.
2.9. Switching frequency determination
Cultures were grown to the OD600 ~0.7 or ~1.7 in LB medium, serially diluted in LB, and plated on 0.5× LB, 0.8% agar. Plates were incubated at 37°C overnight. CFU per milliliter and number of variant colonies were determined for each strain and variant.
2.10. RNA isolation
Cultures of A. baumannii strain AB5075 and the ΔarpR mutant were grown in LB medium at 37°C with shaking to an OD600 ~0.7. Cultures of A. baumannii strain AB5075 harboring pWH1266 or parpR/WH1266 were grown in LB medium supplemented with tetracycline (5 μg/ml) at 37°C with shaking to an OD600 ~0.5. Cells were harvested from cultures by centrifugation and RNA was isolated with the MasterPure RNA Purification Kit according to the manufacturer's protocol (Epicentre). Contaminating DNA was removed by treatment with TURBO DNA‐free according to the manufacturer's protocol (Ambion). DNA contamination was evaluated by PCR with purified RNA as template and RNA concentration was quantified with a NanoDrop ND‐1000 spectrophotometer.
2.11. Quantitative real‐time PCR
Total RNA (1 μg) purified from strains AB5075 with or without plasmid and ΔarpR were converted into cDNA by using the iScript cDNA synthesis kit (Bio‐Rad) with random primers and SuperScript III reverse transcriptase (Invitrogen). Cycling parameters for cDNA synthesis were as follows: 25°C for 5 min, 42°C for 45 min, and 85°C for 5 min. cDNA reactions were then diluted 1:10 with sterile H2O supplemented with 10 μg/ml yeast t‐RNA (Roche). Diluted cDNA was used as template for experimental reactions. Oligonucleotide primer pairs for quantitative real‐time PCR (qRT‐PCR) were generated by the Primer‐BLAST program available at www.ncbi.nlm.nih.gov/tools/primer-blast/. Primers were designed to amplify a 145‐bp fragment of clpX (clpX qRT For 5′‐GCGTTTGAAAGTCGGGCAAT‐3′, clpX qRT Rev 5′‐CCATTGCAAACGGCACATCT‐3′) and a 151‐bp fragment of arpA (arpA qRT 1 5′‐TCGCGTACATATCCGGCAAA‐3′, arpA qRT 2 5′‐GGCAAGCGGCTTATCAACTG‐3′). qRT‐PCR was performed using iQ SYBR Green Supermix (Bio‐Rad) with Bio‐Rad CFX Connect cycler. The following cycle parameters were utilized to amplify and quantify fragments: 95°C for 3 min and then 95°C for 10 s, 55°C for 10 s, and 72°C for 20 s, repeated 40 times. Melt curve data were collected to ensure proper amplification of target genes. Data were generated from three separate RNA isolation and cDNA preparations and at least two technical replicates for each primer set. Relative expression of each gene was determined by comparing target gene expression with control gene (clpX) expression using the Pfaffl method (Pfaffl, 2001).
2.12. Antimicrobial resistance assays
Strains to be tested were grown to an OD600 of 0.3 and then diluted to an OD600 of 0.05 with sterile LB. The minimum inhibitory concentration for various antibiotics was determined using E‐test strips on LB agar plates according to the manufacturer's instructions (Biomerieux). The MICs for each antibiotic were determined after 12 hr of growth at 37°C. All susceptibility tests were done in duplicate.
3. Results
3.1. An RND‐type efflux system is required for the opaque to translucent colony opacity phase variation
Previously, our laboratory reported on a phase variable mechanism in A. baumannii that results in the interconversion between opaque and translucent colonies (Tipton et al., 2015). This phase variation was stimulated at high density and when opaque colonies are grown for 36–48 hr, they become highly mottled in appearance due to translucent variants arising at high frequency within the colony (Figure 1a). This mottled appearance formed the basis for a genetic screen to identify mutants with a reduced frequency of phase variation in the opaque to translucent direction, as these mutants would have a reduction or absence in the mottled appearance of the colony. A library of EZ‐Tn5 <Tet‐1> insertions in AB5075 was screened for colonies that did not exhibit the mottled appearance after 36 hr of growth and mutants with this phenotype were isolated and confirmed by subsequent replating. One mutant 5075.8B (Figure 1a) was characterized further and the transposon insertion was mapped to nucleotide 64 of a 1,101 nucleotide open reading frame (ABUW_0034) encoding a putative periplasmic membrane fusion component of RND‐type efflux systems (Figure 1b). This protein exhibited the highest degree of identity to annotated RND‐type transporters from Pseudomonas mendocina (47% identity/69% similarity) and Pseudomonas pseudoalcaligenes (47% identity/68% similarity). A second open reading frame (ABUW_0035) was present beginning 3 bp downstream of ABUW_0034 that encoded a putative inner membrane transporter component of RND‐type systems. This protein exhibited the highest identity to an annotated RND‐type transporter from Bacillus mycoides (85% identity/93% similarity) and to Alkanindiges illinoisensis (75% identity/86% similarity). Given their close proximity, these two genes likely formed an operon and were previously identified in A. baumannii as a locus upregulated in the presence of farnesol and were designated as acrAB (Kostoulias et al., 2016). However, the overall similarity of these proteins to the E. coli AcrA (23% identity/39 similarity) and AcrB (25% identity/43% similarity) was low and there are other RND‐type systems in E. coli that exhibit greater identity to the A. baumannii proteins. In addition, as outlined below, this RND system confers aminoglycoside resistance, therefore, these genes were designated arpA and arpB (aminoglycoside resistance pump). The frequency of phase variation from opaque to translucent was quantitated in individual colonies of the 5075.8B mutant and wild‐type cells after 48 hr of growth, which revealed an average 55‐fold decrease in the opaque to translucent switch in the arpA:: EZ‐Tn5 <Tet‐1> mutant 5075.8B. While this analysis was being conducted, it was observed that there was significant colony to colony variation within the same strain with respect to phase variation frequencies, leading to large standard deviations. As a result of this, broth grown cells were used for subsequent experiments, which did reduce variability, although significant variability still existed. In broth cultures, the phase variation rate of the 5075.B mutant was 77‐fold lower than wild type (Table 1).
Figure 1.
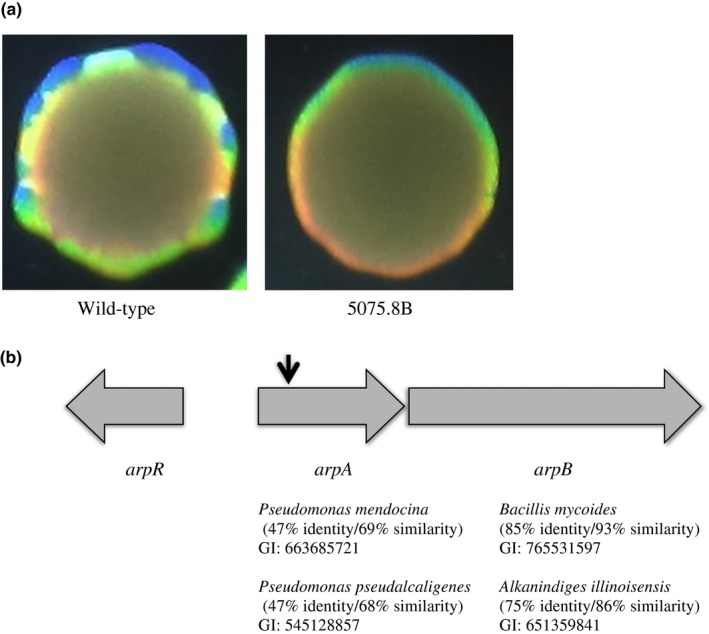
Decreased phase variation in an arpA mutant. (a) A typical wild‐type opaque colony variant of AB5075 is shown compared to 5075.8B arpA:: EZ‐Tn5 <Tet‐1> after 36 hr of growth on a 0.5× LB, 0.8% agar plate. The mottled appearance at the outside edge of the wild type is due to translucent variants arising within the opaque colony. (b) The organization of the arpAB region and the site of the arpA:: EZ‐Tn5 <Tet‐1> insertion that blocks phase variation is depicted by an arrow. Proteins exhibiting the closest match to ArpA and ArpB are shown below each gene
Table 1.
Phase variation frequencies
| Strain | Relative O to T phase variation frequencya |
|---|---|
| AB5075 wild type | 1b |
| 5075.8B arpA1::EZTn‐5Tc | 0.013 ± 0.02 (77‐fold decrease) |
| KT2 ∆arpA2 | 0.014 ± 0.03 (71‐fold decrease) |
| AB00075 arpB::Tc | 0.0013 ± 0.001 (769‐fold decrease) |
| KT3 ∆arpR | 0.0009 ± 0.0002 (1,916‐fold decrease) |
| Strain | Relative T to O phase variation frequencya |
|---|---|
| AB5075 wild type | 1c |
| KT2 ∆arpA2 | 1.5 ± 0.6 |
| AB00075 arpB::Tc | 1.2 ± 0.5 |
| KT3 ∆arpR | N.D. |
Determined in cultures at an OD600 of 1.85–1.9.
Phase variation frequency from O to T was typically 25%–35%.
Phase variation frequency from T to O was typically 40%–50%.
To verify the role of the arpA gene in phase variation and to avoid polar effects on the downstream arpB gene that likely occurred from the arpA:: EZ‐Tn5 <Tet‐1> insertion, an in‐frame deletion in arpA (∆arpA2) was constructed. This mutation had a similar effect on phase variation as the original arpA1::EZTN<Tc> mutant, where the frequency of switching from opaque to translucent was reduced 71‐fold in cultures at an OD600 = 1.9 (Table 1). This ∆arpA2 mutant strain was designated KT2.
Complementation analysis was performed to determine if the loss of arpA was responsible for the phase variation defect. When a plasmid containing the arpA gene was introduced into the arpA deletion mutant (KT2), colonies regained the ability to form the mottled phenotype due to translucent variants arising at high frequency (data not shown). In addition, the frequency of phase variation in 24 hr colonies was increased by 3,600‐fold in the arpA deletion mutant containing the cloned arpA gene (parpA), relative to the arpA deletion mutant containing the pWH1266 vector alone (Figure S1, panel A).
3.2. An arpB mutation also decreases the opaque to translucent phase variation
The gene located immediately downstream from arpA is predicted to encode a putative inner membrane transporter of RND efflux systems and likely functions with ArpA in this process. This gene was designated arpB. To determine if arpB also regulates phase variation, two separate transposon insertions in arpB (AB00075, AB00076) were obtained from the A. baumannii AB5075 transposon insertion library at the University of Washington (Gallagher et al., 2015). Like the arpA mutant, opaque colonies of the arpB mutants did not produce mottled colonies at 48 hr. The frequency of phase variation for AB00075 arpB::Tc in broth grown cells at an OD600 of 1.9 was reduced an average of 769‐fold compared to wild‐type cells (Table 1).
3.3. arpA and arpB mutations do not alter the translucent to opaque switch
The effect of the ∆arpA mutation on the reciprocal opacity switch, from translucent to opaque, was measured in KT2 grown to an OD of 1.9, where the rate of phase variation was increased 1.5‐fold over wild type (Table 1). The arpB::Tc mutant AB00075 exhibited a phase variation rate for translucent to opaque that was 1.2‐fold greater than wild type.
3.4. An arpB mutant still produces an extracellular signal that stimulates phase variation
Previous work demonstrated that the increase in phase variation at high cell density is mediated, in part, by the accumulation of a secreted signal (Tipton et al., 2015). If this signal is dependent on ArpAB for secretion and sensed at the cell surface or the cytoplasmic membrane, then this could explain the reduction in phase variation in the arpA mutant. To test this possibility, conditioned media were prepared from opaque variants of wild‐type and the ∆arpB mutant at an optical density of 1.7 and tested for the ability to stimulate phase variation in the opaque to translucent direction. Conditioned media from wild‐type cells stimulated the opaque to translucent conversion 11‐fold and the conditioned media from the arpB mutant stimulated phase variation ninefold (Table 2). There was not a statistically significant difference between these values (p = .39) indicating that the arpB mutant produced a similar level of extracellular signal activity as the wild‐type parent.
Table 2.
Effect of conditioned media on relative phase variation frequency
| Growth conditiona | Opaque to translucent switching frequency |
|---|---|
| LB broth | 0.35% ± 0.29 |
| Wild‐type opaque conditioned media | 3.9% ± 2.8 (11‐fold increase) |
| ∆arpB opaque conditioned media | 3.2% ± 2.7 (9‐fold increase) |
Determined in cells grown to an optical density A600 of 1.0.
3.5. Role of ArpAB in antimicrobial resistance
It is well established that RND‐type systems can export antimicrobials and confer higher levels of resistance (Alvarez‐Ortega et al., 2013; Anes et al., 2015; Coyne et al., 2011; Magnet et al., 2001; Rosenfeld et al., 2012; Routh et al., 2011; Venter et al., 2015). Therefore, both the arpA and arpB mutants were tested for levels of resistance to various antibiotics. For this analysis, both the opaque and translucent variants of each mutant and wild‐type cells were tested (Table 3). Due to the multiple resistances that already exist in AB5075, the number of antibiotics that could be tested was limited. There were no significant differences between wild‐type and the arpA mutant for any of the antibiotics tested (Table 3), although there were subtle differences between opaque and translucent variants of wild type as reported previously (Tipton et al., 2015). However, the arpB mutant AB00075 was more sensitive to the two aminoglycosides that were tested, amikacin and tobramycin (Table 3). This sensitivity was seen in both the opaque and translucent variants of AB00075 arpB::Tc.
Table 3.
Minimum inhibitory concentrations for antibiotics
| Strain | MIC (μg/ml) | |||||
|---|---|---|---|---|---|---|
| COL | TET | AK | RIF | TIG | TOB | |
| WT opaque | 0.38 | 3 | 128 | 3 | 3 | 96 |
| ∆arpA opaque | 0.38 | 3 | 128 | 3 | 3 | 96 |
| arpB::Tc opaque | 0.38 | ND | 64 | 3 | 3 | 24 |
| WT translucent | 0.38 | 3 | 64 | 4 | 3 | 24 |
| ∆arpA translucent | 0.38 | 3 | 64 | 4 | 3 | 24 |
| arpB::Tc translucent | 0.38 | ND | 16 | 4 | 3 | 8 |
COL, colistin; TET, tetracycline; AK, amikacin; RIF, rifampicin; TIG, tigecycline; TOB, tobramycin.
3.6. arpA and arpB mutations selectively increase surface motility in the opaque variants
Both the arpA and arpB mutants formed irregular, slightly spreading colonies on 0.8% agar plates that were distinct from wild‐type colonies that contained smooth rounded edges. This observation suggested that motility may be enhanced by the arpA and arpB mutations. To test this hypothesis, the motility of both opaque and translucent arpA and arpB mutants were compared to opaque and translucent wild‐type AB5075 on 0.3% Eiken agar plates. In opaque colonies, both the arpA and arpB mutants exhibited increased motility, 53 mm and 75 mm, respectively, over wild‐type value of 33 mm (Figure 2). However, in the translucent form, there was no significant difference in motility between wild‐type and either the arpA or arpB mutants (Figure 2). The increased motility observed in the arpA mutant was restored back to wild‐type levels by parpA containing the cloned arpA gene relative to cells containing the pWH1266 vector alone (Figure S1, panel B).
Figure 2.
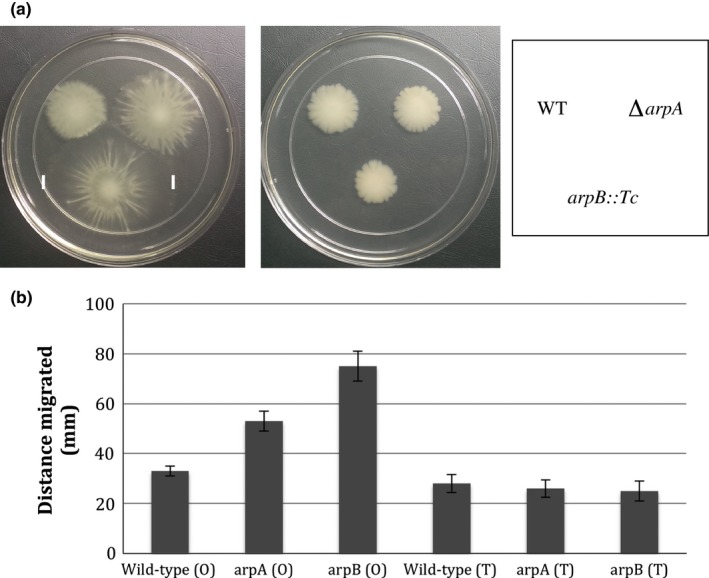
Surface motility. (a) The motility of opaque (left panel) and translucent (right panel) variants of wild‐type AB5075, the ∆arpA mutant KT2, and arpB::Tc mutant AB00075 are shown after 12 hr of growth on 0.35% Eiken agar plates incubated at 37°C. (b) Quantitation of surface motility. The values shown represent the average of three separate motility assays for each strain. Conditions for the motility assays were the same as shown in A
3.7. ArpAB is required for virulence in Galleria mellonella
RND‐type efflux systems have been shown to have roles in virulence in a number of bacterial pathogens (Alvarez‐Ortega et al., 2013; Delmar et al., 2014; Routh et al., 2011; Taylor, Bina, & Bina, 2012; Venter et al., 2015; Warner et al., 2007; Yoon et al., 2015). Therefore, the impact of arpA and arpB mutations on virulence were examined in a Galleria mellonella waxworm model that has previously been shown to be a useful model of virulence in A. baumannii (Esterly et al., 2014; Gaddy et al., 2012; Heindorf, Kadari, Heider, Skiebe, & Wilharm, 2014; Iwashkiw et al., 2012; Jacobs et al., 2014; Nwugo et al., 2012; Peleg et al., 2009; Repizo et al., 2015; Stahl, Bergmann, Gottig, Ebersberger, & Averhoff, 2015; Tipton et al., 2015). Since previous work indicated that the opaque form was more virulent, these studies were done with the opaque forms of wild‐type AB5075, KT2 ∆arpA and the arpB::Tc mutant (Tipton et al., 2015). Relative to wild‐type AB5075, both the ∆arpA and arpB mutations substantially reduced the ability of A. baumannii to kill G. mellonella waxworms (Figure 3a). The decreased virulence exhibited by the arpA mutant was restored by parpA containing the cloned arpA gene relative to cells containing the pWH1266 vector alone (Figure S1, panel C).
Figure 3.
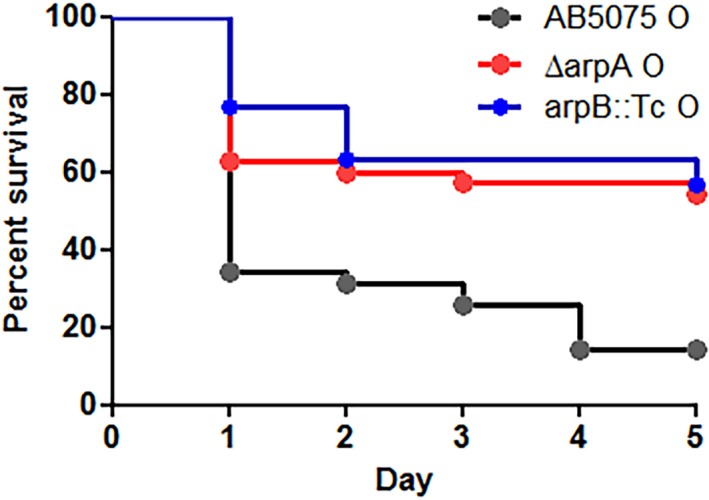
arpA and arpB mutants exhibit decreased virulence in a Galleria mellonella model. The ability of wild‐type AB5075 and the isogenic ∆arpA and arpB::Tc mutants to kill G. mellonella waxworms is shown on the Kaplan–Meier plots. The data shown represent the average of three independent experiments with a total of 30 worms per strain
3.8. A divergently transcribed gene encoding a TetR‐type regulator represses arpAB and is required for the opaque to translucent switch
Adjacent to the arpAB genes in A. baumannii was a divergently transcribed gene encoding a predicted TetR‐type repressor of the AcrR family (Figure 1). In E. coli, the AcrR protein acts to repress the acrAB operon, which is organized in a similar manner as arpAB. To determine if a similar function was present in A. baumannii, an in‐frame deletion was constructed in this gene, designated arpR. The ∆arpR deletion did not have a significant impact on arpAB expression, with the levels of expression 1.1‐fold higher in the mutant (Figure 4). However, the arpR mutation reduced the frequency of the opaque to translucent conversion by 1,916‐fold (Table 1).
Figure 4.
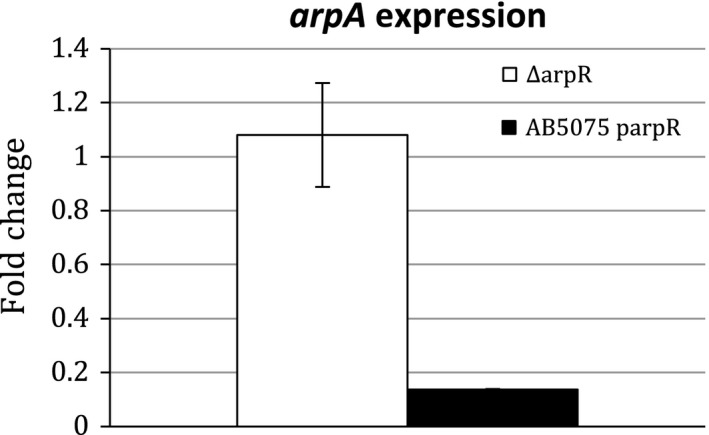
Effect of ArpR on arpA expression. The levels of arpA expression were determined by quantitative RT‐PCR and the values shown are relative to the control gene clpX. The values for the left side represent the levels of arpA expression in the arpR mutant relative to wild type. The values on the right side represent the levels of arpA expression in cells overexpressing arpR from a plasmid relative to cells containing the vector alone
As a second method to determine if ArpR could function as a repressor, the arpR gene was overexpressed by cloning the gene into pWH1266, where expression was driven from the promoter for the β‐lactamase gene. When this plasmid was introduced into wild‐type AB5075, the colony morphology was altered with irregular slightly spreading colonies that were similar to arpA and arpB mutants, suggesting that expression of both arpA and arpB were reduced when ArpR was overexpressed. To investigate this possibility, expression of the first gene arpA was examined by qRT‐PCR and was found to be 7.3‐fold lower in cells with parpR versus the pWH1266 vector alone (Figure 4). Next, we investigated if the reduced ArpAB expression resulting from ArpR overexpression impacted the opaque to translucent switch. In cells overexpressing the arpR gene, the frequency of phase variation was reduced threefold, relative to cells containing the vector only (data not shown).
4. Discussion
In this study, a previously undescribed RND‐type efflux system in A. baumannii was found to regulate phase variation in the opaque to translucent direction. The original mutation defining this phenotype was a transposon insertion in arpA, a gene encoding a membrane fusion component of a putative RND‐type efflux system. This arpA mutation resulted in a 77‐fold decrease in the frequency of phase variation from the opaque to translucent colony opacity phenotype (Table 1). The role of arpA in this process was confirmed by the construction of a nonpolar, in‐frame deletion within arpA, which decreased the conversion to translucent colonies by 71‐fold, and by complementation analysis with the wild‐type arpA gene. In addition, a transposon insertion in the downstream arpB gene obtained from the A. baumannii AB5075 transposon library at the University of Washington (Gallagher et al., 2015) exhibited a stronger effect with a 769‐fold reduction in phase variation (Table 1). Interestingly, neither the arpA or arpB mutations had a significant effect on phase variation in the translucent to opaque direction. This provides strong evidence that a distinct mechanism regulates phase variation in each direction.
Two additional phenotypes were altered by the loss of arpA and arpB. First, surface motility of the opaque variants on 0.3% agar plates was increased, with the arpB mutant exhibiting a more substantial increase than the arpA mutant (Figure 2). Interestingly, in translucent variants, the motility of both the arpA and arpB mutants was similar to wild‐type. The basis for the selective motility increase in opaque variants is unknown, but previously it was noted that in wild‐type cells, the opaque variants were more motile (Tipton et al., 2015). Although the actual mechanism responsible for motility in AB5075 has not been identified, if this mechanism was only expressed in opaque cells and also repressed by ArpAB, it could explain the selective increase in motility seen in arpA or arpB mutants in the opaque background. Second, the arpA mutation decreased the ability of A. baumannii AB5075 to kill G. mellonella waxworms. Additional examples exist where the loss of an RND‐type system decreases virulence (Alvarez‐Ortega et al., 2013; Delmar et al., 2014; Routh et al., 2011; Taylor et al., 2012; Venter et al., 2015; Warner et al., 2007; Yoon et al., 2015). The decreased virulence may be due to a role for ArpAB in efflux of host antimicrobial peptides or other antibacterial compounds present inside G. mellonella.
The arpB mutant consistently exhibited stronger effects than the arpA mutant for phase variation (Table 1), surface motility (Figure 2), and antibiotic sensitivity (Table 3). It is unlikely that this results from the arpA mutation being leaky, as the in‐frame deletion removed most of the coding region. The weaker phenotype of the arpA mutation may indicate that a periplasmic fusion protein from another RND‐type system can partially substitute for ArpA. The stronger effect of a arpB mutation suggests that ArpB is unable to be substituted for by another RND component and has a more critical role than ArpA. However, this hypothesis is speculative at the current time. In addition, the outer membrane channel that works with ArpAB is unknown, but AbuO, a TolC‐like protein is a possible candidate (Srinivasan, Vaidyanathan, & Rajamohan, 2015).
A TetR‐type transcriptional regulator was encoded adjacent to the arpAB genes and divergently transcribed, a genetic organization similar to that of acrR‐acrAB in E. coli and mexR‐mexAB in P. aeruginosa. A ∆arpR mutation did not alter arpAB expression, however, overexpressing the arpR gene decreased arpAB expression by 7.3‐fold. In addition, cells overexpressing arpR exhibited phenotypes that at least partially mimicked the arpAB mutations, as the colonies formed the irregular spreading appearance and the frequency of the opaque to translucent conversion was reduced threefold. It is unclear why the effects of ArpR on arpAB expression are only seen when it is overexpressed, but under the laboratory growth conditions we tested, it does not have a role in arpAB regulation, possibly because it is not expressed at high enough levels to mediate repression. If certain environmental conditions increased arpR expression, this condition would be predicted to repress arpAB and reduce the opaque to translucent conversion.
To our knowledge, this represents the first report of an RND‐type efflux system regulating bacterial phase variation. A direct role for ArpAB in this process is unlikely and loss of ArpAB may indirectly modulate a regulatory pathway controlling phase variation. At the present time, the regulatory mechanism regulating the interconversion between opaque and translucent colonies is unknown. However, whole genome sequencing suggests that nucleotide changes are not responsible. This information, together with the observation that phase variation between colony opacity phenotypes is essentially undetectable in cells at low density, but is sharply activated at high density and reaches frequencies above 10% (Table 1) suggests a nonmutational mechanism is involved.
A possible mechanism to control phase variation is via a bistable switch involving one or more regulatory proteins (Casadesus & Low, 2013; Chang et al., 2010; Dubnau & Losick, 2006; Ferrell, 2002; Maamar & Dubnau, 2005; Mitrophanov & Groisman, 2008; Piggot, 2010; Turner, Vallet‐Gely, & Dove, 2009; Veening, Smits, & Kuipers, 2008). Bistability has previously been shown to regulate colony opacity in Pseudomonas fluorescens (Gallie et al., 2015). Based on this information, there are several possibilities for how ArpAB might regulate phase variation. First, the extracellular signal that stimulates the opaque to translucent phase variation might require ArpAB for secretion. This would imply that the signal works via a sensing mechanism that operates at the cell surface or at the cytoplasmic membrane and then regulates the bistable switch controlling the phase variation. However, spent culture supernatants from an arpB mutant and wild‐type cells appear to activate the opaque to translucent switch at equal frequencies (Table 2). A second possibility is that a metabolite normally secreted by the ArpAB system accumulates in arpAB mutants and this alters a regulatory pathway that controls phase variation. The isolation of both extragenic and high‐copy suppressors that restore phase variation to an arpA or arpB mutant should help uncover the role of this efflux system in regulating the process of colony opacity phase variation in A. baumannii and these studies are in progress.
Conflict of Interest
None declared.
Supporting information
Acknowledgments
This work was funded by grant 1R21AI115183 from the National Institutes of Health. P. N. R. is also supported by grants from the Merit Review program and a Research Career Scientist award, both from the Department of Veterans Affairs. We are grateful to Dr. Colin Manoil for providing mutants from the University of Washington A. baumannii transposon library. In addition, we thank Chui‐Yoke Chin for help with the Kaplan–Meier plots..
Tipton, K. A. , Farokhyfar, M. and Rather, P. N. Multiple roles for a novel RND‐type efflux system in Acinetobacter baumannii AB5075. MicrobiologyOpen. 2017;6:e00418 https://doi.org/10.1002/mbo3.418
Funding Information
This work was funded by grant 1R21AI115183 from the National Institutes of Health. P. N. R. is also supported by grants from the Merit Review program and a Research Career Scientist award, both from the Department of Veterans Affairs.
References
- Alvarez‐Ortega, C. , Olivares, J. , & Martinez, J. L. (2013). RND multidrug efflux pumps: What are they good for? Frontiers in Microbiology, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anes, J. , McCusker, M. P. , Fanning, S. , & Martins, M. (2015). The ins and outs of RND efflux pumps in Escherichia coli . Frontiers in Microbiology, 6, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes, L. C. , Visca, P. , & Towner, K. J. (2014). Acinetobacter baumannii: Evolution of a global pathogen. Pathogens and Disease, 71, 292–301. [DOI] [PubMed] [Google Scholar]
- Bergogne‐Berezin, E. , & Towner, K. J. (1996). Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clinical Microbiology Reviews, 9, 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesus, J. , & Low, D. A. (2013). Programmed heterogeneity: Epigenetic mechanisms in bacteria. The Journal of Biological Chemistry, 288, 13929–13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D. E. , Leung, S. , Atkinson, M. R. , Reifler, A. , Forger, D. , & Ninfa, A. J. (2010). Building biological memory by linking positive feedback loops. Proceedings of the National Academy of Sciences of the United States of America, 107, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnot‐Katsikas, A. , Dorafshar, A. H. , Aycock, J. K. , David, M. Z. , Weber, S. G. , & Frank, K. M. (2009). Two cases of necrotizing fasciitis due to Acinetobacter baumannii . Journal of Clinical Microbiology, 47, 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K. H. , & Schweizer, H. P. (2006). mini‐Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa . Nature Protocols, 1, 153–161. [DOI] [PubMed] [Google Scholar]
- Coyne, S. , Courvalin, P. , & Perichon, B. (2011). Efflux‐mediated antibiotic resistance in Acinetobacter spp. Antimicrobial Agents and Chemotherapy, 55, 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damier‐Piolle, L. , Magnet, S. , Bremont, S. , Lambert, T. , & Courvalin, P. (2008). AdeIJK, a resistance‐nodulation‐cell division pump effluxing multiple antibiotics in Acinetobacter baumannii . Antimicrobial Agents and Chemotherapy, 52, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmar, J. A. , Su, C. C. , & Yu, E. W. (2014). Bacterial multidrug efflux transporters. Annual Review of Biophysics, 43, 93–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau, D. , & Losick, R. (2006). Bistability in bacteria. Molecular Microbiology, 61, 564–572. [DOI] [PubMed] [Google Scholar]
- Esterly, J. S. , McLaughlin, M. M. , Malczynski, M. , Qi, C. , Zembower, T. R. , & Scheetz, M. H. (2014). Pathogenicity of clinical Acinetobacter baumannii isolates in a Galleria mellonella host model according to bla(OXA‐40) gene and epidemiological outbreak status. Antimicrobial Agents and Chemotherapy, 58, 1240–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell, J. E. Jr (2002). Self‐perpetuating states in signal transduction: Positive feedback, double‐negative feedback and bistability. Current Opinion in Cell Biology, 14, 140–148. [DOI] [PubMed] [Google Scholar]
- Gaddy, J. A. , Arivett, B. A. , McConnell, M. J. , Lopez‐Rojas, R. , Pachon, J. , & Actis, L. A. (2012). Role of acinetobactin‐mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infection and Immunity, 80, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, L. A. , Ramage, E. , Weiss, E. J. , Radey, M. , Hayden, H. S. , Held, K. G. , … Manoil, C. (2015). Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii . Journal of Bacteriology, 197, 2027–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, J. , Libby, E. , Bertels, F. , Remigi, P. , Jendresen, C. B. , Ferguson, G. C. , … Rainey, P. B. (2015). Bistability in a metabolic network underpins the de novo evolution of colony switching in Pseudomonas fluorescens . PLoS Biology, 13, e1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootz, T. D. , & Marra, A. (2008). Acinetobacter baumannii: An emerging multidrug‐resistant threat. Expert Review of Anti‐Infective Therapy, 6, 309–325. [DOI] [PubMed] [Google Scholar]
- Guerrero, D. M. , Perez, F. , Conger, N. G. , Solomkin, J. S. , Adams, M. D. , Rather, P. N. , & Bonomo, R. A. (2010). Acinetobacter baumannii‐associated skin and soft tissue infections: Recognizing a broadening spectrum of disease. Surgical Infections, 11, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindorf, M. , Kadari, M. , Heider, C. , Skiebe, E. , & Wilharm, G. (2014). Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PLoS ONE, 9, e101033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling, R. B. , Janes, B. K. , Kimball, H. , Tran, T. , Bundesmann, M. , Check, P. , … Miller, C. (2002). Toxic waste disposal in Escherichia coli . Journal of Bacteriology, 184, 3699–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, T. T. , Karkhoff‐Schweizer, R. R. , Kutchma, A. J. , & Schweizer, H. P. (1998). A broad‐host‐range Flp‐FRT recombination system for site‐specific excision of chromosomally‐located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene, 212, 77–86. [DOI] [PubMed] [Google Scholar]
- Hujer, K. M. , Hujer, A. M. , Hulten, E. A. , Bajaksouzian, S. , Adams, J. M. , Donskey, C. J. , … Bonomo, R. A. (2006). Analysis of antibiotic resistance genes in multidrug‐resistant Acinetobacter sp. isolates from military and civilian patients treated at the walter reed army medical center. Antimicrobial Agents and Chemotherapy, 50, 4114–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger, M. , Schmucker, R. , Kishan, V. , & Hillen, W. (1990). Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene, 87, 45–51. [DOI] [PubMed] [Google Scholar]
- Iwashkiw, J. A. , Seper, A. , Weber, B. S. , Scott, N. E. , Vinogradov, E. , Stratilo, C. , … Feldman, M. F. (2012). Identification of a general O‐linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathogens, 8, e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, A. C. , Thompson, M.G. , Black, C. C. , Kessler, J. L. , Clark, L. P. , McQueary, C. N. , … Zurawski, D. V. , (2014). AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio, 5, e01076–01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly‐Guillou, M. L. (2005). Clinical impact and pathogenicity of Acinetobacter. Clinical Microbiology and Infection, 11, 868–873. [DOI] [PubMed] [Google Scholar]
- Kostoulias, X. , Murray, G. L. , Cerqueira, G. M. , Kong, J. B. , Bantun, F. , Mylonakis, E. , … Peleg, A. Y. (2016). Impact of a Cross‐kingdom signaling molecule of Candida albicans on Acinetobacter baumannii physiology. Antimicrobial Agents and Chemotherapy, 60, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. Y. , Chen, C. L. , Wu, S. R. , Huang, C. W. , & Chiu, C. H. (2014). Risk factors and outcome analysis of Acinetobacter baumannii complex bacteremia in critical patients. Critical Care Medicine, 42, 1081–1088. [DOI] [PubMed] [Google Scholar]
- Lin, M. F. , Lin, Y. Y. , & Lan, C. Y. (2015). The Role of the two‐component system baesr in disposing chemicals through regulating transporter systems in Acinetobacter baumannii . PLoS ONE, 10, e0132843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman, W. , Kalk, T. , Menezes, C. N. , John, M. A. , & Grobusch, M. P. (2008). A case of community‐acquired Acinetobacter baumannii meningitis ‐ has the threat moved beyond the hospital? Journal of Medical Microbiology, 57, 676–678. [DOI] [PubMed] [Google Scholar]
- Maamar, H. , & Dubnau, D. (2005). Bistability in the Bacillus subtilis K‐state (competence) system requires a positive feedback loop. Molecular Microbiology, 56, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet, S. , Courvalin, P. , & Lambert, T. (2001). Resistance‐nodulation‐cell division‐type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrobial Agents and Chemotherapy, 45, 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand, I. , Damier‐Piolle, L. , Courvalin, P. , & Lambert, T. (2004). Expression of the RND‐type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two‐component system. Antimicrobial Agents and Chemotherapy, 48, 3298–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov, A.Y. , & Groisman, E. A . (2008). Positive feedback in cellular control systems. BioEssays, 30, 542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwugo, C. C. , Arivett, B. A. , Zimbler, D. L. , Gaddy, J. A. , Richards, A. M. , & Actis, L. A. (2012). Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii . PLoS ONE, 7, e51936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg, A. Y. , Jara, S. , Monga, D. , Eliopoulos, G. M. , Moellering, R. C. Jr , & Mylonakis, E. (2009). Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrobial Agents and Chemotherapy, 53, 2605–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg, A. Y. , Seifert, H. , & Paterson, D. L. (2008). Acinetobacter baumannii: Emergence of a successful pathogen. Clinical Microbiology Reviews, 21, 538–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research, 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot, P. (2010). Epigenetic switching: Bacteria hedge bets about staying or moving. Current Biology, 20, R480–R482. [DOI] [PubMed] [Google Scholar]
- Repizo, G. D. , Gagne, S. , Foucault‐Grunenwald, M. L. , Borges, V. , Charpentier, X. , Limansky, A. S. , … Salcedo, S. P. (2015). Differential Role of the T6SS in Acinetobacter baumannii Virulence. PLoS ONE, 10, e0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld, N. , Bouchier, C. , Courvalin, P. , & Perichon, B. (2012). Expression of the resistance‐nodulation‐cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR‐type regulator. Antimicrobial Agents and Chemotherapy, 56, 2504–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh, M. D. , Zalucki, Y. , Su, C. C. , Zhang, Q. , Shafer, W. M. , & Yu, E. W. (2011). Efflux pumps of the resistance‐nodulation‐division family: A perspective of their structure, function, and regulation in gram‐negative bacteria. Advances in Enzymology and Related Areas of Molecular Biology, 77, 109–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, P. , Deye, G. , Srinivasan, A. , Murray, C. , Moran, K. , Hulten, E. , … Petruccelli, B. (2007). An outbreak of multidrug‐resistant Acinetobacter baumannii‐calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clinical Infectious Diseases, 44, 1577–1584. [DOI] [PubMed] [Google Scholar]
- Srinivasan, V. B. , Vaidyanathan, V. , & Rajamohan, G. (2015). AbuO, a TolC‐like outer membrane protein of Acinetobacter baumannii, is involved in antimicrobial and oxidative stress resistance. Antimicrobial Agents and Chemotherapy, 59, 1236–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, J. , Bergmann, H. , Gottig, S. , Ebersberger, I. , & Averhoff, B. (2015). Acinetobacter baumannii virulence is mediated by the concerted action of three phospholipases d. PLoS ONE, 10, e0138360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D. L. , Bina, X. R. , & Bina, J. E. (2012). Vibrio cholerae VexH encodes a multiple drug efflux pump that contributes to the production of cholera toxin and the toxin co‐regulated pilus. PLoS ONE, 7, e38208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton, K. A. , Dimitrova, D. , & Rather, P. N. (2015). Phase‐variable control of multiple phenotypes in Acinetobacter baumannii Strain AB5075. Journal of Bacteriology, 197, 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, K. H. , Vallet‐Gely, I. , & Dove, S. L. (2009). Epigenetic control of virulence gene expression in Pseudomonas aeruginosa by a LysR‐type transcription regulator. PLoS Genetics, 5, e1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening, J. W. , Smits, W. K. , & Kuipers, O. P. (2008). Bistability, epigenetics, and bet‐hedging in bacteria. Annual Review of Microbiology, 62, 193–210. [DOI] [PubMed] [Google Scholar]
- Venter, H. , Mowla, R. , Ohene‐Agyei, T. , & Ma, S. (2015). RND‐type drug efflux pumps from Gram‐negative bacteria: Molecular mechanism and inhibition. Frontiers in Microbiology, 6, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, D. M. , Folster, J. P. , Shafer, W. M. , & Jerse, A. E. (2007). Regulation of the MtrC‐MtrD‐MtrE efflux‐pump system modulates the in vivo fitness of Neisseria gonorrhoeae. The Journal of Infectious Diseases, 196, 1804–1812. [DOI] [PubMed] [Google Scholar]
- Yoon, E. J. , Chabane, Y. N. , Goussard, S. , Snesrud, E. , Courvalin, P. , De, E. , & Grillot‐Courvalin, C . (2015). Contribution of resistance‐nodulation‐cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii . MBio, 6, e00309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


