Abstract
The recombinant retrovirus, MoFe2-MuLV (MoFe2), was constructed by replacing the U3 region of Moloney murine leukemia virus (M-MuLV) with homologous sequences from the FeLV-945 LTR. NIH/Swiss mice neonatally inoculated with MoFe2 developed T-cell lymphomas of immature thymocyte surface phenotype. MoFe2 integrated infrequently (0 to 9%) near common insertion sites (CISs) previously identified for either parent virus. Using three different strategies, CISs in MoFe2-induced tumors were identified at six loci, none of which had been previously reported as CISs in tumors induced by either parent virus in wild-type animals. Two of the newly identified CISs had not previously been implicated in lymphoma in any retrovirus model. One of these, designated 3-19, encodes the p101 regulatory subunit of phosphoinositide-3-kinase-gamma. The other, designated Rw1, is predicted to encode a protein that functions in the immune response to virus infection. Thus, substitution of FeLV-945 U3 sequences into the M-MuLV long terminal repeat (LTR) did not alter the target tissue for M-MuLV transformation but significantly altered the pattern of CIS utilization in the induction of T-cell lymphoma. These observations support a growing body of evidence that the distinctive sequence and/or structure of the retroviral LTR determines its pattern of insertional activation. The findings also demonstrate the oligoclonal nature of retrovirus-induced lymphomas by demonstrating proviral insertions at CISs in subdominant populations in the tumor mass. Finally, the findings demonstrate the utility of novel recombinant retroviruses such as MoFe2 to contribute new genes potentially relevant to the induction of lymphoid malignancy.
The gammaretroviruses represent a group of mammalian oncogenic retroviruses typically associated with the induction of leukemia and lymphoma in the natural host. Moloney murine leukemia virus (M-MuLV), a prototype gammaretrovirus, induces a T-lymphoblastic lymphoma of the thymus in virtually 100% of susceptible neonatal mice, with a latency of 3 to 4 months. The major determinant of tumorigenic potential and T-cell disease specificity resides in the M-MuLV long terminal repeat (LTR), particularly within the tandemly duplicated, 75-bp transcriptional enhancers (13). The M-MuLV LTR functions preferentially in T cells, a consequence of the cell type-specific binding of nuclear proteins to sites in the transcriptional enhancers. The cell type-specific activity of the M-MuLV enhancers has been correlated positively with the efficient induction of T lymphoma by the retrovirus (41). Feline leukemia virus (FeLV) is a naturally occurring gammaretrovirus that infects the domestic cat. Unlike M-MuLV, infection with FeLV in the natural host is associated with a variable and rather unpredictable disease outcome. Natural FeLV infections in the domestic cat are associated with malignant and proliferative diseases, including lymphomas and leukemias of lymphoid, myeloid, or erythroid origin, as well as degenerative diseases, including anemia (40).
We previously described a natural isolate of FeLV, termed FeLV-945, from non-T-cell malignant, proliferative, and degenerative diseases that occurred in a geographic cohort. Although FeLV-945 was not identified in T-cell lymphomas in the cohort, it was identified in non-T-cell multicentric lymphomas of uncertain phenotype, as well as in cases of myeloproliferative disease and anemia (1, 7). The LTR of FeLV-945 is characterized by a unique repeat motif not previously described from any other natural FeLV isolate. The U3 region of the FeLV-945 LTR contains only a single copy of the transcriptional enhancer, unusual for tumor-derived proviruses, followed 25 bp downstream by the tandem triplication of a 21-bp sequence. Our previous studies showed that the 21-bp triplication contributes enhancer function to the LTR, that it functions preferentially in a cell type-specific manner, and that it confers a replicative advantage to the virus in those cells (2, 24, 38). Recent studies showed that the activity of the 21-bp triplication depends on the specific binding of the c-Myb transcription factor to sites that cross the repeat junctions and the consequent recruitment of the coactivator CBP (15). To examine the influence of the triplication-containing FeLV-945 LTR on disease induction, we previously created an MuLV-FeLV recombinant retrovirus in which the U3 region of the M-MuLV LTR was substituted with homologous sequences from FeLV-945. The recombinant virus was termed MoFe2-MuLV (MoFe2). MoFe2 was shown to be infectious and tumorigenic in NIH/Swiss mice. Specifically, when neonatal animals were inoculated intraperitoneally with MoFe2, lymphomas of T-cell origin arose uniformly with a latency of 4 to 10 months (43).
The LTR of M-MuLV or FeLV, like those of other gammaretroviruses, functions in the malignant process by directing high levels of virus expression in relevant target tissues and also by insertionally activating oncogenes at the sites of proviral integration. When the same genetic locus is observed to be interrupted by proviral integration in multiple independent tumors, it is inferred that the commonly interrupted locus encodes an oncogene whose activation is relevant to tumor induction (22, 44). Such a locus is then referred to as a common insertion site (CIS). Indeed, the CISs used by M-MuLV and FeLV in the induction of lymphoma have been well characterized. Approximately 80% of M-MuLV-induced thymic lymphomas contain proviral insertions within at least one of three cellular loci: c-myc, pim-1, or pvt-1 (13). The c-myc and pim-1 loci are also involved in thymic lymphomas induced by FeLV, along with other loci, including bmi-1 and fit-1 (45). In contrast, in the non-T-cell multicentric lymphomas from which FeLV-945 was identified, none of these loci is targeted as a CIS. Rather, a locus in feline DNA of unknown function, termed flvi-1, is a common target of FeLV-945 proviral integration. Presumably, the unique structure of the FeLV-945 LTR is relevant to activation of a gene sequence within or near flvi-1 (1, 24). Considering that the MoFe2 LTR contains sequence elements typical of both M-MuLV and FeLV-945, it was of interest to evaluate the patterns of oncogene activation by the recombinant virus and to determine whether it resembles either parent virus. Initial studies suggested that the oncogenes known to be targeted by the parental viruses are not targets for integration by MoFe2. Specifically, no integrations at c-myc, pim-1, pvt-1, or flvi-1 loci were detected in an initial analysis of 12 animals (43). Thus, the pattern of oncogene involvement in MoFe2-induced tumors is apparently distinct from both M-MuLV and FeLV-945. In the present study, multiple approaches were used to identify CISs in a large set of MoFe2-induced lymphomas. The results revealed infrequent utilization of c-myc, pim-1, pvt-1, or flvi-1. CISs were identified at six loci, none of which had previously been reported as a CIS in tumors induced by either parent virus in wild-type animals and two of which had not previously been reported in any model. These findings demonstrate that the substitution of FeLV-945 LTR sequences into M-MuLV significantly altered the pattern of oncogene utilization in tumors, thus supporting a growing body of evidence that the distinctive sequence and/or structure of the retroviral LTR determines its pattern of insertional activation (5, 8, 12, 31, 32, 34, 35, 39). In addition, the data demonstrate the oligoclonal nature of retrovirus-induced lymphomas with respect to proviral integration at CISs and indicate the utility of recombinant retroviruses such as MoFe2 to identify novel oncogenes associated with lymphoma.
MATERIALS AND METHODS
Virus inoculations and tumor collection.
Newborn NIH/Swiss mice (<24 h of age) were inoculated intraperitoneally with 106 infectious units of MoFe2-MuLV in a 0.1-ml volume. Animals were monitored regularly and sacrificed by carbon dioxide asphyxiation when signs of morbidity were apparent. Necropsy was performed and tissues were collected for storage at −80°C.
Flow cytometry.
Thymocytes from diseased animals (∼106 cells) were suspended in 100 μl of standard azide buffer (SAB; pH 7.2) (containing [per liter] 10 g of FA Bacto buffer [Difco], 1 g of sodium azide, and 10 ml of heat-inactivated fetal bovine serum). Antibody was added (0.5 μg/106 cells) and was incubated for 30 min on ice in the dark. Cells were washed and resuspended in 100 μl of SAB without fetal bovine serum. Flow cytometry was performed on a Becton Dickinson FACSCalibur flow cytometer and analyzed with Becton Dickinson CellQuest Pro software. The antibodies used, obtained from BD Biosciences (San Jose, Calif.), were as follows: phycoerythrin-conjugated rat anti-mouse CD8a (Ly-2) monoclonal antibody (53-6.7), fluorescein isothiocyanate-conjugated rat anti-mouse CD4 (L3T4) monoclonal antibody (RM4-5), phycoerythrin-conjugated rat immunoglobulin G2a(κ) [IgG2a(κ)] monoclonal immunoglobulin isotype control (R35-95), and fluorescein isothiocyanate-conjugated rat IgG2a(κ) monoclonal immunoglobulin isotype control (R35-95).
Southern blot analysis.
Large-molecular-weight DNA was isolated from tumors, and Southern blot analysis was performed as previously described (1). The T-cell-receptor β (TCRβ) locus was examined by using 86T5, a 600-bp EcoRI fragment from the murine TCRβ cDNA (18). Proviral insertion patterns were analyzed by using a hybridization probe specific for the FeLV portion of the MoFe2 LTR. The probe, rLTR1, contains the FeLV-derived portion of the MoFe2 LTR between EcoRV and HincII restriction sites. The flvi-1 locus was examined by using probe A, a 1.2-kb PstI-EcoRI fragment, and probe D, a 0.9-kb BamHI-SacI fragment, cloned from feline flvi-1 flanking the domain of common retroviral insertion (25). The c-myc locus was examined by using pSVcmyc1 (ATCC 41029), a clone of mouse genomic DNA representing c-myc exons 2 and 3. The pim-1 locus was examined by using a 0.9-kb BamHI fragment of murine pim-1 genomic DNA (a gift from Anton Berns, The Netherlands Cancer Institute). The pvt-1 locus was examined by using a 2.2-kb XbaI fragment of murine pvt-1 (9). Genomic DNA was digested with EcoRI, BamHI, or HindIII for analysis of the flvi-1, c-myc, pim-1, and pvt-1 loci.
Construction and screening of libraries for identification of CISs.
Genomic DNA from the thymic tumor of animal MF8 was digested with EcoRI and size fractionated on a 16.5 ml of 5 to 20% potassium acetate gradient by centrifugation in an SW28 rotor at 20,000 rpm for 15.5 h at 20°C. Fractions containing DNA of the appropriate size were pooled and cloned into the bacteriophage λ vector λ ZapII (Stratagene, La Jolla, Calif.). The fragment of interest containing the host-virus junction was then isolated by screening the resulting clones with the rLTR1 probe. The fragment was then submitted for automated sequence analysis. A 301-bp region of host DNA near the site of integration was PCR amplified with the primers MF8T-1 (5′-TACTTGACACCAGCCTGAGTACC-3′) and MF8T-2 (5′-CTCTGTCTAGCATCTCACAG-3′). The resulting amplification product, MF8T-HVJ, was radiolabeled and used as a probe for Southern blot analysis of DNA from other MoFe2-induced tumors in the collection. A recombinant library of genomic DNA from MoFe2-induced tumor 493-3 (43) was prepared in a bacteriophage λ vector, λGEM-II (Stratagene), as previously described (26). Recombinants containing integrated MoFe2-MuLV proviral DNA were then isolated from the library using radiolabeled rLTR1 as a probe. From one of the selected recombinants, λ3-19, a 500-bp EcoRI-HindIII fragment was isolated from host DNA immediately flanking the integrated provirus. That fragment, termed 3-19HR0.5, was used as a probe for Southern blot analysis of DNA from other MoFe2-induced tumors in the collection.
Proviral integrations in the MF8T or 3-19 loci were PCR amplified with one primer specific for the proviral LTR and the other specific for host sequences near the site of insertion. Oligonucleotide primers used in PCR to amplify host-virus junction fragments from tumor DNA included A, B, C, and D from the MoFe2 LTR sequence; E, F, and G from the MF8T sequence; and H, I, and J from the 3-19 sequence. Primer sequences were as follows: A, 5′-CTCTTGCAGTTGCATCCGACTTGT; B, 5′-GCTGTTCCATCTGTTCCTGACCTTGA; C, 5′-AGGCTTAACTGTTCTTGGCCTCAAGC; D, CGCAAGTCTATGCTGAGACTTGAC; E, 5′-ACAATGAACAGCCAGCGAG; F, 5′-GTCTTCAATGCCCCTGTGTAGTG; G, 5′-GTCTTCAATGCCCCTGTGTAGTG; H, 5′-ATGCAAGGCTTCCAATGC; I, 5′-ATGACCTTGTGGTCGGT; and J, 5′-GGCTGGCCTTAAACTTGTGTATAGTCGGA. Amplification products were examined by agarose gel electrophoresis, extracted from the gel by using Qiaquik gel extraction kit (Qiagen, Inc., Valencia, Calif.), and subjected to automated sequence analysis. The resulting sequence from each integration site was compared to the Mouse Genome Database by using Ensemble (http://www.ensemble.org/Mus_musculus/) and the NCBI Mouse Genome Resources (http://www.ncbi.nlm.nih.gov/genome/guide/mouse/) assemblies. Each proviral integration was thereby positioned precisely in the mouse genome.
PCR-based approach for identification of CISs.
Host-virus junctions were PCR amplified from tumor DNA by using the Universal genome walker kit (BD Biosciences) as described in the user manual, with the modifications described below. High-molecular-weight DNA was digested with DraI or StuI, and libraries were constructed as described in the manual. PCRs were performed by using the Advantage cDNA polymerase mix (BD Biosciences) in a PE Biosystems GeneAmp PCR 2400. Amplification conditions for primary and secondary reactions were as follows: 1 cycle of 94°C for 15 s, 7 cycles of 94°C for 2 s and 70°C for 3 min, and 32 cycles of 94°C for 2 s and 65°C for 3 min, followed by an elongation step at 68°C for 10 min. Primary amplification was performed with the primers AP1 (BD Biosciences) and FeLV.307m (5′-TAAATGAGGCGGAAGGTCGGACTCTG-3′). Secondary PCR was performed with a 1:50 dilution of the primary reaction with primers AP2 (BD Biosciences) and MoFe.206m (5′-GCTATATCCGAGGCTTAACTGTTCTTGGC-3′). Secondary amplification products were examined by agarose gel electrophoresis, extracted from the gel by using QiaQuik gel extraction kit (Qiagen), and subjected for automated sequence analysis. The resulting sequence from each integration site was compared to the Mouse Genome Database by using Ensemble and the NCBI Mouse Genome Resources (http://www.ncbi.nlm.nih.gov/genome/guide/mouse/) assemblies. Each proviral integration was thereby positioned precisely in the mouse genome. Each integration site was then compared to the Mouse Retroviral Tagged Cancer Gene Database (RTCGD; http://rtcgd.ncifcrd.gov) to determine whether it had been previously reported as a proviral insertion site.
Northern blot analysis.
Total cellular RNA was isolated from tissues by using TRIzol reagent (Invitrogen Corp., Carlsbad, Calif.) according to the instructions of the manufacturer or by guanidinium isothiocyanate extraction and cesium chloride density gradient ultracentrifugation as described previously (27). Northern blot analysis was performed with 10 μg of total cellular RNA as previously described (27). Hybridization probes for Northern blot analysis were developed by reverse transcriptase-PCR (RT-PCR) with tumor-derived RNA that had been treated with DNA-free (Ambion, Inc., Austin, Tex.). RT-PCR was performed with Superscript One-Step RT-PCR reagents with Platinum Taq (Invitrogen) according to the manufacturer's instructions. Oligonucleotide primers used in RT-PCR to amplify probes for Northern blot analysis included Rasgrp.F (5′-GCAGAAACTATGACAACTACAGGCG), Rasgrp.B (5′-CGGAGGTGAAGTTCTGGTTTTG), P101.F (5′-ACATCCTACAGGAAGTCCTTCTC), P101.B (5′-GCTGGTGCTCTTTAAGAGTTTATAG), Spred1.F (5′-CAAGTCAGCCAGATACCATTCAGTC), and Spred1.B (5′-CGTTCCCCATCCTCTTTTCTTC). Amplification products were examined by gel electrophoresis, cloned into pGEM-T plasmid vector (Promega Corp., Madison, Wis.), and verified by nucleotide sequence analysis.
RESULTS AND DISCUSSION
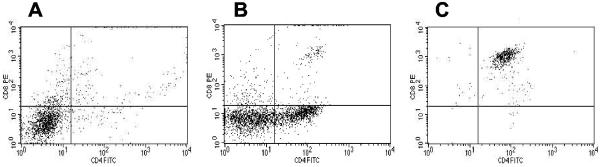
Our original report of the MuLV-FeLV recombinant retrovirus, MoFe2, described the induction of T-cell lymphoma in 12 infected animals after a latency of 15 to 41 weeks (average of 24 weeks) (43). In the present study, an additional 51 NIH/Swiss mice were inoculated as neonates with MoFe2, and tumors were collected after a latency of 2 to 8 months (average of 16 weeks). The more rapid kinetics of tumor induction are probably a reflection of the inoculum size used in the present study (106 infectious units) compared to the previous study (260 PFU as measured on XC cells [43]). Tumors were detected at multiple sites in individual animals, typically involving the thymus, spleen, and lymph nodes. As reported in the previous study (43), analysis of TCRβ gene rearrangement confirmed the tumors as lymphomas of T-cell origin and demonstrated that the multiple tumors within an individual animal typically represented the same clonal expansion (data not shown). Flow cytometric analysis indicated that MoFe2-induced lymphomas represented phenotypes of immature and maturing thymocytes, including CD4− CD8−, CD4+ CD8+, and CD4+ CD8− cells (Fig. 1). These findings indicated that MoFe2-induced lymphomas apparently originate in the thymus, as do tumors induced by M-MuLV (13). Thus, the target tissue for M-MuLV transformation was not altered by the introduction of FeLV-945 U3 sequences. Southern blot analysis was used to detect clonal MoFe2 insertions at loci commonly targeted for integration in tumors induced by either M-MuLV or FeLV-945. Previous studies have shown that c-myc, pvt-1, and pim-1 and are targets of M-MuLV integration in ∼50, ∼10, and ∼60% of lymphomas examined, respectively (10, 14, 47). FeLV-945 was not found to integrate at any of these loci but was identified at a locus termed flvi-1 in 36% of tumors examined (1, 24). In the present study, the c-myc, pvt-1, pim-1, and flvi-1 loci were found to be targeted infrequently by MoFe2 integration in lymphomas (6, 0, 9, and 0%, respectively). These observations confirmed our previous finding that the pattern of oncogene activation by the recombinant virus is distinct from either parent virus and suggested that novel targets of common integration might be identified in MoFe2-induced tumors.
FIG. 1.
Flow cytometric analysis of MoFe2-induced thymic lymphomas. Surface expression of CD4 and CD8 on three tumors was examined by flow cytometry. The results demonstrate CD4− CD8− (A), mixed tumor (CD4− CD8− and CD4+ CD8−) (B), and CD4+ CD8+ (C) surface phenotypes.
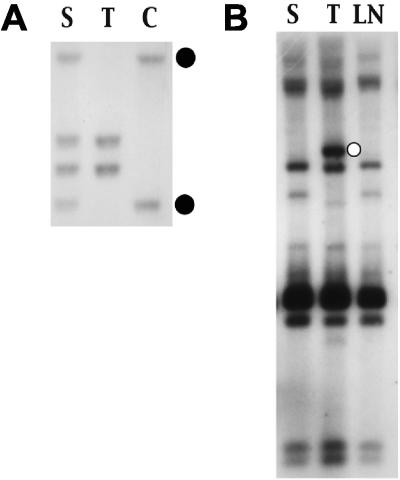
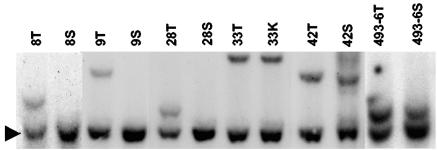
Several approaches were used to identify CISs in MoFe2-induced lymphomas. First, when proviral integration patterns were examined in the multiple tumors of individual animals, clonal integrations were observed in 8 of 44 cases that distinguished the tumor in one tissue from other tissues in the same animal. For example, animal MF8 developed tumors in the thymus, spleen, and lymph nodes that appeared by examination of TCRβ gene rearrangement to represent the same clonal expansion (Fig. 2A). The tumors also exhibited nearly identical patterns of MoFe2 integration; however, the thymic tumor contained a single clonal proviral insertion that was not apparent in tumors from other anatomic sites (Fig. 2B). This observation indicated that clonal insertion occurred after tumor spread and only in the thymic lymphoma, suggesting that integration in that locus might be involved in tumor maintenance or progression in the thymus. To localize the insertion and analyze it further, the unique host-virus junction fragment (Fig. 2B) was isolated by size selection on a potassium acetate gradient, molecularly cloned into a bacteriophage λ vector, and isolated by screening with a probe specific for the MoFe2 LTR. A restriction fragment of host DNA adjacent to the proviral insertion site was then used as a probe in Southern blot analysis to screen other MoFe2-induced tumors for insertion in the same locus. The results demonstrated clonal interruption of the locus not only in the thymic tumor from animal MF8 but also in thymic lymphomas from 5 other animals of 62 examined (Fig. 3). Interruption of the locus in 6 of 62 MoFe2-induced tumors thus defines it as a CIS. The locus was termed MF8T. Southern blot analysis further demonstrated that, in three animals, insertion at MF8T occurred exclusively in the thymic tumor and not in tumors at other anatomic sites. In three other animals, interruption of MF8T was identified in thymus, as well as in tumors of spleen or kidney (Fig. 3). Insertion at MF8T exclusively in the thymic tumor in three of six cases supports the hypothesis that MF8T insertion may be related to tumor maintenance or progression in the thymus.
FIG. 2.
Southern blot analysis of DNA from MoFe2-induced lymphoma of animal MF8. (A) Genomic DNA (8 μg) from lymphoma in the spleen (S), in the thymus (T), and from uninvolved control tissue of the same animal (C) was digested with HpaI and hybridized to radiolabeled probe for the TCRβ locus. The closed circles indicate the germ line configuration of the TCRβ locus. (B) Genomic DNA samples (8 μg) from lymphomas in the spleen (S), thymus (T), and lymph node (LN) of animal MF8 were digested with KpnI and hybridized to rLTR1, a probe specific for the integrated provirus of MoFe2-MuLV. The open circle indicates a clonal host-virus junction fragment that distinguishes the thymic tumor from tumors in other tissue sites of the same animal.
FIG. 3.
Southern blot analysis of the MF8T locus in MoFe2-induced lymphomas. DNA was digested with HindIII and hybridized to the MF8T-HVJ probe. Shown are DNA samples from six tumors with insertions in the MF8T locus, indicated by the animal number and tissue origin of the tumor (T, thymus; S, spleen; K, kidney). The arrow indicates the germ line configuration of the locus.
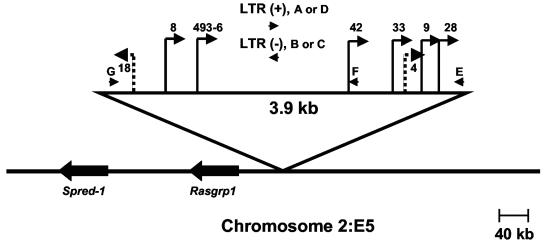
The identity of the MF8T locus was obtained by isolating it in unintegrated form from a genomic library of NIH/Swiss mouse DNA. Once the Mouse Genome Database became publicly available, comparison with sequence information from the unintegrated locus rapidly localized MF8T to mouse chromosome 2:E5. A PCR-based strategy was then developed to precisely position the clonal proviral integrations that had occurred at the MF8T locus in multiple independent lymphomas. Specifically, Southern blot analysis was first used with probes from the unintegrated locus to narrow the region within which each insertion occurred. Each host-virus junction was then PCR amplified with one primer specific for the proviral LTR (primer A, B, C, or D) and the other specific for host sequences near the site of insertion (primer E, F, or G). Amplification products were then sequenced to determine the precise position and transcriptional direction of each integrated provirus. The results demonstrated that proviral insertions at MF8T occurred within a domain of 3.9-kb of genomic DNA and that all were in the same transcriptional direction (Fig. 4). The PCR-based strategy was then used again to evaluate whether proviral integrations may have occurred at MF8T in other tumors in which insertion was not detectable by Southern blot analysis. In fact, host-virus junction fragments were amplified by using this strategy from 2 of 20 tumors examined. In both cases, proviral integration occurred within the previously identified 3.9-kb domain, although the insertion in animal MF18 was observed to be in opposite transcriptional orientation from the others (Fig. 4). These results indicate that ∼10% of MoFe2-lymphomas contain subdominant populations with insertions at MF8T. It is noteworthy that flvi-1, a CIS previously identified in FeLV-945-induced lymphomas, was originally localized to mouse chromosome 2:E1-E5 by in situ hybridization to metaphase chromosomes (25). Comparison of the flvi-1 sequence to the mouse genome database more precisely localized the CIS to mouse chromosome 2:E1, ca. 30 Mbp from MF8T.
FIG. 4.
Physical map of the MF8T locus. Depicted is the 3.9-kb domain of common proviral insertion. Vertical lines represent the positions of proviral integrations (indicated by animal number) with the transcriptional orientation of each provirus depicted by the direction of the arrow. Solid lines represent insertions detectable by Southern blot analysis. Checkered lines represent insertions undetectable by Southern blot analysis but identified by PCR amplification. Oligonucleotide primers used in PCR to amplify host-virus junctions are illustrated as horizontal arrowheads. The positions and transcriptional directions of the Spred-1 and Rasgrp1 genes are indicated.
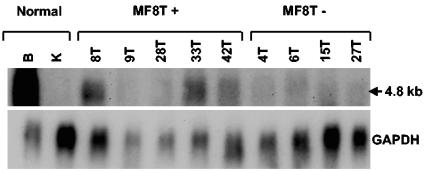
Use of the Mouse Genome Database predicted several genes in the region of chromosome 2:E5 surrounding the proviral insertions, including Spred-1 located 282 kb away and Rasgrp1 located 58 kb away from the domain of common insertion (Fig. 4). The majority of proviral insertions at MF8T were located upstream and in a transcriptional orientation opposite that of Spred-1 and Rasgrp1, a finding consistent with potential activation through enhancer insertion (Fig. 4). Others have reported that proviral insertion can affect the expression of multiple genes near the integration site, that insertion does not always affect the expression of the closest gene, and that insertion can activate the expression of cellular genes over distances as great as 300 kb (19, 23). With this in mind, expression of Spred-1 and Rasgrp1 were analyzed by Northern blot analysis in normal mouse tissues, in tumors containing proviral insertions, and in those without detectable insertions at MF8T. Although Spred-1 transcripts of 5.1 and 3.8 kb were readily detected in brain of uninfected animals, little or no expression was detected in MoFe2-induced tumors with or without insertions at MF8T (data not shown). Thus, proviral insertion did not activate expression from Spred-1. A 4.8-kb Rasgrp1 transcript was detected in the brain of uninfected NIH/Swiss mice and was also expressed to variable levels in tumors with MF8T insertions. In contrast, Rasgrp1 expression was low or undetectable in tumors from animals without MF8T proviral insertion (Fig. 5). Thus, proviral insertion at MF8T may activate expression of Rasgrp1 but does not do so uniformly.
FIG. 5.
Northern blot analysis of Rasgrp1 expression in tumors containing (MF8T+) or lacking (MF8T−) proviral insertion at MF8T. Total RNA (10 μg) was examined from lymphomas indicated by animal number and by the anatomic origin of the tumor (T, thymus) and also from normal tissues of uninfected NIH/Swiss mice (B, brain; K, kidney). Northern blots were hybridized to a probe specific for Rasgrp1 exons 9 to 16 and were subsequently hybridized to a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe as a loading control. Size of the observed transcript is indicated.
Rasgrp1 encodes a guanine nucleotide exchange factor required for T-cell development (11). Examination of the RTCGD revealed that Rasgrp1 had not previously been identified as a target for common insertion in tumors induced by M-MuLV in wild-type mice. The locus was previously identified as a CIS in genetically modified animals infected with M-MuLV (20, 33), in SL3-3-induced T-cell lymphomas (22), and in AKXD and BXH2 tumor models (44) (Table 1). Proviral insertions near Rasgrp1 have been reported to cluster in four domains of the locus, within intron 2 or 3, or in clusters from 0 to 80-kb upstream of the gene (17, 22, 33, 44). The CIS identified in MoFe2-induced tumors occurs within the previously identified cluster 50 to 63 kb upstream of the gene (Fig. 4). Although expression of Rasgrp1 was not uniformly upregulated in MoFe-2-induced end-stage tumors (Fig. 5), it is possible that an effect of proviral insertion on Rasgrp1 expression may occur earlier in the disease process, a possibility that has not yet been examined. Expression of Rasgrp1 has not been examined in other studies in which it was identified as a CIS.
TABLE 1.
Sites of proviral insertion identified in MoFe2-induced lymphomas in NIH/Swiss mice
| Locus (chromosome) | Full gene name | CIS in MoFe2-induced tumorsa | Previously identified insertion siteb | Modelsc
|
|
|---|---|---|---|---|---|
| Mouse strain (virus) (reference) | Tumor type | ||||
| MF8T (2:E5) | Ras guanyl releasing protein 1 | Y | Y (CIS) | BXH2 (Akv) (44) | Myeloid |
| EμMyc (M-MuLV) (33) | B cell, T cell | ||||
| p27Kip−/+ (M-MuLV) (20) | B cell, T cell | ||||
| p27Kip−/− (M-MuLV) (20) | T cell | ||||
| AKXD (Akv) (44) | B cell, T/B mix | ||||
| NIH/Swiss (SL3-3) (22) | T cell | ||||
| EμMyc;Pim1(−);Pim2(−)(M-MuLV)(33) | B cell, T cell | ||||
| 3-19 (11:B3) | P101 regulatory subunit of PI3Kγ | Y | N | ||
| Jundm2 (12:D1) | Jun dimerization protein 2 (predicted) | Y | Y (CIS) | p27Kip−/+ (M-MuLV) (20) | T cell |
| p27Kip−/− (M-MuLV) (20) | T cell | ||||
| EμMyc (M-MuLV) (33) | B cell | ||||
| NMRI (SL3-3) (42) | T cell | ||||
| Cas-Br-M (Cas-Br-M MuLV) (21) | Myeloid | ||||
| Ahi1 (10:A3) | Abelson helper integration site | Y | Y (CIS) | SIM.S, NIH/Swiss (A-MuLV) (37) | B cell |
| Cdkn2a−/− (M-MuLV) (29) | B cell | ||||
| p27Kip−/+ (M-MuLV) (20) | T cell | ||||
| p27Kip−/− (M-MuLV) (20) | T cell | ||||
| NIH/Swiss (SL3-3) (22) | T cell | ||||
| AKXD (Akv) (44) | B cell | ||||
| Cbfb-MYH11 (MLV-Ampho4070A) (6) | Myeloid | ||||
| EμMyc; Pim1(−);Pim2(−)(M-MuLV) (33) | B cell, T cell, T/B mix | ||||
| EμMyc (M-MuLV) (33) | B cell | ||||
| BXH2 (Akv) (44) | Myeloid | ||||
| Rras2 (7:E3) | Ras-related protein 2 | Y | Y (CIS) | Cdkn2a−/− (M-MuLV) (29) | Lymphoma (unspecified) |
| p27Kip−/+ (M-MuLV) (20) | T cell | ||||
| p27Kip−/− (M-MuLV) (20) | T cell | ||||
| NIH/Swiss (SL3-3) (22) | T cell | ||||
| AKXD (Akv) (44) | B cell, T/B mix | ||||
| AKXD (17) | Myeloid, T cell | ||||
| EμMyc; Pim1(−); Pim2(−) (M-MuLV) (33) | B cell, T cell | ||||
| Rw1 (1:B) | RW1 protein (predicted) | Y | N | − | − |
| Rin2 (2:G1) | Ras and Rab interactor 2 | N | Y (CIS) | BXH2 (Akv) (44) | Unspecified |
| Irs2 (8:A1.1) | Similar to insulin receptor substrate 2 (predicted) | N | Y (CIS) | EμMyc; Pim1(−); Pim2(−) (M-MuLV) (33) | B cell |
| AKXD (Akv) (44) | B cell, T/B mix | ||||
| Traip (9:F1) | TRAF-interacting protein | N | N | − | − |
| Grf2 (2:A3) | Guanine nucleotide releasing factor 2 | N | N | − | − |
| Lamr1 (9:F3) | C laminin receptor | N | N | − | − |
| Kpnb1 (11:D) | Karyopherin (importin) beta 1 | N | N | − | − |
To determine whether a proviral integration site was a CIS in MoFe2-induced lymphomas, Southern blot analysis of multiple tumors was performed by using a region of each integration site as a probe. Y, yes; N, no.
To determine whether a proviral integration site in MoFe2-induced lymphomas had been previously reported, integration sites were compared to the RTCGD. Y, yes; N, no. Previously identified CISs are indicated in parentheses.
When proviral integration sites were previously identified on the RTCGD, the strain of mouse, virus infection, and tumor type from each study are indicated. T/B mix, mix of T and B cells.
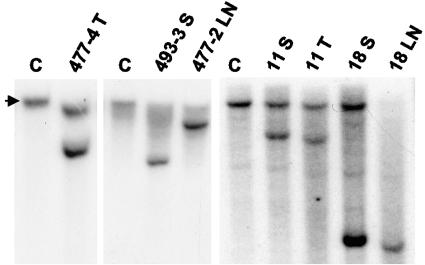
A second approach to identify CISs in MoFe2-induced lymphomas involved the construction and screening of a recombinant library of tumor DNA. A genomic library was constructed in a bacteriophage λ vector from a MoFe2-induced tumor shown by Southern blot analysis to contain only three clonal proviral insertions (data not shown). Proviral integrations were selected from the library by hybridization with a probe specific for the FeLV portion of the MoFe2 LTR. Host DNA sequences adjacent to the proviral insertions were then isolated and used as probes in Southern blot analysis to screen other MoFe2-induced tumors for interruptions of the same loci. Two of the clonal proviral insertions were characterized by this approach. One integration site was identified by sequence analysis as Kpnb1, a locus not previously identified as an integration site in any retrovirus model. This locus was not shown to be a CIS in MoFe2-induced tumors (Table 1). The other integration site was found to be interrupted in 5 of 64 tumors examined, thus defining it as a CIS (Fig. 6). This locus has been termed 3-19. Insertions at 3-19 were identified in tumors at multiple anatomic sites, including the spleens, thymuses, and lymph nodes of infected animals (Fig. 6). Thus, unlike MF8T, there does not appear to be a selective advantage for insertions in 3-19 in any specific tissue. Once the Mouse Genome Database became publicly available, the 3-19 locus was readily mapped by sequence comparison to mouse chromosome 11:B3. Proviral insertions at 3-19 were precisely positioned by PCR with one primer specific for the proviral LTR (primer A, B, C, or D) and the other specific for host sequences near the site of insertion (primer H, I, or J). Amplification products were then sequenced to determine the precise position and transcriptional direction of each integrated provirus. The results demonstrated that proviral insertions at 3-19 occurred within a domain of 5.5 kb of host DNA and that all were in the same transcriptional direction (Fig. 7). When the same PCR-based strategy was used to identify possible proviral integrations in 3-19 that were not detectable by Southern blot analysis, host-virus junction fragments were amplified from 2 of 18 tumors examined. In both cases, insertions occurred within the previously identified 5.5-kb domain and in the same transcriptional orientation as others in the cluster (Fig. 7). Thus, ∼11% of MoFe2-induced lymphomas contain subdominant populations with insertions at 3-19.
FIG. 6.
Southern blot analysis of the 3-19 locus in MoFe2-induced lymphomas. Shown are DNA samples from five lymphomas indicated by the animal number and anatomic origin of the tumor (T, thymus; S, spleen; LN, lymph node) and from an uninfected NIH/Swiss mouse (C). DNA was digested with EcoRI (477-4, -11, and -18) or KpnI (493-3 and 477-2) and hybridized to a radiolabeled probe representing the 3-19 locus. The arrow represents the germ line configuration of the locus.
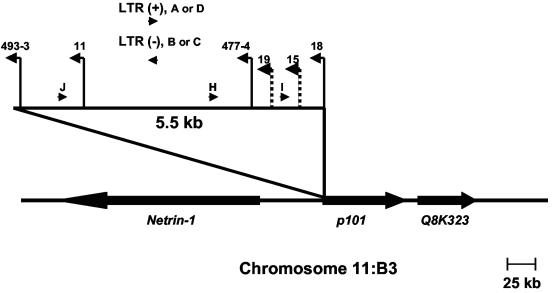
FIG. 7.
Physical map of the 3-19 locus. Depicted is the 5.5-kb domain of common proviral insertion. Vertical lines represent the positions of proviral integrations (indicated by animal number) with the transcriptional orientation of each provirus depicted by the direction of the arrow. Solid lines represent insertions detectable by Southern blot analysis. Checkered lines represent insertions undetectable by Southern blot analysis but identified by PCR amplification. Oligonucleotide primers used in PCR to amplify host-virus junctions are illustrated as horizontal arrowheads. The positions and transcriptional directions of the Netrin-1, p101, and Q8K323 genes are indicated.
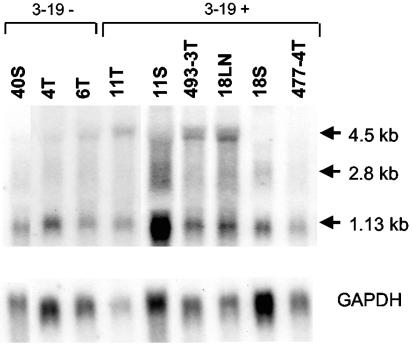
Use of the Mouse Genome Database further facilitated the identification of genes in the vicinity of 3-19 (Fig. 7). The cluster of insertions was located immediately upstream of a recently identified gene that encodes the p101 regulatory subunit of phosphoinositide-3-kinase-gamma (PI3Kγ) (3, 4). (The predicted gene, designated F730038I15RIK on the Mouse Genome Database, is referred to below as p101.) One proviral insertion occurred 273 bp downstream of the p101 transcription start site. The remainder occurred within 5.5 kb upstream of the transcriptional start, all in opposite transcriptional orientation. Analysis of p101 expression revealed three transcripts of 4.5, 2.8, and 1.13 kb (Fig. 8). By comparison, the Ensemble database predicts a 2.8-kb transcript that encodes p101. Because p101 is a recently identified gene about which little is known, neither the coding capacity nor the relationship between the multiple transcripts is understood. In general, lymphomas that contained proviral insertion at 3-19 were observed to express higher levels of one or more of the transcripts. For example, the 4.5-kb transcript was detected in three of four animals with insertions in 3-19 but was observed at low or undetectable levels in tumors lacking that insertion. It was noteworthy, however, that the 4.5-kb transcript was not uniformly detected in tumors from all anatomic sites of the same animal. For example, the transcript was readily detectable in the lymph node tumor of animal MF18 but not in the tumor from the spleen, although both contained the proviral insertion (Fig. 6). Similarly, tumors of the spleen and thymus of animal MF11 demonstrated apparently clonal proviral insertion at 3-19 (Fig. 6), but the patterns of p101 expression were distinct (Fig. 8). These observations suggest that the microenvironment of the tumor may influence expression of p101. PI3Kγ, of which p101 is the regulatory subunit, is known to be activated by G-protein-coupled receptors through direct interaction with Gβγ. Further, it is an activator of mitogen-activated protein kinase signaling and is expressed preferentially in cells of hematopoietic lineage (3, 4, 28). Thus, PI3Kγ function may be relevant to growth regulation in the target cell for transformation by MoFe2. Examination of the RTCGD revealed that the 3-19 locus has not previously been identified as a locus of common insertion in M-MuLV-induced tumors or in any other model (Table 1).
FIG. 8.
Northern blot analysis of p101 expression in tumors containing (3-19+) or lacking (3-19−) proviral insertion at 3-19. Total RNA (10 μg) was examined from lymphomas indicated by animal number and by the anatomic origin of the tumor (T, thymus; S, spleen; LN, lymph node). Northern blots were hybridized to a probe specific for p101 exon 10. The membranes were subsequently hybridized to a GAPDH probe as a loading control. The sizes of the observed transcripts are indicated.
In our previous analysis of MoFe2-induced lymphoma, variations in LTR sequence were observed that correlated with time to disease induction. Specifically, LTRs in relatively early appearing tumors (19 to 23 weeks) retained the original MoFe2 LTR intact and unaltered, but LTRs in late-appearing tumors (29 to 41 weeks) had acquired various duplications of the enhancer (43). In the present study, the sequences of LTRs were examined from five insertions at MF8T and from three insertions at 3-19. This analysis demonstrated that all LTRs retained the original sequence intact and unaltered (data not shown). Thus, the LTR sequence variation observed in the previous study was not recapitulated. This may reflect the generally shorter latency to tumor induction in the present study (16 weeks versus 24 weeks; (43).
A third approach to identifying CISs in MoFe2-induced tumors was PCR based with commercially available reagents designed to amplify DNA sequences for which little information is available (Universal Genome Walker Kit; BD Biosciences); thus, the reagents were ideal for amplifying host-virus junction fragments from tumor DNA. For this approach, tumor DNA was digested with a restriction enzyme, ligated to adaptor molecules, and PCR amplified with primer AP1 specific for the adaptor and primer FeLV.307m specific for the MoFe2 LTR. To increase the specificity, secondary amplification was performed with primer AP2 specific for the adaptor and primer MoFe.206m specific for the MoFe2-LTR. Secondary amplification products representing host-virus junctions were then sequenced, and the exact site of proviral insertion was mapped by comparison to the mouse genome database. Insertion sites were then compared to the RTCGD to evaluate which ones had been previously identified as targets of integration. Probes were developed from the host sequence at each integration site for Southern blot analysis of other tumors in the collection to determine whether any integration site represented a CIS. Using this PCR-based approach, three tumors have been examined and nine clonal proviral integrations have been characterized. Of these, four were found to be CISs in MoFe2-induced lymphomas (Table 1). First, the Jundm2 locus on mouse chromosome 12:D1 was found to be interrupted by proviral insertion in 2 of the 41 tumors examined. Two insertions were identified in the locus in a single tumor, one located 58-kb upstream from the predicted Jundm2 gene and the other located within the Jundm2 coding region between exons 2 and 3. The insertion located upstream of the gene was undetectable by Southern blot analysis and thus represents a subdominant population in the tumor mass (data not shown). The Jundm2 gene is predicted to encode a member of the AP-1 family of transcription factors that functions as a Jun dimerization partner. The second CIS identified by the PCR-based approach was localized to mouse chromosome 10:A3 and identified as Ahi-1 (Table 1). Ahi-1, shown to be interrupted by proviral insertion in 2 of 32 MoFe2-induced tumors examined, was originally described as a common target of integration for the helper virus of Abelson-MuLV in pre-B-cell lymphomas (37). As determined by Southern blot analysis, the insertions at Ahi-1 were subclonal in one case and clonal in the other (data not shown). Third, the Rras2 locus on mouse chromosome 7:E3 was also identified as a CIS, clonally interrupted in 2 of 41 MoFe2-induced lymphomas examined (Table 1). Rras2 is a member of the superfamily of Ras-related proteins previously shown to possess oncogenic potential (16). None of these loci, i.e., Jundm2, Ahi-1, or Rras2, has been previously been described as a CIS in M-MuLV-induced tumors in wild-type mice; however, they were previously identified as a CIS or a single insertion site in M-MuLV-induced lymphomas in genetically modified animals, including EμMyc mice that do not express pim-1 and pim-2 (33), p27Kip1 knockout mice (20), or Cdkn2a knockout mice (29). The fourth CIS identified in MoFe2-induced lymphomas by the PCR-based approach was identified as the Rw1 locus on mouse chromosome 1:B, shown to be clonally interrupted in 2 of 32 tumors examined (Table 1). Little is known about the Rw1 protein, although it is thought to function in the immune response to virus infection (46). The Rw1 locus has not previously been reported as a target of retroviral insertion in lymphomas induced by M-MuLV or in any other retrovirus model. Of the remaining insertions isolated by the PCR-based approach, none was identified as a CIS in MoFe2-induced lymphomas. Two of the insertion sites, Rin2 and Irs2, have been previously identified as CISs in other model systems, including EμMyc mice that do not express pim-1 and pim-2 (33) and the BXH2 and AKXD models (44). The remaining insertion sites—Traip, Grf2, and Lamr1—have not previously been reported (Table 1).
Taken together, the studies described above show that the substitution of FeLV-945 U3 sequences into the M-MuLV LTR did not alter the target tissue for M-MuLV transformation (Fig. 1) but significantly altered the pattern of CIS utilization in the induction of T-cell lymphoma. CISs in MoFe2-induced tumors were identified at six loci, none of which had been previously reported as CISs in tumors induced by either parent virus in wild-type animals. Two of the CISs had not previously been implicated in lymphoma in any retrovirus model (Table 1). These observations support a growing body of evidence that the distinctive sequence and/or structure of the retroviral LTR determines its pattern of insertional activation (5, 8, 12, 31, 32, 34, 35, 39). It is noteworthy that the CIS pattern of the recombinant virus, although distinct from either parent virus, resembled that of SL3-3 MuLV. SL3-3 has been reported to integrate near c-myc and pim-1 with a frequency like that of MoFe2 (∼10%) and also near Rasgrp1, Rras2, and Jundm2 (22, 42). In fact, Rras2 was identified as the most common target for insertion of SL3-3 in lymphomas induced in NIH/Swiss mice (22). The transcriptional enhancer element in the MoFe2 LTR contains a core binding site distinct from M-MuLV (TGTGGTAAG) but identical to SL3-3 (TGTGGTTAA) (36, 43). Unlike M-MuLV or SL3-3, the U3 LTR sequence of MoFe2 contains the unique 21-bp element triplicated in tandem downstream of the enhancer (43). Recent studies showed that the 21-bp triplication binds the transcription factor c-Myb, which then recruits the coactivator CBP to the LTR (15). Assembly of this transcription factor complex may be important for modulating expression of a distinctive set of target oncogenes. The present findings further demonstrate the oligoclonal nature of retrovirus-induced lymphomas that appear by Southern blot analysis to represent clonal expansions. MoFe2-induced lymphomas were apparently clonal, as evidenced by Southern blot analysis of TCRβ gene rearrangement and proviral integration pattern (representative example in Fig. 2). Nonetheless, ca. 10% of tumors were shown to contain subdominant populations with insertions at MF8T or 3-19 loci (Fig. 4 and 7), and another tumor contained subdominant populations with insertions near Jundm2 and Ahi-1 (data not shown). The presence of subdominant populations in an apparently clonal tumor have also been reported in Akv, M-MuLV, and type B leukemogenic virus tumor models (5, 30). Finally, the studies reported here demonstrate the utility of novel recombinant retroviruses such as MoFe2 for the identification of new oncogenes involved in lymphoma. The search for CISs in the present study was by no means exhaustive; nonetheless, new CISs were identified whose potential impact on tumor induction is intriguing.
Acknowledgments
We gratefully acknowledge the valuable technical contributions of Joshua Kayser, Jared Kushner, Courtney Hanby, and Catherine DiGiorgio.
This study was supported by PHS grant CA83823 from the National Cancer Institute, by a grant from the Ladies Leukemia League, and by Development Funds of the Tulane Cancer Center. C.J. was supported in part by a grant from Cancer Association of Greater New Orleans.
REFERENCES
- 1.Athas, G. B., B. Choi, S. Prabhu, P. A. Lobelle-Rich, and L. S. Levy. 1995. Genetic determinants of feline leukemia virus-induced multicentric lymphomas. Virology 214:431-438. [DOI] [PubMed] [Google Scholar]
- 2.Athas, G. B., P. Lobelle-Rich, and L. S. Levy. 1995. Function of a unique sequence motif in the long terminal repeat of feline leukemia virus isolated from an unusual set of naturally occurring tumors. J. Virol. 69:3324-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baier, R., T. Bondeva, R. Klinger, A. Bondev, and R. Wetzker. 1999. Retinoic acid induces selective expression of phosphoinositide 3-kinase gamma in myelomonocytic U937 cells. Cell Growth Differ. 10:447-456. [PubMed] [Google Scholar]
- 4.Brock, C., M. Schaefer, H. P. Reusch, C. Czupalla, M. Michalke, K. Spicher, G. Schultz, and B. Nurnberg. 2003. Roles of G beta gamma in membrane recruitment and activation of p110 gamma/p101 phosphoinositide 3-kinase gamma. J. Cell Biol. 160:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broussard, D. R., J. A. Mertz, M. Lozano, and J. P. Dudley. 2002. Selection for c-myc integration sites in polyclonal T-cell lymphomas. J. Virol. 76:2087-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castilla, L. H., P. Perrat, N. J. Martinez, S. F. Landrette, R. Keys, S. Oikemus, J. Flanegan, S. Heilman, L. Garrett, A. Dutra, S. Anderson, G. A. Pihan, L. Wolff, and P. P. Liu. 2004. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 101:4924-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandhasin, C., P. A. Lobelle-Rich, and L. S. Levy. 2004.. Feline leukemia virus LTR variation and disease association in a geographic and temporal cluster. J. Gen. Virol. 85:2937-2942. [DOI] [PubMed]
- 8.Chen, H., and F. K. Yoshimura. 1994. Identification of a region of a murine leukemia virus long terminal repeat with novel transcriptional regulatory activities. J. Virol. 68:3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cory, S., M. Graham, E. Webb, L. Corcoran, and J. M. Adams. 1985. Variant (6;15) translocations in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c-myc oncogene. EMBO J. 4:675-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuypers, H. T., G. Selten, W. Quint, M. Zijlstra, E. R. Maandag, W. Boelens, P. van Wezenbeek, C. Melief, and A. Berns. 1984. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell 37:141-150. [DOI] [PubMed] [Google Scholar]
- 11.Dower, N. A., S. L. Stang, D. A. Bottorff, J. O. Ebinu, P. Dickie, H. L. Ostergaard, and J. C. Stone. 2000. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat. Immunol. 1:317-321. [DOI] [PubMed] [Google Scholar]
- 12.Ethelberg, S., J. Lovmand, J. Schmidt, A. Luz, and F. S. Pedersen. 1997. Increased lymphomagenicity and restored disease specificity of AML1 site (core) mutant SL3-3 murine leukemia virus by a second-site enhancer variant evolved in vivo. J. Virol. 71:7273-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan, H. 1997. Leukemogenesis by Moloney murine leukemia virus: a multistep process. Trends Microbiol. 5:74-82. [DOI] [PubMed] [Google Scholar]
- 14.Fan, H., H. Chute, E. Chao, and P. K. Pattengale. 1988. Leukemogenicity of Moloney murine leukemia viruses carrying polyoma enhancer sequences in the long terminal repeat is dependent on the nature of the inserted polyoma sequences. Virology 166:58-65. [DOI] [PubMed] [Google Scholar]
- 15.Finstad, S. L., S. Prabhu, K. R. Rulli, and L. S. Levy. 3 September 2004, posting date. Regulation of FeLV-945 by c-Myb binding and CBP recruitment to the LTR. Virol. J. 1:3. [Online] http://www.virologyj.com/content/1/1/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, S. M., A. D. Cox, G. Drivas, M. G. Rush, P. D'Eustachio, and C. J. Der. 1994. Aberrant function of the Ras-related protein TC21/R-Ras2 triggers malignant transformation. Mol. Cell. Biol. 14:4108-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, G. M., D. Skapura, and M. J. Justice. 2000. Genetic profile of insertion mutations in mouse leukemias and lymphomas. Genome Res. 10:237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedrick, S. M., D. I. Cohen, E. A. Nielsen, and M. M. Davis. 1984. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature 308:149-153. [DOI] [PubMed] [Google Scholar]
- 19.Himmel, K. L., F. Bi, H. Shen, N. A. Jenkins, N. G. Copeland, Y. Zheng, and D. A. Largaespada. 2002. Activation of clg, a novel dbl family guanine nucleotide exchange factor gene, by proviral insertion at evi24, a common integration site in B-cell and myeloid leukemias. J. Biol. Chem. 277:13463-13472. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, H. C., C. P. Martins, Y. Bronkhorst, E. Randel, A. Berns, M. Fero, and B. E. Clurman. 2002. Identification of oncogenes collaborating with p27Kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proc. Natl. Acad. Sci. USA 99:11293-11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joosten, M., Y. Vankan-Berkhoudt, M. Tas, M. Lunghi, Y. Jenniskens, E. Parganas, P. J. Valk, B. Lowenberg, E. van den Akker, and R. Delwel. 2002. Large-scale identification of novel potential disease loci in mouse leukemia applying an improved strategy for cloning common virus integration sites. Oncogene 21:7247-7255. [DOI] [PubMed] [Google Scholar]
- 22.Kim, R., A. Trubetskoy, T. Suzuki, N. A. Jenkins, N. G. Copeland, and J. Lenz. 2003. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J. Virol. 77:2056-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazo, P. A., A. J. Klein-Szanto, and P. N. Tsichlis. 1990. T-cell lymphoma lines derived from rat thymomas induced by Moloney murine leukemia virus: phenotypic diversity and its implications. J. Virol. 64:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levesque, K. S., L. Bonham, and L. S. Levy. 1990. flvi-1, a common integration domain of feline leukemia virus in naturally occurring lymphomas of a particular type. J. Virol. 64:3455-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levesque, K. S., M. G. Mattei, and L. S. Levy. 1991. Evolutionary conservation and chromosomal localization of flvi-1. Oncogene 6:1377-1379. [PubMed] [Google Scholar]
- 26.Levy, L. S., and P. A. Lobelle-Rich. 1992. Insertional mutagenesis of flvi-2 in tumors induced by infection with LC-FeLV, a myc-containing strain of feline leukemia virus. J. Virol. 66:2885-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy, L. S., P. A. Lobelle-Rich, and J. Overbaugh. 1993. flvi-2, a target of retroviral insertional mutagenesis in feline thymic lymphosarcomas, encodes bmi-1. Oncogene 8:1833-1838. [PubMed] [Google Scholar]
- 28.Lopez-Ilasaca, M., P. Crespo, P. G. Pellici, J. S. Gutkind, and R. Wetzker. 1997. Linkage of G protein-coupled receptors to the MAPK signaling pathway through PI 3-kinase gamma. Science 275:394-397. [DOI] [PubMed] [Google Scholar]
- 29.Lund, A. H., G. Turner, A. Trubetskoy, E. Verhoeven, E. Wientjens, D. Hulsman, R. Russell, R. A. DePinho, J. Lenz, and M. Van Lohuizen. 2002. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat. Genet. 32:160-165. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Hernandez, J., A. B. Sorensen, and F. S. Pedersen. 2001. Murine leukemia virus proviral insertions between the N-ras and unr genes in B-cell lymphoma DNA affect the expression of N-ras only. J. Virol. 75:11907-11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martiney, M. J., L. S. Levy, and J. Lenz. 1999. Suppressor mutations within the core binding factor (CBF/AML1) binding site of a T-cell lymphomagenic retrovirus. J. Virol. 73:2143-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto, Y., Y. Momoi, T. Watari, R. Goitsuka, H. Tsujimoto, and A. Hasegawa. 1992. Detection of enhancer repeats in the long terminal repeats of feline leukemia viruses from cats with spontaneous neoplastic and nonneoplastic diseases. Virology 189:745-749. [DOI] [PubMed] [Google Scholar]
- 33.Mikkers, H., J. Allen, P. Knipscheer, L. Romeijn, A. Hart, E. Vink, A. Berns, and L. Romeyn. 2002. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Genet. 32:153-159. [DOI] [PubMed] [Google Scholar]
- 34.Miura, T., M. Shibuya, H. Tsujimoto, M. Fukasawa, and M. Hayami. 1989. Molecular cloning of a feline leukemia provirus integrated adjacent to the c-myc gene in a feline T-cell leukemia cell line and the unique structure of its long terminal repeat. Virology 169:458-461. [DOI] [PubMed] [Google Scholar]
- 35.Morrison, H. L., B. Soni, and J. Lenz. 1995. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J. Virol. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pantginis, J., R. M. Beaty, L. S. Levy, and J. Lenz. 1997. The feline leukemia virus long terminal repeat contains a potent genetic determinant of T-cell lymphomagenicity. J. Virol. 71:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poirier, Y., C. Kozak, and P. Jolicoeur. 1988. Identification of a common helper provirus integration site in Abelson murine leukemia virus-induced lymphoma DNA. J. Virol. 62:3985-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prabhu, S., P. A. Lobelle-Rich, and L. S. Levy. 1999. The FeLV-945 LTR confers a replicative advantage dependent on the presence of a tandem triplication. Virology 263:460-470. [DOI] [PubMed] [Google Scholar]
- 39.Rajan, L., D. Broussard, M. Lozano, C. G. Lee, C. A. Kozak, and J. P. Dudley. 2000. The c-myc locus is a common integration site in type B retrovirus-induced T-cell lymphomas. J. Virol. 74:2466-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rezanka, L. J., J. L. Rojko, and J. C. Neil. 1992. Feline leukemia virus: pathogenesis of neoplastic disease. Cancer Investig. 10:371-389. [DOI] [PubMed] [Google Scholar]
- 41.Short, M. K., S. A. Okenquist, and J. Lenz. 1987. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J. Virol. 61:1067-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorensen, A. B., M. Duch, H. W. Amtoft, P. Jorgensen, and F. S. Pedersen. 1996. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J. Virol. 70:4063-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starkey, C. R., P. A. Lobelle-Rich, S. Granger, B. K. Brightman, H. Fan, and L. S. Levy. 1998. Tumorigenic potential of a recombinant retrovirus containing sequences from Moloney murine leukemia virus and feline leukemia virus. J. Virol. 72:1078-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, T., H. Shen, K. Akagi, H. C. Morse, J. D. Malley, D. Q. Naiman, N. A. Jenkins, and N. G. Copeland. 2002. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32:166-174. [DOI] [PubMed] [Google Scholar]
- 45.Tsatsanis, C., R. Fulton, K. Nishigaki, H. Tsujimoto, L. Levy, A. Terry, D. Spandidos, D. Onions, and J. C. Neil. 1994. Genetic determinants of feline leukemia virus-induced lymphoid tumors: patterns of proviral insertion and gene rearrangement. J. Virol. 68:8296-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tscharke, D. C., R. Wilkinson, and A. Simmons. 2000. Use of mRNA differential display to study the action of lymphocyte subsets in vivo and application to a murine model of herpes simplex virus infection. Immunol. Lett. 74:127-132. [DOI] [PubMed] [Google Scholar]
- 47.Wirschubsky, Z., P. Tsichlis, G. Klein, and J. Sumegi. 1986. Rearrangement of c-myc, pim-1 and Mlvi-1 and trisomy of chromosome 15 in MCF- and Moloney-MuLV-induced murine T-cell leukemias. Int. J. Cancer 38:739-745. [DOI] [PubMed] [Google Scholar]