Abstract
Multiple amino acid changes within herpes simplex virus type 1 (HSV-1) gB and gK cause extensive virus-induced cell fusion and the formation of multinucleated cells (syncytia). Early reports established that syncytial mutations in gK could not cause cell-to-cell fusion in the absence of gB. To investigate the interdependence of gB, gK, and UL20p in virus-induced cell fusion and virion de-envelopment from perinuclear spaces as well as to compare the ultrastructural phenotypes of the different mutant viruses in a syngeneic HSV-1 (F) genetic background, gB-null, gK-null, UL20-null, gB/gK double-null, and gB/UL20 double-null viruses were constructed with the HSV-1 (F) bacterial artificial chromosome pYEBac102. The gK/gB double-null virus YEbacΔgBΔgK was used to isolate the recombinant viruses gBsyn3ΔgK and gBamb1511ΔgK, which lack the gK gene and carry the gBsyn3 or gBamb1511 syncytial mutation, respectively. Both viruses formed small nonsyncytial plaques on noncomplementing Vero cells and large syncytial plaques on gK-complementing cells, indicating that gK expression was necessary for gBsyn3- and gBamb1511-induced cell fusion. Lack of virus-induced cell fusion was not due to defects in virion egress, since recombinant viruses specifying the gBsyn3 or gKsyn20 mutation in the UL19/UL20 double-null genetic background caused extensive cell fusion on UL20-complementing cells. As expected, the gB-null virus failed to produce infectious virus, but enveloped virion particles egressed efficiently out of infected cells. The gK-null and UL20-null viruses exhibited cytoplasmic defects in virion morphogenesis like those of the corresponding HSV-1 (KOS) mutant viruses. Similarly, the gB/gK double-null and gB/UL20 double-null viruses accumulated capsids in the cytoplasm, indicating that gB, gK, and UL20p do not function redundantly in membrane fusion during virion de-envelopment at the outer nuclear lamellae.
Herpes simplex viruses specify at least 11 virally encoded glycoproteins as well as several nonglycosylated membrane-associated proteins that function in several important roles, including virus entry via fusion of the viral envelope with cellular membranes, intracellular virion morphogenesis and egress, cell-to-cell spread, and virus-induced cell fusion (42, 43, 53, 58, 60). Mutations that cause extensive virus-induced cell-to-cell fusion have been mapped to at least four regions of the viral genome: the UL20 gene (3, 40, 41), the UL24 gene (32, 57), the UL27 gene encoding glycoprotein B (gB) (5, 47), and the UL53 gene coding for gK (4, 13, 29, 49, 50, 55).
The UL20 and UL53 (gK) genes encode multipass transmembrane proteins of 222 and 338 amino acids, respectively, which are conserved in all alphaherpesviruses (13, 40, 51). UL20p assumes a membrane topology that constitutes a mirror image of gK with both the amino and carboxyl termini of UL20p located intracellularly, while both the amino and carboxyl termini of gK are located extracellularly (17, 21, 41). gK is known to be posttranslationally modified by N-linked carbohydrate addition, while UL20p is not glycosylated (13, 29, 51). UL20p and gK localize to trans-Golgi network membranes after endocytosis from cell surfaces (21). UL20p and gK are essential for cytoplasmic virion morphogenesis, since mutant viruses lacking either gK or UL20p accumulate capsids within the cytoplasm that are unable to acquire envelopes by budding into trans-Golgi network-associated membranes (3, 18, 19, 24, 31, 33, 41). Furthermore, UL20p is essential for virus-induced cell fusion caused by either gB or gK syncytial mutations (19), and it is necessary for gK cell surface expression (17, 19, 21), strongly suggesting that direct or indirect interactions between gK and UL20 are necessary for intracellular protein trafficking, virus egress, and virus-mediated cell-to-cell fusion (15, 17, 19, 24, 41).
Glycoprotein B (gB) is highly conserved across all subfamilies of herpesviruses. It exists within cells as a homodimeric type I integral membrane protein that is N-glycosylated at multiple sites (10, 11, 27, 37, 53, 59). A variety of evidence indicates that gB plays important roles in membrane fusion phenomena during virus entry and virus-induced cell fusion: herpes simplex virus type 1 (HSV-1) mutants lacking gB are not able to enter cells (6) due to a postattachment defect that can be resolved by polyethylene glycol-mediated fusion of viral envelopes with cellular membranes (7); single-amino-acid substitutions and truncations of the carboxyl terminus of gB cause extensive virus-induced cell fusion (2, 5, 8, 25); and transient coexpression of gB with gD, gH, and gL causes cell-to-cell fusion, which is substantially increased by carboxyl-terminal truncations of gB (20, 26, 34, 48). These results suggest a direct role for gB in membrane fusion and suggest that perturbations of the carboxyl-terminal domains of gB facilitate gB-mediated cell-to-cell fusion.
It has been shown that syncytial mutations in gK failed to cause virus-induced cell fusion in the absence of gB, indicating that gB was essential for gK-associated cell-to-cell fusion (7). In this paper, we used the HSV-1 (F) bacterial artificial chromosome BAC system (62) to generate multiple mutant viruses lacking either gK, UL20, or gB or combinations of gK and gB or UL20 and gB. Comparison of the BAC-generated insertion-deletion mutants in the HSV-1 (F) genetic background revealed subtle but important differences in individual virus phenotypes in comparison to the laboratory-adapted virus strain KOS, most likely due to the limited passage of these viruses in cell culture. In addition, mutant viruses carrying syncytial mutations in gB in the absence of either gK or UL20 were generated. Characterization of these viruses revealed that gK is necessary for gB-mediated virus-induced cell fusion. Concurrent deletion of either gB and gK or gB and UL20p did not prevent perinuclear virion de-envelopment, indicating that gB and gK or gB and UL20p do not function redundantly in mediating fusion between the viral envelopes and the outer nuclear membrane.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells, CV-1 cells, and 293 cells were obtained from the American Type Culture Collection (Rockville, Md.). Cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL; Grand Island, N.Y.), supplemented with 10% fetal calf serum and antibiotics. Fd20-1 cells and D6 cells were maintained as described previously (6, 41). The UL19- and UL20-complementing cell line G5 was a gift of P. Desai (Johns Hopkins Medical Center) (14). gKDIV5, gKsyn20DIV5, and tsB5 were described previously (17, 23). Flp-In-CV-1 cells were maintained as directed by the manufacturer (Invitrogen, Carlsbad, Calif.).
Plasmids.
Plasmid pcDNA5/FRT/gK was constructed by inserting the gK gene, PCR amplified from KOS viral DNA, into pcDNA5/FRT/V5-His-TOPO (Invitrogen). Plasmid p20F2 contains a 7,185-bp HSV-1 (KOS) genomic fragment spanning the region comprising UL19, UL20, UL21, and UL22 and was described previously (41). Primers 5′CMVEGFP_20.5_BclI (5′-TTCTTCTTCTTCTGATCATGCTGACGTAAGTAGTTATTAATAGTAATCAATTACGGGG-3′) and 3′CMVEGFP_BclI (5′-CTCTTCCTCTCCTGATCAATGAGTTTGGACAAACCACAACTAGA-3′) were used to PCR amplify a cytomegalovirus-enhanced green fluorescent protein (EGFP) gene cassette from pTF9200 (22). The resultant PCR product was digested with BclI and ligated into p20F2 that was restricted the BamHI and treated with calf intestinal phosphatase. The resultant plasmid, designated pΔ1920-EGFP, contained a cytomegalovirus-EGFP gene cassette which replaced the HSV-1 region in p20F2 contained within the BamHI sites at genomic positions 39234 and 41549 (GenBank accession number NC_001806), removing the entire UL20 open reading frame (ORF) as well as the first 1,293 bp of the UL19 ORF.
Construction of transformed Flp-In-CV-1 cell lines.
Generation of stable Flp-In-CV-1 expression cell lines was performed essentially as directed by the manufacturer (Invitrogen) and as described previously (41). Confluent Flp-In-CV-1 monolayers in six-well plates were transfected with 0.3 μg of pcDNA5/FRT/gK and 2.7 μg of pOG44 (Invitrogen), which expresses Flp recombinase, with Lipofectamine 2000. After 2 weeks, hygromycin B (125 μg/ml)-resistant colonies were tested for the ability to complement the growth of YEbacΔgK virus. All colonies tested complemented YEbacΔgK virus. Isolate FcgK-1, derived from pcDNA5/FRT/gK-transfected cells, was chosen for further use. FcgK-1 cells were used in generating YEbacΔgK virus stocks.
Construction of a replication-deficient adenovirus expressing gB.
pAd5-Blue is a replication-defective, E1- and E3-deleted adenovirus type 5 (Ad5)-containing plasmid (44). gB was PCR amplified from HSV-1 KOS viral DNA with primers gB5′ClaI (5′-TCCTCCATCGATATGCGCCAGGGCGCCCCCG-3′) and gB3′XbaI (5′-CTTCTTTCTAGATCACAGGTCGTCCTCGTCGGC-3′) and transferred into the unique ClaI and XbaI sites located in the E1-deleted region of pAd5-Blue, immediately downstream of the cytomegalovirus promoter, to generate pAd5-gB. pAd5-gB was transfected into 293 cells in a T-25 flask with Lipofectamine 2000, according to the manufacturer's directions (Invitrogen); 72 h posttransfection, cell lysates were prepared by multiple freeze-thaw cycles. A small portion of the cell lysate was then used to infect a subsequent flask of 293 cells. After four passages, a high-titer stock of Ad5-gB was obtained, which was deficient for replication on Vero cells. Ad5-gB virus at different multiplicities of infection (MOIs) was tested for its ability to complement YEbacΔgB on Vero cells, and an MOI of 10 was found to be optimal.
Construction of HSV-1 mutants with deletions of the UL20, gB, and/or gK genes (pYEbac102, pYEbacΔgB, pYEbacΔUL20, pYEbacΔgK, pYEbacΔgBΔUL20, and pYEbacΔgBΔgK).
Insertion-deletion mutagenesis of pYEbac102 DNA was accomplished in Escherichia coli with the λ gam recE recT (GET) recombination system (45, 46), as described previously for mutagenesis of the KSHV genome (39). Electrocompetent YEbac102 Escherichia coli DH10B cells were transformed with plasmid pGETrec, which contains the genes encoding recE, recT, and bacteriophage λ gam, grown on plates containing chloramphenicol (12.5 μg/ml) and ampicillin (100 μg/ml). Individual colonies were picked and grown overnight in Luria-Bertani (LB) medium containing chloramphenicol and ampicillin. The next day, the culture was inoculated into 250 ml of LB containing chloramphenicol and ampicillin until an optical density at 600 nm of 0.4 was reached. Addition of l-arabinose to a final concentration of 0.2% (wt/vol) and further incubation for 40 min induced expression of the recE, recT and λ gam genes from plasmid pGETrec. The cells were then harvested and made electrocompetent.
For the ΔUL20-GFPzeo mutation, a PCR fragment containing a GFP-zeocin resistance (GFP-zeo) gene cassette flanked by ≈50 bp of viral sequences on both sides was used for recombination to construct pYEbacΔUL20, containing the GFP-zeo gene cassette within the targeted UL20 genomic region. For the ΔgB-Kan mutation, a PCR fragment containing a kanamycin resistance gene cassette flanked by ≈50 bp of viral sequences on both sides was used for recombination to construct pYEbacΔgB, which contained the kanamycin resistance gene cassette within the targeted UL27 genomic region. For ΔgK-GFPzeo, a PCR fragment containing a GFP-zeo gene cassette flanked by ≈50 bp of viral sequences on both sides was used for recombination to construct pYEbacΔgK, containing the GFP-zeo gene cassette within the targeted UL53 genomic region. Briefly, 40 μl of electrocompetent DH10B cells harboring both pYEbac102 and pGETrec were electroporated with 200 ng of each PCR product to delete the target gene (gB, UL20, or gK) with standard electroporation parameters (1.8 kV/cm, 200 Ω, and 25 μF). Following electroporation, cells were grown in 1 ml of LB for 60 min and subsequently streaked onto LB agar plates containing chloramphenicol (12.5 μg/ml) as well as either kanamycin (50 μg/ml) or zeocin (25 μg/ml). Mutant pYEbac102 DNA containing a deletion in the gB, UL20, or gK gene was isolated from bacterial colonies, and a second round of electroporation was performed to remove plasmid pGETrec. Following electroporation, cells were grown on agar plates containing chloramphenicol as well as either kanamycin or zeocin. Similar procedures were followed, starting with electrocompetent DH10B E. coli cells harboring pYEbacΔgB, in order to generate pYEbacΔgBΔgK and pYEbacΔgBΔUL20.
Confirmation of the targeted mutations in pYEbac102 DNA.
HSV-1 BAC DNAs (pYEbac102, pYEbacΔgB, pYEbacΔUL20, pYEbacΔgK, pYEbacΔgBΔUL20, and pYEbacΔgBΔgK) were purified from 500 ml of BAC cultures with the Qiagen large-construct kit (Qiagen; Valencia, Calif.). BAC DNA was digested with KpnI, run on a 0.7% agarose gel, and transferred to charged nylon membranes (Bio-Rad; Richmond, Calif.). Southern blot hybridization was performed with either a 1.1-kb kanamycin or 800-bp GFP-zeocin PCR product labeled with biotin (New England Biolabs; Boston, Mass.). Chemiluminescence detection of the DNA was performed with the North2South chemiluminescent hybridization and detection kit as described by the manufacturer (Pierce Inc.; Rockford, Ill.). All mutations were sequenced to verify the presence of the desired mutations in the BACs. To verify that no spurious mutations were introduced into the viral genome of the individual BAC mutants during mutagenesis or during the transfection procedure, each null mutation was rescued by homologous recombination with DNA fragments spanning each gene. Each rescue virus was verified to have the phenotypic and replication characteristics of the wild-type BAC YEbac102.
Transfection of HSV-1 BAC DNAs.
Transient transfection of cells with BAC DNAs was performed with Lipofectamine 2000 (Invitrogen). Complementing cell lines were grown to 95% confluency in six-well plates. Cells were transfected with BAC DNA mixed with Lipofectamine 2000 in Opti-MEM medium as recommended by the manufacturer (Invitrogen). After 6 h of incubation at 37°C, the medium was removed from the transfected cells, cells were washed with phosphate-buffered saline, and subsequent fresh Dulbecco's modified Eagle's medium with 10% fetal calf serum was added. At 72 h posttransfection, virus stocks were collected.
Plaque morphology of YEbac102 mutants.
For plaque morphology, Vero, Fd20-1, FcgK-1, and D6 cells were infected at MOIs of 0.01 and 0.001 with the YEbac102, YEbacΔgB, YEbacΔUL20, YEbacΔgK, YEbacΔgBΔgK, and YEbacΔgBΔUL20 viruses and visualized by immunohistochemistry at 48 h postinfection with horseradish peroxidase-conjugated anti-HSV antibodies (Dako) and Novared (VectorLabs) substrate development. For complementation of YEbacΔgBΔgK and YEbacΔgBΔUL20 viruses, Fd20-1 and FcgK-1 were infected with Ad5-gB at an MOI of 10, 1 h prior to HSV-1 infection. Due to the constitutive expression of gB in all cells, plaques were visualized by immunohistochemistry at 48 h postinfection with an anti-gD monoclonal antibody (RG-1103; Rumbaugh-Goodwin Institute; Plantation, Fla.) with the Vectastain Elite ABC kit and Novared substrate development kit (VectorLabs; Burlingame, Calif.) according to the manufacturer's directions and as described previously (17, 19).
One-step growth kinetics of YEbac102 mutants.
Analysis of one-step growth kinetics was as described previously (17, 23). Briefly, each virus at an MOI of 5 was adsorbed to approximately 6 × 105 Vero cells at 4°C for 1 h. Thereafter, warm medium was added, and virus was allowed to penetrate for 2 h at 37°C. Any remaining extracellular virus was inactivated by low-pH treatment (0.1 M glycine, pH 3.0). Cells and supernatants were harvested immediately thereafter (0 h) or after 4, 9, 16, 20, or 24 h of incubation at 37°C. Virus titers were determined by endpoint titration of virus stocks on a complementing cell line.
Generation of recombinant UL20-null and gK-null viruses specifying mutant gB.
In order to generate recombinant viruses specifying mutations in gB, PCR amplification of the UL27 region from purified viral DNA of YEbac102 (wild-type gB), tsb5 (gBsyn3), or amb1511 (gBamb1511) was performed. Each PCR product was used for homologous recombination with both the YEbacΔgBΔUL20 and YEbacΔgBΔgK viruses. Virus plaques that formed on Fd20-1 (UL20 complementing) or FcgK-1 (gK complementing) cells were picked and sequentially plaque purified at least five times. Replacement of the gB-null resident kanamycin resistance cassette and concomitant insertion of the specific mutant gB gene were confirmed by DNA sequencing.
Generation of UL19 and UL20 double-null viruses.
Viruses that contained a cytomegalovirus immediate-early promoter-driven EGFP gene cassette in place of a UL19/UL20 gene deletion were generated by homologous recombination with plasmid pΔ1920-EGFP and the parental virus gKDIV5, gKsyn20DIV5, or tsB5 (gBsyn3). G5 cells were transfected with pΔ1920-EGFP and at 6 h posttransfection were infected at an MOI of 0.1 with either gKDIV5, gKsyn20DIV5, or tsB5 (gBsyn3) to generate UL19 and UL20 double-null viruses that were designated Δ1920, Δ1920-syn20, and Δ1920-syn3, respectively. Viruses were subsequently plaque isolated on G5 cells by fluorescence microscopy and sequentially plaque purified at least five times. Insertion of the cytomegalovirus-EGFP gene cassette and the concomitant deletion of the UL19 and UL20 genes were confirmed by diagnostic PCR and DNA sequencing.
Electron microscopy.
Cell monolayers were infected with the indicated virus at an MOI of 3. All cells were prepared for transmission electron microscopy (TEM) examination 24 h postinfection. Infected cells were fixed in a mixture of 2% paraformaldehyde and 1.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.3. Following treatment with 1% OsO4 and dehydration in an ethanol series, the samples were embedded in Epon-Araldyte resin and polymerized at 70°C. Thin sections were made on an MTXL ultratome (RMC Products), stained with 5% uranyl acetate and citrate/nitrate/acetate lead, and observed with a Zeiss 10 transmission electron microscope as described previously (19, 41).
RESULTS
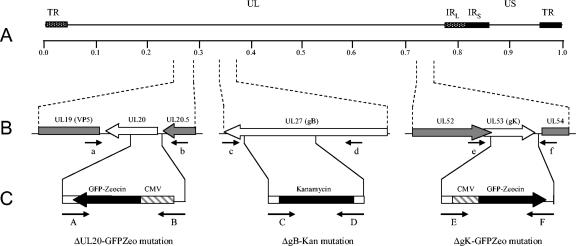
Construction of HSV-1 BACs pYEbacΔgB, pYEbacΔUL20, pYEbacΔgK, pYEbacΔgBΔUL20, and pYEbacΔgBΔgK.
The complete HSV-1 (F) genome was recently cloned into a bacterial artificial chromosome (pYEbac102), enabling genetic manipulation of the HSV-1 genome in E. coli (62). The BAC-based GET homologous recombination system was used to construct deletions within the gB, UL20, and gK genes in E. coli. Deletion of each gene was accomplished by targeting the gB, UL20, or gK open reading frame (ORF) with specific primers as detailed in the Materials and Methods section. For construction of the ΔUL20-GFPzeo mutation, primers A and B were used for homologous recombination and positioned to remove a 353-bp region of the UL20 gene upon insertion of a GFP-zeocin resistance gene cassette (Table 1; Fig. 1B and C), extending from the UL20 ATG to an HpaI site inside the UL20 ORF.
TABLE 1.
Synthetic oligonucleotide primers
| Primer designation | Name | Sequencea | Purpose and product size (bp) |
|---|---|---|---|
| A | 3′ UL20-GFPZeo | 5′-CACGGACATCCCCCAAAACACGGGCGCCGACAACGGCAGACGATCCCTCTTGATGTTagacatgataagatacattgatgagtttgg-3′ | UL20 GET recombination (2,025) |
| B | 5′ UL20-GFPZeo | 5′-CTGACGTAAGCGACCCTTTGCGGTTTCGGTCTCCCCACCTCCACCGCACACCCCgtccgttacataacttacggtaaatgg-3′ | |
| C | 3′ gB-Kan | 5′-GTTGGGAACTTGGGTGTAGTTGGTGTTGCGGCGCTTGCGCATGACCATGTCcggttgatgagagctttgttgtaggtggac-3′ | gB GET recombination (1,209) |
| D | 5′ gB-Kan | 5′-CTGACCCTGTGGAACGAGGCCCGCAAGCTGAACCCCAACGCCATCGCCTCGTGAccacgttgtgtctcaaaatctctgatgtta-3′ | |
| E | 5′ gK-GFPZeo | 5′-AGCACCTCTCAACCGTCGTCTTGATAACGGCGTACGGCCTCGTGCTCGTGTGGTGAgtccgttacataacttacggtaaatgg-3′ | gK GET recombination (2,022) |
| F | 3′ gK-GFPZeo | 5′-CAGTTTCCAATTTGCATATGCCGTTACGGTTTCCGCCGGCCTGGATGTGACGagacatgataagatacattgatgagtttgg-3′ | |
| a | UL20-Rev | 5′-GAAGGCTGCGGCCTCGTTCCAG-3′ | Diagnostic PCR for wt UL20 (909) or ΔUL20-GFPZeo (2,469) |
| b | UL20-For | 5′-ACGGCCTTATCAAAACACTCGCCTC-3′ | |
| c | gB-Rev | 5′-TCACAGGTCGTCCTCGTCGGC-3′ | Diagnostic PCR for wt gB (2,127) or ΔgB-Kan (2,261) |
| d | gB-For | 5′-GTGATCGACAAGATCAACGCCAAGG-3′ | |
| e | gK-For | 5′-GGCACGCCATGGTCGCCGTCCGTTCCCTGC-3′ | Diagnostic PCR for wt gK (1,265) or ΔgK-GFPZeo (2,238) |
| f | gK-Rev | 5′-ATCACATACCCCGTTCCGCTTCCG-3′ |
For GET recombination primers, the region homologous to HSV-1 DNA is in uppercase and the region which binds to the marker gene is in lowercase letters.
FIG. 1.
Schematic of the strategy for the construction of pYEbac102 mutant BACs. (A) The top line represents the prototypic arrangement of the HSV-1 genome, with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (B) Shown below are the expanded genomic regions of the UL20, UL27, and UL53 ORFs as well as the approximate locations of the sites to which insertion of the marker genes was targeted and the primers used in diagnostic PCR to confirm the presence of each mutation. (C) PCR fragments containing the kanamycin resistance or GFP-zeocin resistance gene cassette flanked by ≈50 bp of viral sequences on both sides were used for targeted GET recombination in E. coli to construct pYEbac102 mutant BACs with insertion-deletion mutations in the UL20, UL27, and/or UL53 ORFs. The approximate locations of the primers used in amplification of each PCR fragment are also shown.
Inactivation of the gB gene was achieved by insertion of a kanamycin resistance gene cassette within gB (ΔgB-Kan mutation). Specifically, primers C and D were designed to remove a 970-bp fragment starting with gB ORF nucleotide 1655 upon homologous recombination with pYEbac102 (Table 1; Fig. 1B and C). This gB deletion does not disrupt the UL27.5 ORF overlapping the 5′ end of gB (9) and is similar to that of the KΔT gB-null virus (6), with the exception that the ΔgB-Kan mutation was constructed to include a stop codon at the 5′ end of the kanamycin resistance cassette ensuring that no aberrant proteins could be made via a translational readthrough of the kanamycin resistance gene cassette.
For construction of the ΔgK-GFPzeo mutation, primers E and F were positioned to delete almost the entire gK gene (941 bp) without affecting the proximal UL52 gene, which partially overlaps the gK ORF (Table 1; Fig. 1B and C). Each pYEbac102 mutant construct contains one or more of the insertion-deletion mutations mentioned above: pYEbacΔgB contains the ΔgB-Kan mutation; pYEbacΔUL20 contains the ΔUL20-GFPzeo mutation; pYEbacΔgK contains the ΔgK-GFPzeo mutation; pYEbacΔgBΔUL20 contains both the ΔgB-Kan and the ΔUL20-GFPzeo mutations; and pYEbacΔgBΔgK contains both the ΔgB-Kan and the ΔgK-GFPzeo mutations.
The YEbac102 viral constructs were tested for the presence of the engineered insertion-deletion mutation via diagnostic PCR. Primers a and b (Table 1; Fig. 1A), located outside of the UL20 gene, amplified the predicted 909-bp diagnostic UL20 DNA fragments from BACs that contain wild-type UL20, while a predicted 2,469-bp product was produced from BACs that contain the ΔUL20-GFPzeo mutation (Fig. 2A, panel i). Primers c and d (Table 1; Fig. 1A) located within gB but outside the deleted region amplified the predicted 2,127-bp diagnostic gB DNA fragment from BACs that contain wild-type gB, while a predicted 2,261-bp product was produced from BACs that contain the ΔgB-Kan mutation (Fig. 2A, panel iii). Primers e and f (Table 1; Fig. 1A) located outside of the gK deletion region amplified a 1,265-bp diagnostic gK DNA fragment from BACs that contain wild-type UL20, while a predicted 2,238-bp product was produced from BACs that contain the ΔgK-GFPzeo mutation (Fig. 2A, panel ii).
FIG. 2.
Genomic analysis of pYEbac102 mutants. (A) Diagnostic PCR to confirm the insertion of the relevant gene cassettes in place of the deleted genomic regions in UL20 (A, panel i) UL27 (gB) (A, panel ii), and UL53 (gK) (A, panel iii). (B) KpnI restriction fragment analysis of pYEbac102 mutant BACs in comparison to wild-type pYEbac102. The black arrow demarcates the position of the KpnI DNA fragment altered by the ΔgB-Kan mutation, while the gray arrow points to the missing 9,432-bp fragment resulting from the ΔUL20-GFPzeo mutation. (C and D) Southern blot analysis of pYEbac102 mutant BACs. The KpnI-restricted BACs from panel B were hybridized with either a kanamycin resistance gene (C) or GFP-zeocin resistance gene (D) biotinylated probe. The biotinylated kanamycin resistance gene probe hybridized to an estimated 4,835-bp DNA fragment in pYEbacΔgB, pYEbacΔgBΔgUL20, and pYEbacΔgBΔgK DNA, which was absent in the wild-type pYEbac102 DNA (indicated by a black arrow in panel C). The biotinylated GFP-zeocin probe identified a predicted 10,992-bp DNA fragment (gray arrow) in pYEbacΔUL20 and pYEbacΔgBΔgUL20 DNA corresponding to the ΔUL20-GFPzeo mutation, and a predicted 8,956-bp fragment (white arrow) in pYEbacΔgK and pYEbacΔgBΔgK DNA corresponding to the ΔgK-GFPzeo mutation (D).
Further confirmation of the genetic content of the pYEbac102 mutant constructs was obtained via restriction endonuclease fragment analysis (Fig. 2B) and Southern blotting (Fig. 2C and D). Restriction enzyme analysis of pYebac102, pYEbacΔgB, pYEbacΔUL20, pYEbacΔgK, pYEbacΔgBΔUL20 and pYEbacΔgBΔgK DNA with KpnI produced a similar genomic DNA restriction pattern with a few exceptions caused by the specified alterations of gB, UL20, and gK containing KpnI DNA fragments. First, the 4,701-bp band containing the UL27 (gB) ORF increased in size by 134 bp to 4,835 bp for pYEbacΔgB, pYEbacΔgBΔUL20, and pYEbacΔgBΔgK as a result of the ΔgB-Kan mutation. This 4,835-bp KpnI DNA fragment (demarcated by a black arrow) was visible in the lanes labeled with the above-mentioned constructs appearing between the predicted 4,680-bp and 4959-bp KpnI DNA fragments (Fig. 2B). In addition, Southern blot analysis revealed that a kanamycin resistance gene cassette probe hybridized to a single 4,835-bp KpnI DNA fragment contained only in pYEbacΔgB, pYEbacΔgBΔUL20, and pYEbacΔgBΔgK restriction digests (indicated by a black arrow) (Fig. 2C), indicating that the ΔgB-Kan mutation had been successfully transferred into a single genomic site.
Second, the wild-type 9,432-bp KpnI DNA fragment containing the UL20 ORF increased in size by 1,560 bp to 10,992 bp for pYEbacΔUL20 and pYEbacΔgBΔUL20. It is apparent that the 9,432-bp band is missing in pYEbacΔUL20 and pYEbacΔgBΔUL20, as indicated by a gray arrow (Fig. 2B). However, the 10,992-bp KpnI DNA fragment is not easily identifiable due to the presence of KpnI genomic fragments with predicted sizes of 10,837 and 11,024 bp. The 10,992-bp DNA fragment contains the ΔUL20-GFPzeo mutation, which was confirmed by probing the corresponding blot with a GFP-zeocin probe (Fig. 2D, gray arrow). The GFP-zeocin probe hybridized to unique DNA fragments of the predicted sizes for both pYEbacΔUL20 and pYEbacΔgBΔUL20. Finally, the ΔgK-GFPzeo mutation is predicted to increase the size of the wild-type 7,983-bp DNA fragment containing the UL53 (gK) ORF by 973 to 8,956 bp. These DNA fragments were not readily discernible in the ethidium bromide-stained restriction profile (Fig. 2A). However, the GFP-zeocin probe hybridized to the 8,956-bp DNA fragment (white arrow) in pYEbacΔgK and pYEbacΔgBΔgK, confirming the creation of the ΔgK-GFPzeo mutation (Fig. 2C). Again, the presence of a single DNA fragment detected by the GFP-zeocin probe indicates the absence of any spurious insertions.
Generation of infectious virus from pYEbac102-based constructs.
To generate virus stocks from the mutant pYEbac102 constructs, transient transfection of individual BAC DNAs into specific cell lines transformed with either the gB, UL20, or gK gene and capable of complementing the resultant viruses was performed. Specifically, pYEbac102 was transfected into Vero cells, pYEbacΔgB into gB-complementing D6 cells (6), pYEbacΔUL20 into UL20-complementing Fd20-1 cells (42), and pYEbacΔgK into FcgK-1 cells. The FcgK-1 cell line constitutively expresses the gK gene under the cytomegalovirus immediate-early promoter and was constructed to efficiently complement the ΔgK-GFPzeo mutation and minimize the chance for generation of wild-type viruses via recombination with the endogenous gK gene (see Materials and Methods). For all transfection experiments, virus plaques became visible 72 h posttransfection, and virus stocks exhibiting maximum cytopathic effects were collected at appropriate points.
For complementation of the double-null viruses produced by transfection of pYEbacΔgBΔUL20 and pYEbacΔgBΔgK, the replication-deficient adenovirus Ad5-gB was constructed, which constitutively expresses gB under the cytomegalovirus immediate-early promoter (see Materials and Methods). A series of experiments were performed to assess the optimum MOI of this virus that could maximize complementation of gB-null viruses. These experiments showed that Ad5-gB at an MOI of 10 efficiently complemented gB-deficient viruses (not shown). To complement double-null viruses having the gB gene deletion, UL20- or gK-complementing cell lines were infected with Ad5-gB 1 h prior to transfection with pYEbacΔgBΔUL20 or pYEbacΔgBΔgK. The same strategy was used to amplify virus stocks of the double-null viruses.
Plaque morphology and growth kinetics of HSV-1 YEbac102 mutants.
HSV-1 viruses containing deletions in UL20, gB, and gK have been described previously (3, 6, 19, 30, 33). These viruses were produced by homologous recombination of plasmid constructs with viral DNA in cell culture experiments. Typically, isolation of these viruses required multiple plaque purifications to ensure that the mutant virus stocks did not contain contaminating wild-type virus. In contrast, the YEbac102 UL20, gB, and gK null viruses were constructed in E. coli with the GET recombination system (45, 46). In this case, production of mutant viruses did not require any plaque purification, since the progeny viruses did not contain any contaminating wild-type genomes.
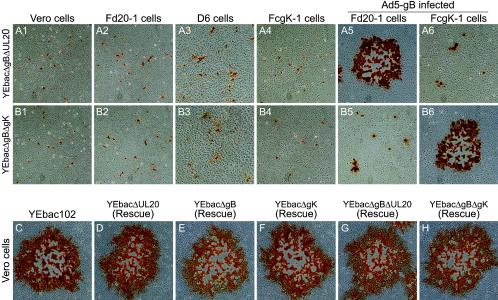
To assess the effect of the deletion of the various genes on cell-to-cell spread, the plaque morphologies of the YEbac102, YEbacΔgB, YEbacΔUL20, and YEbacΔgK viruses were examined in Vero, Fd20-1, D6, and FcgK-1 cells at 48 h postinfection (Fig. 3). The wild type YEbac102 produced large plaques of a similar size on all infected cells (Fig. 3A). Infection of noncomplementing cells with either YEbacΔUL20 or YEbacΔgK resulted in small plaques containing on average three to five cells (Fig. 3B1, B3, B4, D1, D3, and D3, respectively), while complementation of the UL20 and gK gene deletions resulted in large plaques comparable in size to the YEbac102 plaques (Fig. 3B2 and D4, respectively). Similarly, infection of noncomplementing cells with the YEbacΔgB virus produced only single infected cells (Fig. 3C1, C2, and C4), indicating that the virus was unable to spread beyond the primary infected cell, while infection of complementing D6 cells resulted in large plaques (Fig. 3C3). The YEbacΔgBΔUL20 and YEbacΔgBΔgK double-null viruses formed large plaques when plated onto Ad5-gB-infected Fd20-1 or FcgK-1 cells, respectively (Fig. 4A5 and B6), single infected cells on Vero cells, or cells able to complement for only gK or UL20 (Fig. 4A1, A2, A4, B1, B2, and B4), or very small plaques in cells able to complement gB (Fig. 4A3, A6, B3, and B5).
FIG. 3.
Plaque phenotypes of gB-, gK-, and UL20-null viruses. The plaque phenotypes of YEbac102, YEbacΔUL20, YEbacΔgB, and YEbacΔgK were observed on Vero (A1, B1, C1, and D1), Fd20-1 (A2, B2, C2, and D2), D6 (A3, B3, C3, and D3), and FcgK-1 (A4, B4, C4, and D4) cells. Confluent cell monolayers were infected with wild-type YEbac102 (A1, A2, A3, and A4), YEbacΔUL20 (B1, B2, B3, and B4), YEbacΔgB (C1, C2, C3, and C4), or YEbacΔgK (D1, D2, D3, and D4) at an MOI of 0.001, and viral plaques were visualized at 48 h postinfection by immunohistochemistry.
FIG. 4.
Plaque phenotypes of double-null viruses and rescued viruses. The plaque phenotypes of YEbacΔgBΔgUL20 and YEbacΔgBΔgK on Vero (A1 and B1), Fd20-1 (A2 and B2), D6 (A3 and B3), and FcgK-1 (A4 and B4) cells or Ad5-gB infected Fd20-1 (A5 and B5) and FcgK-1 (A6 and B6) cells are shown. Confluent cell monolayers were infected with YEbacΔgBΔgUL20 (A1, A2, A3, A4, A5, and A6) or YEbacΔgBΔgK (B1, B2, B3, B4, B5, and B6) at an MOI of 0.001. The plaque phenotypes of the rescued gB-, gK-, and UL20-null (D, E, and F) and double-null (G and H) viruses in comparison to the wild-type YEbac102 (C) on Vero cells are shown. Viral plaques were visualized at 48 h postinfection by immunohistochemistry.
Rescue of the individual mutations from each BAC-derived virus was performed by homologous recombination with wild-type DNA fragments spanning each gene. Double-null mutations were rescued sequentially with the gB-null mutation rescued first followed by rescue of either the gK-null or UL20-null mutations. Importantly, all rescued viruses formed large plaques similar to the wild-type YEbac102 virus when plated on noncomplementing Vero cells, indicating that there were no spurious mutations in the parental mutant BAC viruses affecting virus replication and egress (Fig. 4C, D, E, F, G, and H).
To examine the effect of the various mutations on virus replication, Vero cells were infected at an MOI of 5 with either the wild-type or a mutant virus. Virus stocks were prepared at 0, 4, 9, 16, 20, or 24 h postinfection and titrated in triplicate onto complementing cells (Fig. 5). The kinetics of both YEbacΔUL20 and YEbacΔgK viral replication was substantially slower than that of the YEbac102 virus. Maximum titers for the YEbacΔUL20 and YEbacΔgK mutants viruses were 105 PFU/ml in comparison to 108 PFU/ml for the YEbac102 virus at 24 h postinfection (Fig. 5). In contrast, the YEbacΔgB, YEbacΔgBΔUL20, and YEbacΔgBΔgK viruses failed to produce infectious virus in Vero cells (Fig. 5).
FIG. 5.
Viral replication kinetics. Comparison of the viral replication characteristics of YEbac102 (•),YEbacΔUL20 (▪),YEbacΔgK (▴),YEbacΔgB (○),YEbacΔgBΔUL20 (□), and YEbacΔgBΔgK (▵) on Vero cells. One-step kinetics of infectious virus production were calculated after infection at an MOI of 5 followed by incubation at 37°C.
Ultrastructural characterization of the YEbacΔgBΔUL20 and YEbacΔgBΔgK double-null mutant viruses.
Previous reports indicated that gB was not essential for virion egress (6, 7), while both gK and UL20p were found to be essential for cytoplasmic virion morphogenesis and egress (19, 33). To ascertain whether gB functioned redundantly with either gK or UL20p in virion de-envelopment from perinuclear spaces, the ultrastructural phenotypes of each YEbac102-based mutant virus carrying UL20, gB, or gK deletions relative to the wild-type YEbac102 virus were determined by transmission electron microscopy at 24 h postinfection.
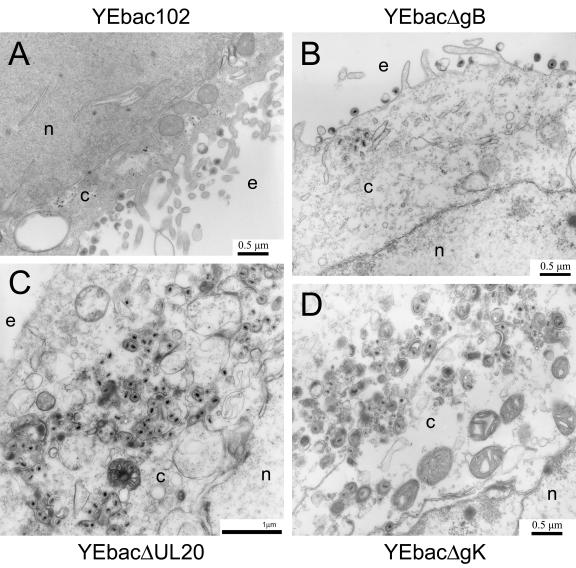
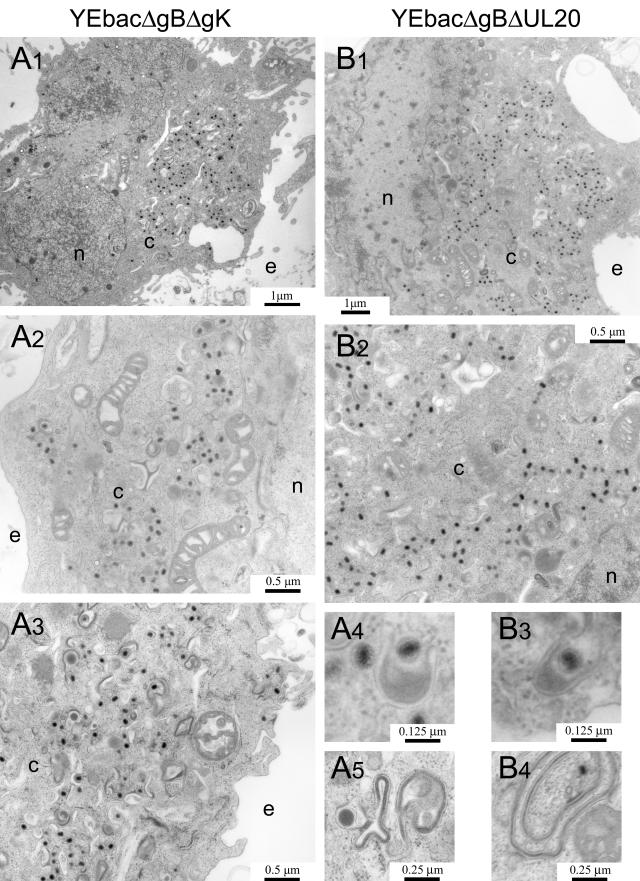
As expected, the YEbac102 and YEbacΔgB viruses exhibited no apparent defects in virion egress, exemplified by the presence of fully enveloped virions extracellularly (Fig. 6A and B) as well as the presence of fully enveloped virions intracellularly (not shown). Unlike the wild-type virus, ultrastructural visualization of YEbacΔUL20- and YEbacΔgK-infected Vero cells revealed a cytoplasmic defect in virion egress, characterized by the presence of unenveloped capsids in the cytoplasm as well as aberrantly enveloped virions (Fig. 6C and D). These results are consistent with previous findings (19, 33). In general, ultrastructural examination of Vero cells infected with the YEbacΔgBΔgK (Fig. 7A) and YEbacΔgBΔUL20 (Fig. 7B) double-null viruses revealed virion morphogenetic defects similar to those of the gK- and UL20-null viruses, exhibiting large numbers of unenveloped capsids in the cytoplasm. In addition, the observed ultrastructural phenotypes revealed specific defects in cytoplasmic envelopment: (i) multiple capsids appeared proximal to membranes apparently unable to complete the budding process into cytoplasmic vesicles (Fig. 7A4 and B3) and (ii) cytoplasmic vesicular membranes appeared to contain electron-dense material consistent with the accumulation of tegument proteins in the presence or absence of proximal capsids (Fig. 7A5 and B4).
FIG. 6.
Ultrastructural morphology of gB-, gK-, and UL20-null viruses. Electron micrographs of Vero cells infected with YEbac102 (A), YEbacΔgB (B), YEbacΔUL20 (C), or YEbacΔgK (D). Confluent cell monolayers were infected at an MOI of 5, incubated at 37°C for 24 h, and prepared for transmission electron microscopy. Nuclear (n), cytoplasmic (c), and extracellular (e) spaces are marked.
FIG.7.
Ultrastructural morphology of gB/gK and gB/UL20 double-null viruses. Electron micrographs of Vero cells infected with YEbacΔgBΔgUL20 (A1, A2, A3, A4, and A5) or YEbacΔgBΔgK (B1, B2, B3, and B4). Confluent cell monolayers were infected at an MOI of 5, incubated at 37°C for 24 h, and prepared for transmission electron microscopy. Panels A4 and B3 show partially enveloped capsids. Panels A5 and B4 show a tegument-like accumulation on membranes that are folded irregularly. Nuclear (n), cytoplasmic (c), and extracellular (e) spaces are marked.
Generation and characterization of recombinant gK-null and UL20-null viruses containing the gBsyn3 or gBamb1511 mutation.
It was previously determined that gB was required for expression of the syn20 (gK) syncytial phenotype (7). To determine whether gK was required for virus-induced cell fusion due to syncytial mutations in gB, YEbac102-based recombinant viruses carrying either the gBsyn3 or the gBamb1511 syncytial mutation in the absence of the gK gene were constructed. These viruses were constructed by rescuing the YEbacΔgBΔgK virus with either wild-type gB, gBsyn3, or gBamb1511 (see Materials and Methods). The same strategy was used to isolate recombinant viruses carrying the gBsyn3 or gBamb1511 mutation in the absence of the UL20 gene by rescuing the gB deletion in the YEbacΔgBΔUL20 virus.
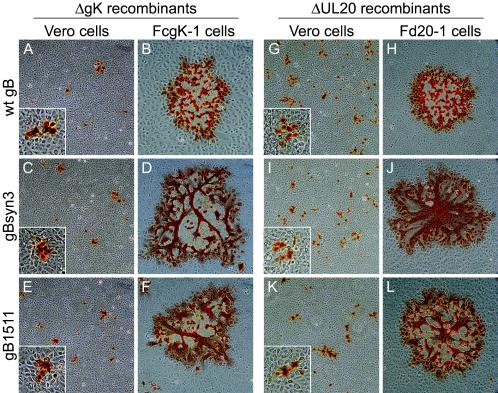
To assess the effect of each gB mutation on virus-induced cell fusion, Vero cells as well as either gK- or UL20-complementing cells were infected with each mutant virus at an MOI of 0.001. Virus plaques were visualized at 48 h postinfection by immunohistochemistry as described previously (19, 41). Viruses specifying the wild-type, syn3, or amb1511 gB in the gK-null background formed small, nonsyncytial plaques on Vero cells (Fig. 8A, C, and E). As expected, the virus containing the wild-type gB formed large, nonsyncytial plaques on FcgK-1 cells (Fig. 8B), while gK-null viruses specifying either the gBsyn3 or gBamb1511 mutation formed large, syncytial plaques on FcgK-1 cells (Fig. 8D and F). Similarly, UL20-null viruses specifying wild-type, syn3, or amb1511 gB formed small, nonsyncytial plaques on Vero cells (Fig. 8G, I, and K). The UL20-null virus containing wild-type gB formed large, nonsyncytial plaques on Fd20-1 cells (Fig. 8H), and the UL20-null viruses specifying either the gBsyn3 or gBamb1511 mutation formed large, syncytial plaques on Fd20-1 cells (Fig. 8J and L).
FIG. 8.
Plaque morphology of gK- and UL20-null viruses carrying syncytial mutations in gB. Plaque phenotypes of gK-null (A, B, C, D, E, and F) and UL20-null (G, H, I, J, K, and L) recombinant viruses containing specific gB mutations on Vero (A, C, E, G, I, and K) and FcgK-1 (B, D, and F), or Fd20-1 (H, J, and L) cells. Confluent cell monolayers were infected with the gK-null viruses containing wild-type gB (A and B), gBsyn3 (C and D), or gBamb1511 (E and F) or UL20-null viruses containing wild-type gB (G and H), gBsyn3 (I and J), or gBamb1511 (K and L) at an MOI of 0.001, and viral plaques were visualized at 48 h postinfection by immunohistochemistry. Insets are shown at 2.4× magnification to facilitate visualization of the nonsyncytial phenotype of the small plaques.
Virion assembly and egress are not required for either gB- or gK-mediated virus-induced cell-to-cell fusion.
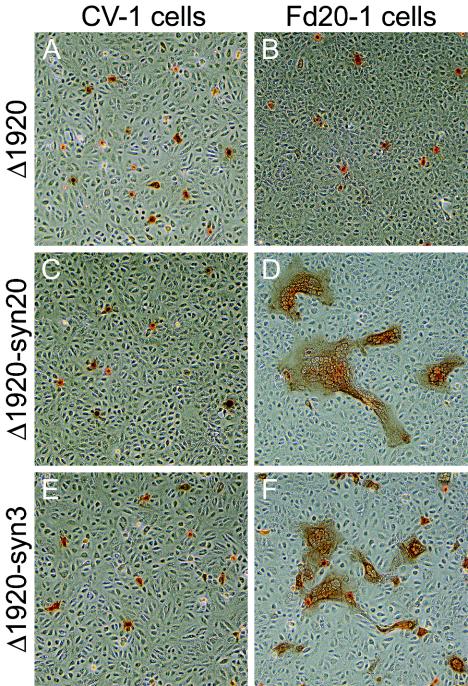
To investigate the potential requirement for virion egress in virus-induced cell fusion, recombinant viruses carrying a UL19 and UL20 deletion in an otherwise wild-type, syn20, or tsB5 genetic background were constructed. UL19 encodes the major capsid protein (VP5), and capsid formation cannot occur in its absence (14). Briefly, a cytomegalovirus-EGFP gene cassette was inserted into the viral genome, effectively deleting a sizeable portion of the UL19 ORF and all of the UL20 ORF. Recombinant viruses were isolated via fluorescence microscopy and plaque purified on G5 cells, which complement the UL19/UL20-null defect (14), and characterized as described in Materials and Methods. CV-1 or Fd20-1 cells were infected with Δ1920, Δ1920-syn20, and Δ1920-syn3 viruses at an MOI of 0.005. As expected, these infections did not cause virus-induced cell fusion in noncomplementing CV-1 cells (Fig. 9A, C, and E). However, the Δ1920-syn20 and Δ1920-syn3 viruses formed syncytial plaques containing a range of 3 to 100 nuclei per syncytium in Fd20-1 cells, which complement only UL20 (Fig. 9D and F), while the Δ1920 virus failed to cause virus-induced cell fusion (Fig. 9B).
FIG. 9.
Plaque phenotypes of Δ1920, Δ1920-syn20, and Δ1920-syn3 viruses on CV-1 (A, C, and E) and FcgK-1 (B, D, and F) cells. Confluent cell monolayers were infected with the Δ1920 (A and B), Δ1920-syn20 (C and D), or Δ1920-syn3 (E and F) at an MOI of 0.001, and viral plaques were visualized at 48 h postinfection by immunohistochemistry.
DISCUSSION
HSV-1 BAC-generated recombinant viruses.
Investigation of the structure and function of individual viral proteins has been greatly facilitated through the use of recombinant viruses specifying mutated proteins. Typically, recombinant viruses have been produced by homologous recombination between a mutated DNA fragment and viral genomes. In most instances, the resultant recombinant viruses have to be extensively purified due to the presence of a large amount of contaminating wild-type viruses. Recently, the HSV-1 (F) genome was cloned into a bacterial artificial chromosome (BAC) by inserting the BAC into the intergenic region between UL3 and UL4 without deletion of any viral genes (62), unlike previously constructed HSV-1 BACs, which contained deletions of one or more viral genes (28, 56, 61). Transfection of the resultant pYEbac102 plasmid into cells resulted in production of the YK304 infectious virus, which was indistinguishable from wild-type HSV-1 in vitro and in vivo, exhibiting wild-type virulence in mice after intracerebral inoculation (62).
Taking advantage of the HSV-1 pYEbac102 system, we generated recombinant viruses lacking either the gK, gB, or UL20 gene as well as viruses having deletions in both gK and gB or gB and UL20. As expected, the gB/gK and gB/UL20 double-null viruses failed to produce infectious virus or spread beyond the primary infected Vero cells. Generally, the gK-, gB-, and UL20-null viruses exhibited defects in virus replication and plaque morphologies similar to those of corresponding viruses constructed in the KOS genetic background. However, it was noted that the HSV-1 (F) gK- and UL20-null viruses exhibited up to 10-fold greater defects in virus replication, while virus plaques were typically smaller than those produced by the KOS mutant viruses. These results may be attributed to genetic differences between the F and KOS virus strains, although no apparent differences were noted between the wild-type viruses with regard to their replication kinetics and plaque morphology. Alternatively, it is possible that the requirement for extensive passage of mutant viruses in the KOS background during plaque purification may have resulted in improved adaptation of these viruses for growth in Vero cells. In this regard, generation of mutant viruses via the pYEbac102 system provides a unique advantage in assessment of the structure and function of individual viral proteins without the potential complications arising from extensive cell culture passage of virus stocks.
gB, UL20p, and gK are not required for virion de-envelopment.
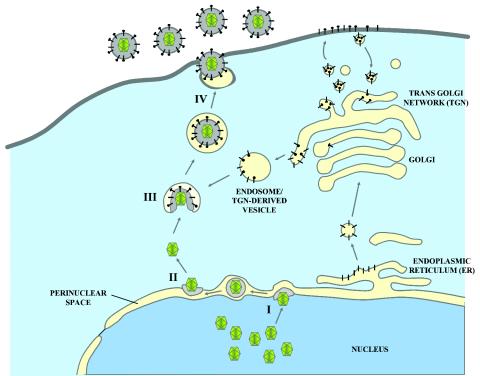
HSV-1 is thought to acquire an initial envelope when capsids formed in the nucleus of cells bud into the perinuclear space. Subsequently, a putative fusion event between viral envelopes and the outer nuclear membranes results in capsid release into the cytoplasm. Final envelopment is thought to occur by budding into cytoplasmic vesicles derived from the trans-Golgi network or endosomes that facilitate egress of enveloped virions to extracellular spaces (Fig. 10). Currently, it is not known whether viral glycoproteins involved in virus entry or virus-induced cell fusion also functions in this de-envelopment step. The results obtained in this study clearly show that gK, UL20, and gB are not essential for virion de-envelopment.
FIG. 10.
Schematic of HSV-1 virion morphogenesis and egress. (I) Mature capsids budding through the inner nuclear membrane into the perinuclear space. (II) De-envelopment of perinuclear virions at the outer nuclear membrane. (III) Re-envelopment of cytoplasmic capsids by budding into cytoplasmic vesicles. (IV) Final egress to the extracellular space.
In the case of gB, previous studies suggested that lack of gB did not adversely influence egress of virion particles to supernatants of infected cells, although these virions lacked gB and were rendered noninfectious (6, 7). Similar results have been reported for gH, gL, and gD, which are thought to be required for virus-induced and virus-free (transient expression) membrane fusion (16, 38, 54). Ultrastructural examination of the gB-null virus confirmed initial findings and clearly showed that lack of gB did not adversely affect the production of enveloped virions in extracellular spaces. Investigations with recombinant viruses specifying single gene deletions in either gB, gK, or UL20p did not exclude the possibility that gB and gK may function in a redundant manner to facilitate virion de-envelopment. This possibility can be discounted on the basis that the gB/gK double-null virus efficiently produced unenveloped capsids in the cytoplasm of infected cells. Similar results were obtained for the gB/UL20 double-null virus. Furthermore, it is improbable that UL20p or gK alone could mediate virion de-envelopment because these two proteins act in a cooperative manner for intracellular transport and virus-induced cell fusion. Construction of a triple-null virus lacking gB, gK, and UL20p could resolve this possibility. Alternatively, other viral glycoproteins such as gH/gL may function redundantly with gK, UL20p, or gB in the de-envelopment process.
Virions found in perinuclear spaces are known to contain the UL34 and UL31 tegument proteins, which are absent from extracellular enveloped virions (52). Thus, it is possible that the de-envelopment process involves tegument-membrane protein interactions different from those occurring during virus entry. In this regard, virion de-envelopment from perinuclear spaces seems to bear little similarity to either virus entry or virus-induced cell fusion, suggesting that this membrane fusion event may be regulated by a fundamentally different mechanism.
Role of gK in gB-mediated cell-to-cell fusion.
Previous reports showed that gK syncytial mutations could not cause virus-induced cell fusion in the absence of gB, indicating that gB served functions essential for cell-to-cell fusion (7). Both the gBsyn3 and gBamb1511 mutations cause extensive virus-induced cell fusion (2, 5). However, unlike the gBsyn3 single-amino-acid mutation, which does not enhance gB-mediated fusion in the transient coexpression system, the gBamb1511 truncation causes greatly enhanced fusion in the transient expression system (20). These findings suggest that the gBamb1511 truncation may disengage gB from other viral or cellular functions responsible for downregulation of the fusogenic potential of gB. However, recombinant viruses containing the gB syncytial mutation gBsyn3 or gBamb1511 but lacking the gK or the UL20 gene failed to cause virus-induced cell fusion, indicating that expression of both gK and UL20p is required for gB-mediated membrane fusion.
In addition, deletion of the carboxyl-terminal 28 amino acids of gB specified by the amb1511 mutation does not abolish the requirement for expression of both gK and UL20p in gB-mediated virus-induced cell fusion, suggesting that gB amino acid sequences other than the carboxyl-terminal 28 amino acids are required for gK or UL20p regulatory interactions with gB. Recently, gK expression was reported to inhibit membrane fusion caused by coexpression of gB, gD, gH, and gL (1). These findings seem to contradict results obtained herein with the recombinant viruses lacking gK. However, a similar result was reported for gM, which inhibited membrane fusion in the transient coexpression system (34, 36) but was required for virus-induced cell fusion caused by syncytial mutations in gB (12). These results suggest that gM and gK may regulate virus-induced cell fusion both positively and negatively depending on the presence of different sets of viral proteins.
Virion egress is not required in virus-induced cell fusion.
Virus-induced cell fusion starts at approximately 5 h postinfection at 37°C, at which time infectious virions are produced and egress out of infected cells. Therefore, it is possible that egressing virions may contribute directly or indirectly to the virus-induced membrane fusion phenomena mediated by cell surface-expressed gB and gK carrying syncytial mutations. It has been shown that cell surface expression of gB is not affected in a gK-null or UL20-null virus (19, 33). Therefore, the requirement for the presence of UL20p and gK for gB-mediated virus-induced cell fusion may be due to defects in virion egress. Earlier reports indicated that production of virion particles may not be necessary for virus-induced cell fusion, inasmuch as syncytial mutants caused fusion in the presence of the replication inhibitor phosphonoacetic acid (35). These early findings were confirmed herein by the phenotype of the UL19/UL20 double-null viruses containing either the gBsyn3 or gBamb1511 mutation. These viruses are unable to form capsids because of the deletion in the UL19 gene, coding for the major capsid protein VP5. Both viruses caused virus-induced cell fusion in the UL20-complementing cell line, indicating that virion formation and egress are not necessary for virus-induced cell fusion.
The rapid generation of recombinant viruses with the YEbac102 system greatly facilitates the characterization of the function of individual viral proteins while helping to discern potentially complex interactions among multiple proteins through the introduction of multiple mutations into an identical genetic background. In this report, we have demonstrated the utility of this system in investigating the role of gB, gK, and UL20p in virus-induced cell fusion and morphogenesis. The existence of an intimate relationship between gK/UL20p and gB in virus-induced cell fusion suggests that both gK and UL20p act to regulate the fusogenic properties of gB. Future studies will investigate direct physical interactions among gB, gK, and UL20p and their role in the regulation of membrane fusion phenomena in the HSV-1 life cycle.
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (AI4300) to K.G.K. J.M.M. was supported by a Louisiana Economic Development fellowship.
We thank Olga Borkshsenious for expert technical assistance with electron microscopy. We are indebted to Dr. Y. Kawaguchi for providing pYEbac102.
REFERENCES
- 1.Avitabile, E., G. Lombardi, and G. Campadelli-Fiume. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 77:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baghian, A., L. Huang, S. Newman, S. Jayachandra, and K. G. Kousoulas. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond, V. C., and S. Person. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132:368-376. [DOI] [PubMed] [Google Scholar]
- 5.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 6.Cai, W., S. Person, S. C. Warner, J. Zhou, and N. A. DeLuca. 1987. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J. Virol. 61:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W. Z., S. Person, C. DebRoy, and B. H. Gu. 1988. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J. Mol. Biol. 201:575-588. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y. E., L. Menotti, F. Filatov, G. Campadelli-Fiume, and B. Roizman. 1998. UL27.5 is a novel gamma2 gene antisense to the herpes simplex virus 1 gene encoding glycoprotein B. J. Virol. 72:6056-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claesson-Welsh, L., and P. G. Spear. 1987. Amino-terminal sequence, synthesis, and membrane insertion of glycoprotein B of herpes simplex virus type 1. J. Virol. 61:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claesson-Welsh, L., and P. G. Spear. 1986. Oligomerization of herpes simplex virus glycoprotein B. J. Virol. 60:803-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis-Poynter, N., S. Bell, T. Minson, and H. Browne. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 68:7586-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debroy, C., N. Pederson, and S. Person. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36-48. [DOI] [PubMed] [Google Scholar]
- 14.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietz, P., B. G. Klupp, W. Fuchs, B. Kollner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, T. P., X. Alvarez, and K. G. Kousoulas. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, T. P., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 78:5347-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster, T. P., J. M. Melancon, and K. G. Kousoulas. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 21.Foster, T. P., J. M. Melancon, T. L. Olivier, and K. G. Kousoulas. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 78:13262-13277. [DOI] [PMC free article] [PubMed]
- 22.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 1998. Expression of the enhanced green fluorescent protein by herpes simplex virus type 1 (HSV-1) as an in vitro or in vivo marker for virus entry and replication. J. Virol. Methods 75:151-160. [DOI] [PubMed] [Google Scholar]
- 23.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75:12431-12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gage, P. J., M. Levine, and J. C. Glorioso. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol. 67:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 27.Highlander, S. L., W. F. Goins, S. Person, T. C. Holland, M. Levine, and J. C. Glorioso. 1991. Oligomer formation of the gB glycoprotein of herpes simplex virus type 1. J. Virol. 65:4275-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horsburgh, B. C., M. M. Hubinette, D. Qiang, M. L. MacDonald, and F. Tufaro. 1999. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 6:922-930. [DOI] [PubMed] [Google Scholar]
- 29.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchinson, L., F. L. Graham, W. Cai, C. Debroy, S. Person, and D. C. Johnson. 1993. Herpes simplex virus (HSV) glycoproteins B and K inhibit cell fusion induced by HSV syncytial mutants. Virology 196:514-531. [DOI] [PubMed] [Google Scholar]
- 31.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69:5401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 33.Jayachandra, S., A. Baghian, and K. G. Kousoulas. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 71:5012-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kousoulas, K. G., S. Person, and T. C. Holland. 1978. Timing of some of the molecular events required for cell fusion induced by herpes simplex virus type 1. J. Virol. 27:505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyano, S., E. C. Mar, F. R. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485-1491. [DOI] [PubMed] [Google Scholar]
- 37.Laquerre, S., S. Person, and J. C. Glorioso. 1996. Glycoprotein B of herpes simplex virus type 1 oligomerizes through the intermolecular interaction of a 28-amino-acid domain. J. Virol. 70:1640-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luna, R. E., F. Zhou, A. Baghian, V. Chouljenko, B. Forghani, S. J. Gao, and K. G. Kousoulas. 2004. Kaposi's sarcoma-associated herpesvirus glycoprotein K8.1 is dispensable for virus entry. J. Virol. 78:6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLean, C. A., S. Efstathiou, M. L. Elliott, F. E. Jamieson, and D. J. McGeoch. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72:897-906. [DOI] [PubMed] [Google Scholar]
- 41.Melancon, J. M., T. P. Foster, and K. G. Kousoulas. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 78:7329-7343. [DOI] [PMC free article] [PubMed]
- 42.Mettenleiter, T. C. 2002. Brief overview on cellular virus receptors. Virus Res. 82:3-8. [DOI] [PubMed] [Google Scholar]
- 43.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moraes, M. P., G. A. Mayr, and M. J. Grubman. 2001. pAd5-Blue: direct ligation system for engineering recombinant adenovirus constructs. BioTechniques 31:1050. [DOI] [PubMed] [Google Scholar]
- 45.Narayanan, K., R. Williamson, Y. Zhang, A. F. Stewart, and P. A. Ioannou. 1999. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 6:442-447. [DOI] [PubMed] [Google Scholar]
- 46.Orford, M., M. Nefedov, J. Vadolas, F. Zaibak, R. Williamson, and P. A. Ioannou. 2000. Engineering EGFP reporter constructs into a 200 kb human beta-globin BAC clone using GET recombination. Nucleic Acids Res. 28:E84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pertel, P. E. 2002. Hum. herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pogue-Geile, K. L., G. T. Lee, S. K. Shapira, and P. G. Spear. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136:100-109. [DOI] [PubMed] [Google Scholar]
- 50.Pogue-Geile, K. L., and P. G. Spear. 1987. The single base pair substitution responsible for the syn phenotype of herpes simplex virus type 1, strain MP. Virology 157:67-74. [DOI] [PubMed] [Google Scholar]
- 51.Ramaswamy, R., and T. C. Holland. 1992. In vitro characterization of the HSV-1 UL53 gene product. Virology 186:579-587. [DOI] [PubMed] [Google Scholar]
- 52.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roizman, B., and A. E. Sears. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 54.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryechan, W. T., L. S. Morse, D. M. Knipe, and B. Roizman. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29:677-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saeki, Y., T. Ichikawa, A. Saeki, E. A. Chiocca, K. Tobler, M. Ackermann, X. O. Breakefield, and C. Fraefel. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum. Gene Ther. 9:2787-2794. [DOI] [PubMed] [Google Scholar]
- 57.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277-295. [DOI] [PubMed] [Google Scholar]
- 58.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell. Microbiol. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 59.Spear, P. G. 1993. Membrane fusion induced by herpes simplex virus, p. 201-232. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press., Boca Raton, Fla.
- 60.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stavropoulos, T. A., and C. A. Strathdee. 1998. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J. Virol. 72:7137-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]