Abstract
We established a system for propagation of Sindbis virus (SIN)-based replicons in tissue culture in the form of a tricomponent genome virus. Three RNA fragments containing complementing genetic information required for virus replication are packaged into separate viral particles, and each cell produces at least 1,000 packaged replicons and the number of packaged helpers sufficient to perform the next passage. This system can be used to generate large stocks of packaged replicons. The formation of infectious recombinant SIN virus was not detected in any experiments. These features make multicomponent genome SIN an attractive system for a variety of research and biotechnology applications.
During the last decade, significant progress in understanding the mechanisms of replication and virus-host interactions has led to the wide use of viruses with RNA genomes for the delivery and expression of heterologous genes in vivo and in vitro (2, 7, 12, 13, 18-20). Alphaviruses were among the first to be adapted for application as vector systems (16, 28), and presently they are widely used in recombinant vaccine development and large-scale protein production (23, 29).
The cDNAs encoding infectious genomes have been developed for a number of alphaviruses, including Sindbis virus (SIN) (24), Semliki Forest virus (SFV) (17), Venezuelan equine encephalitis (VEE) (3), eastern equine encephalitis (EEE) (26), and others. Cloning of these genomes facilitated in vitro manipulations of viral genetic material and reverse genetics experiments. These studies strongly promoted the application of alphaviruses as gene delivery vehicles.
The alphavirus genome consists of a single-stranded RNA of positive polarity that is about 12 kb in length (27). It is capped at the 5′ end and contains a 3′-terminal poly(A) tail. The genome serves as a template for synthesis of viral nonstructural proteins (nsPs) forming the replicase-transcriptase complex (RdRp) required for synthesis of a genome-length, minus-strand intermediate that is a template for transcription of both genome-length, plus-strand RNA and the subgenomic 26S RNA. The latter RNA, encoded by the 3′ one-third of the genome, accumulates in infected cells to high levels and is translated into viral structural proteins. The structural proteins are dispensable for viral RNA replication and transcription, and their coding sequence can be replaced by heterologous genes (11, 28). The deletion of the structural genes makes these RNAs (replicons) unable to spread, but they can be packaged into viral particles by cotransfecting the cells with in vitro-synthesized replicon RNAs and one or two helper RNAs (1, 5, 11, 16). These helpers contain 5′ and 3′ cis-acting RNA elements that serve as replication promoters utilized by the viral replicative enzymes provided by replicons in trans and the subgenomic promoters that drive the expression of viral structural proteins. The required structural proteins can be expressed as a single polyprotein, or the capsid- and glycoproteins-coding genes can be separated between two helpers. Traditionally, helpers are designed so as not to contain packaging signals and, thus, package themselves inefficiently. This cotransfection-based technology meets the requirements of research needs, but it limits the large-scale production of replicon-containing particles necessary for recombinant protein manufacturing and other applications.
In an effort to devise alternative strategies that allow production of large stocks of packaged SIN replicons, we established a system for their propagation at an escalating scale in the form of a tricomponent genome virus. This approach can facilitate further application of Sindbis virus-based vectors for multiple needs.
The general scheme for creating and passaging the tricomponent genome viruses is demonstrated in Fig. 1A. Initially, all three in vitro-synthesized RNAs are delivered into the cells by transfection, initiating productive replication and release of particles with all of the RNA segments packaged. These samples can then be further passaged to make the larger stocks required for heterologous protein production and/or immunization. The advantage of using not one, but two helpers is in the reduced formation of full-length infectious recombinant viral genomes.
FIG. 1.
Replication of SIN replicons in BHK-21 cells using helpers with different 5′ ends. (A) Experimental protocol proposed for tricomponent genome SIN propagation. Transfection of BHK-21 cells by replicon and two-helper RNAs results in the release of viral particles with three different genomes. These particles can be used for the next round of infection. (B) Schematic representation of replicon and helper genomes and helper 5′ UTRs (based on predicted secondary structure). DH-BB- and DH-BBdelSL2-based helpers had the 5′ tRNAAsp sequence derived from the naturally occurring SIN DI RNA (see reference 1 for details); BB- and BBdelSL2-based helpers had natural SIN 5′ UTRs. All of the helpers contained a deletion of nucleotides 426 to 7334 of the SIN genome, and delSL2 indicates an additional deletion of nucleotides 47 to 152 of the SIN genome. A total of 107 BHK-21 cells were cotransfected with SINrep/LacZ or SINrep/GFP replicons and indicated pairs of helper RNAs. Harvested virus particles were used for the next round of infection of 3 × 106 BHK-21 cells at the indicated MOIs (measured in infectious units [inf.u]/cell) in 100-mm-diameter dishes. Viruses were harvested at 40 h postinfection and were used further without concentrating. Titers refer to unconcentrated harvested virus-containing media. N.A. stands for not applicable; the passage could not be performed at an MOI of 10 or 1 infectious units/cell due to low titers of packaged replicons harvested after the previous passage. All of the experiments were performed multiple times and generated very reproducible titers. (C) Replication of replicon and helper RNAs in the cells after their cotransfection or infection (passage 2 [P2]) with tricomponent genome viruses. Autoradiographs of dried gels with RNAs metabolically labeled with [3H]uridine. RNAs were transfected using the conditions described in the text or were infected at an MOI of 3 or 30 infectious units/cell for the samples of replicons packaged with 5′ tRNA UTR- or SIN 5′ UTR-containing helpers, respectively. RNA labeling was performed between 4 and 8 h posttransfection or 12 to 16 h postinfection in the presence of ActD and [3H]uridine (20 mCi/ml). RNAs were isolated from the cells by using TRIzol reagent, denatured with glyoxal in dimethyl sulfoxide, and analyzed by agarose gel electrophoresis using previously described conditions (1). SINrep/LacZ and BB/C plus BB/Gl helpers, lanes 1 and 5; SINrep/LacZ and BBdelSL2/C plus BBdelSL2/Gl helpers, lanes 2 and 6; SINrep/GFP and BB/C plus BB/Gl helpers, lanes 3 and 7; SINrep/GFP and BBdelSL2/C plus BBdelSL2/Gl helpers, lanes 4 and 8; SINrep/LacZ and DH-BB/C plus DH-BB/Gl helpers, lanes 9 and 13; SINrep/LacZ and DH-BBdelSL2/C plus DH-BBdelSL2/Gl helpers, lanes 10 and 14; SINrep/GFP and DH-BB/C plus DH-BB/Gl helpers, lanes 11 and 15; SINrep/GFP and DH-BBdelSL2/C plus DH-BBdelSL2/Gl helpers, lanes 12 and 16. Co-transf., cotransfection.
The first step in separating the coding material of Sindbis virus among the three complementing genomes was to find cis-acting RNA elements that would promote not only the efficient replication of all of the genome fragments and synthesis of the viral structural proteins but also packaging of helper RNAs into viral particles. The previously published data concerning replication of SIN defective interfering (DI) RNAs (22) demonstrated that these small RNAs could accumulate in virus samples to very high concentrations, an effect that is achieved by a variety of modifications, including extended deletions in the viral genome, generating heterologous 5′-untranslated regions (UTRs), and exploiting the so-called packaging signals (RNA sequences that elevate encapsidation efficiency) (10). The 5′ tRNAAsp-derived structure present in the naturally occurring SIN DI RNA was shown to increase helper RNA replication by two- to threefold (1, 6). The tRNA-containing DI RNAs and the helper genomes that were designed later were also more efficiently packaged into viral particles, because the heterologous, tRNA-containing 5′ UTR, besides being a promoter for RNA replication, was capable of functioning as a packaging signal (10). Thus, we designed a series of helpers (Fig. 1B) with predicted differences in replication levels and compared their efficiencies in packaging replicons and their own genomes into viral particles. They contained (i) either the 5′-terminal tRNAAsp sequence or the natural SIN 5′ UTR and (ii) the deletions of the downstream sequences (delSL2) that were previously shown to have a stimulatory effect on RNA replication. Using previously described conditions (5), we electroporated 5 μg of in vitro-synthesized replicon RNAs and pairs of helper RNAs encoding capsid or glycoprotein genes (7 μg of each) into BHK-21 cells. The subgenomic RNA of the glycoprotein-coding helper also contained the Ross River virus capsid gene (CRRV) with an intact translational enhancer and protease domain but with a deleted RNA-binding domain. The presence of this gene in the cassette was previously shown to strongly increase the expression of glycoproteins (9). The replicon RNAs carried green fluorescent protein (GFP) or β-galactosidase (LacZ) genes under control of the subgenomic promoters (SINrep/GFP and SINrep/LacZ replicons, respectively) (1, 4).
Virus-containing media were usually harvested within 24 h postelectroporation, after the development of a cytopathic effect (CPE). Titers of packaged replicons (in infectious units/ml) were measured by infecting 5 × 105 BHK-21 cells with harvested samples (after they were diluted 1,000- to 10,000-fold), followed by an evaluation of the numbers of GFP- and β-galactosidase-positive cells at 6 to 8 h postinfection. Helpers with a natural SIN 5′ UTR demonstrated a more efficient packaging of replicons, and titers of replicon-containing particles packaged with either BB/C and BB/Gl or BBdelSL2/C and BBdelSL2/Gl pairs (Fig. 1B) of helpers were 5 - to 10-fold higher than those of the samples generated with tRNA-containing helpers, and they approached 2 × 109 to 5 × 109 infectious units/ml (Fig. 1B). This higher efficiency of replicon packaging was not a result of differences in helper RNA replication (Fig. 1C), because 5′ tRNA UTR helpers replicated more efficiently.
Next, the naïve BHK-21 cells were infected with samples of replicons packaged with different helpers at a multiplicity of infection (MOI) of 10 infectious units/cell. Particles were harvested at 30 to 40 h postinfection, after development of profound CPE. After this passage, titers of the replicons packaged with BB/C plus BB/Gl and BBdelSL2/C plus BBdelSL2/Gl helpers became 4 to 5 orders of magnitude lower (data not shown), and even after repeating this passage at an MOI of 100 infectious units/cell, the titers of replicon-containing viral particles were still 100- to 1,000-fold below the levels found in the samples after RNA transfection (Fig. 1B). This was an indication that after electroporation (that we usually refer to as passage 1), helpers with SIN 5′ UTRs were packaged to a concentration insufficient to infect all of the cells and to support the next round of packaging and productive virus replication. In these samples, syntheses of helper genomic and subgenomic RNAs (Fig. 1C lanes 5 to 8) and SIN structural proteins (data not shown) were barely detectable in the infected cells. The deletion of SL2 in BBdelSL2/C and BBdelSL2/Gl helpers, which was previously shown to cause an increase in DI RNA replication (6), was expected to increase their self packaging because of their accumulation at higher concentrations in the cells. However, our results indicated that the more efficient replication was not sufficient to strongly promote the helpers' self packaging. The use of these helpers made titers of the packaged replicons after passage 2 noticeably higher than those of stocks generated with BB/C and BB/Gl helpers (Fig. 1B). However, they remained 100-fold lower than those in the samples harvested after electroporation.
The use of the tRNA 5′ UTR had a strong positive effect on packaging of helper RNAs into viral particles. When a DH-BB/C and DH-BB/Gl pair (Fig. 1B) of helper RNAs was used, the titers of reporter replicon-containing particles remained the same after performing two, three, or more passages (Fig. 1B and data not shown). Syntheses of the replicon and helper RNAs as well as SIN structural proteins were readily detectable in the cells during passaging (Fig. 1C and data not shown). Virus samples prepared by using DH-BB/C and DH-BB/Gl or DH-BBdelSL2/C and DH-BBdelSL2/Gl helpers were incapable of forming large plaques in the same manner that wild-type SIN does. The probability of copackaging of all three RNA genome components (replicon and two helper RNAs) was very low. In the additional experiments, we evaluated the concentration of SINrep/LacZ replicon-containing particles, capable of infecting single cells and particles containing all three genomes (replicon and both DH-BB/C and DH-BB/Gl helpers). BHK-21 cells were infected with samples of packaged replicons at different dilutions, and after 2 to 3 days of incubation under agarose cover, we detected 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-stained foci that formed aroundthe cells initially infected with all three genomes essential for productive replication. The ratio of single SINrep/LacZ-infected cells to these foci was found to be 1,000:1. It was consistent with the 20:1 ratio previously detected for replicon and bipartite genome-containing particles (1). To further evaluate the ratio of replicon and helper genomes in the population of viral particles, packaged viruses were pelleted by ultracentrifugation and concentration of virus-specific RNAs was analyzed by Northern hybridization. In different samples, the ratio of packaged replicons to packaged helpers was found to be in a range between 1:1 and 1:5 (data not shown). Thus, as with some plant viruses, these SIN-derived viruses could be characterized as containing multicomponent genomes with the segments predominantly packaged into separate virions (14).
In all of the experiments, there was no evidence of recombinant infectious SIN formation even after five passages of viral particles in cell culture. The sensitivity of the method of recombinant virus detection used was addressed in an additional experiment in which 104 PFU of wild-type SIN (an equivalent of the amount of virus produced by a single cell) was added to a 10-ml stock of tricomponent genome virus. A 0.1-ml sample was then used to make the next passage at an MOI of 10 infectious units of replicon/cell, as in other experiments. After 40 h of incubation, wild-type SIN was easily detected by its plaque-forming activity (data not shown). The concentration of infectious virus rapidly increased during further passaging.
The deletion of SL2 in the helpers with a tRNA sequence in the 5′ ends (the DH-BBdelSL2/C and DH-BBdelSL2/Gl pair) did not notably change helper RNA replication and packaging. Both replication and packaging efficiencies were found to be similar to those detected for the DH-BB-based helpers without deletions in the 5′ end (Fig. 1C). However, in some experiments, we detected lower levels of replicating helper RNAs and their subgenomes in the cells during passaging of SIN replicons (Fig. 1C, lane 16, and data not shown). The lower helper self-packaging efficiency was more likely due to alterations in packaging than in RNA replication.
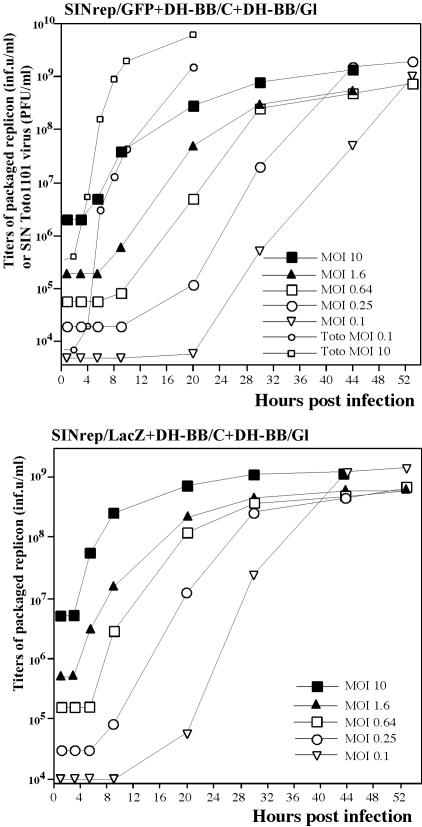
To further investigate the characteristics of our genetically engineered multicomponent genome viruses, we infected BHK-21 cells with stocks of SINrep/GFP and SINrep/LacZ replicons packaged with DH-BB/C and DH-BB/Gl helpers (that were additionally amplified by passaging once at an MOI of 10 infectious units/cell on BHK-21 cells). Infections were performed at various MOIs, and titers of replicon-containing particles were assessed at different times postinfection. After infection at MOIs between 0.1 and 10 infectious units/cell (Fig. 2A and B), viruses caused CPE and titers of particles with replicons approached 109 infectious units/ml. In the range of 0.1 to 0.25 infectious units/cell, replication of the multicomponent genome viruses was considerably slower, but 44 h postinfection the titers reached the same values as those during passaging at an MOI of 10 infectious units/cell. At an MOI below 0.1 infectious units/cell, the CPE did not develop, and final titers of packaged replicons were 3 to 4 orders of magnitude lower (data not shown).
FIG. 2.
Growth curves of the tricomponent genome SIN. BHK-21 cells were infected with tricomponent genome viruses or wild-type SIN Toto1101 virus at indicated MOIs in infectious units/cell. At the indicated times, media were replaced and titers of packaged replicons were determined as described in the text. Titers of SIN Toto1101 virus were determined using a standard plaque assay. One of the highly reproducible experiments is demonstrated. Wild-type SIN (Toto1101) virus was included in the experiment to illustrate the difference in growth rates between normal and tricomponent genome viruses. inf.u, infectious units.
To assess the possibility of DI RNA accumulation (21), the SINrep/GFP and SINrep/LacZ replicons were packaged by DH-BB/C and DH-BB/Gl helpers and were serially passaged at two different MOIs (2 and 10 infectious units/cell). By passage 5, SINrep/LacZ passaged at an MOI of 10 infectious units/cell accumulated DI RNAs that were readily detectable on RNA gel (data not shown) and by a decrease in the titer of replicon-containing particles (Table 1). Similar results were obtained in other experiments: DI RNAs accumulated by passage 5 when passaging was performed at an MOI of ∼10 infectious units/cell, but they did not accumulate at lower MOIs.
TABLE 1.
Titers of packaged replicons after passaging at different MOIsa
| Replicon (MOI) | RNA cotransfection | Passage no.:
|
|||
|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | ||
| SINrep/LacZ (10 inf.u/cell) | 3.5 × 108 | 6.2 × 108 | 2.7 × 108 | 3.2 × 108 | 1.1 × 107 |
| SINrep/LacZ (2 inf.u/cell) | 3.5 × 108 | 1.3 × 109 | 1.5 × 109 | 3.3 × 108 | 3.4 × 108 |
| SINrep/GFP (10 inf.u/cell) | 3.6 × 108 | 3.7 × 108 | 2.5 × 108 | 3.3 × 108 | 3.6 × 108 |
| SINrep/GFP (2 inf.u/cell) | 3.6 × 108 | 1.1 × 109 | 2.3 × 109 | 4.0 × 108 | 2.1 × 109 |
inf.u, infectious unit.
One of the concerns in the development of the multicomponent genome SIN was the possibility of interference between replicating helper RNAs and replicons expressing the genes of interest in the infected cells. To test if the replication of DH-BB helpers and expression of structural proteins affect the expression of the heterologous proteins, we packaged three different SIN replicons by cotransfecting them with either BB/C and BB/Gl helpers (that had normal SIN 5′ ends and were packaged into viral particles very inefficiently) or by DH-BB/C and DH-BB/Gl helpers (that demonstrated highly efficient self packaging, replication, and expression of SIN structural proteins). Replicons contained either no translational enhancer (SINrep/GFP), part of the translational enhancer (SINrep/Cdel/GFP), or the entire SIN capsid-coding sequence with an unmodified translational enhancer (SINrep/C/GFP) (Fig. 3A). The naïve BHK-21 cells were infected at the same MOI of 20 infectious units/cell, and GFP expression was assayed 18 h postinfection by flow cytometry. The results shown in Fig. 3B demonstrate that the presence of helper RNAs capable of efficient replication had only a minor effect on GFP expression. The presence of the 5′ tRNA-containing helpers in the cells caused a noticeably wider distribution of cell fluorescence, but the median values remained very similar for both pairs of helpers. In an additional experiment, those three replicons and similar SIN replicons expressing β-galactosidase (Fig. 3A) were all packaged with DH-BB/C and DH-BB/Gl helpers. BHK-21 cells infected with these tricomponent genome viruses demonstrated an accumulation of GFP and β-galactosidase, which were readily detectable on Coomassie-stained gels (Fig. 3C and D). Expression of both proteins depended on the integrity of the translational enhancer, indicating that this structure helps the replicon subgenomic RNA to efficiently compete with the helpers' subgenomic RNAs for the translational machinery.
FIG. 3.
Expression of heterologous proteins by SIN replicons in the presence of replicating helper RNAs. (A) Schematic representation of SIN replicons without translational enhancer, SINrep/GFP and SINrep/LacZ; with a partial translational enhancer (SIN capsid-coding sequence with deleted amino acids 59 to 113), SINrep/Cdel/GFP, SINrep/Cdel/LacZ; and with complete capsid-coding sequence, SINrep/C/GFP and SINrep/C/LacZ. Both complete capsid and capsid with deletions were capable of cleaving themselves co- and posttranslationally. (B) BHK-21 cells were infected with GFP-expressing replicons packaged with 5′ tRNA UTR- or SIN 5′UTR-containing helpers at an MOI of 20 infectious units (inf.u)/cell. At 18 h postinfection, the expression of GFP was analyzed by flow cytometry on a Vantage fluorescence-activated cell sorter (Becton Dickinson). (C) A total of 5 × 105 BHK-21 cells were infected with GFP-expressing replicons packaged with DH-BB/C and DH-BB/Gl helpers (lanes 1, 2, and 3) or BB/C and BB/Gl helpers (lane 4) at an MOI of 20 infectious units/cell. Cells were harvested 30 h postinfection, and cell lysates equivalent to 105 cells were analyzed by gel electrophoresis. The gel was stained with Coomassie brilliant blue R-250. Lysate of cells infected with packaged SINrep/GFP replicon, lanes 1 and 4; SINrep/Cdel/GFP replicon, lane 2; SINrep/C/GFP replicon, lane 3. (D) BHK-21 cells were infected with β-galactosidase (β-gal.)-expressing replicons packaged with DH-BB/C and DH-BB/Gl helpers at an MOI of 20 infectious units/cell. Cells were harvested 30 h postinfection, and cell lysates were analyzed by gel electrophoresis. Accumulation of β-galactosidase was also tested using a Galacto-light Plus System (Applied Biosystems). The lysate of 5 × 104 cells was loaded on the gel. The gel was stained by Coomassie brilliant blue R-250. Lysate of uninfected cells, lane 1; cells infected with packaged SINrep/LacZ replicon, lane 2; SINrep/Cdel/LacZ replicon, lane 3; SINrep/C/LacZ replicon, lane 4.
Taken together, the results of the study suggest that the single-stranded RNA genome of SIN virus can be drastically modified. It can be divided into three segments that not only encode proteins with complementing functions but are also encapsidated into separate viral particles. This is not unusual; for example, many plant viruses have segmented genomes, with each segment packaged into distinct particles. Such genomes are referred to as multicomponent genomes (14). It is difficult to expect the existence of similar animal viruses in vivo under natural conditions, because the development of productive infection implies the presence of all of the genomes with complementing functions in the same cell. However, it is possible to create them (at least for alphaviruses) for in vitro manipulations and biotechnology needs. Moreover, particles with different genomes have been previously recognized for Aura virus, but their biological significance remains unclear (25).
In contrast to the original SIN virus genome, the tricomponent genome variant does not cause plaque formation in tissue culture, because all three genomes can be combined in the same cell only at a reasonably high MOI. In this case, all of the surrounding cells in the monolayers are infected with replicon-containing particles (with a small percentage of cells infected with all three genomes), and this presence of replicons in all of the cells in the monolayers makes development of plaques an impossible event. It was previously demonstrated that replicons and one helper (encoding all of the viral structural proteins) could form bipartite genome particles that are capable of plaque formation (11). However, we estimated the ratio of virions with replicon genome and both replicon and helper genome at between 20:1 and 40:1 (1). Thus, in the two-helper system described here, the concentration of released virions containing all three genomes together does not exceed 0.1% of the virus sample.
Nevertheless, the multicomponent genome virus still can be considered cytopathic, because cells infected, even with replicons only, eventually die due to apoptosis development and translational/transcriptional shutoff (8, 15).
We demonstrated that this unusual SIN could be easily propagated at an escalating scale, and passaging is a reasonable alternative to making repeated electroporations. Infection by the multicomponent SIN genome at an MOI of 0.1 to 10 infectious units/cell caused infection to spread in tissue culture. Using this method, the amount of packaged replicons can be increased within one or two passages after electroporation by between 100- and 10,000-fold, respectively. During passaging of this tricomponent genome virus, each cell produces at least 1,000 packaged replicons and the amount of the packaged helpers sufficient to perform the next passage. Thus, 1013- to 1014-infectious-unit samples of packaged replicons can be generated on 1010 to 1011 cells. Importantly, the formation of infectious recombinant SIN virus was not detected in any experiments, indicating that the probability of this event is very low. The low level of recombination and the possibility of preparing large amounts of packaged replicons make SIN with multicomponent genomes an attractive system for a variety of research and biotechnology needs.
In addition to SIN, similar packaging and expression systems can likely be created for other alphaviruses. However, additional studies of the RNA promoters and RNA packaging signals may be required.
Acknowledgments
We thank Peter Mason for critical reading of the manuscript.
This work was supported by Public Health Service grant AI053135.
REFERENCES
- 1.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crotty, S., and R. Andino. 2004. Poliovirus vaccine strains as mucosal vaccine vectors and their potential use to develop an AIDS vaccine. Adv. Drug Deliv. Rev. 56:835-852. [DOI] [PubMed] [Google Scholar]
- 3.Davis, N. L., L. V. Willis, J. F. Smith, and R. E. Johnston. 1989. In vitro synthesis of infectious Venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology 171:189-204. [DOI] [PubMed] [Google Scholar]
- 4.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Prágai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frolov, I., E. Frolova, and S. Schlesinger. 1997. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 71:2819-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frolov, I., T. A. Hoffman, B. M. Prágai, S. A. Dryga, H. V. Huang, S. Schlesinger, and C. M. Rice. 1996. Alphavirus-based expression systems: strategies and applications. Proc. Natl. Acad. Sci. USA 93:11371-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolov, I., and S. Schlesinger. 1994. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 68:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolov, I., and S. Schlesinger. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolova, E., I. Frolov, and S. Schlesinger. 1997. Packaging signals in alphaviruses. J. Virol. 71:248-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geigenmuller-Gnirke, U., B. Weiss, R. Wright, and S. Schlesinger. 1991. Complementation between Sindbis viral RNAs produces infectious particles with a bipartite genome. Proc. Natl. Acad. Sci. USA 88:3253-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarowitz, S. G. 2001. Plant viruses, p. 533-998. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 15.Levine, B., J. E. Goldman, H. H. Jiang, D. E. Griffin, and J. M. Hardwick. 1996. Bcl-2 protects mice against fatal alphavirus encephalitis. Proc. Natl. Acad. Sci. USA 93:4810-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liljeström, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. BioTechnology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 17.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundstrom, K. 2001. Alphavirus vectors for gene therapy applications. Curr. Gene Ther. 1:19-29. [DOI] [PubMed] [Google Scholar]
- 19.McKenna, P. M., J. P. McGettigan, R. J. Pomerantz, B. Dietzschold, and M. J. Schnell. 2003. Recombinant rhabdoviruses as potential vaccines for HIV-1 and other diseases. Curr. HIV Res. 1:229-237. [DOI] [PubMed] [Google Scholar]
- 20.Miller, M. A., C. L. Lavine, S. D. Klas, L. M. Pfeffer, and M. A. Whitt. 2004. Recombinant replication-restricted VSV as an expression vector for murine cytokines. Protein Expr. Purif. 33:92-103. [DOI] [PubMed] [Google Scholar]
- 21.Monroe, S. S., J.-H. Ou, C. M. Rice, S. Schlesinger, E. G. Strauss, and J. H. Strauss. 1982. Sequence analysis of cDNAs derived from Sindbis virions and of defective interfering particles. J. Virol. 41:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monroe, S. S., and S. Schlesinger. 1983. RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5′ ends. Proc. Natl. Acad. Sci. USA 80:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polo, J. M., J. P. Gardner, Y. Ji, B. A. Belli, D. A. Driver, S. Sherrill, S. Perri, M. A. Liu, and T. W. Dubensky, Jr. 2000. Alphavirus DNA and particle replicons for vaccines and gene therapy. Dev. Biol. (Basel) 104:181-185. [PubMed] [Google Scholar]
- 24.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rumenapf, T., D. T. Brown, E. G. Strauss, M. Konig, R. Rameriz-Mitchel, and J. H. Strauss. 1995. Aura alphavirus subgenomic RNA is packaged into virions of two sizes. J. Virol. 69:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoepp, R. J., J. F. Smith, and M. D. Parker. 2002. Recombinant chimeric western and eastern equine encephalitis viruses as potential vaccine candidates. Virology 302:299-309. [DOI] [PubMed] [Google Scholar]
- 27.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 28.Xiong, C., R. Levis, P. Shen, S. Schlesinger, C. Rice, and H. V. Huang. 1989. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science 243:1188-1191. [DOI] [PubMed] [Google Scholar]
- 29.Yamanaka, R. 2004. Alphavirus vectors for cancer gene therapy (review). Int. J. Oncol. 24:919-923. [PubMed] [Google Scholar]