Abstract
Adeno-associated virus (AAV) and other parvoviruses inhibit proliferation of nonpermissive cells. The mechanism of this inhibition is not thoroughly understood. To learn how AAV interacts with host cells, we investigated AAV's interaction with adenovirus (Ad), AAV's most efficient helper virus. Coinfection with Ad and AAV results in an AAV-mediated inhibition of Ad5 gene expression and replication. The AAV replication proteins (Rep) activate and repress gene expression from AAV and heterologous transcription promoters. To investigate the role of Rep proteins in the suppression of Ad propagation, we performed chromatin immunoprecipitation analyses that demonstrated in vivo AAV Rep protein interaction with the Ad E2a gene promoter. In vitro binding of purified AAV Rep68 protein to the Ad E2a promoter was characterized by electrophoretic mobility shift assays (Kd = 200 ± 25 nM). A 38 bp, Rep68-protected region (5′-TAAGAGTCAGCGCGCAGTATTTACTGAAGAGAGCCT-3′) was identified by DNase I footprint analysis. The 38-bp protected region contains the weak E2a TATA box, sequence elements that resemble the Rep binding sites identified by random sequence oligonucleotide selection, and the transcription start site. These results suggest that Rep binding to the E2a promoter contributes to the inhibition of E2a gene expression from the Ad E2a promoter and may affect Ad replication.
Adeno-associated virus (AAV) is a nonpathogenic human parvovirus that normally depends on a helper virus to efficiently complete its replication cycle (reviewed in reference 43). The 4.7-kb single-stranded linear genome has inverted terminal repeat (ITRs) hairpin structures that serve as replication origins. Two open reading frames encode four replication proteins (Rep78, -68, -52, and -40) and three structural capsid proteins (VP1 to -3). The mRNAs for Rep78 and Rep68 proteins are transcribed from the p5 promoter, whereas the mRNAs for Rep52 and Rep40 are transcribed from the p19 promoter. Rep68 and Rep40 differ from Rep78 and Rep52 as a result of splicing that replaces 92 amino acid residues at the carboxyl terminus with nine residues. Rep proteins are pleiotropic effectors of viral replication and gene expression. Rep78 and Rep68 are involved in viral replication, integration into chromosome 19, and regulation of AAV and heterologous gene expression (43). Rep52 and Rep40 proteins are not essential for viral replication but are important for packaging viral DNA into preformed viral capsids (10, 30). Rep78 and Rep68 are site-specific DNA-binding proteins that recognize a 16-bp element in the A stem of the AAV inverted terminal repeat (ITR) (13, 26, 47, 56). Rep78 and Rep68 have endonuclease activity; all four Rep proteins possess helicase and ATPase activities (14). Rep78 also has a ligase activity (55). The enzymatic activities of the larger Rep proteins are required for viral DNA replication and establishment of the provirus state.
Productive AAV infection requires coinfection with a helper virus; infection without a helper virus results in integration into chromosome 19. Adenovirus (Ad) is the most efficient helper for AAV, but human papillomavirus (HPV), cytomegalovirus (CMV), vaccinia virus, Epstein-Barr virus, and herpes simplex virus (HSV) can provide helper functions (5, 40, 58). Productive infection may also be achieved by the use of genotoxic agents, synchronized cells or infection of differentiating keratinocytes (41, 68, 69). Expression of a subset of Ad early genes establishes a permissive AAV replication environment. E1a activates AAV and Ad transcription (9, 18, 27, 35, 52). The E1b and E4 proteins form a complex associated with the transport of AAV mRNA to the cytoplasm and the conversion of single-stranded to double-stranded AAV vector DNA (16, 17). E2a encodes a single-stranded DNA-binding protein that stimulates viral DNA replication and gene transcription (8, 27). The E3 gene increases the efficiency of Ad-induced cell lysis and the release of Ad after a productive infection but is not required for AAV replication (59, 60).
Although AAV is considered nonpathogenic, it has profound effects on the proliferation of the host cell, the replication of helper viruses, and cellular transformation. AAV inhibits proliferation of nonpermissive cells, but the mechanism of this inhibition is not thoroughly understood. It should be noted that the phenomenon of inhibition of proliferation under nonpermissive conditions by AAV has only been examined in vitro in cultured cells. It is not known whether this is also an in vivo phenomenon. AAV type 2 (AAV2) infection of primary human fibroblasts transactivates p21WAF1 gene expression, causing cell cycle arrest by suppressing phosphorylation of pRB family proteins (21). Rep78 downregulates the human c-fos, ras, and c-myc proto-oncogene promoters (22, 24, 62). AAV inhibits Ad and papillomavirus propagation (6, 7, 23). Expression of Rep protein inhibits the replication of HSV, bovine papillomavirus, HPV, and human immunodeficiency virus (1, 2, 22, 23, 53). AAV inhibits cellular transformation associated with HSV and Ad (46). Rep proteins block E1a- and ras-mediated transformation of cultured cells (29). The Rep proteins inhibit HPV transformation in cell culture perhaps by suppressing the papillomavirus p97 core promoter through disruption of the interaction between the TATA-binding protein and the TATA box (57).
To understand how AAV interacts with host cells, we investigated its relationship with Ad, AAV's most efficient helper virus. The Rep proteins play important roles in the inhibition. Inhibition of Ad replication has been observed after plasmid transfection of AAV Rep gene plasmids (65). Our laboratory has observed that AAV and its Rep proteins inhibit Ad replication and decreases Ad E2a mRNA and protein expression (28). Cotransfection of HeLa cells with reporter constructs containing the Ad early reporters and a plasmid expressing Rep78 demonstrated that Rep78 repressed the E2a promoter (28). Rep78 has no effect on E2a mRNA stability, suggesting that the Rep78 mediated decrease in Ad E2a mRNA results from inhibition of transcription (44).
To study Rep interaction with the Ad early promoters in vivo, we performed chromatin immunoprecipitation (CHIP) analysis on HeLa cells coinfected with AAV2 and Ad5 and infected with Ad and transfected with a Rep78 expression plasmid. We determined that AAV2 Rep proteins could be cross-linked to the Ad5 E2a promoter. The binding of AAV Rep protein to the Ad E2a promoter was characterized by electrophoretic mobility shift assay (EMSA). The Kd was determined to be 200 ± 25 nmol. A 38-bp protected region from nucleotides (nt) 27057 to 27085 in the Ad5 genome was identified by DNase I footprint analysis. The 38-bp protected region contains a weak TATA box, a region with homology to the canonical Rep binding motifs, and the transcription start site. These results suggest that Rep binding to the E2a promoter contributes to the inhibition of transcription from the Ad E2a promoter and Ad replication.
MATERIALS AND METHODS
Cells, plasmids, virus, antibodies, and protein expression.
HeLa cells from the American Type Culture Association were grown in Eagle minimum essential medium supplemented with 2 mM glutamine, 50 mg of penicillin/ml, 50 mg of streptomycin/ml, 100 μg of gentamicin/ml, 2.5 μg of amphotericin B/ml, and 10% fetal bovine serum. Cells were maintained as monolayer cultures at 37°C in a 5% CO2 atmosphere.
The pCDM series plasmids contain a variation of the rep gene inserted into pCDM8 (Invitrogen, Inc.) under the control of the CMV early promoter (70). Plasmids pCDMRep78 and pCDMRep78G contain the rep gene from the AAV2 genome (nt 263 to 2233) (70). The G refers to the replacement of the Rep52/40 methionine initiation codon with a glycine codon (10). Plasmid pCDMRep68G contains the spliced, carboxyl terminus-encoding region of Rep68 from pNTC28 inserted into pCDMRep78G. Plasmid pNTC28 lacks the major p40 mRNA intron and does not express Rep78 or Rep52 (10). Plasmid pE2aLUC contains the Ad2 E2a gene early promoter element inserted into the pGL3-Basic luciferase reporter vector (Promega, Inc.) (28). Plasmid pJM17 contains a mutant Ad5 genome lacking the E3 gene and 4.3-kb insertion of prokaryotic DNA at nt 1330 (39).
AAV2 was obtained by transfecting 25 μg of pNTC244 (10) containing the complete AAV genome onto each of 15 15-cm dishes of Ad-infected HeLa cells with Lipofectamine (Invitrogen Corp.) according to the manufacturer's recommendations. At 48 to 72 h after transfection, cells were scraped from the plates, pelleted by low-speed centrifugation, and resuspended in 40 ml of phosphate-buffered saline (PBS). Cells were lysed by three freeze-thaw cycles and then sonicated for three 15-s cycles at level 3 with a Fisher Sonic Dismembranator. Cellular debris was pelleted at 2,000 × g for 10 min, and the supernatant was brought to 1% sodium deoxycholate and 0.025% trypsin. The crude virus was incubated at 37°C for 60 min and centrifuged as described above. A 2.5-ml heparin agarose (H-6508; Sigma) column equilibrated with PBS at 4°C. The crude virus preparation was passed twice over the column, washed with 7.5 ml of PBS containing 250 mM NaCl, and eluted with 7.5 ml of PBS containing 750 mM NaCl. The eluted virus was brought to 10% glycerol and stored at −80°C. Titers of the purified virus were determined by using indirect immunofluorescence or a modified infectious center assay as described previously (6, 30). Ad5 was obtained by infecting HeLa cells; the Ad5 was then purified, and the titers were determined as described previously (66). Frank Graham kindly provided AdlacZ5, which contains the Escherichia coli β-galactosidase (β-Gal) reporter gene under control of the Ad5 E3 transcription promoter (42).
Polyclonal antibody was affinity purified from serum of rabbits immunized with recombinant E. coli Rep protein (61). B6 cl.10 monoclonal antibody to the E2A protein was kindly provided by Arnold Berk. Anti-rabbit immunoglobulin G (IgG; A-4914; Sigma) and anti-mouse IgG (A-2554; Sigma) peroxidase conjugates were diluted 1:104 prior to use.
Purification of Rep68.
Rep was expressed in E. coli (M15pRep4) harboring pStump68 in pQE70 (71). Cells were grown in Luria-Bertani medium, M9 salts, and 1% glucose at 37°C to an A600 of 0.8 to 1. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 0.2 mM, and cells were harvested after 3 h and stored frozen at −70°C. Preparation of the extract and Ni-nitrilotriacetic acid (NTA) chromatography were performed at 0 to 4°C. Partially thawed cell pellets (40 to 50 g) were suspended in 6 volumes of 50 mM Tris-HCl (pH 7.5), 10% (vol/vol) glycerol, 0.1% (vol/vol) Triton X-100, 2 mM EDTA, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride (PMSF) with a Dounce homogenizer. NaCl and MgCl2 were added with stirring to 0.5 and 5 mM, respectively, and the extract was stirred for 30 min. The extract was subject to centrifugation for 30 min at 20,000 × g. Polyethylene glycol 8000 (PEG 8000; 0.25 volumes of a 50% [wt/vol] solution) was added with stirring to the supernatant, followed by further stirring for 30 min. The precipitate was collected by centrifugation (20,000 × g, 30 min) and dissolved in 30 to 40 ml of 20 mM Tris-HCl (pH 7.5), 20% glycerol, and 0.5 M NaCl with a Dounce homogenizer. The sample was applied to 5-ml column of Ni-NTA Superflow (Qiagen) equilibrated in the same buffer, washed with starting buffer, and eluted successively with buffer containing 0.05 and 0.4 M imidazole; Rep68 eluted in the 0.4 M imidazole eluate. Fractions containing Rep68 were pooled, dithiothreitol (to 0.5 mM) and EDTA (to 0.2 mM) were added, and the material was concentrated to 2 to 3 ml with Vivaspin 6-ml concentrators (10,000 molecular weight cutoff). The concentrated material was applied as 0.5-ml aliquots to a column (1 by 32 cm) of Superose 12 (Pharmacia) equilibrated in 20 mM Tris-HCl (pH 7.5), 10% (vol/vol) glycerol, 0.5 M NaCl, 0.5 mM dithiothreitol, 2 mM MgCl2, and 0.2 mM EDTA. Chromatography was performed at room temperature at 0.5 ml per min; fractions were collected and immediately placed on ice. Rep68 containing fractions were combined and stored frozen at −70°C. Rep68PNB, which has a lysine-to-histidine change in the purine nucleotide binding site (11), was expressed and purified in a similar manner.
Purification of RepNT.
DNA sequences encoding the first 225 residues of Rep78/68 were cloned into pET16b (pRepNT), and the protein was expressed in E. coli BL21D3star (Invitrogen). RepNT was purified by essentially the same protocol except that the gel filtration step was performed at 0 to 4°C on a open column (1.5 by 100 cm) of Sephacryl S200 (Pharmacia).
Purification of Rep40.
Rep40 was cloned into pET9a and expressed in E. coli BL21D3star as an untagged protein. Steps prior to Superose 12 chromatography were performed at 0 to 4°C. The extract (from 130 g of cells) was prepared as for Rep68 and brought to 7.5% (wt/vol) PEG by the addition of 0.175 volumes 50% (wt/vol) of PEG 8000 and subjected to centrifugation (20,000 × g, 30 min). The supernatant was brought to 20% PEG (by addition of 0.48 volumes (relative to the original extract) of 50% (wt/vol) PEG. The precipitate was collected by centrifugation and dissolved in 20 mM Tris-HCl (pH 7.5), 20% (vol/vol) glycerol, 40 mM NaCl, 0.5 mM dithiothreitol, and 0.5 mM EDTA. The material was applied to a 70-ml column of Q-Sepharose (Pharmacia) equilibrated in the same buffer. The column was eluted in the same buffer and Rep40 eluted in the unbound material. Fractions were pooled (200 ml) and concentrated in Amicon ultrafiltration cell using a PM10 to membrane to 25 ml. During concentration, Rep40 precipitated selectively and was collected by centrifugation. The protein was dissolved in 20 mM Tris-HCl (pH 7.5), 10% (vol/vol) glycerol, 200 mM NaCl, 0.5 mM dithiothreitol, and 0.5 mM EDTA, and aliquots of 0.5 ml were subjected to chromatography at room temperature on Superose 12 (Pharmacia) equilibrated in 20 mM Tris-HCl (pH 7.5), 10% (vol/vol) glycerol, 200 mM NaCl, 0.5 mM dithiothreitol, and 0.2 mM EDTA. Rep40-containing fractions collected during Superose 12 chromatography were immediately placed on ice, pooled, concentrated, and stored at −70°C.
β-Gal assays.
HeLa cells at ∼80% confluency in 24-well plates were transfected with 3 μl of Lipofectamine 2000 (Invitrogen) and 800 ng of pCDMRep78G (or pCDMRep68G) per well according to the manufacturer's instructions with 0.1% bovine serum albumin (BSA) included in 400 μl of serum-free medium during transfection. After 4 h, the cells were infected at a multiplicity of infection (MOI) of 1 with AdlacZ5. After 1 h, the medium was replaced with 500 μl of complete medium. Adherent cells were released by scraping 48 h postinfection and separated into two equal aliquots. One aliquot was assayed for AdlacZ-induced β-Gal activity using the Galacto-Star (Tropix System) according to the manufacturer's instructions. Cells were pelleted by low-speed centrifugation, resuspended in 100 μl of GalactoStar lysis buffer, incubated 10 min at room temperature, and clarified by centrifugation for 10 min at 8,000 × g. Then, 30 μl of supernatant was incubated with 100 μl of substrate, and luminescence was measured on a Lumat LB 9600 (Berthold Technologies). The other aliquot was lysed by freeze-thawing in complete medium and used for second infections. HeLa cells at ∼80% confluency were infected in 24-well plates in 500 μl of serum-free medium. After 1 h the medium was replaced with complete medium, and the cells were assayed for β-Gal activity after 24 h as described above.
Immunoblot analysis.
Samples containing equal amounts of protein were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and transferred to nitrocellulose membranes. Blots were incubated with primary antibodies at a 1:103 dilution and incubated overnight at 4°C. Blots were incubated with secondary horseradish peroxidase conjugates diluted at 1:104 for 1 h at room temperature. The secondary antibody was detected with 100 mM Tris-HCl (pH 8.5), 0.015% (vol/vol) H2O2, 1.25 mM luminol, and 0.2 mM coumaric acid.
CHIP analysis.
HeLa cells in duplicate 15-cm plates at 60% confluence were transfected with 25 μg of pCDMRep78 or pCDM8 and 100 μl of Lipofectamine (Invitrogen) in serum-free medium containing 0.1% BSA. The medium was replaced after 4 h with complete medium and, after 24 h, the medium was replaced with complete medium containing Ad5 at an MOI of 5. Cells were harvested at 48 h posttransfection. For AAV2 infections, cells in duplicate 15-cm plates at 80% confluency were infected with AAV2 at an MOI of 100 for 1 h in serum-free medium. The medium was replaced with complete medium containing Ad5 at an MOI of 5, and the cells were harvested 24 h postinfection.
Cells were harvested by trypsinization, pelleted by low-speed centrifugation, and washed twice with PBS containing 2 mM CaCl2 and 2 mM MgCl2. The cells were resuspended on ice in buffer A (10 mM HEPES [pH 7.8], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, 1 μM leupeptin, 1 μg of pepstatin A/ml, 1 mM PMSF, and 1 mM benzamidine) and 0.4% (wt/vol) Triton X-100. The cells were incubated on ice for 10 min, and the nuclei were pelleted by centrifugation. All subsequent steps were performed at 4°C, and centrifugations were 4 min at 1,300 × g. Nuclei were prepared by differential centrifugation, and the preparation of formaldehyde cross-linked chromatin was based on a protocol described by Ritzi et al. (54). The nuclei from two 15-cm plates were resuspended in 1.0 ml of buffer A containing 200 mM NaCl. The nuclei were added to 9 ml of prewarmed buffer A containing 1.1% (vol/vol) formaldehyde (37°C) and incubated at 37°C water bath for 10 min, and the reaction was quenched by the addition of glycine to 0.125 M. Nuclei were centrifuged and washed twice with PBS containing 2 mM CaCl2, 2 mM MgCl2, and 0.5% (wt/vol) NP-40. Nuclei were resuspended in 8 ml of IPP buffer (50 mM Tris-HCl [pH 7.9], 150 mM NaCl, 20 mM EDTA, 0.5% NP-40, 1 mM DTT, 1 μM leupeptin, 1 μg of pepstatin A/ml, 1 mM PMSF, and 1 mM benzamidine) and incubated for 30 min on a rotary incubator. Chromatin was pelleted at 2,000 × g for 10 min and resuspended in 1 ml of IPP buffer. Chromatin was subjected to centrifugation through a 35-ml cushion of IPP buffer containing 100 mM sucrose at 5,400 rpm in a SW28 rotor for 10 min, and the chromatin pellet was resuspended in 2.0 ml of TE (10 mM Tris-HCl [pH 7.5], 1 mM EDTA). Chromatin was sonicated by using a Sonic 550 Dismembrator and a microtip (Fisher Scientific) for 2 min at setting 5 with 1-s pulses on ice. Absorbance at 260 nm was used to estimate chromatin concentration. Approximately 1 mg of chromatin was isolated from two 15-cm plates.
Then, 50 μl of Immunopure protein A-agarose beads (Pierce) was washed with 1 ml of IPP buffer, vortexed, and pelleted by low-speed centrifugation. The beads were resuspended in 930 μl of IPP buffer containing 200 μg of calf thymus DNA/ml and incubated for 3 h at 4°C on a rotary mixer. Cross-linked chromatin was denatured by the addition of 1% SDS and 40 mM β-mercaptoethanol for 10 min at 37°C. The denatured cross-linked chromatin was then incubated at 4°C for 1 h with 50 μl of the treated beads to minimize nonspecific binding in subsequent incubations. The beads were removed by low-speed centrifugation, and the supernatant containing precleared cross-linked chromatin was incubated overnight at 4°C with 50 μl of IPP-washed beads, 730 μl of IPP buffer containing 200 μg of calf thymus DNA/ml and 200 μl of affinity-purified Rep antibody (∼1 μg). The beads were pelleted by centrifugation (1 min at 3,000 × g at 4°C) and transferred to new 1.7-ml Eppendorf tubes after each of the following washes (1.0 ml): eight times with radioimmunoprecipitation assay buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1% [wt/vol] NP-40, 1 mM EDTA, 0.5% [wt/vol] sodium deoxycholate, 1 μM leupeptin, 1 μg of pepstatin A/ml, 1 mM PMSF, and 1 mM benzamidine), three times with LiCl buffer (10 mM Tris-Cl [pH 8.0], 250 mM LiCl, 0.5% [wt/vol] NP-40, 1 mM EDTA, 1 μM leupeptin, 1 μg of pepstatin A/ml, 1 mM PMSF, and 1 mM benzamidine) and five times with TE. Beads were resuspended after the final wash in 200 μl of 50 mM glycine pH 3 containing 1% SDS (wt/vol) and incubated for 10 min at 37°C. After centrifugation the supernatant was incubated with 10.0 μl of RNase A and RNase T1 (Ambion RNase Cocktail) for 1 h at 37°C. Proteinase K (1.0 μg/ml) was added to 150 μl of the supernatant and incubated at 37°C for an additional 6 h to reverse the protein-DNA cross-links. The DNA was extracted twice with phenol-chloroform (1:1), precipitated with 2.5 volumes of ethanol, and dissolved in 50 μl of TE for PCR analysis. Cross-links were also reversed on 100 μg of the chromatin/protein before immunoprecipitation as a control for immunoprecipitation specificity. PCR was performed on 1 μl of the DNA in a 25-μl reaction volume with 5.0 U of Taq polymerase (Promega). The samples were denatured at 95°C for 5 min and then subjected to 40 cycles of 94°C for 1 min, 45°C for 30 s, and 72°C for 1 min with a final incubation at 72°C for 5 min. The samples (8 μl) were electrophoresed by using 2% (wt/vol) agarose in Tris-acetate-EDTA buffer. The sizes of the amplified products and the primers used for the Ad early promoters were as follows: E1a, 532 bp, CCTCGAGCATCATCATAATATACCTTA and AGAAGCTTGGAGGAGAAAACTCTACTCG; E1b, 261 bp, TTCTCGAGTCTGGGCAACCTTTGGA and GCAAGCTTGAGGTCAGATGTAAACAAGA; E2a, 286 bp, GACTCGAGATATCATGTGGGGTCC, and AAAAGCTTAGATGAGCTTCGGCGCAC; E3, 311 bp, ATCTCGAGTCAACGGAATCCGCGCC and CCAAGCTTGGAGCTCACCGACTCGTC; and E4, 360 bp CCTCGAGCATCATCATAATATACCTTA and AAAAGCTTTCGACACGGCACCAGCTCA. Primers for amplifying the 150-bp E2a fragment used in gel shift analyses were as follows: AAGATCAGCTTCGGCGCACGCTGGAAGACG and CAGGTGCTGGCGCCGGGTGTGGCCGCTGGA.
EMSA.
All binding reactions were prepared on ice in 20 μl of 10 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 250 mM NaCl, 4% glycerol (vol/vol), 0.5 mM DTT, 0.5 mM EDTA, 250 μg of poly(dI-dC)-poly(dI-dC) (Sigma)/ml, and 50 μg of BSA/ml and then shifted to 30°C for 20 min. Tris-borate-EDTA nondenaturing 4% acrylamide-bisacrylamide (40:1) electrophoresis was performed at 100 V at 4°C.
DNase I footprint analysis.
Nuclease protection assays were performed according to the method of Galas and Schmitz with modifications (19). pE2aLUC was linearized with either HindIII and XhoI, end labeled with [γ-32P]ATP and polynucleotide kinase, and digested with XhoI (after HindIII linearization, “upper strand”) and HindIII (after XhoI linearization, “lower strand”) to yield a 303-bp fragment. DNA binding reactions were performed in 20 μl of 20 mM Tris-HCl (pH 7.9), 2 mM MgCl2, 50 mM NaCl, 10% (wt/vol) glycerol, 0.1 mM DTT, 5 μg of poly(dI-dC)/ml, and 50 μg of BSA/ml for 20 min at 30°C. DNase I (RQ1; Promega) was added to 20 U/ml, and the reactions were incubated for 2 min at room temperature. The DNase I digestion was stopped by the addition of 50 μl of 200 mM NaCl, 40 mM EDTA, 1% (wt/vol) SDS, and 125 μg of tRNA/ml. The samples were digested with 1 μg proteinase K/μl for 1 h at 37°C, extracted twice with phenol-chloroform, ethanol precipitated, and resuspended in 6 μl of loading buffer containing 90% (vol/vol) formamide and bromophenol blue and xylene cyanol dyes. Samples were heated to 95°C, chilled on ice, and separated by using a 6% acrylamide electrophoresis gel containing 8 M urea.
RESULTS
The Rep78 and Rep68 inhibit Ad replication and are preferentially cross-linked at the Ad5 E2a promoter.
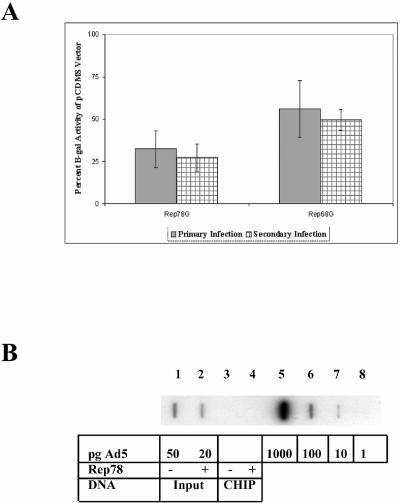
To investigate interactions between AAV2, its encoded Rep proteins, and Ad, we established a model system of AAV-mediated inhibition of Ad replication by transfecting Rep-expressing plasmids into cells that were subsequently infected with Ad. We hypothesized that strong expression of Rep proteins will inhibit Ad replication by interacting with the Ad genome in the vicinity of early transcription promoters. To confirm that Rep proteins alone can inhibit Ad replication, Rep-expressing plasmids were transfected onto HeLa cells and subsequently infected with AdlacZ5. The AdlacZ5 vector is an E3 deletion mutant that contains the E. coli β-Gal gene containing an intact E3 promoter (42). However, it has not been proven that β-Gal expression is indeed directed by the viral promoter (F. L. Graham, unpublished data). At 48 h after infection, the cultures were harvested and lysed, and aliquots were used to infect another plate of HeLa cells, or to measure β-Gal activity. The secondary infections were harvested 24 h later, and the β-Gal activity was determined. Diminished β-Gal activity in the first infection is indicative of inhibition of E3 promoter activity and/or Ad DNA replication The β-Gal activity in the second infection is indicative of the amount of Ad produced in the first infection. Control experiments indicate that the amount of β-Gal activity in infected cells is directly proportional to the amount of virus in the inoculum (not shown). Figure 1A demonstrates that Rep78G inhibited Ad replication by 65% and that Rep68G inhibited Ad replication by 35%. Similar levels of inhibition were observed in the second infection, which is indicative of the amount of virus production from the first infection. The apparent difference between Rep78G and Rep68G may be due to differences in protein expression; the expression of Rep68G is typically lower than Rep78G. These results indicate that Rep78/68 expression, in the absence of other AAV components, suppresses Ad replication.
FIG. 1.
AAV2 Rep proteins inhibit Ad5 replication. (A) β-Gal activity was measured from HeLa cell extracts obtained after transfection with pCDM8 plasmids expressing Rep proteins and infected with Ad5lacZ5. β-Gal activity was normalized to the pCDM8 vector control (not shown). Error bars represent the standard deviations from four experiments performed in triplicate (n = 12). (B) Slot blot hybridization analysis on nuclear extracts from HeLa cells with radiolabeled Ad5 probe. HeLa cells were transfected with either pCDM8 (slots 1 and 3) or pCDMRep78 (slots 2 and 4). Input slots contain equal amounts of chromatin, as determined by the absorbance at 260 nm. CHIP slots contain chromatin-immunoprecipitated DNA. Slots 5 to 8 contain decreasing amounts of the Ad5 genome excised from plasmid pJM17. The radioactive signal in slots 5 to 8 was measured with a phosphorimager, and the amount of DNA in slots 1 to 4 was determined by comparison to the known amounts of DNA in slots 5 to 8.
Previous work from our lab suggested that the Rep proteins inhibit Ad E2a mRNA transcription in AAV-Ad-coinfected cultures (28, 44). To determine whether the Rep proteins interact directly with Ad DNA in vivo, we tried to cross-link Rep proteins to Ad DNA by using formaldehyde. Formaldehyde is a high-resolution (2-Å) cross-linking agent that produces protein-DNA, protein-RNA, and protein-protein cross-links in vivo (45). Amino and imino groups of proteins (primarily lysine and arginine) and nucleic acids (primarily cytosine) react with the dipolar formaldehyde to form a Schiff base that forms a reversible reaction with a second amino group and condenses (37, 38). Cells were transfected with pCDMRep78 or pCDM8 and infected with Ad5 24 h posttransfection, and nuclei were prepared at 24 h postinfection. The nuclei were subjected to formaldehyde cross-linking, and chromatin was isolated. The cross-links were reversed, and equal amounts of chromatin were analyzed by slot blot and Southern hybridization with radiolabeled Ad5 DNA. Figure 1B demonstrates that Ad5 genomic DNA was present in the chromatin fraction and suggests transfection with a Rep78-expressing plasmid resulted in a 60% lower yield of Ad5 DNA (slot 2) compared to transfection of pCDM8 (slot 1). This reduced yield of Ad DNA is consistent with the comparable level of inhibition of β-Gal activity in Fig. 1A, suggesting that inhibition of DNA replication accounts for the reduced activity. These results confirm that Rep78 inhibits Ad DNA synthesis and that the diminished Ad production observed in Fig. 1A is likely due to the lower level of DNA synthesis. No Ad5 DNA was detected by hybridization after DNA was obtained by CHIP analysis with Rep antibody (slots 3 and 4). Therefore, we performed PCR analyses on the immunoprecipitated DNA to detect specific Ad DNA sequences.
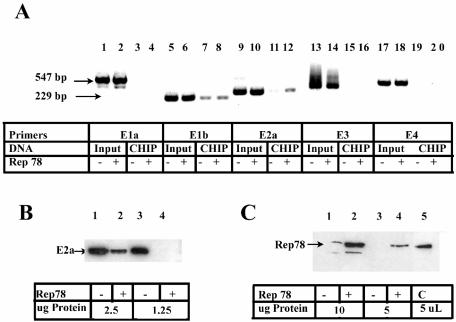
To identify sites in the 35,938-bp Ad5 genome where Rep might regulate gene expression, we used CHIP, followed by PCR analysis with primers that amplify Ad5 promoter regions. The cross-linked, anti-Rep immunoprecipitated samples used in Fig. 1B were subjected to PCR. Figure 2A shows that Ad5 genomic DNA from all five promoter regions was present in the chromatin-protein pellet after formaldehyde cross-linking and could be amplified efficiently with all five primer sets (input lanes). After immunoprecipitation with Rep antibody, the Ad5 E2a primers (lanes 11 to 12) clearly amplified more DNA from cells transfected with the pCDMRep78 plasmid (lane 12) than the vector control (lane 11). Typically, investigators use an antibody of the same isotype to control for nonspecific interactions. We were able to use the same antibody, since no Rep protein was present after transfection of the vector control plasmid. Quantitation of the slot blot by radiolabeled Ad5 DNA hybridization analysis (Fig. 1B) revealed that the Rep immunoprecipitated samples (+) contained 60% fewer genomic copies of Ad5 than the input samples, further magnifying the difference observed between lanes 11 and 12.
FIG. 2.
Rep78 protein is preferentially cross-linked at the Ad5 E2a promoter region. (A) CHIP assay from nuclear lysates of Ad5-infected cells transfected with pCDMRep78. Input DNA (before immunoprecipitation, diluted 1:1,000) and CHIP DNA (after immunoprecipitation, 1 μl) were amplified by using Ad5 promoter region primers. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. Lanes labeled “+” were from cells transfected with pCDMRep78; lanes labeled “−” were from cells transfected with pCDM8. The sizes of the largest and smallest products are indicated at the left side of the gel. Equal amounts of protein were compared from nuclear lysates for the expression of E2a (B) and Rep78 (C). Lysates were from Ad-infected HeLa cells transfected with pCDM8 (lanes 1 and 3) or pCDMRep78 (lanes 3 and 4). Lane 5 in panel C contains a positive control for the Rep78 protein.
Infection with AAV2 inhibits Ad5 E2a mRNA and protein expression (44). To verify that Rep78 inhibits E2a in our plasmid transfection-infection model, we transfected HeLa cells with pCDMRep78 and infected the cultures with Ad5. The expression of the Ad5 E2a protein was inhibited (Fig. 2B), indicating that Rep78 inhibits expression from the E2a promoter in plasmid transfection. Rep78 protein expression was verified Western analysis (Fig. 2C).
The in vivo association of Rep78 with the E2a promoter region, coupled with our observations that Rep78 inhibits E2a protein and mRNA expression, strengthens our hypothesis that Rep mediates its effects on Ad5 through protein-DNA interaction. Although our data indicate that Rep78 is preferentially cross-linked at or near the Ad5 E2a promoter, it does not rule out that Rep is bound near other Ad5 promoter sites or elsewhere in the Ad5 genome.
Rep proteins are cross-linked to the Ad5 E2a promoter region during AAV2 coinfection with Ad5.
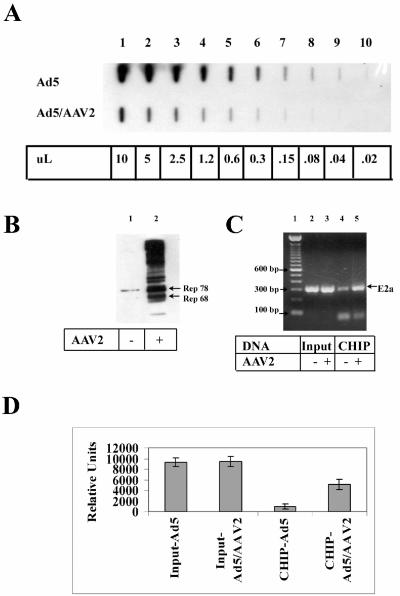
Since Rep78 expression from the CMV promoter after transfection results in a higher level of protein expression than during an infection with AAV2, we examined cross-linking of Rep to the E2a promoter region after AAV2 infection, where the Rep proteins are expressed at physiologically relevant levels. HeLa cells were infected with Ad5 or coinfected with AAV2 and Ad5 and nuclei harvested 24 h postinfection. The nuclei were subjected to formaldehyde cross-linking, and chromatin was isolated and quantitated. The relative amount of Ad5 in equal amounts of isolated chromatin was quantitated by Southern analysis (Fig. 3A). Densitometry analysis revealed that infection with AAV2 reduced the number of copies of Ad5 by a factor of 8, suggesting that AAV infection is more effective than transient transfection in inhibiting Ad replication. The more effective inhibition of Ad replication by infection with AAV, compared to transfection of the rep gene, may also be due to the known evidence that AAV and Ad DNA replication colocalize to cellular replication centers and that AAV genomes presumably compete directly with Ad genomes for the replication apparatus (65). CHIP analysis was performed on equivalent amounts of chromatin isolated from the Ad-infected HeLa nuclei. Western analysis was performed on 25% of the final supernatant before the reversal of cross-links to ensure that the amplification would be in the linear range of response. Immunoblot analysis showed that both Rep78 and Rep68 were immunoprecipitated with affinity-purified Rep antibody after cross-linking to chromatin (Fig. 3B). The presence of higher-molecular-weight protein bands most likely results from Rep proteins becoming covalently bound to DNA replication intermediates during an AAV infection (51). Equivalent genomic copies of Ad Input (before immunoprecipitation) and CHIP DNA were assayed by PCR with the Ad5 E2a primers. Figure 3C demonstrates that, whereas the input DNA was amplified to a similar level (lanes 1 and 2), the amount of CHIP DNA amplified from AAV2-infected cells was enriched by a factor of 4. The PCR was performed in triplicate and quantitated (Fig. 3D). These results show that Rep proteins are preferentially cross-linked at or near the E2a promoter during an AAV2 coinfection with Ad5.
FIG. 3.
Rep proteins are cross-linked to the Ad5 E2a promoter region during AAV2 coinfection. (A) Equal amounts of DNA from formaldehyde-cross-linked nuclei from Ad5-infected or Ad5- and AAV-coinfected HeLa cells were analyzed by slot blot hybridization with an Ad5 probe. The numbers at the bottom of the panel refer to the amount of purified DNA from the original 50 μl obtained after chromatin isolation. (B) The presence of the Rep proteins was verified by SDS-PAGE and immunoblot analysis after CHIP analyses. Lane 1 is from Ad-infected cells, and lane 2 is from Ad-AAV-coinfected cells. (C) CHIP analysis was performed with primers that amplify the Ad5 E2a region. Equivalent amounts of input (lanes 2 and 3) and CHIP (lanes 4 and 5) Ad DNA (as determined in panel A) were amplified and separated by agarose gel electrophoresis. Lane 1 contains 1.0 μg of 100-bp DNA ladder separated. (D) The ethidium-bromide stained bands from panel C were quantitated by using a Kodak Image Station 440 from PCRs performed in triplicate. Error bars represent standard deviations.
Characterization of Rep68 binding to the E2a promoter region by EMSA.
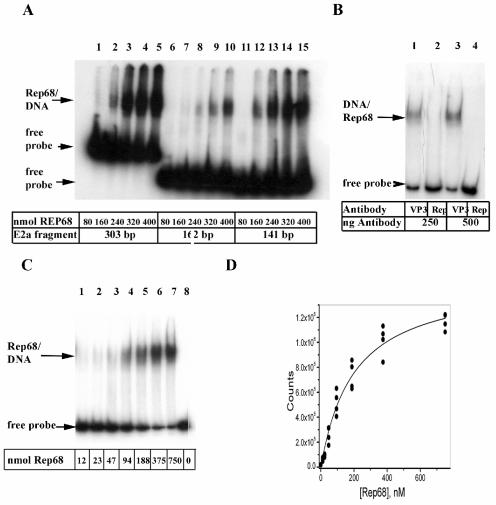
Because formaldehyde cross-links proteins to DNA and proteins cross-link to other proteins, we cannot conclude from CHIP analysis that Rep is bound directly to DNA. Sonication results in DNA fragments with an average size of ∼1 kb. Therefore, the 303-bp amplified fragment observed in Fig. 2A and 3B may have resulted from a DNA fragment of ≥1 kb. Conceivably, the Rep protein is cross-linked over 1 kb from the E2a promoter. To determine whether Rep binds the amplified region directly, we tested the binding of purified Rep68 to the 303-bp fragment by EMSA. We used Rep68 purified from E. coli instead of Rep78. Rep68 contains the same DNA-binding domain as Rep68, and both proteins interact with the binding site in the AAV ITR. Rep68 also inhibits Ad replication (Fig. 1A). Therefore, we anticipate that the two proteins will behave similarly in DNA-binding experiments with the Ad E2a promoter. As observed in Fig. 4B (lanes 1 to 5), the addition of Rep68 to the 303-bp fragment containing the Ad5 E2a promoter region results in a retarded band that is saturable. Rep68PNB (which lacks a functional purine nucleotide binding site) bound to the 303-bp fragment as efficiently as wild-type Rep68. An N-terminal 225-amino-acid fragment of Rep68, RepNT, and Rep40 did not bind to the fragment (data not shown). The 303-bp fragment contains a weak TATA motif, an ATF binding site, two E2F binding sites, three Rep partial binding motifs (GAGC), and the transcription start site. Chiorini et al. present evidence that tandem repeats of GAGC are preferred sites for Rep binding (13). Therefore, we refer to a single GAGC as a Rep partial binding motif. The 303-bp fragment was digested with NciI, yielding 141- and 162-bp fragments. The 141-bp fragment contains two Rep partial binding motifs; the ATF, E2F, and TBP transcription factor binding sites; and a transcription start site. The 162-bp fragment contains the remaining sequences starting 16 bp upstream of the ATF site and contains one Rep partial binding motif. Figure 4A shows that binding affinity of Rep68 to the 141-bp fragment (lanes 12 to 15) is stronger than the 162-bp fragment (lanes 6 to 10). The results suggest that Rep68 binds with some sequence specificity to the 141-bp fragment containing the transcription factor binding sites, the principal transcription start site, and two Rep partial binding motifs (GAGC and GCGC).
FIG. 4.
Characterization of Rep68 binding to the E2a promoter region by EMSA. (A) The 303-bp promoter element from pE2aLUC was obtained by digestion with HindIII and XhoI and end labeled. The 162- and 141-bp promoter fragments were obtained by digestion of the 303-bp element with NciI. EMSA was performed by using ∼10 fmol of each DNA fragment with increasing amounts of Rep68 protein. (B) The 303-bp fragment was used as a template to amplify a 150-bp fragment containing sequences essentially identical to the 141-bp element in panel A. Primers corresponding to the ends of the 141-bp fragment (with an additional 9 nt) were used to amplify the 150-bp fragment in a reaction containing [α-32P]dATP. EMSA was performed by using both affinity purified VP3 and Rep antibody. Totals of 250 and 500 ng of antibody were used. Antibodies contained ∼50 μg of IgG/ml. (C) Increasing amounts of Rep68 were added to 7.5 fmol of the 150-bp fragment. (D) The dissociation constant was determined after performing the titration in panel C in quadruplicate. The dried gel was exposed to a phosphorimager cassette, and densitometry was performed with ImageQuant software.
Although recombinant Rep68 was purified to near homogeneity, the preparation may contain an unrelated DNA-binding protein that binds to the labeled DNA. To test this possibility, affinity-purified Rep antibody was added to the binding reaction. The DNA target for these assays was a 150-bp fragment amplified by PCR containing all of the sequence elements found in the 141-bp fragment described above. The addition of the Rep antibody did not result in a supershift but rather disrupted the Rep68/DNA complex (Fig. 4B, lanes 2 and 4), whereas affinity-purified antibody to the AAV capsid protein (VP3) did not affect the complex (Fig. 4B, lanes 1 and 3). The 150-bp fragment (7.5 fmol) was titrated with increasing concentrations of Rep68 in quadruplicate, and a representative result is shown in Fig. 4C. Free probe and the Rep68/DNA complex were quantitated, and the results were plotted in Fig. 4D. A hyperbolic {B = N[L]/(K&double_tag;n + [L])} binding isotherm and the Hill equation {B = N[L]n/(Kn + [L]n)} were used as fitting functions for the data shown in Fig. 4D, where B is bound ligand, L is the free ligand concentration, N is the number of total sites, n is the Hill coefficient, and K is the dissociation constant. Based on application of the F-test, a fit to the Hill equation could not be justified statistically over a fit to a simple hyperbola. It is possible that cooperative binding would be revealed if a true equilibrium binding method with better precision were used. The estimate for the binding constant based on a simple binding model is 200 ± 25; however, this analysis is a simplification since the binding model is likely to be complex. Dissociation constants have been reported for Rep68 binding to the AAV ITR, A-stem, and chromosome 19 S1 integration sites (Table 1) (12, 20, 36). Each of these sequences contain three or four GAGC repeats that may lead to tighter binding
TABLE 1.
Kd estimates of known Rep68 target sitesa
| DNA | Kd (nM) | Source or reference |
|---|---|---|
| Chromosome 19, S1 site | 5.5 | 20 |
| AAV hairpin ITR | 0.9 | 12 |
| AAV hairpin ITR | 6.3 | 36 |
| AAV A-stem of ITR | 0.8 | 12 |
| AAV A-stem of ITR | 20 | 36 |
| Ad5 E2a promoter | 200 | This study |
Dissociation constants were estimated by using EMSA assays similar to the methods described in the text.
Rep68 protects a region of the Ad5 E2a promoter from DNase I digestion.
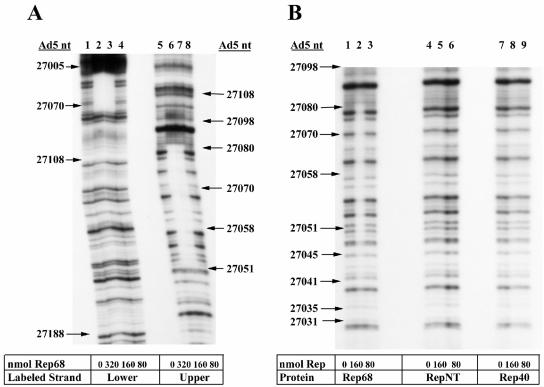
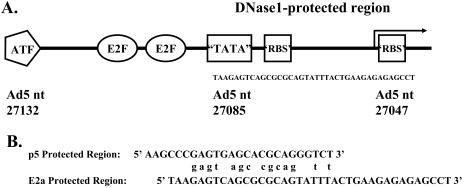
Rep68 binds specifically to a 141-bp region of Ad5 DNA containing cis elements of the E2a promoter. To locate the Rep binding site in this 141-bp region, we utilized DNase I footprinting. The 303-bp fragment used in Fig. 4 labeled on the upper (antisense) or lower (sense) strands was incubated with Rep68 and digested with DNase I, and the DNA fragments were separated by denaturing electrophoresis. Figure 5 shows autoradiograms of DNase I protection assays with variable amounts of Rep68. The protected region extends from nt 27047 to 27085 in the Ad5 genome (lanes 2, 4, 6, and 7). The same protected region is observed on both strands. Wild-type Rep68 was used in the experiment shown in Fig. 5A; the same region was protected when the mutant Rep68PNB protein was used (data not shown). An additional protected region at approximately nt 26835 was observed by using the highest concentration of Rep68 (data not shown). This protected region may account for the binding of Rep68 to the 162-bp fragment observed in Fig. 4B. Consistent with the EMSAs, neither RepNT nor Rep40 provided protection from nuclease digestion (Fig. 5B), whereas Rep68 (lanes 3 to 5) protected a region from nt 27057 to 27085. These studies localize the Rep68-bound region between the E2a TATA box and the transcription start site. Figure 6A illustrates the location of the protected region. There is nucleotide sequence similarity between the AAV p5 promoter, which contains a similar Rep-protected region, and the E2a promoter. The similarities occur in the putative Rep binding motifs of both promoters.
FIG. 5.
Rep68 protects a region of the Ad5 E2a promoter from DNase I digestion. (A) DNase I protection assays were performed on the 303-bp fragment (∼600 pmol) containing labeled lower strand (lanes 1 to 4) or labeled upper strand (lanes 5 to 8) with decreasing concentrations of Rep68. (B) DNase I protection assays were performed on labeled upper strand in the presence of Rep68, RepNT, and Rep40. Radiolabeled DNA size markers (not shown) were used to determine the corresponding nucleotide position in the Ad5 genome.
FIG. 6.
Ad5 E2a and AAV p5 promoter regions protected by Rep68. (A) Schematic diagram of the Ad5 E2a promoter showing principal transcription factor binding sites and the transcription start site (arrow). The DNase I-protected region is bracketed. (B) Sequences between the TATA box (underlined) and transcription start sites (arrows) for the AAV p5 and Ad E2a transcription promoters. A total of 14 identical nucleotides that are found in the DNase I-protected regions are indicated.
DISCUSSION
AAV has profound effects on the replication of Ad, its most efficient helper virus. AAV inhibits Ad production from severalfold to >100-fold (7, 15, 28). Earlier studies implicated Rep proteins in the AAV-mediated inhibition of Ad replication (28, 65). We confirmed these observations by demonstrating that both Rep78 and Rep68 inhibit the replication of AdlacZ5. AAV Rep proteins inhibit gene expression from a variety of viral and cellular transcription promoters (reviewed in reference 43), but the majority of the assays were performed in transient-transfection assays with reporter plasmids. We demonstrated that Rep78 expressed in a cell line inhibits the expression of Ad E2a protein and the accumulation of its mRNA (28). We have demonstrated an in vivo interaction of Rep78 with the Ad5 E2a promoter region by using CHIP and PCR analyses. This association was demonstrated in Ad-infected cells transfected with a Rep78 expression plasmid and in cells coinfected with wild-type AAV and Ad5. That the Ad5 E2a promoter is a target of the larger Rep proteins complements previous work from our laboratory demonstrating that AAV Rep expression alone mediates reduction of E2a protein and mRNA levels (28, 44). The interaction observed between Rep78 and the E2a promoter likely has physiological relevance. The Rep protein may bind to other Ad promoter regions or other sites in the Ad genome. However, we focused on the E2a promoter to characterize Rep protein binding.
CHIP analysis does not necessarily identify DNA-binding proteins since binding and cross-linking can be mediated through other proteins. Therefore, we examined Rep68 protein binding to the Ad E2a promoter fragment by EMSA. In the presence of nonspecific competitor, Rep68 bound specifically to a 303-bp fragment containing the E2a promoter region. Splitting this fragment revealed that Rep68 preferentially bound to a 141-bp fragment containing the major cis-acting regulatory elements of the E2a promoter: an ATF, two tandem E2F sites, and a weak TATA box (TTAAGA). The Rep protein displayed limited binding to the 162-bp fragment containing two Rep binding partial motifs separated by ∼90 bp. We focused on the 141-bp fragment because of Rep68's stronger affinity for it. We determined a binding constant for the binding of Rep68 protein to the 141-bp fragment to be 200 nmol. Binding constants have been determined for the AAV ITR element and the isolated A-stem of the ITR, and these are presented in Table 1. The AAV ITR and S1 DNA referred to in Table 1 contain three or four tandem copies of GAGC. The Rep68 Kd for the E2a promoter is much weaker than for the ITR element. Similarly, the affinity of Rep68 for the p40 transcription promoter is reportedly 100 times lower than for the ITR (49). It should be noted that these values for binding constants are not based on more complex models that will likely be required to account for the association of Rep68/Rep78 with DNA binding elements and should be interpreted cautiously.
Analysis of the Rep68-E2a promoter complex by nuclease protection revealed that 38 bp were protected by Rep68. The protected region is located from the E2a TTAAGA box to several base pairs downstream of the transcription initiation site (Fig. 6). This region contains sequence elements that resemble the Rep binding sites identified by Chiorini et al. by random sequence oligonucleotide selection (13). Rep binds to elements in the p5, p19, c-H-ras, the HIV long-terminal-repeat TAR region, HPV p97, cellular E2F, and the CMV immediate-early promoter region (1, 3, 24, 32, 33, 57). A variety of other cellular DNA sequences have been identified that have Rep binding sites that more closely resemble the ITR element and many of these interact with Rep68 (67). DNase I footprinting of Rep68 binding to the AAV p5 promoter identified a 26-bp region from the TATA box to slightly downstream of the initiation site that was protected by Rep68 (48). Rep protein affinity for the other transcription promoters cited has not been determined. The DNA sequence between the AAV p5 TATA element and the start site for transcription is homologous to the A-stem of the AAV ITR. This arrangement is also found in the E2a promoter where the DNase I protected region extends from the E2a TATA region to the mRNA start site and contains a region homologous to the ITR (Fig. 6B). Of 16 identical nucleotides that contain the Rep binding motif, 14 are shared between the two promoters. Rep protein binding to the AAV p5 promoter results in transcription repression (32, 33, 48). Rep protein binding to the E2a promoter may explain how the Rep proteins inhibit E2a gene expression in Ad-infected cells.
The presence of Rep protein binding site homology in the E2a promoter suggests that the DNA-binding domains of Rep78 and Rep68 are required for the interaction. This suggestion was corroborated experimentally in the inability of Rep40 or RepNT to bind to the E2a promoter fragment. We did not observe Rep40 or RepNT interaction with the E2a promoter in either DNase I footprinting (Fig. 5) or EMSA analyses (data not shown). The 225-amino-acid RepNT protein binds to the AAV ITR but with reduced affinity (data not shown). Therefore, it is not surprising that it does not interact with the weak Rep binding element in the E2a promoter. The absence of multiple Rep binding motifs and the relatively large Kd suggest that Rep might interact with other proteins to mediate its effect on the Ad E2a promoter. Rep68 containing a mutant purine nucleotide binding domain (Rep68PNB) binds to the E2a promoter with affinity equal to that of the wild-type protein. This mutant Rep protein does not bind ATP; therefore, nucleotide binding is not necessary for E2a promoter interactions. Rep68PNB also binds to the AAV p5 Rep binding site but repression of transcription at the p5 promoter requires functional Rep ATPase activity (32, 33).
The AAV Rep78/68 proteins positively and negatively regulate viral transcription due to the presence of a Rep binding element within the p5 promoter, facilitating interactions with transcription factors bound to the p5 promoter. Rep78 and -68 proteins repress the AAV p5 and p19 promoters in the absence of Ad infection, and the promoters are differentially regulated in the presence of helper virus (34). Activation of the AAV p19 promoter requires interaction between proximal p19 elements and Rep bound at the p5 promoter. An interaction between Rep bound at the p5 promoter and Sp1 bound at the p19 promoter is mediated by DNA looping that results in a scaffold positioning the p5 YY1 complex near the p19 promoter (49). Regulation of the p19 and p40 promoters by Rep is related to the spacing between the Sp1 site and either the TATA box or the start of transcription (49). The NS1 protein, a homologue of Rep from the minute virus of mice, transactivates the p38 promoter by a direct interaction interacting with Sp1 (31). In addition to the transcription factor Sp1 the Rep protein has also been shown to interact with TATA-binding protein, PC4, Topors, c-Jun, and E2F-1 and may prevent the assembly of TFIID complexes on DNA (4, 25, 50, 57, 63, 64). The Ad E2a promoter has two binding sites for the E2F transcription factor. Rep binds to the cellular E2F-1 promoter, which also has E2F binding sites, and represses Ad-induced transcription of E2F-1 (4). The interaction between Rep78 and the p97 promoter contributes to the inhibition of HPV replication presumably by disrupting the interaction between TBP and the TATA box (57). The recognition of DNA secondary structure by Rep78/68 might also be important for the regulation of transcription by the Rep protein (3). Rep78/68 interaction with other transcription promoters has not been thoroughly investigated beyond EMSAs. However, a theme runs through these studies in which Rep78/68 binds to the transcription promoter even though it lacks a canonical Rep binding site. The affinity of the Rep protein for these other promoters may also be weak, as we have observed here with the E2a promoter. Since Rep interacts with other transcription factors in binding to AAV and other promoters, interaction with cellular transcription factors may be required to stabilize Rep binding and to maximize its effects in transcription regulation.
Acknowledgments
This study was supported by the National Institutes of Health grants GM64765 and AI51471.
We thank Susan Dignam for preparation of the Rep68 proteins, Roy Collaco for the preparation of Rep and Cap protein antibodies, and Kristin Verrill for preparation of AAV2.
REFERENCES
- 1.Antoni, B. A., A. B. Rabson, I. L. Miller, J. P. Trempe, N. Chejanovsky, and B. J. Carter. 1991. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J. Virol. 65:396-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bantel-Schaal, U., and H. zur Hausen. 1988. Adeno-associated viruses inhibit SV40 DNA amplification and replication of herpes simplex virus in SV40-transformed hamster cells. Virology 164:64-74. [DOI] [PubMed] [Google Scholar]
- 3.Batchu, R. B., and P. L. Hermonat. 1995. The trans-inhibitory Rep78 protein of adeno-associated virus binds to TAR region DNA of the human immunodeficiency virus type 1 long terminal repeat. FEBS Lett. 367:267-271. [DOI] [PubMed] [Google Scholar]
- 4.Batchu, R. B., M. A. Shammas, J. Y. Wang, and N. C. Munshi. 2001. Dual level inhibition of E2F-1 activity by adeno-associated virus Rep78. J. Biol. Chem. 276:24315-24322. [DOI] [PubMed] [Google Scholar]
- 5.Buller, R. M., J. E. Janik, E. D. Sebring, and J. A. Rose. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, B. J., C. A. Laughlin, L. M. de la Maza, and M. Myers. 1979. Adeno-associated virus autointerference. Virology 92:449-462. [DOI] [PubMed] [Google Scholar]
- 7.Casto, B. C., J. A. Armstrong, R. W. Atchison, and W. M. Hammon. 1967. Studies on the relationship between adeno-associated virus type 1 (AAV-1) and adenoviruses. II. Inhibition of adenovirus plaques by AAV; its nature and specificity. Virology 33:452-458. [DOI] [PubMed] [Google Scholar]
- 8.Chang, L. S., and T. Shenk. 1990. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J. Virol. 64:2103-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, L. S., Y. Shi, and T. Shenk. 1989. Adeno-associated virus P5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J. Virol. 63:3479-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chejanovsky, N., and B. J. Carter. 1989. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173:120-128. [DOI] [PubMed] [Google Scholar]
- 11.Chejanovsky, N., and B. J. Carter. 1990. Mutation of a consensus purine nucleotide binding site in the adeno-associated virus rep gene generates a dominant negative phenotype for DNA replication. J. Virol. 64:1764-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiorini, J. A., S. M. Wiener, R. A. Owens, S. R. Kyostio, R. M. Kotin, and B. Safer. 1994. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J. Virol. 68:7448-7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiorini, J. A., L. Yang, B. Safer, and R. M. Kotin. 1995. Determination of adeno-associated virus Rep68 and Rep78 binding sites by random sequence oligonucleotide selection. J. Virol. 69:7334-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collaco, R., V. Kalman-Maltese, A. D. Smith, J. D. Dignam, and J. P. Trempe. 2003. A biochemical characterization of the adeno-associated virus Rep40 helicase. J. Biol. Chem. 278:34011-34017. [DOI] [PubMed] [Google Scholar]
- 15.Di Pasquale, G., and J. A. Chiorini. 2003. PKA/PrKX activity is a modulator of AAV/adenovirus interaction. EMBO J. 22:1716-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher, K. J., W. M. Kelley, J. F. Burda, and J. M. Wilson. 1996. A novel adenovirus-adeno-associated virus hybrid vector that displays efficient rescue and delivery of the AAV genome. Hum. Gene Ther. 7:2079-2087. [DOI] [PubMed] [Google Scholar]
- 18.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31:177-212. [DOI] [PubMed] [Google Scholar]
- 19.Galas, D. J., and A. Schmitz. 1978. DNase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5:3157-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, S. I., M.-A. Kawano, K.-I. Ishizu, H. Watanabe, M. Hasegawa, S.-N. Kanesashi, Y.-S. Kim, A. Nakanishi, K. Kataoka, and H. Handa. 2004. Rep68 protein of adeno-associated virus type 2 interacts with 14-3-3 proteins depending on phosphorylation at serine 535. Virology 320:144-155. [DOI] [PubMed] [Google Scholar]
- 21.Hermanns, J., A. Schulze, P. Jansen-Db1urr, J. A. Kleinschmidt, R. Schmidt, and H. zur Hausen. 1997. Infection of primary cells by adeno-associated virus type 2 results in a modulation of cell cycle-regulating proteins. J. Virol. 71:6020-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermonat, P. L. 1994. Down-regulation of the human c-fos and c-myc proto-oncogene promoters by adeno-associated virus Rep78. Cancer Lett. 81:129-136. [DOI] [PubMed] [Google Scholar]
- 23.Hermonat, P. L. 1992. Inhibition of bovine papillomavirus plasmid DNA replication by adeno-associated virus. Virology 189:329-333. [DOI] [PubMed] [Google Scholar]
- 24.Hermonat, P. L. 1991. Inhibition of H-ras expression by the adeno-associated virus Rep78 transformation suppressor gene product. Cancer Res. 51:3373-3377. [PubMed] [Google Scholar]
- 25.Hermonat, P. L., A. D. Santin, R. B. Batchu, and D. Zhan. 1998. The adeno-associated virus Rep78 major regulatory protein binds the cellular TATA-binding protein in vitro and in vivo. Virology 245:120-127. [DOI] [PubMed] [Google Scholar]
- 26.Im, D. S., and N. Muzyczka. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 27.Janik, J. E., M. M. Huston, and J. A. Rose. 1981. Locations of adenovirus genes required for the replication of adenovirus-associated virus. Proc. Natl. Acad. Sci. USA 78:1925-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jing, X. J., V. Kalman-Maltese, X. Cao, Q. Yang, and J. P. Trempe. 2001. Inhibition of adenovirus cytotoxicity, replication, and E2a gene expression by adeno-associated virus. Virology 291:140-151. [DOI] [PubMed] [Google Scholar]
- 29.Khleif, S. N., T. Myers, B. J. Carter, and J. P. Trempe. 1991. Inhibition of cellular transformation by the adeno-associated virus rep gene. Virology 181:738-741. [DOI] [PubMed] [Google Scholar]
- 30.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krady, J. K., and D. C. Ward. 1995. Transcriptional activation by the parvoviral nonstructural protein NS-1 is mediated via a direct interaction with Sp1. Mol. Cell. Biol. 15:524-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyostio, S. R., R. A. Owens, M. D. Weitzman, B. A. Antoni, N. Chejanovsky, and B. J. Carter. 1994. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. J. Virol. 68:2947-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyostio, S. R., R. S. Wonderling, and R. A. Owens. 1995. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J. Virol. 69:6787-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lackner, D. F., and N. Muzyczka. 2002. Studies of the mechanism of transactivation of the adeno-associated virus p19 promoter by Rep protein. J. Virol. 76:8225-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laughlin, C. A., N. Jones, and B. J. Carter. 1982. Effect of deletions in adenovirus early region 1 genes upon replication of adeno-associated virus. J. Virol. 41:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarty, D. M., D. Pereira, I. Zolotukhin, X. Zhou, J. Ryan, and N. Muzyczka. 1994. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J. Virol. 68:4988-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGhee, J. D., and P. H. von Hippel. 1975. Formaldehyde as a probe of DNA structure. I. Reaction with exocyclic amino groups of DNA bases. Biochemistry 14:1281-1296. [DOI] [PubMed] [Google Scholar]
- 38.McGhee, J. D., and P. H. von Hippel. 1975. Formaldehyde as a probe of DNA structure. II. Reaction with endocyclic imino groups of DNA bases. Biochemistry 14:1297-1303. [DOI] [PubMed] [Google Scholar]
- 39.McGrory, W. J., D. S. Bautista, and F. L. Graham. 1988. A simple technique for the rescue of early region I mutations into infectious human adenovirus type 5. Virology 163:614-617. [DOI] [PubMed] [Google Scholar]
- 40.McPherson, R. A., L. J. Rosenthal, and J. A. Rose. 1985. Human cytomegalovirus completely helps adeno-associated virus replication. Virology 147:217-222. [DOI] [PubMed] [Google Scholar]
- 41.Meyers, C., M. Mane, N. Kokorina, S. Alam, and P. L. Hermonat. 2000. Ubiquitous human adeno-associated virus type 2 autonomously replicates in differentiating keratinocytes of a normal skin model. Virology 272:338-346. [DOI] [PubMed] [Google Scholar]
- 42.Mittal, S. K., A. J. Bett, L. Prevec, and F. L. Graham. 1995. Foreign gene expression by human adenovirus type 5-based vectors studied using firefly luciferase and bacterial beta-galactosidase genes as reporters. Virology 210:226-230. [DOI] [PubMed] [Google Scholar]
- 43.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2359. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa. [Google Scholar]
- 44.Nada, S., and J. P. Trempe. 2002. Characterization of adeno-associated virus rep protein inhibition of adenovirus E2a gene expression. Virology 293:345-355. [DOI] [PubMed] [Google Scholar]
- 45.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 46.Ostrove, J. M., D. H. Duckworth, and K. I. Berns. 1981. Inhibition of adenovirus-transformed cell oncogenicity by adeno-associated virus. Virology 113:521-533. [DOI] [PubMed] [Google Scholar]
- 47.Owens, R. A., J. P. Trempe, N. Chejanovsky, and B. J. Carter. 1991. Adeno-associated virus rep proteins produced in insect and mammalian expression systems: wild-type and dominant-negative mutant proteins bind to the viral replication origin. Virology 184:14-22. [DOI] [PubMed] [Google Scholar]
- 48.Pereira, D. J., D. M. McCarty, and N. Muzyczka. 1997. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J. Virol. 71:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pereira, D. J., and N. Muzyczka. 1997. The cellular transcription factor SP1 and an unknown cellular protein are required to mediate Rep protein activation of the adeno-associated virus p19 promoter. J. Virol. 71:1747-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prasad, C. K., C. Meyers, D. J. Zhan, H. You, M. Chiriva-Internati, J. L. Mehta, Y. Liu, and P. L. Hermonat. 2003. The adeno-associated virus major regulatory protein Rep78-c-Jun-DNA motif complex modulates AP-1 activity. Virology 314:423-431. [DOI] [PubMed] [Google Scholar]
- 51.Prasad, K. M. R., C. Zhou, and J. P. Trempe. 1997. Characterization of the Rep78/adeno-associated virus complex. Virology 229:183-192. [DOI] [PubMed] [Google Scholar]
- 52.Richardson, W. D., and H. Westphal. 1981. A cascade of adenovirus early functions is required for expression of adeno-associated virus. Cell 27:133-141. [DOI] [PubMed] [Google Scholar]
- 53.Rittner, K., R. Heilbronn, J. A. Kleinschmidt, and G. Sczakiel. 1992. Adeno-associated virus type 2-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) replication: involvement of p78rep/p68rep and the HIV-1 long terminal repeat. J. Gen. Virol. 73(Pt. 11):2977-2981. [DOI] [PubMed] [Google Scholar]
- 54.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971-3984. [DOI] [PubMed] [Google Scholar]
- 55.Smith, R. H., and R. M. Kotin. 2000. An adeno-associated virus (AAV) initiator protein, Rep78, catalyzes the cleavage and ligation of single-stranded AAV ori DNA. J. Virol. 74:3122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Snyder, R. O., D. S. Im, T. Ni, X. Xiao, R. J. Samulski, and N. Muzyczka. 1993. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J. Virol. 67:6096-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su, P. F., S. Y. Chiang, C. W. Wu, and F. Y. Wu. 2000. Adeno-associated virus major Rep78 protein disrupts binding of TATA-binding protein to the p97 promoter of human papillomavirus type 16. J. Virol. 74:2459-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomson, B. J., F. W. Weindler, D. Gray, V. Schwaab, and R. Heilbronn. 1994. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology 204:304-311. [DOI] [PubMed] [Google Scholar]
- 59.Tollefson, A. E., J. S. Ryerse, A. Scaria, T. W. Hermiston, and W. S. Wold. 1996. The E3-11.6-kDa adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology 220:152-162. [DOI] [PubMed] [Google Scholar]
- 60.Tollefson, A. E., A. Scaria, T. W. Hermiston, J. S. Ryerse, L. J. Wold, and W. S. Wold. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 70:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trempe, J. P., E. Mendelson, and B. J. Carter. 1987. Characterization of adeno-associated virus Rep proteins in human cells by antibodies raised against Rep expressed in Escherichia coli. Virology 161:18-28. [DOI] [PubMed] [Google Scholar]
- 62.Walz, C., and J. R. Schlehofer. 1992. Modification of some biological properties of HeLa cells containing adeno-associated virus DNA integrated into chromosome 17. J. Virol. 66:2990-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weger, S., E. Hammer, and R. Heilbronn. 2002. Topors, a p53 and topoisomerase I binding protein, interacts with the adeno-associated virus (AAV-2) Rep78/68 proteins and enhances AAV-2 gene expression. J. Gen. Virol. 83:511-516. [DOI] [PubMed] [Google Scholar]
- 64.Weger, S., M. Wendland, J. A. Kleinschmidt, and R. Heilbronn. 1999. The adeno-associated virus type 2 regulatory proteins rep78 and rep68 interact with the transcriptional coactivator PC4. J. Virol. 73:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weitzman, M. D., K. J. Fisher, and J. M. Wilson. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winters, W. D., and W. C. Russell. 1971. Studies on the assembly of adenovirus in vitro. J. Gen. Virol. 10:181-194. [DOI] [PubMed] [Google Scholar]
- 67.Wonderling, R. S., and R. A. Owens. 1997. Binding sites for adeno-associated virus rep proteins within the human genome. J. Virol. 71:2528-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yakobson, B., T. A. Hrynko, M. J. Peak, and E. Winocour. 1989. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J. Virol. 63:1023-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yakobson, B., T. Koch, and E. Winocour. 1987. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J. Virol. 61:972-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang, Q., and J. P. Trempe. 1993. Analysis of the terminal repeat binding abilities of mutant adeno-associated virus replication proteins. J. Virol. 67:4442-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young, S. M., D. M. McCarty, N. Degtyareva, and R. J. Samulski. 2000. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 74:3953-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhan, D., A. D. Santin, Y. Liu, G. P. Parham, C. Li, C. Meyers, and P. L. Hermonat. 1999. Binding of the human papillomavirus type 16 p97 promoter by the adeno-associated virus Rep78 major regulatory protein correlates with inhibition. J. Biol. Chem. 274:31619-31624. [DOI] [PubMed] [Google Scholar]