Abstract
Both beta- and gammaherpesviruses encode G protein-coupled receptors (GPCRs) with unique pharmacological phenotypes and important biological functions. An example is ORF74, the γ2-herpesvirus Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded GPCR, which is highly constitutively active and considered the key oncogene in Kaposi's sarcoma pathogenesis. In contrast, the current annotation of the Epstein-Barr virus (EBV) genome does not reveal any GPCR homolog encoded by this human oncogenic γ1-herpesvirus. However, by employing bioinformatics, we recognized that the previously established EBV open reading frame BILF1 indeed encodes a GPCR. Additionally, BILF1 is a member of a new family of related GPCRs exclusively encoded by γ1-herpesviruses. Expression of hemagglutinin-tagged BILF1 in the HEK293 epithelial cell line revealed that BILF1 is expressed as an approximately 50-kDa glycosylated protein. Immunocytochemistry and confocal microscopy revealed that BILF1 localizes predominantly to the plasma membrane, similar to the localization of KSHV ORF74. Using chimeric G proteins, we found that human and rhesus EBV-encoded BILF1 are highly potent constitutively active receptors, activating Gαi. Furthermore, BILF1 is able to inhibit forskolin-triggered CREB activation via stimulation of endogenous G proteins in a pertussis toxin-sensitive manner, verifying that BILF1 signals constitutively through Gαi. We suggest that EBV may use BILF1 to regulate Gαi-activated pathways during viral lytic replication, thereby affecting disease progression.
Many beta- and gammaherpesviruses have acquired G protein-coupled receptors (GPCRs), some of which are functional chemokine receptors (30). While all members of the betaherpesvirus family, e.g., cytomegalovirus (CMV), encode GPCR homologs, it is the general notion that γ1-herpesviruses, e.g., Epstein-Barr virus (EBV), unlike γ2-herpesviruses, e.g., Kaposi's sarcoma-associated herpesvirus (KSHV), do not encode GPCR homologs (46). Most virus-encoded GPCRs, including UL33, M33, R33, UL78, M78, R78, US27, and US28 (6, 7, 10, 18, 41, 42, 60), are dispensable for viral growth in tissue culture. However, several in vivo studies have revealed that viral GPCRs are highly significant for viral replication and for virus-induced pathogenesis in the natural hosts (6, 7, 18, 42, 49, 50). For example, M33-deficient murine CMV does not disseminate to the salivary glands of infected mice (18), R33-deficient rat CMV is less virulent for immunocompromised rats than wild-type virus, and in parallel to M33 mutant murine CMV, R33 mutant rat CMV does not replicate efficiently in the salivary glands of infected rats (7). Furthermore, R78-deficient rat CMV is less virulent than wild-type rat CMV (6), and M78 deletion mutant murine CMV replicates and/or disseminates to a lower level in the salivary glands, spleen, and liver in both immunocompetent and immunocompromised mice (42).
γ2-Herpesvirus-encoded GPCRs also have important functions. Murine gammaherpesvirus ORF74 knockout virus suffers from decreased efficiency of reactivation from latency both in vitro and in vivo compared to wild-type virus (32, 39). This is surprising because ORF74 is regarded as an early lytic gene and is not expressed in the vast majority of otherwise latently infected cells (14, 29). This suggests that lytic replication can transform latently infected cells, presumably through paracrine mechanisms. The activities and biological effects of the KSHV-encoded chemokine receptor ORF74 are well characterized. ORF74 is highly constitutively active (1, 3, 22, 48) and mediates its signals through several different Gα proteins and by activating βγ subunits. As a result, ORF74 triggers several of the major signaling transduction pathways, including the phospholipase C and protein kinase C pathways (1, 48, 54), the phosphoinositol-3′-kinase-AKT/protein kinase B pathway (38, 54), and the mitogen-activated protein kinase pathways JNK, p38 (3), and p44/p42 MAPK (52, 54). Consequently, ORF74 induces various growth factors and angiogenic and proinflammatory cytokines (38, 44, 45, 52, 53). Through activation of vascular endothelial growth factor, ORF74 stimulates the proliferation of transfected cells (1, 3) and induce angiogenesis of human umbilical vein endothelial cells (3, 57). Injection of ORF74-expressing mouse fibroblasts into the flank of nude mice causes vascularized tumors (3), and most significantly, transgenic mice expressing ORF74 ubiquitously or within hematopoietic or endothelial cells develop Kaposi's sarcoma-like lesions in multiple organs (24, 26, 37, 63). Therefore, ORF74 is considered the key viral oncogene in Kaposi's sarcoma pathogenesis.
In this paper we describe that the EBV-encoded open reading frame (ORF) BILF1 encodes a functional GPCR. The EBV genome was sequenced and annotated 20 years ago (2). The EBV annotation is based on BamHI fragments, and the BILF1 ORF is localized to the small BamHI fragment I. At least two lytic transcripts, a 1.1-kb and a 1.4-kb transcript, are known to be expressed from this region (27). We discovered that BILF1 contained several hallmarks of GPCRs, including seven hydrophobic transmembrane domains, conserved cysteines in the amino terminus and in the extracellular loops, amino-terminal glycosylation sites, and intracellular phosphorylation sites. We therefore set out to test whether BILF1 was a functional GPCR. We show that BILF1 is expressed as a heavily glycosylated membrane protein and that BILF1 is a highly potent GPCR, constitutively signaling through Gαi. Given the importance of ORF74-mediated activity for γ2-herpesvirus replication and for the development of Kaposi's sarcoma, it is intriguing that the oncogenic γ1-herpesviruses also encode constitutively active GPCRs. Because of this new GPCR similarity between the two human oncogenic gammaherpesviruses, further research into the activities and functions of BILF1 is important. Not only will it result in a better understanding of the significance of lytic gene products for gammaherpesvirus replication and reactivation, it may also give clues to why both γ1- and γ2-herpesviruses express GPCRs with similar pharmacological characteristics during lytic replication.
MATERIALS AND METHODS
Bioinformatics.
The BLAST program available at http://www.ncbi.nlm.nih.gov/BLAST/ was used to identify EBV-encoded sequences with similarities to known GPCRs. The program TMHMM was used to predict transmembrane helices, NetNGlyc and NetOGlyc were used to predict N-linked and O-GalNAc glycosylation sites, respectively, and NetPhos was used to predict phosphorylation sites. All prediction programs are available at http://www.cbs.dtu.dk/services/.
Cell culture, reagents, and plasmids.
Human EBV DNA (strain B95.8) and rhesus monkey EBV DNA were kindly provided by Fred Wang, Harvard University, Cambridge, Mass. The glucose-dependent insulinotropic peptide (GIP) receptor, the κ-opiate receptor, the wild-type Gαq expression construct, GIP, and dynorphin were kindly provided by Christian Elling, 7TM-pharma, Hoersholm, Denmark. The recombinant G proteins Gαqs5 and GαΔ6qi4myr were kindly provided by Evi Kostenis, 7TM-pharma, Hoersholm, Denmark. Gαqs5 and GαΔ6qi4myr have switched receptor specificity from wild-type Gαq-interacting receptors to Gαs- and Gαi-interacting receptors, respectively; Gαqs5 has replaced the five C-terminal amino acids of wild-type Gαq with the five corresponding C-terminal amino acids from Gαs, and GαΔ6qi4myr lacks the first six N-terminal amino acids, replacing the four C-terminal amino acids of wild-type Gαq with the corresponding four C-terminal amino acids from Gαi, and including an N-terminal myristoylation site (15, 31, 51).
The T-REx-293 cell line (Invitrogen R710-07) was grown in Dulbecco's modified Eagle's medium (Invitrogen 31966-021) plus 10% fetal bovine serum and penicillin-streptomycin (Invitrogen 15140-022) with the addition of 15 μg of blasticidin S (Invitrogen R210-01) per ml and 100 μg of zeocin (Invitrogen R250-01) per ml or 150 μg of hygromycin B (Invitrogen 10687-010) per ml at 37°C and 5% CO2. COS-7 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen 21885-025), 10% fetal bovine serum, and penicillin-streptomycin at 37°C and 10% CO2.
Receptor cloning.
Cloning of human and rhesus EBV BILF1 and receptor hemagglutinin tagging was done with standard PCR techniques with PFU polymerase (Stratagene) with, for human EBV BILF1, forward primer 5′-TAGAAGCTTATGTACCCATACGACGTACCAGACTACGCACTCTCCACCATGGCCCCC and reverse primer 5′-TAGCTCGAGTCAGGTGGACTGGCTAGGC and, for rhesus EBV BILF1, forward primer 5′-TAGAAGCTTATGTACCCATACGACGTACCAGACTACGCACTCTCCACCCTGGCCCC and reverse primer 5′-TAGCTCGAGTCAGGTGGACTGGGTGGAC). Boldface type indicates the start codon for methionine; italic type indicates the part of the primer that overlaps the template gene. Human and rhesus EBV BILF1 were inserted into pcDNA5/FRT/TO (Invitrogen V6520-20) by cohesive end ligation. The resulting human and rhesus EBV BILF1 constructs (BILF1-pcDNA5/FRT/TO) are hereafter called HuBILF1 and RhBILF1, respectively. The constructs also contain the hygromycin resistance gene with an flp recombination target (FRT) site embedded in the 5′ coding region. The hygromycin resistance gene lacks a promoter and the ATG initiation codon
Generation of stable, BILF1-inducible cell lines.
Stable, tetracycline-inducible BILF1 cell lines were generated with the Flp-In T-Rex Core kit (Invitrogen K6500-01) by flp recombinase-mediated integration. The 293 T-REx cell line contains one integrated FRT site originally encoded by the pFRT/lacZeo vector (Invitrogen V6015-20) and expresses the tetracycline repressor originally encoded by the pcDNA6/TR vector (Invitrogen V1025-01). The FRT site is maintained by selection for zeocin resistance, and the tetracycline repressor is maintained by selection for blasticidin resistance. The integrated FRT site is contained just downstream of the ATG initiation codon of the lacZ-zeo fusion gene. Briefly, T-REx-293 cells were transfected with HuBILF1 or RhBILF1 and pOG44 (Invitrogen V6005-20) for transient expression of the Flp recombinase with the Fugene-6 transfection reagent (Roche 1814433) according to the manufacturer's protocol. Upon cotransfection, the Flp recombinase mediates homologous recombination between the FRT site, so that BILF1 is inserted into the genome at the already integrated FRT site. This insertion brings the simian virus 40 promoter and the ATG initiation codon (from the lacZ-zeo fusion gene) into proximity and in frame with the ATG minus hygromycin resistance gene, and at the same time inactivates the lacZ-zeo fusion gene. Stable BILF1 clones were selected for blasticidin and hygromycin resistance and controlled for zeocin sensitivity and screened for the lack of β-galactosidase activity. The resulting HuBILF1 and RhBILF1 cell lines therefore contain the BILF1 ORF under the control of a tetracycline-regulated hybrid CMV/tetO2 promoter. The CMV/tetO2 promoter is inactive in cells expressing the tetracycline repressor (T-REx-293 cell line), where it is activated by the addition of tetracycline. It should be noted that the CMV/tetO2 promoter is fully active in cells not expressing the tetracycline repressor.
Immunoblotting and receptor deglycosylation.
For receptor studies, cells were stimulated with tetracycline (Invitrogen Q100-19) to induce receptor expression. For Western blotting, cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate, 0.1% sodium dodecyl sulfate with the addition of protease inhibitors [phenylmethylsulfonyl fluoride, aprotinin, leupeptin, and pepstatin] and phosphatase inhibitors [activated Na3VO4 and NaF]). For receptor glycosylation studies, cells were lysed in 0.1 M Tris-HCl-10 mM EDTA-1% Triton X-114-1 mM phenylmethylsulfonyl fluoride. Triton X-114 is soluble in aqueous solutions at 0°C but not at 37°C. This property makes is suitable for separating membrane fractions (and the embedded receptors) from water-soluble proteins. Briefly, cells were lysed on ice for 15 min and centrifuged for 30 min at 5,500 rpm at 4°C. The supernatant was incubated at 37°C until unclear and spun for 10 min at 4,000 rpm at room temperature to separate the detergent phase from the aqueous phase (upper phase). The detergent phase was supplemented with 0.1 M Tris-HCl (pH 8.1) to starting volume and incubated on ice until clarified, and the phase separation was repeated. The detergent phase was again supplemented with 0.1 M Tris-HCl (pH 8.1) to starting volume and clarified with 2.5 μl of 10% CHAPS. The supernatant detergent phase was separated from the aqueous phase by centrifugation for 15 min at 4,000 rpm at 4°C. Receptor deglycosylation was done with glycopeptidase F (Sigma G5166) according to the manufacturer's protocol, and proteins were analyzed by Western blotting with antihemagglutinin antibody (Biosite HA.11). For detection of the internal control, glyceraldehyde-3-phosphate dehydrogenase, blots were stripped in Restore Western blot stripping buffer (Pierce 21059) for 20 min at room temperature and analyzed with mouse anti-rabbit glyceraldehyde-3-phosphate dehydrogenase antibody (Research Diagnostics RDI-TRK5G4-6C5).
Immunocytochemistry and confocal microscopy.
For immune staining, HuBILF1 and RhBILF1 cell lines were grown on CC2-treated Lab-Tek chamber glass slides (Nalge Nunc International 154917). One day after seeding into the chamber glass slides, the cells were transfected with CXCR4-GFP20, US28-GFP20 or ORF74-GFP35 with Fugene 6 transfection reagent (Roche 1 814 443) according to the manufacturer's recommendations. The following day, cells were stimulated with tetracycline (0.1 μg/ml); 24 h after tetracycline stimulation, cells were fixed in 3.7% formaldehyde, washed in phosphate-buffered saline, and permeabilized in 0.2% Triton X-100 for 20 min on ice. Cells were blocked in phosphate-buffered saline containing 1% bovine serum albumin and 5% goat serum for 30 min at room temperature and incubated with antihemagglutinin antibody (Biosite HA.11) for 1 h at room temperature. Cells were washed and incubated with Alexa Fluor 594-conjugated goat anti-mouse immunoglobulin antibody (Molecular Probes A-11005) for 1 h at room temperature. Cells were mounted with Fluoromount-G (Southern Biotechnology Associates 0100-01). Confocal fluorescence microscopy was done on an inverse Zeiss Axiovert 200 fluorescence microscope equipped with a Coolsnap charge-coupled device camera. Digital images were transferred to Adobe Photoshop and used without further processing.
Inositol phosphate production.
For inositol phosphate turnover, COS-7 cells were transfected with the calcium phosphate precipitation method (48). Briefly, 20 μl of 2 M CaCl2, DNA, and Tris-EDTA buffer to a total volume of 160 μl were mixed and added dropwise to 160 μl of 2× HEPES-buffered saline buffer. After 45 min of incubation, the mixture was added to the cells and incubated with the addition of 100 μM chloroquine for 5 h. One day after transfection, the cells were transferred to six-well plates (5 × 105 cells/well) and incubated for 24 h with 4 μCi of myo-[3H]inositol (Amersham TRK911) in 0.8 ml of complete medium/well. Cells were washed twice in immunoprecipitation buffer (20 mM HEPES, pH 7.4, supplemented with 140 mM NaCl, 5 mM KCl, 1 mM MgSO4, 1 mM CaCl2, 10 mM glucose, and 0.05% bovine serum albumin) and incubated in 1.0 ml of immunoprecipitation buffer supplemented with 10 mM LiCl at 37°C for 90 min. GIP and dynorphin were added to a final concentration of 10−7 M after 15 min of incubation. After incubation, the buffer was removed, and accumulated inositol phosphates were extracted for 30 min on ice in 1.0 ml of 10 mM formic acid. The generated [3H]inositol phosphates were purified on Dowex 1X8 anion-exchange resin, and radioactivity was counted in a scintillation counter (48). Radioactivity values are given as counts per minute.
CREB reporter assay.
For luciferase reporter assays, COS-7 cells (35,000 cells/well) were seeded in 96-well plates and transfected with a reporter-cDNA cocktail consisting of 6 ng of pFA2-CREB and 50 ng of pFR-Luc reporter plasmids with the PathDetect cyclic AMP response element binding protein (CREB) system (Stratagene) and various amounts of receptor and G protein DNA with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Following transfections, cells were treated with 10 μM forskolin or 10 μM forskolin plus 0.1 μg of pertussis toxin per ml in an assay volume of 100 μl for 24 h. The assay was terminated by washing the cells twice with phosphate-buffered saline and adding 100 μl of phosphate-buffered saline supplemented with 1 mM MgCl2 and CaCl2 and 100 μl of luciferase assay reagent (LucLite; Packard). Luminescence was measured in a TopCounter (Top Count NXT; Packard) for 5 s. Luminescence values are given as relative light units.
RESULTS
EBV encodes a GPCR homolog.
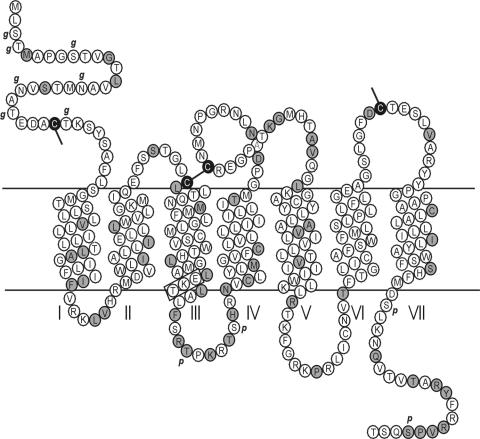
Initially, BLAST analysis revealed that the equine herpesvirus 2-encoded GPCR homolog, ORF E6, contained limited sequence homology (17% amino acid identity, PAM250 similarity matrix) to EBV ORF BILF1. Transmembrane helix analysis clearly demonstrated that BILF1 contains seven hydrophobic helices (Fig. 1), a hallmark of all GPCRs. In addition, BILF1 has several additional characteristics of GPCRs. These include conserved cysteines in the amino-terminal end and in the extracellular loops (Fig. 1), which are known to form structurally and functionally important disulfide bonds in GPCRs (19). Furthermore, BILF1 is predicted to contain seven N-terminal glycosylation sites, which are important for GPCR-ligand interactions, receptor expression, and cellular localization (33), as well as four intracellular phosphorylation sites (Fig. 1), which are important for GPCR-mediated signaling, receptor regulation, and intracellular targeting (36). Interestingly, the DRY (aspartic acid, arginine, tyrosine) motif at the intracellular end of TM-III, which is conserved in most rhodopsin-like GPCRs, is replaced with an EKT (glutamic acid, lysine, threonine) motif in BILF1. However, this conservative amino acid substitution (same order of acidic, basic, and polar side chains) can be considered an alternative DRY motif (Fig. 1 and 2). It should be noted that alternative DRY box motifs are present in numerous viral GPCRs.
FIG. 1.
Serpentine diagram of the human EBV BILF1 receptor, showing the seven transmembrane helices. Differences between human EBV and rhesus EBV are indicated in black on grey, and identical amino acids are indicated in black on white. Computer-predicted glycosylation sites are indicated with g, and predicted phosphorylation sites are indicated with p. The alternative DRY box, EKT, is marked with a rectangle.
FIG. 2.
Multiple sequence alignment of BILF1-related sequences identified with BLAST. The alignment was done with ClustalW version 1.8, and shading was done with Boxshade version 3.21, available at http://www.ch.embnet.org/software/BOX_form.html. Abbreviations used in this figure: PLHV, porcine lymphotropic herpesvirus; CHV3, callitrichine herpesvirus 3 (also known as marmoset EBV); AHV, alcelaphine herpesvirus; EHV, equine herpesvirus. The rectangle indicates the DRY box. Positions sharing 50% amino acid identity or more are indicated in white on black. Positions sharing 50% amino acid homology or more are indicated in black on gray. Positions with less than 50% amino acid identity or homology are indicated in black on white. The consensus line asterisk indicates 100% conserved positions, and dots indicate positions with 50% or better homology or identity.
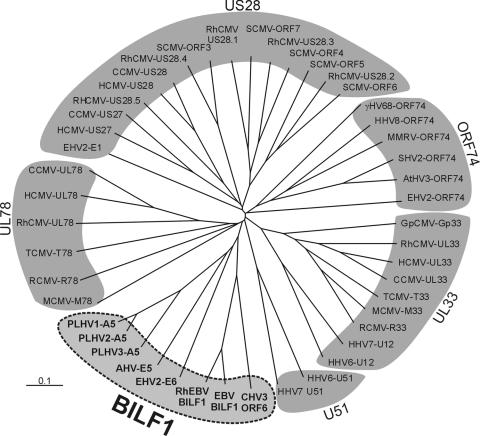
BILF1 belongs to a novel receptor subfamily.
Further BLAST analysis revealed that BILF1 is encoded not only by human EBV but also by other γ1-herpesviruses. Together, this group of BILF1-related sequences constitute a new family of related GPCRs, exclusively encoded by and a trait for γ1-herpesviruses (Fig. 2). Indeed, BILF1 is one of just six genes present only in γ1-herpesviruses (47). This genomic evidence suggests that BILF1 plays an important role in the life cycle of γ1-herpesviruses. Multiple sequence alignment of eight BILF1-related sequences with a wide range of viral GPCRs belonging to the US28, UL33, UL78, UL51, and ORF74 subfamilies revealed that all BILF1 sequences group together and constitute a separate viral GPCR subfamily (Fig. 3)
FIG. 3.
Dendrogram of herpesvirus-encoded G protein-coupled receptors based on their amino acid identities. Receptor alignment was generated in Clustal X (1.81), and the phylogenetic tree was visualized with TreeView. The length of each branch reflects amino acid discrepancies. Each entry consists of the abbreviated virus name followed by the name of the reading frame encoding the receptor. The BILF1 receptor family is highlighted in bold and consists of porcine lymphotrophic herpesvirus (PLHV) types 1, 2 and 3 ORF A5, alcelaphine herpesvirus 1 (AHV) ORF E5, equine herpesvirus 2 (EHV2) ORF E6, rhesus Epstein-Barr virus (RhEBV) ORF BILF1, human Epstein-Barr virus (EBV) ORF BILF1, and callitrichine herpesvirus 3 (CHV3) ORF 6.
Cloning and generation of cell lines.
We cloned human EBV and rhesus EBV BILF1 and inserted the receptor reading frames into pCDNA5/FRT/TO, generating HuBILF1 and RhBILF1, respectively. The BILF1 sequences were confirmed by sequence analysis and corresponded to nucleotides 152161 to 153099 in the human herpesvirus 4 complete genome (GenBank accession no. NC_001345) and to nucleotides 147746 to 148684 in the rhesus EBV (cercopithicine herpesvirus 15) complete genome (GenBank accession no. AY037858). We generated several clonal cell lines of both HuBILF1 and RhBILF1. For our expression studies, we selected clones with the lowest BILF1 background expression that could be induced to express high levels of BILF1 after tetracycline induction. Tetracycline dose experiments showed that 0.1 μg of tetracycline/ml of medium was sufficient to induce maximal BILF1 expression 24 h poststimulation (data not shown).
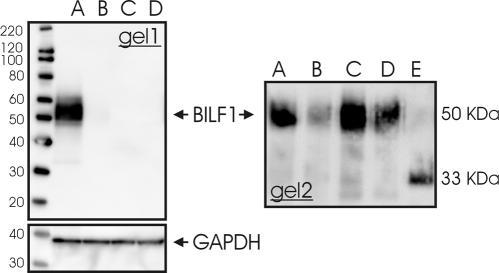
BILF1 is heavily glycosylated.
To study receptor glycosylation, membrane samples from tetracycline-induced HuBILF1-expressing cell lines were either untreated or treated with glycopeptidase F to remove N-linked glycosylation sites prior to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) separation and Western blotting. Staining with antihemagglutinin antibody showed that BILF1 is expressed as a heterogeneous approximately 50 kDa heavily glycosylated protein since glycopeptidase F treatment reduced the apparent mass to approximately 33 kDa, which is close to the calculated mass of 34.5 kDa (Fig. 4). These data support the computer-based prediction of N-terminal glycosylation sites in BILF1.
FIG. 4.
Western blot of tetracycline-induced HuBILF1 cell lines. Gel 1: Total radioimmunoprecipitation-extracted protein were boiled and reduced with 100 mM dithiothreitol prior to SDS-PAGE separation and Western blotting. A) Tetracycline-stimulated HuBILF1 cells, B) unstimulated HuBILF1 cells, C) tetracycline-stimulated parental HEK293 Flp In T-Rex cells, and D) unstimulated parental HEK293 Flp In T-Rex cells. The lower image shows the same plot after antibody stripping and restaining with an anti-glyceraldehhyde-3-phosphate dehydrogenase (GAPDH) antibody for loading control. Gel 2: Detergent phase samples were either (A) untreated, (B) boiled, (C) reduced with 100 mM dithiothreitol (DTT), or boiled and reduced (D) without or (E) with glycopeptidase F treatment prior to SDS-PAGE separation and Western blotting. Nondeglycosylated BILF1 is expressed as an approximately 50-kDa protein. Glycopeptidase F treatment reduced the apparent mass to approximately 33 kDa.
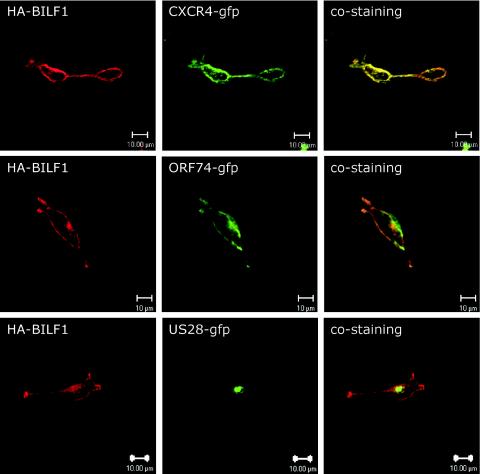
BILF1 is localized to the plasma membrane.
Tetracycline-induced HuBILF1 and RhBILF1 cell lines were stained with an antihemagglutinin antibody, and receptor localization was studied by confocal microscopy. As shown in Fig. 5, BILF1 localizes mainly to the plasma membrane. This expression pattern is concurrent with the expression pattern of most GPCRs, but is in contrast to the predominantly intracellular localization of many viral GPCRs (30). BILF1 colocalizes with the human CXC chemokine receptor CXCR4 and with the KSHV ORF74 to the cell plasma membrane, whereas BILF1 localizes differently than US28, which has a distinct intracellular accumulation. These data support our prediction that BILF1 is indeed a membrane protein and implies that our cell lines express properly folded hemagglutinin-tagged BILF1.
FIG. 5.
Cellular localization of human and rhesus EBV BILF1. Representative images of HuBILF1-expressing 293 cells cotransfected with either A) CXCR4-GFP, B) ORF74-GFP, or C) US28-GFP. Cells were fixed, permeabilized, and stained with an antihemagglutinin antibody. The images were taken with a 60X oil immersion objective. The images show that BILF1 has a distinct receptor membrane association similar to that of CXCR4 and ORF74 and different from the predominantly intracellular localization of US28.
BILF1 signals constitutively through Gαi.
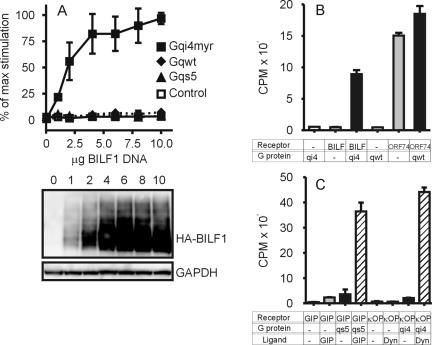
To study whether human EBV BILF1 is a functional GPCR, we tested the activation of a variety of Gα proteins. To facilitate these studies of constitutive, i.e., ligand-independent, BILF1-mediated signaling, we used chimeric Gαq proteins comprising the three major types of Gα subunits; Gαq, Gαs and Gαi. Both the wild-type and chimeric Gαq proteins Gαqs5 and GαΔ6qi4myr signal via activation of phospholipase C, leading to accumulation of [3H]inositol phosphates. COS-7 cells were transfected with an increasing amount of HuBILF1 DNA together with a constant quantity of different Gα DNA or empty expression vector DNA. The Western blot in Fig. 6 show that an increase in BILF1 DNA corresponds to an increase in BILF1 protein expression. Figure 6A shows that BILF1 did not activate phospholipase C through endogenous Gαq. As a positive control for endogenous Gαq function, we transfected COS-7 cells with KSHV ORF74, which constitutively activates Gαq. As shown in Fig. 6B, transfected COS-7 cells were fully capable of activating phospholipase C through endogenous Gαq. Cotransfection with Gαqs5 revealed that BILF1 does not signal via activation of G protein α subunits of the s class (Fig. 6A). As a positive control for Gαs function, we cotransfected COS-7 cells with Gαqs5 and the GIP receptor (which activates Gαs upon ligand engagement) and stimulated transfected cells with GIP. As shown in Fig. 6C, Gαqs5 was fully capable of activating phospholipase C after GIP-receptor activation. Finally, we investigated whether BILF1 could signal through G protein α subunits of the i class. Cotransfection with GαΔ6qi4myr revealed that BILF1 indeed signals through G protein α subunits of the i class. Figure 6A shows that increasing doses of BILF1 increase phospholipase C activity only when cotransfected with chimeric G protein with specificity towards Gαi-activating receptors. As an additional positive control for Gαi function, we cotransfected COS-7 cells with the κ-opiate receptor and GαΔ6qi4myr and stimulated transfected cells with dynorphin. As seen in Fig. 6C, GαΔ6qi4myr was also fully capable of activating phospholipase C after κ-opiate receptor activation. Importantly, none of the chimeric and endogenous G proteins were active without concurrent receptor expression (Fig. 6A, B, and C). To further test the specificity of the system and to ensure that our results were not merely a result of G protein overexpression, we cotransfected BILF1 with wild-type Gαq. Transfection with Gαq did not result in an increased basic level of phospholipase C activity, nor did it cause any measurable BILF1 activity (Fig. 6A and B), whereas overexpression of Gαq slightly increased ORF74-mediated signaling (Fig. 6B). Overall, these data show that BILF1 is constitutively active and mediates its signal through G protein α subunits of the i class.
FIG. 6.
HuBILF1 induction of inositol phosphate accumulation. COS-7 cells were transfected with an increasing amount of HuBILF1 DNA together with 5 μg of different chimeric Gα protein DNA or empty expression vector DNA. The recombinant G proteins Gαqs5 and GαΔ6qi4myr have switched receptor specificity from wild-type Gαq interacting with receptors to Gαs- and Gαi-interacting receptors, respectively, but all signal via activation of phospholipase C, leading to accumulation of inositol phosphates (IP3). Panel A shows the activity as a percentage of maximum activity obtained. HuBILF1 plus control DNA (open squares), HuBILF1 plus wild-type Gαq DNA (solid diamonds), HuBILF1 plus Gαqs5 DNA (solid triangles), and HuBILF1 plus GαΔ6qi4myr DNA (solid squares). Data are means and standard deviations of four independent experiments, each experiment carried out in duplicate. The Western blot shows the expression levels of BILF1 in transfected COS-7 cells, with the amount of DNA given above each lane. The amount of DNA in the Western blot corresponds to the amount of DNA used in the [3H]inositol phosphate assay shown above. The lower Western blot image shows the same plot after antibody stripping and restaining with an anti-glyceraldehhyde-3-phosphate dehydrogenase (GAPDH) antibody as a loading control. To illustrate both the magnitude and specificity of the BILF1-mediated response, panels B and C show the actual counts from one representative experiment out of four. Cells were transfected with 10 μg of receptor DNA and 5 μg of G protein DNA and stimulated with ligands as indicated below the graphs. Bars illustrate means and error bars illustrate standard deviations of one experiment carried out in duplicate. BILF1 is HuBILF1, ORF74 is the KSHV-encoded GPCR, qi4 is GαΔ6qi4myr, qwt is wild-type Gαq, qs5 is Gαqs5, GIP (receptor) is the glucose-dependent insulinotropic peptide receptor, κOP (receptor) is the κ-opiate receptor, GIP (ligand) is the glucose-dependent insulinotropic peptide, and Dyn (ligand) is dynorphin. GIP and dynorphin were added to a final concentration of 10−7 M.
We further tested whether rhesus EBV-encoded BILF1 was also constitutively active and whether rhesus EBV BILF1 activated the same G protein as the human EBV BILF1. Cotransfection of RhBILF1 with the chimeric G proteins described above revealed that rhesus EBV BILF1 is also constitutively active and, like the human EBV homolog, mediates its signal through activation of Gαi. Figure 7 shows a representative example of four independent experiments. Besides revealing homologous Gα subunit activation, the RhBILF1 activity level was very similar to the activity of HuBILF1 (compare Fig. 6B and 7). Thus, both human and rhesus EBV BILF1 signal constitutively via Gαi with similar efficacies.
FIG. 7.
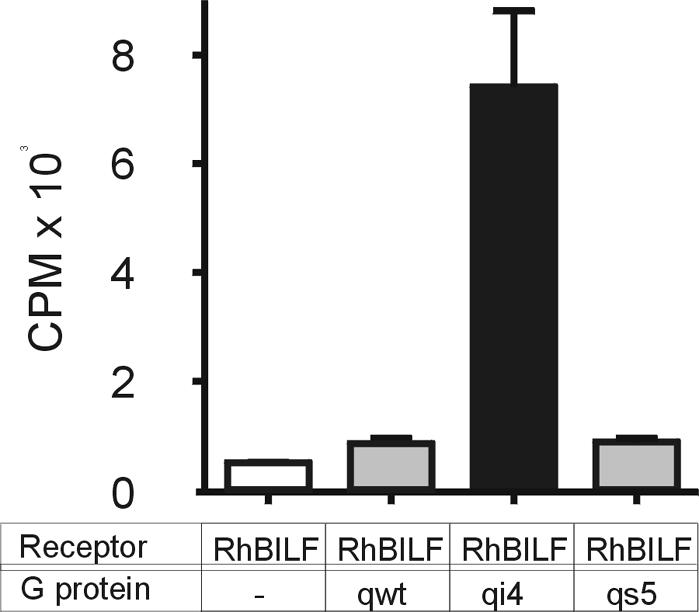
RhBILF1 induction of inositol phosphate accumulation. COS-7 cells were transfected with 6 μg of BILF1 DNA together with 5 μg of different chimeric Gα protein DNA or empty expression vector DNA as indicated below the graph. The figure shows actual counts from one representative experiment out of four. Bars illustrate means and error bars illustrate standard deviations of one experiment carried out in duplicate.
As an additional readout for BILF1-driven Gαi activity, we measured the generation of CREB-stimulated luciferase reporter activity. COS-7 cells were transfected with an increasing amount of HuBILF1 DNA together with a constant quantity of GαΔ6qi4myr DNA or empty expression vector and a reporter-cDNA cocktail consisting of 6 ng of pFA2-CREB and 50 ng of pFR-Luc reporter plasmids. Figure 8A shows that BILF1 activates CREB through GαΔ6qi4myr in a dose-dependent manner. Furthermore, it is evident that BILF1 does not activate CREB without cotransfection with GαΔ6qi4myr. This result therefore confirms the inositol phosphate data, validating that BILF1 signals constitutively through Gαi.
FIG. 8.
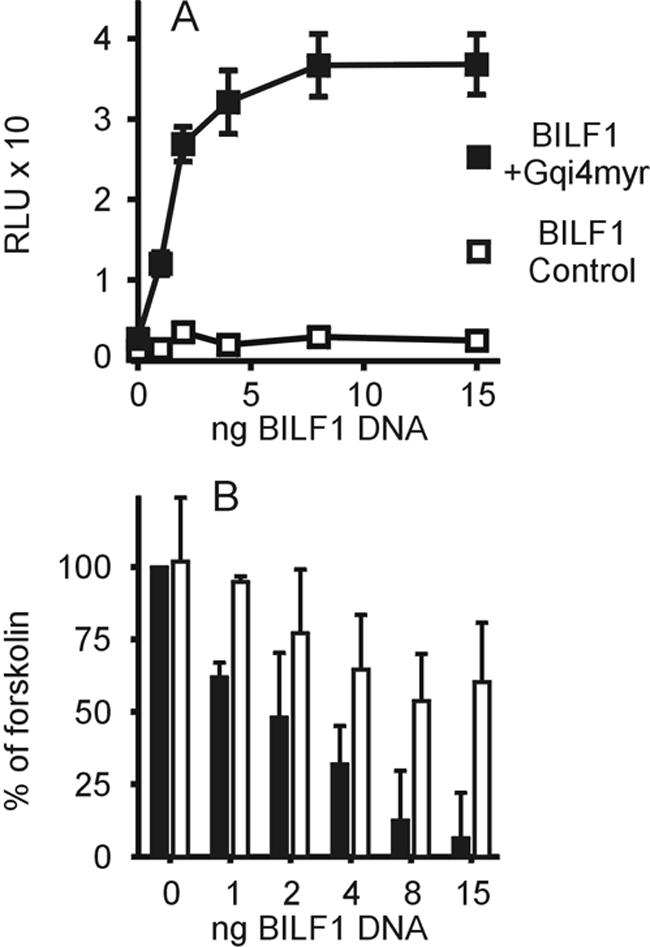
CREB-mediated transcription regulation. (A) COS-7 cells were seeded in 96-well plates and cotransfected with a reporter-cDNA cocktail consisting of 6 ng of pFA2-CREB and 50 ng of pFR-Luc reporter plasmids and increasing amounts of HuBILF1 DNA with (solid squares) or without (open squares) 30 ng of GαΔ6qi4myr DNA. Data points illustrate means and error bars illustrate standard deviations of one experiment out of four, each carried out in triplicate. Luciferase activity is presented as relative light units (RLU). (B) COS-7 cells were seeded in 96-well plates and cotransfected with a reporter-cDNA cocktail consisting of 6 ng of pFA2-CREB and 50 ng of pFR-Luc reporter plasmids and increasing amounts of receptor DNA and incubated in the presence of 10 μM forskolin to stimulate the adenylyl cyclase, with (white bars) or without (solid bars) the addition of 0.1 μg of pertussis toxin per ml to inhibit endogenous Gαi. Data are presented as a percentage of the CREB activity generated by 10 μM forskolin alone above the values obtained for unstimulated HuBILF1-transfected cells.
BILF1 activates endogenous i-class G proteins.
To study whether BILF1 could activate endogenous Gαi, we tested the ability of BILF1 to inhibit forskolin-stimulated CREB activity. COS-7 cells were transfected with increasing amounts of HuBILF1 DNA together with a reporter-cDNA cocktail consisting of 6 ng of pFA2-CREB and 50 ng of pFR-Luc reporter plasmids. After transfection, cells were stimulated with 10 μM forskolin with or without the addition of PTx. Figure 8B show that BILF1 almost completely inhibited forskolin-induced CREB activity in a dose-dependent manner. Furthermore, BILF1-mediated signaling can be inhibited by addition of PTx, a potent inhibitor of Gαi. The addition of PTx restored CREB activity to approximately 60% of maximum forskolin-stimulated CREB activity. These data confirms that BILF1 constitutively activates endogenous G proteins of the i class.
DISCUSSION
Several large DNA viruses encode viral GPCRs with unusual pharmacological and cellular properties and significant biological functions. We have identified a new subfamily of viral GPCRs, the BILF1 receptors, encoded by EBV and other γ1-herpesviruses. Our studies of human EBV BILF1 and the closely related homolog from rhesus EBV show that these receptors are functional GPCRs, constitutively signaling through Gαi. Additionally we show that BILF1 is heavily glycosylated and is localized to the plasma membrane in stably expressing epithelial cell lines. It is intriguing that EBV, a major human oncogenic herpesvirus, encodes a constitutively active GPCR, considering the significance of the KSHV-encoded GPCR, ORF74, for the development of Kaposi's sarcoma. However, even though the significance of ORF74 for Kaposi's sarcoma pathogenesis is well established, many questions regarding its significance for viral replication still remain unanswered.
One interesting function of ORF74 may be to increase the efficiency of KSHV reactivation (39). This observation is intriguing because ORF74 is regarded as an early lytic gene and is not expressed in the vast majority of otherwise latently infected cells (13), which suggests that some level of lytic replication can activate latently infected cells, presumably through paracrine mechanisms. Intriguingly, it has been proposed that dysregulation of the KSHV gene program caused by human immunodeficiency virus type 1 Tat, inflammation, or aborted lytic replication may indeed lead to nonlytic expression of ORF74 (55). Like ORF74, BILF1 is a lytic gene. BILF1 is expressed from 8 h postinduction of lytic replication and throughout infection as an approximately 1.1-kb transcript. Furthermore, BILF1 expression is dependent on viral DNA replication, since expression in the Akata B-cell line is inhibited by phosphonoacetic acid (T. Kledal, unpublished data).
Primary EBV infection is the most common cause of infectious mononucleosis, and by adulthood, nearly all humans are asymptomatic carriers of EBV. Besides causing mononucleosis, in a small percentage of individuals EBV infection is associated with endemic Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, gastric carcinoma, leiomyosarcoma, and AIDS- and transplant-associated B-cell lymphomas (25, 46). The viral and host factors implicated in the development of EBV-associated malignancies are poorly understood. The degree of immunosuppression is one risk factor for some malignancies (16, 28), but immune suppression alone is not sufficient for the development of other types of malignancies. Indeed both Burkitt's lymphoma and Hodgkin's lymphoma occur in patients without immunosuppression (46). Additionally, chromosomal abnormalities and somatic hypermutation of cellular oncogenes have been hypothesized to act in concert with EBV infection to cause malignancies (5, 43).
Like those associated with KSHV, EBV-associated malignancies are characterized by a mainly latently infected cell population, and several latently expressed genes have been shown to cause cell transformation in vitro. Nevertheless, in a population of latently EBV-infected cells, a small fraction of cells are spontaneously permissive for lytic replication (46). It is therefore not impossible that BILF1 could contribute both to control the level of reactivation and be involved in EBV tumorigenesis by acting on uninfected or latently infected cells through paracrine mechanisms.
Many viral GPCR (e.g., UL33, US27, US28, and M78) are characterized by an unusual cellular localization patterns (20, 21, 30). Detailed analysis of US28 showed that it underwent constitutive endocytosis and recycling to the plasma membrane (20, 21). It has been suggested that the constitutive endocytosis and recycling of US28 could be a mechanism for sequestering host CC chemokines, providing a sink for clearing proinflammatory CC chemokines from the tissue surrounding the CMV-infected cell (10), thereby antagonizing the recruitment of cells involved in the immune response against CMV. Furthermore, it has been speculated that the intracellular localization of many viral GPCRs could facilitate incorporation of the viral GPCRs into the maturing virion envelopes. Indeed, UL33, M78, and US27 have all been identified on viral particles (21, 34, 42). The localization of BILF1 to the plasma membrane distinguishes BILF1 from the UL33, US27, US28, and M78 viral GPCR subfamilies. However, BILF1 localizes similarly to ORF74 (52). This parallel could indicate that BILF1 and ORF74 have similar functions during gammaherpesvirus replication.
Several receptors from the US28, UL33, and ORF74 receptor subgroups have been characterized as being constitutively active, a trait of viral GPCRs. As described in the introduction, the KSHV-encoded oncogene ORF74 is highly constitutively active and responds to ligand engagement. Additionally, UL33, M33, and R33 all constitutively stimulate phospholipase C, whereas only the rodent CMV receptors M33 and R33 activate NF-κB-driven transcription (23, 62). UL33 and M33 activate CREB through activation of Gαs and the mitogen-activated protein kinase p38 (62), whereas R33 inhibits forskolin-stimulated CREB through Gαi (23). US28 also activates several different signal transduction pathways, both constitutively and ligand mediated (8, 12, 58, 59, 61, 62).
Even though a lot is known about viral GPCR pharmacology in general and about what signaling pathways that are activated, little is known about the significance of the constitutive activity for viral replication. Besides regulating downstream second-messenger systems, controling viral and host gene transcription, and directing cell morphology and motility, the constitutive activity of viral GPCRs may directly regulate the signaling mediated by other GPCRs (4). If this is indeed the case, BILF1 could function as a regulatory switch for other GPCRs expressed in EBV-infected cells. EBV also controls the transcriptional regulation of host GPCRs. Originally EBV was shown to induce the expression of two orphan GPCRs, EBV-induced (EBI) 1 and 2 (9). EBI-1 is now known as chemokine receptor CCR7, whereas EBI-2 is still an orphan. In addition, it has been shown that EBV-immortalized B cells have altered chemokine receptor expression pattern, which could be responsible for the distorted migration of infected B cells to germinal centers (40). Therefore, EBV seems to control the GPCR settings on many levels, emphasizing the importance of both endogenous GPCRs and viral GPCR for viral replication, dissemination, and immune evasion. It should be noted that BILF1 itself does not display any particular similarities to know chemokine receptors.
Since many receptors activate overlapping second-messenger systems, it is difficult to predict which viral GPCR-activated pathways are responsible for cellular transformation. It was recently shown that the Akt signaling pathway plays a central role in ORF74-mediated oncogenesis (56). Furthermore, it has been shown that ORF74 constitutively activates Akt via Gαi and phospholipase C (11, 17, 54). It is therefore likely that Gαi activity is directly involved in KSHV-mediated cell transformation, though it is unlikely to be the only signaling pathway involved. Other nontransforming viral GPCRs (e.g., R33 and US28) also activate Gαi, and ORF74 activates several additional pathways. Indeed, the fact that ORF74 stimulates a broad range of signaling pathways has been taken into account for the transforming properties of this receptor, and it has been suggested that the transforming effects of ORF74 may be mediated cooperatively by Gαq and Gαi signaling (11, 54).
In this paper we show that the EBV-encoded GPCR BILF1 constitutively activates Gαi. Given the many similarities between BILF1 and ORF74, it is attractive to speculate that BILF1 may play a role in EBV-associated cell transformation. Additionally, the selective and highly constitutive signal triggered by BILF1 suggests that BILF1 plays a significant role during the life cycle of EBV and possibly during the life cycle of all γ1-herpesviruses.
Acknowledgments
This project is supported by Danish Cancer Society grant DP-03-027 and by Danish Medical Research council grant 22-03-0181 to T. Kledal. The Danish Cancer Society and the Novo-Nordisk Foundation supported M. M. Rosenkilde. We also thank Jens Ole Nielsen, Hvidovre Hospital, for generous financial support to the project.
We thank Christian E. Elling and Evi Kostenis, 7TM-pharma, for providing essential reagents and for helpful discussions during this project. We thank Lisbeth Elbak and Inger Smith Simonsen for excellent technical assistance and Ulrik Gether lab members for technical help with confocal microscopy.
REFERENCES
- 1.Arvanitakis, L., E. Geras-Raaka, A. Varma, M. C. Gershengorn, and E. Cesarman. 1997. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature 385:347-350. [DOI] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, and C. Seguin, and. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 3.Bais, C., B. Santomasso, O. Coso, L. Arvanitakis, E. G. Raaka, J. S. Gutkind, A. S. Asch, E. Cesarman, M. C. Gershengorn, E. A. Mesri, and M. C. Gerhengorn. 1998. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86-89. [DOI] [PubMed] [Google Scholar]
- 4.Bakker, R. A., P. Casarosa, H. Timmerman, M. J. Smit, and R. Leurs. 2004. Constitutively active Gq/11-coupled receptors enable signaling by co-expressed G(i/o)-coupled receptors. J. Biol. Chem. 279:5152-5161. [DOI] [PubMed] [Google Scholar]
- 5.Ballerini, P., G. Gaidano, J. Z. Gong, V. Tassi, G. Saglio, D. M. Knowles, and R. la-Favera. 1993. Multiple genetic lesions in acquired immunodeficiency syndrome-related non-Hodgkin's lymphoma. Blood 81:166-176. [PubMed] [Google Scholar]
- 6.Beisser, P. S., G. Grauls, C. A. Bruggeman, and C. Vink. 1999. Deletion of the R78 G protein-coupled receptor gene from rat cytomegalovirus results in an attenuated, syncytium-inducing mutant strain. J. Virol. 73:7218-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beisser, P. S., C. Vink, J. G. Van Dam, G. Grauls, S. J. Vanherle, and C. A. Bruggeman. 1998. The R33 G protein-coupled receptor gene of rat cytomegalovirus plays an essential role in the pathogenesis of viral infection. J. Virol. 72:2352-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billstrom, M. A., G. L. Johnson, N. J. Avdi, and G. S. Worthen. 1998. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J. Virol. 72:5535-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birkenbach, M., K. Josefsen, R. Yalamanchili, G. Lenoir, and E. Kieff. 1993. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J. Virol. 67:2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodaghi, B., T. R. Jones, D. Zipeto, C. Vita, L. Sun, L. Laurent, F. renzana-Seisdedos, J. L. Virelizier, and S. Michelson. 1998. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J. Exp. Med. 188:855-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannon, M. L., and E. Cesarman. 2004. The KSHV G protein-coupled receptor signals via multiple pathways to induce transcription factor activation in primary effusion lymphoma cells. Oncogene 23:514-523. [DOI] [PubMed] [Google Scholar]
- 12.Casarosa, P., R. A. Bakker, D. Verzijl, M. Navis, H. Timmerman, R. Leurs, and M. J. Smit. 2001. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J. Biol. Chem. 276:1133-1137. [DOI] [PubMed] [Google Scholar]
- 13.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J. Virol. 70:8218-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiou, C. J., L. J. Poole, P. S. Kim, D. M. Ciufo, J. S. Cannon, C. M. ap Rhys, D. J. Alcendor, J. C. Zong, R. F. Ambinder, and G. S. Hayward. 2002. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3421-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conklin, B. R., Z. Farfel, K. D. Lustig, D. Julius, and H. R. Bourne. 1993. Substitution of three amino acids switches receptor specificity of Gq alpha to that of Gi alpha. Nature 363:274-276. [DOI] [PubMed] [Google Scholar]
- 16.Curtis, R. E., L. B. Travis, P. A. Rowlings, G. Socie, D. W. Kingma, P. M. Banks, E. S. Jaffe, G. E. Sale, M. M. Horowitz, R. P. Witherspoon, D. A. Shriner, D. J. Weisdorf, H. J. Kolb, K. M. Sullivan, K. A. Sobocinski, R. P. Gale, R. N. Hoover, J. F. Fraumeni, Jr., and H. J. Deeg. 1999. Risk of lymphoproliferative disorders after bone marrow transplantation: a multi-institutional study. Blood 94:2208-2216. [PubMed] [Google Scholar]
- 17.Dadke, D., B. H. Fryer, E. A. Golemis, and J. Field. 2003. Activation of p21-activated kinase 1-nuclear factor kappaB signaling by Kaposi's sarcoma-associated herpes virus G protein-coupled receptor during cellular transformation. Cancer Res. 63:8837-8847. [PubMed] [Google Scholar]
- 18.Davis-Poynter, N. J., D. M. Lynch, H. Vally, G. R. Shellam, W. D. Rawlinson, B. G. Barrell, and H. E. Farrell. 1997. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J. Virol. 71:1521-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elling, C. E., U. Raffetseder, S. M. Nielsen, and T. W. Schwartz. 2000. Disulfide bridge engineering in the tachykinin NK1 receptor. Biochemistry 39:667-675. [DOI] [PubMed] [Google Scholar]
- 20.Fraile-Ramos, A., T. N. Kledal, A. Pelchen-Matthews, K. Bowers, T. W. Schwartz, and M. Marsh. 2001. The human cytomegalovirus US28 protein is located in endocytic vesicles and undergoes constitutive endocytosis and recycling. Mol. Biol. Cell 12:1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraile-Ramos, A., A. Pelchen-Matthews, T. N. Kledal, H. Browne, T. W. Schwartz, and M. Marsh. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218-232. [DOI] [PubMed] [Google Scholar]
- 22.Geras-Raaka, E., L. Arvanitakis, C. Bais, E. Cesarman, E. A. Mesri, and M. C. Gershengorn. 1998. Inhibition of constitutive signaling of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor by protein kinases in mammalian cells in culture. J. Exp. Med. 187:801-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruijthuijsen, Y. K., P. Casarosa, S. J. Kaptein, J. L. Broers, R. Leurs, C. A. Bruggeman, M. J. Smit, and C. Vink. 2002. The rat cytomegalovirus R33-encoded G protein-coupled receptor signals in a constitutive fashion. J. Virol. 76:1328-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, H. G., M. Sadowska, W. Reid, E. Tschachler, G. Hayward, and M. Reitz. 2003. Kaposi's sarcoma-like tumors in a human herpesvirus 8 ORF74 transgenic mouse. J. Virol. 77:2631-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hjalgrim, H., J. Askling, K. Rostgaard, S. Hamilton-Dutoit, M. Frisch, J. S. Zhang, M. Madsen, N. Rosdahl, H. B. Konradsen, H. H. Storm, and M. Melbye. 2003. Characteristics of Hodgkin's lymphoma after infectious mononucleosis. N. Engl. J. Med. 349:1324-1332. [DOI] [PubMed] [Google Scholar]
- 26.Holst, P. J., M. M. Rosenkilde, D. Manfra, S. C. Chen, M. T. Wiekowski, B. Holst, F. Cifire, M. Lipp, T. W. Schwartz, and S. A. Lira. 2001. Tumorigenesis induced by the HHV8-encoded chemokine receptor requires ligand modulation of high constitutive activity. J. Clin. Investig. 108:1789-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hummel, M., and E. Kieff. 1982. Epstein-Barr virus RNA. VIII. Viral RNA in permissively infected B95-8 cells. J. Virol. 43:262-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirk, O., C. Pedersen, A. Cozzi-Lepri, F. Antunes, V. Miller, J. M. Gatell, C. Katlama, A. Lazzarin, P. Skinhoj, and S. E. Barton. 2001. Non-Hodgkin lymphoma in HIV-infected patients in the era of highly active antiretroviral therapy. Blood 98:3406-3412. [DOI] [PubMed] [Google Scholar]
- 29.Kirshner, J. R., K. Staskus, A. Haase, M. Lagunoff, and D. Ganem. 1999. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J. Virol. 73:6006-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kledal, T. N. 2003. Herpesvirus encoded chemokines and chemokine receptors, p. 27-52. In S. Mahalingam (ed.), Chemokines in viral infections. Landes Bioscience, Georgetown, Tex.
- 31.Kostenis, E. 2001. Is Galpha16 the optimal tool for fishing ligands of orphan G-protein-coupled receptors? Trends Pharmacol. Sci. 22:560-564. [DOI] [PubMed] [Google Scholar]
- 32.Lee, B. J., U. H. Koszinowski, S. R. Sarawar, and H. Adler. 2003. A gammaherpesvirus G protein-coupled receptor homologue is required for increased viral replication in response to chemokines and efficient reactivation from latency. J. Immunol. 170:243-251. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig, A., J. E. Ehlert, H. D. Flad, and E. Brandt. 2000. Identification of distinct surface-expressed and intracellular CXC-chemokine receptor 2 glycoforms in neutrophils: N-glycosylation is essential for maintenance of receptor surface expression. J. Immunol. 165:1044-1052. [DOI] [PubMed] [Google Scholar]
- 34.Margulies, B. J., H. Browne, and W. Gibson. 1996. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 225:111-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean, K. A., P. J. Holst, L. Martini, T. W. Schwartz, and M. M. Rosenkilde. 2004. Similar activation of signal transduction pathways by the herpesvirus-encoded chemokine receptors US28 and ORF74. Virology 325:241-251. [DOI] [PubMed] [Google Scholar]
- 36.Mokros, T., A. Rehm, J. Droese, M. Oppermann, M. Lipp, and U. E. Hopken. 2002. Surface expression and endocytosis of the human cytomegalovirus-encoded chemokine receptor US28 is regulated by agonist-independent phosphorylation. J. Biol. Chem. 277:45122-45128. [DOI] [PubMed] [Google Scholar]
- 37.Montaner, S., A. Sodhi, A. Molinolo, T. H. Bugge, E. T. Sawai, Y. He, Y. Li, P. E. Ray, and J. S. Gutkind. 2003. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi's sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell 3:23-36. [DOI] [PubMed] [Google Scholar]
- 38.Montaner, S., A. Sodhi, S. Pece, E. A. Mesri, and J. S. Gutkind. 2001. The Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor promotes endothelial cell survival through the activation of Akt/protein kinase B. Cancer Res. 61:2641-2648. [PubMed] [Google Scholar]
- 39.Moorman, N. J., H. W. Virgin, and S. H. Speck. 2003. Disruption of the gene encoding the gammaHV68 v-GPCR leads to decreased efficiency of reactivation from latency. Virology 307:179-190. [DOI] [PubMed] [Google Scholar]
- 40.Nakayama, T., R. Fujisawa, D. Izawa, K. Hieshima, K. Takada, and O. Yoshie. 2002. Human B cells immortalized with Epstein-Barr virus upregulate CCR6 and CCR10 and downregulate CXCR4 and CXCR5. J. Virol. 76:3072-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neote, K., D. DiGregorio, J. Y. Mak, R. Horuk, and T. J. Schall. 1993. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell 72:415-425. [DOI] [PubMed] [Google Scholar]
- 42.Oliveira, S. A., and T. E. Shenk. 2001. Murine cytomegalovirus M78 protein, a G protein-coupled receptor homologue, is a constituent of the virion and facilitates accumulation of immediate-early viral mRNA. Proc. Natl. Acad. Sci. USA 98:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasqualucci, L., P. Neumeister, T. Goossens, G. Nanjangud, R. S. Chaganti, R. Kuppers, and R. la-Favera. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412:341-346. [DOI] [PubMed] [Google Scholar]
- 44.Pati, S., M. Cavrois, H. G. Guo, J. S. Foulke, Jr., J. Kim, R. A. Feldman, and M. Reitz. 2001. Activation of NF-κB by the human herpesvirus 8 chemokine receptor ORF74: evidence for a paracrine model of Kaposi's sarcoma pathogenesis. J. Virol. 75:8660-8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polson, A. G., D. Wang, J. DeRisi, and D. Ganem. 2002. Modulation of host gene expression by the constitutively active G protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus. Cancer Res. 62:4525-4530. [PubMed] [Google Scholar]
- 46.Rickinson, A., and E. Kieff. 2001. Epstein-Barr virus, p. 2575. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 47.Rivailler, P., Y. G. Cho, and F. Wang. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J. Virol. 76:12055-12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenkilde, M. M., T. N. Kledal, H. Brauner-Osborne, and T. W. Schwartz. 1999. Agonists and inverse agonists for the herpesvirus 8-encoded constitutively active seven-transmembrane oncogene product, ORF-74. J. Biol. Chem. 274:956-961. [DOI] [PubMed] [Google Scholar]
- 49.Saederup, N., S. A. Aguirre, T. E. Sparer, D. M. Bouley, and E. S. Mocarski. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J. Virol. 75:9966-9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saederup, N., Y. C. Lin, D. J. Dairaghi, T. J. Schall, and E. S. Mocarski. 1999. Cytomegalovirus-encoded beta chemokine promotes monocyte-associated viremia in the host. Proc. Natl. Acad. Sci. USA 96:10881-10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulz, A., and T. Schoneberg. 2003. The structural evolution of a P2Y-like G-protein-coupled receptor. J. Biol. Chem. 278:35531-35541. [DOI] [PubMed] [Google Scholar]
- 52.Schwarz, M., and P. M. Murphy. 2001. Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor constitutively activates NF-kappa B and induces proinflammatory cytokine and chemokine production via a C-terminal signaling determinant. J. Immunol. 167:505-513. [DOI] [PubMed] [Google Scholar]
- 53.Shepard, L. W., M. Yang, P. Xie, D. D. Browning, T. Voyno-Yasenetskaya, T. Kozasa, and R. D. Ye. 2001. Constitutive activation of NF-kappa B and secretion of interleukin-8 induced by the G protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus involve G alpha(13) and RhoA. J. Biol. Chem. 276:45979-45987. [DOI] [PubMed] [Google Scholar]
- 54.Smit, M. J., D. Verzijl, P. Casarosa, M. Navis, H. Timmerman, and R. Leurs. 2002. Kaposi's sarcoma-associated herpesvirus-encoded G protein-coupled receptor ORF74 constitutively activates p44/p42 mitogen-activated protein kinase and Akt via Gi- and phospholipase C-dependent signaling pathways. J. Virol. 76:1744-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sodhi, A., S. Montaner, and J. S. Gutkind. 2004. Does dysregulated expression of a deregulated viral GPCR trigger Kaposi's sarcomagenesis? FASEB J. 18:422-427. [DOI] [PubMed] [Google Scholar]
- 56.Sodhi, A., S. Montaner, V. Patel, J. J. Gomez-Roman, Y. Li, E. A. Sausville, E. T. Sawai, and J. S. Gutkind. 2004. Akt plays a central role in sarcomagenesis induced by Kaposi's sarcoma herpesvirus-encoded G protein-coupled receptor. Proc. Natl. Acad. Sci. USA 101:4821-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sodhi, A., S. Montaner, V. Patel, M. Zohar, C. Bais, E. A. Mesri, and J. S. Gutkind. 2000. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res. 60:4873-4880. [PubMed] [Google Scholar]
- 58.Streblow, D. N., C. Soderberg-Naucler, J. Vieira, P. Smith, E. Wakabayashi, F. Ruchti, K. Mattison, Y. Altschuler, and J. A. Nelson. 1999. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell 99:511-520. [DOI] [PubMed] [Google Scholar]
- 59.Streblow, D. N., J. Vomaske, P. Smith, R. Melnychuk, L. Hall, D. Pancheva, M. Smit, P. Casarosa, D. D. Schlaepfer, and J. A. Nelson. 2003. Human cytomegalovirus chemokine receptor US28-induced smooth muscle cell migration is mediated by focal adhesion kinase and Src. J. Biol. Chem. 278:50456-50465. [DOI] [PubMed] [Google Scholar]
- 60.Vieira, J., T. J. Schall, L. Corey, and A. P. Geballe. 1998. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J. Virol. 72:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waldhoer, M., P. Casarosa, M. M. Rosenkilde, M. J. Smit, R. Leurs, J. L. Whistler, and T. W. Schwartz. 2003. The carboxyl terminus of human cytomegalovirus-encoded 7 transmembrane receptor US28 camouflages agonism by mediating constitutive endocytosis. J. Biol. Chem. 278:19473-19482. [DOI] [PubMed] [Google Scholar]
- 62.Waldhoer, M., T. N. Kledal, H. Farrell, and T. W. Schwartz. 2002. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J. Virol. 76:8161-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, T. Y., S. C. Chen, M. W. Leach, D. Manfra, B. Homey, M. Wiekowski, L. Sullivan, C. H. Jenh, S. K. Narula, S. W. Chensue, and S. A. Lira. 2000. Transgenic expression of the chemokine receptor encoded by human herpesvirus 8 induces an angioproliferative disease resembling Kaposi's sarcoma. J. Exp. Med. 191:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]