Abstract
The UL17 protein of herpes simplex virus type 1 is essential for packaging the viral genome into the procapsid, a spherical assembly intermediate, and is present in the mature virus particle. We have examined the distribution of UL17 in various assembly products and virions to determine which component of the virus particle UL17 is associated with and at what stage in capsid assembly UL17 is required. UL17 was present in the procapsid, in the DNA-containing angularized C capsid, and in two other angularized capsid forms, A and B, that lack DNA and are thought to be dead-end products. The results suggest that UL17 is a minor capsid protein which is incorporated into the procapsid during assembly of the particle. UL17 was also found in virions and in noninfectious structures known as light (L) particles, which possess a tegument and envelope but lack a capsid. The level of UL17 in these particles was much greater than the amount that could be attributed to capsid contamination of the purified L-particle preparation, suggesting that UL17 is also a tegument protein. The finding that virions contain approximately twofold more UL17 than do C capsids provided further support for the idea that UL17 is present in two different structural components within the mature virion. The UL25 packaging protein, which is also present in virions, was not found in significant amounts in L particles, indicating that it is associated only with the capsid. UL6, the third virion-associated packaging protein, was present in slightly increased levels in L particles.
The mature herpes simplex virus type 1 (HSV-1) infectious particle has a complex structure consisting of a DNA core inside an icosahedral capsid, a protein layer referred to as the tegument surrounding the capsid, and an outer envelope comprising a lipid bilayer containing the viral glycoproteins (33). The viral genome is a linear double-stranded DNA (dsDNA) molecule, with a terminally redundant region (the a sequence) which contains the cis-acting elements required for cleavage and packaging of the replicated viral DNA. The DNA is synthesized as head-to-tail concatemers in the cell nucleus, and cleavage into unit length monomers is tightly coupled to packaging. The viral genome is inserted into a preformed protein shell known as the procapsid, where it assumes a highly compact, ordered state with liquid crystalline density (2). The procapsid is formed by cocondensation of the three capsid shell proteins, VP5, VP19C, and VP23, together with the viral scaffolding proteins. Initially, a wedge-shaped structure of these proteins forms, enlarging to become the spherical procapsid with an internal scaffold (21, 22). During packaging of the genome, the scaffolding proteins are detached from the inner capsid wall by the viral serine protease and then are lost from the capsid (6, 30, 32, 40). At the same time, the procapsid undergoes a conformational change, forming a stable, angularized capsid, known as the C capsid, which is the precursor of the infectious virus. In the absence of DNA packaging, the internal scaffolding proteins are still cleaved but remain trapped inside the angularized capsid, generating a dead-end product, the B capsid. Empty or A capsids, containing neither DNA nor a protein scaffold, are also produced in the nucleus, and these are thought to result from abortive packaging (11).
Of the seven DNA packaging proteins identified (reviewed in reference 11), only UL6, UL17, and UL25 are present in significant amounts in both the C capsid and the mature virion, although none of the three proteins is essential for assembly of the capsid (8, 18, 27, 35). UL6 is the best characterized of the three capsid-associated DNA packaging proteins. Evidence is accumulating that UL6 forms the portal or channel through which the viral DNA is packaged into the procapsid. Purified UL6 protein forms a ring structure composed of 12 subunits that is strikingly similar to the complexes of the portal protein from dsDNA bacteriophage, such as Φ29, T4, or P22 (23, 41). Furthermore, UL6 has a unique location on the capsid and, like the portal protein of the bacteriophage T4, associates with the scaffolding protein (24). In nonpermissive cells infected with a UL6 or UL17 null mutant, DNA packaging does not take place, whereas in UL25 null mutant-infected cells a low level of viral DNA encapsidation is observed but most of the packaged DNA is not full length (18, 28, 34, 37). On the basis of this observation, it has been suggested that UL25 is required at a later stage in the DNA packaging process than UL6 or UL17 (18, 37). In contrast to UL6, UL25 appears to be present at multiple sites on the capsid, although its precise location has not been determined (26). Because of the phenotype of the UL25 null mutant, it has been suggested that UL25 may be analogous to the head completion proteins of bacteriophages such as P22 (26). To date, very little information is available on the location and function of UL17. Initially it was proposed that UL17 was a tegument protein because it was a constituent of light (L) particles, which are noninfectious particles consisting of a tegument and envelope without a capsid (34). Subsequently it was reported that UL17 was present in B and C capsids (8). We have examined the distribution of UL17 in procapsids; A, B, and C capsids; virions; and L particles to assess the location of UL17 and to gain insight on its incorporation into virions.
MATERIALS AND METHODS
Plasmids: UL6 constructs.
The plasmid pMH54, encoding a NusA-UL6C fusion protein, was generated by ligating the SalI-BamHI UL6 fragment from plasmid pAs30 (28) to pET43.1a (Novagen, Inc.) cleaved with BamHI and SalI. The structure of this plasmid and others described below were confirmed by extensive restriction enzyme analysis. The plasmid pMH42, specifying the maltose binding protein (MBP)-UL6C fusion protein, was produced by ligating the SalI fragment containing UL6 from a cloned HSV-1 strain 17 EcoRI d fragment into the vector pMAL-c2E (New England Biolabs, Inc.) digested with SalI. Both constructs contained only the coding sequences of the C-terminal 298 amino acids of UL6.
UL17 constructs.
The plasmid pGX334 was constructed by ligating the HSV-1 BglII fragment from the plasmid pGX42 (containing the HSV-1 strain 17 EcoRI g fragment) to pUC18Bgl cleaved with BglII (14). The BglII site within pUC18Bgl was created by introducing the BglII linker d(pCAGATCTG) into the HindIII site of pUC18. The 5′ portion and sequences upstream of UL17 were removed by digestion of pGX334 with BamHI and MscI and were replaced with a synthetic oligonucleotide regenerating the UL17 translational start site and amino acid codons up to the first MscI site. The full-length UL17 gene was constructed by the subsequent cloning of the UL17 internal MscI fragment back into the UL17 open reading frame (ORF). A BamHI site was engineered close to the 3′ end of the UL17 ORF by inserting a BamHI linker into the unique NheI site of the construct to create the plasmid pMH19. The UL17 gene, flanked by BamHI sites, was inserted into the BamHI site of the plasmid pAPV (13) for the production of transformed cell lines expressing UL17. The SacI fragment from the UL17 gene in the plasmid pMH19 was cloned into the SacI site of pET43.1b, generating the plasmid pET43.1bUL17frag for expression of the NusA-UL17C fusion protein, containing amino acids 154 to 703 of UL17. The plasmid pMH19 was also digested with Ecl36II, an isoschizomer of SacI, and the UL17 fragment ligated into the vector pMAL-c2X that had been digested with XmnI to create the plasmid pMAL-c2X/UL17frag for the expression of the MBP-UL17C fusion protein.
UL25 constructs.
The EcoRI-EcoRV fragment at the 5′ end of the UL25 gene in the HSV-1 strain 17 genomic BamHI u fragment, inserted in the BamHI site of the vector pUC119, was replaced by an EcoRI-EcoRV PCR fragment to generate the plasmid pGX294. This cloning step recreated the N-terminal sequences of UL25 and removed the sequences upstream of the UL25 ATG start site and replaced them with a BamHI site. Similarly, sequences at the 3′ end and downstream of the UL25 ORF in BamHI u were removed by digestion of the plasmid pGX294 with AflIII and HindIII and were replaced with a double-stranded synthetic oligonucleotide. This cloning step regenerated the 3′ end of the UL25 ORF and introduced a BamHI site immediately after the translational stop codon. The UL25 ORF, flanked by BamHI sites in the plasmid pTA5, was cloned into the BamHI site of the vector pMAL-c2 to form the plasmid pTA3 for the production of a pMAL-UL25 fusion protein. The EcoRI-AatII sequences in the plasmid pTA5 were replaced with a double-stranded synthetic oligonucleotide, resulting in the modification of the UL25 ORF by the insertion of six histidine codons immediately after the translational start codon to create the plasmid pTA8. The modified UL25 gene was inserted as a BamHI fragment into the BamHI site of the baculovirus transfer vector pAcCL29.1 to form the plasmid pTA10.
UL38 constructs.
The plasmid pΔ38YFP contained the yellow fluorescent protein (YFP) ORF flanked by PCR products from the HSV-1 genome. Initially the PCR product, encompassing nucleotides 83010 to 84522 of the HSV-1 genome and flanked by HindIII and NheI sites, respectively, was cloned into the plasmid pGEM-T Easy (Promega) such that the NheI site in the PCR fragment was close to the SacI site in the vector. This plasmid was named pΔ 38-5′. The YFP ORF was released from the pEYFP-C1 plasmid (Clontech) by digestion with NheI and SacI and was ligated into NheI/SacI-digested pΔ 38-5′ to create the plasmid pΔ 38-5′YFP. A second PCR product, extending from nucleotide 85982 to 87572 of the HSV-1 genome and flanked by BglII and SacI sites, respectively, with a stop codon for the YFP ORF, was initially cloned into pGEM-T Easy. The plasmid generated was called pΔ 38-3′. The UL38 sequences were subsequently released from pΔ 38-3 ′as a BglII-SacI fragment and ligated into pΔ 38-5′YFP, digested with the same enzymes, to create the plasmid pΔ38YFP. The plasmid pIM248 was derived from pE38 (31). A BamHI linker was introduced into the HindIII site of the plasmid pE38. The UL38 ORF, now flanked by BamHI sites, was removed from the modified plasmid by cleavage with BamHI and ligated to pAPV digested with BamHI to form pIM248.
Viruses.
HSV-1 wild-type (wt) strain 17, wt strain KOS, ts1201, the UL25 null mutant KUL25NS, the UL6 null mutant lacZ-UL6−, and the UL17 insertional mutant UL17-stop have all been described previously (3, 18, 28, 30, 34, 36). An HSV-1 strain 17 UL38 deletion mutant, vΔ38YFP, was constructed by removing nucleotide sequences between 84522 to 85982 in the HSV-1 strain 17 genome (16) and replacing them with the yellow fluorescent protein ORF. This mutant was produced by recombining the plasmid pΔ38YFP with wt strain 17. The plasmid linearized with HindIII was transfected into a transformed rabbit skin (RS) cell line expressing UL38 (UL38RSC) by using lipofectin PLUS (Life Technologies). Four hours after transfection the cells were infected with 1 PFU of wt strain 17 per cell. The cells were harvested after 48 h at 37°C, the recombinant UL38 deletion virus, which was identified by its ability to form fluorescent plaques, was plaque purified on the UL38RSC cell line, and a virus stock was prepared. lacZ-UL6− was propagated in the transformed RS cell line UL6RSC1, and the UL17 stop mutant was grown in the transformed RS cell line UL17RSC5. The construction of these three transformed cells is described below. The recombinant baculovirus AcUL25 was made by recombining the modified UL25 gene in the plasmid pTA10 into the baculovirus expression vector AcPAK6 as described previously (29).
Cells.
BHK-21 clone 13 cells were cultured in Glasgow minimal essential medium supplemented with 10% tryptose phosphate broth, 10% newborn calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The transformed cell lines described below and the RS cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum and the above antibiotics (1). Sf21 cells were propagated in TC100 containing penicillin and streptomycin as described above and 5% fetal calf serum.
Construction of transformed cell lines.
The UL6RSC1 cell line was constructed by transfecting RS cells with the plasmid pgDUL6 and selecting for G418-resistant cell lines that supported the growth of lacZ-UL6−, essentially as described previously (4, 28). The UL17RSC5 cell line was made in the same way as UL6RSC1, except that two plasmids, pAPVUL17 (containing the HSV-1 UL17 gene under the control of the ribonucleotide reductase promoter) and pSVneo2 (containing a gene encoding neomycin resistance) were transfected into RS cells. The UL38RSC cell line was made essentially as described for the UL17RSC5 cell line, using the plasmids pIM248 and pSVneo2.
Purification of recombinant proteins.
The proteins MBP-UL25 (encoded by pTA3), MBP-UL6C (encoded by pMH42), and MBP-UL17C (encoded by pMAL-UL17C) were expressed in Escherichia coli BL21, and the recombinant proteins were purified by affinity chromatography on amylose resin columns. The NusA-UL6C (encoded by pMH54) and NusA-UL17C (encoded by pET43.1bUL17frag) proteins were each expressed in E. coli BL21-CodonPlus-RP (Stratagene), and the recombinant proteins were purified under native conditions by affinity chromatography on nickel-agarose columns. The eluted UL17 recombinant proteins were denatured with either urea or guanidine HCl, and the denaturant was removed by dialysis in the presence of low concentrations of sodium dodecyl sulfate (SDS). The UL25 histidine-tagged protein was expressed in Sf21 cells infected with AcUL25 and was purified under native conditions by affinity chromatograph on a nickel-agarose column.
Antibodies.
The mouse monoclonal antibodies VP16 (1-21), specific for HSV-1 VP16; 6F10 (21), specific for VP5; and MCA406, specific for UL26.5, were supplied by Autogen Bioclear UK, Ltd., Santa Cruz Biotechnology, Inc., and Serotec, respectively. The rabbit polyclonal antibody R186, specific for VP23, and the mouse monoclonal antibody DM165, specific for VP5, have been described previously (12, 15). Monoclonal antibodies were raised against UL17, UL6, and UL25. Because the solubility of each DNA packaging protein was low when expressed in large amounts in E. coli, the full-length UL25 ORF and the N-terminal truncated UL6 and UL17 ORFs were fused to the MBP ORF to generate more soluble proteins that could be readily purified for immunizing mice. To reduce the possibility of isolating hybridomas excreting antibodies specific for the MBP protein, a different fusion partner, NusA, was used for the final inoculation of UL17 and UL6 proteins. In the case of UL25, a 6× His-tagged UL25 protein was used instead of the NusA fusion partner. A solution of fusion protein (20 μg) was emulsified with an equal volume of Freund's complete adjuvant and was injected subcutaneously into an 8-week-old female BALB/c mouse. Four weeks later, the mouse was boosted subcutaneously with 20 μg of the same protein emulsified with incomplete Freund's adjuvant, followed by subsequent inoculations of the same material every 2 weeks for 6 weeks. Finally, 200 μg of purified NusA-fusion protein (for the production of UL6 or UL17 monoclonal antibodies) or 6× His-tagged-UL25 (for UL25 monoclonal antibodies) was injected intraperitoneally. Four days later the spleen cells were harvested and fused to Sp2/O-Ag14 myeloma cells, and hybridomas secreting antibodies specific for the DNA packaging protein were identified. Antibody was either purified from the hybridoma cell culture supernatant on a protein A-Sepharose column or concentrated in the cell culture medium by using a specialized culture flask, CELLine, from IBS Integra Biosciences.
Purification of L particles and virions of HSV-1.
BHK cells in 850-cm2 roller bottles were infected with approximately 0.003 PFU of wt virus per cell or with 5 PFU of mutant virus per cell and incubated at 37°C for 3 days (wt virus) or for 24 h (mutant virus). Extracellular virus and L particles were concentrated by centrifugation and were purified on 5 to 15% Ficoll gradients as described previously (39). The bands corresponding to virions and L particles were collected using a BioComp fractionator and then diluted in Eagle's medium lacking phenol red, and the material was pelleted at 20,000 rpm at 4°C in a Sorval AH629 rotor. The envelope of L particles was removed by treatment with Nonidet P40 at a final concentration of 1% as described by McLauchlan and Rixon (17).
Purification of capsids.
BHK cells in 850-cm2 roller bottles were infected with 5 PFU of virus per cell, and the infected cells were incubated at 37°C for 16 h. The cells were harvested and the capsids were isolated as described by Zhou et al. (43).
Procapsid isolation.
Procapsids were isolated essentially as described by Sheaffer et al. (35) using UL19 monoclonal antibodies DM165 and 6F10.
Western blot analysis.
Proteins, separated by SDS-polyacrylamide gel electrophoresis (PAGE), were transferred by electrophoresis onto a nitrocellulose membrane, and immunoblot analysis was performed using the enhanced chemiluminescence (ECL) method based on the luminol detection system (Amersham). Antibodies were removed by treating the blot with stripping buffer (100 mM mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.7) at 55°C for 30 min, and the blot was extensively washed with phosphate-buffered saline containing 0.05% Tween 20. When a blot was probed with several primary antibodies, each antibody was used in a separate ECL reaction. Protein bands in exposed film were quantified by densitometric analysis of digital images using Quantity One software (Bio-Rad Laboratories).
Silver staining of proteins.
Proteins, separated by SDS-PAGE, were stained with ammoniacal silver essentially as described by Harlow and Lane (10).
RESULTS
Mouse monoclonal antibodies specific for UL6, UL17, or UL25.
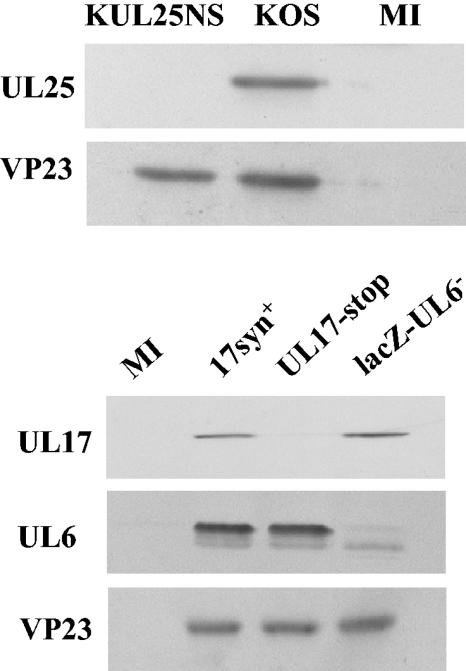
Hybridoma cell lines secreting UL6-, UL17-, or UL25-specific monoclonal antibodies were isolated, and for each DNA packaging protein a monoclonal antibody was selected that reacted strongly in immunoprecipitation and Western blot assays against the full-length unmodified HSV-1 protein in an extract of insect cells that had been infected with the relevant recombinant baculovirus. The specificity of each of the three monoclonal antibodies used in the present study was confirmed by Western blot analysis of extracts of cells infected with wt HSV-1 or a mutant virus that did not express the relevant DNA packaging protein (Fig. 1). As well as recognizing the UL6 protein, the UL6 antibody recognized a protein band in the wt virus-infected cell protein profile that was slightly smaller than that of UL6. Because this protein was also present in similar amounts in cells infected with the UL6 null mutant but was absent from mock-infected cells, it is likely that this protein band represents a cross-reaction between the antibody and a viral protein unrelated to UL6. The blot was also probed with antibodies to VP23 as a control to check that the cells had been efficiently infected with virus. Similar levels of this structural protein were detected in all the samples.
FIG. 1.
Specificity of UL6, UL17, and UL25 monoclonal antibodies. Cells were infected with HSV-1 wt virus strain 17, wt virus strain KOS, UL17-stop virus, lacZ-UL6−, or KUL25NS or were mock infected (MI). At 20 h postinfection the cells were harvested and the proteins were separated by SDS-PAGE and blotted onto a nitrocellulose membrane. The blot was probed sequentially with monoclonal antibodies UL6 Mab175, UL17 Mab203, and UL25 Mab166 as well as VP23-specific rabbit antibodies.
UL17 copurifies with wt virus A, B, and C capsids.
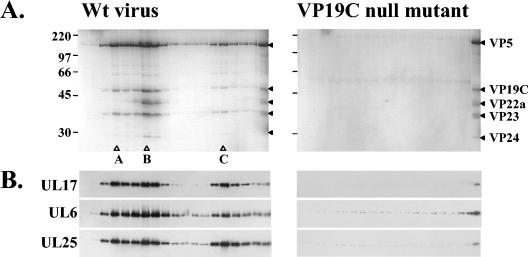
To determine whether UL17 was a capsid protein, BHK cells were infected either with a VP19C null mutant virus, which does not produce capsids, or with wt virus, and the cells were harvested after 18 h of incubation at 37°C. Each virus-infected cell extract was sedimented through a 40% (wt/vol) sucrose cushion, and the resulting pellet was resuspended by brief sonication prior to centrifugation through a 20 to 50% (wt/vol) sucrose gradient. Three distinct capsid bands, corresponding to A, B, and C capsids, were detected only in the gradient of the extract from wt virus-infected cells. Gradient fractions from both samples were collected, and the proteins, separated by SDS-PAGE, were visualized by Coomassie blue staining (Fig. 2). Analysis of proteins in the gradient fractions of the extract from wt virus-infected cells confirmed the identity of the capsid types. B capsids were characterized by the presence of the scaffolding protein VP22a. Peaks of A, B, or C capsid were not discernible on the Coomassie blue-stained gel of the gradient fractions from the extract from VP19C null mutant-infected cells, consistent with the phenotype of the VP19C null mutant (Fig. 2A). The separated proteins were also blotted onto a nitrocellulose membrane, and UL17, UL6, and UL25 proteins were detected by Western blot analysis (Fig. 2B). All three proteins were concentrated in the fractions containing A, B, and C capsids from the wt virus-infected cell extract, whereas no significant peaks of these proteins were observed in the corresponding region of the gradient of the material from the VP19C null mutant-infected cell extract. The results strongly suggested that UL17 associated with all three forms of angularized capsids.
FIG. 2.
Sucrose gradient sedimentation analysis of extracts from BHK cells infected with wt virus strain 17 or the VP19C null mutant vΔ38YFP. The extracts were sedimented through a 10 to 40% sucrose gradient, and 32 successive fractions were collected starting from the top of the gradient. The proteins in fractions 14 to 32 were analyzed by SDS-PAGE and detected by staining with Coomassie brilliant blue (A). Protein size markers (in kilodaltons), indicated by the short lines, are shown at the far left-hand side of the panel. The open triangles show the peak fractions of A, B, and C capsids. The far lane on the right-hand side of each image of the gel shows a wt B-capsid protein profile. The HSV-1 capsid proteins are denoted by the closed triangles. The packaging proteins in fractions 14 to 31 were identified by Western blotting, using UL6, UL17, and UL25 antibodies (B).
Comparison of the amount of UL17 in A, B, and C capsids and virions.
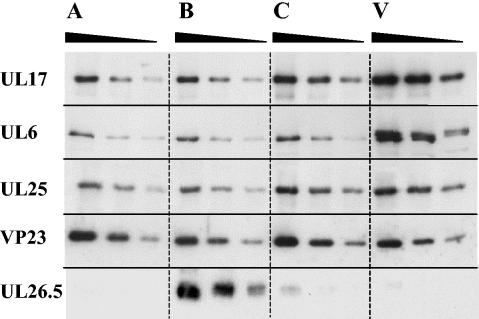
To estimate the levels of UL17 in angularized capsids and virions, purified A, B, and C capsids and virions were standardized with respect to their VP23 content by using Western blot analysis. Twofold serial dilutions were made of each sample, and the proteins were analyzed by Western blotting using UL17, UL6, and UL25 antibodies. The blots were also probed with antibody against the scaffolding protein UL26.5 to confirm the capsid type and with VP23-specific antibody to quantify the particles (Fig. 3). A and C capsids had low amounts of the scaffolding protein compared to that for B capsids, suggesting that the capsids were of high purity. The relative amounts of each DNA packaging protein in capsids and virions that had been standardized with respect to their VP23 content were determined and are shown in Table 1, together with values from an independent experiment. The finding that B capsids had smaller amounts of UL25 than C capsids was consistent with earlier results (26, 35). The level of UL17 was similar in A and B capsids but was slightly higher in C capsids (Fig. 3). Virions had greater amounts of UL17 than C capsids. There was no significant difference in the amount of UL6 in the three forms of angularized capsids. In the virion protein profile, two protein bands close together reacted with the UL6-specific antibody, the lower one of which comigrated with the UL6 band in the capsid samples. Because the bands were not well separated and because the upper band was probably a cross-reacting protein(s), as shown later, it was not possible to determine the relative amount of UL6 in virions in this experiment.
FIG. 3.
Comparison of the relative amount of UL17 in A, B, and C capsids and virions. Serial twofold dilutions were prepared from samples that had been equalized with respect to VP23 content, and the proteins were separated by SDS-PAGE. Structural and packaging proteins were detected by Western blotting using antibodies specific for UL17, UL6, UL25, VP23, and the UL26.5 products.
TABLE 1.
Comparison of the levels of UL6, UL17, and UL25 in A, B, C capsids and virionsa
| Protein | A capsid | B capsid | C capsid | Virion |
|---|---|---|---|---|
| UL6 | 94.0 | 100 | 89.4 | 563.4b |
| 104.2 | 100 | 93.3 | 338.6b | |
| UL17 | 92.8 | 100 | 163.9 | 527.6 |
| 83.1 | 100 | 174.2 | 425.9 | |
| UL25 | 90.1 | 100 | 177.7 | 238.4 |
| 98.4 | 100 | 241.2 | 315.7 |
The relative amount of DNA packaging protein was determined by densitometric analysis of digital images of exposed films of Western blots. The results are shown as a percentage of the protein relative to the levels in B capsids. The two sets of data are derived from independent experiments. Values for VP23 were 100% in all cases.
Values are artificially high because of cross-reacting proteins.
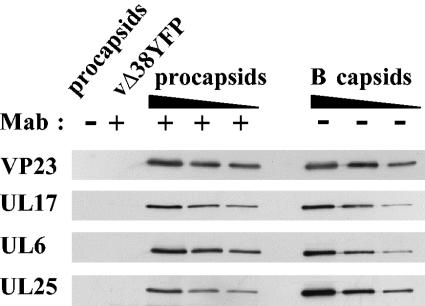
Comparison of the amount of UL17 in purified procapsids and B capsids.
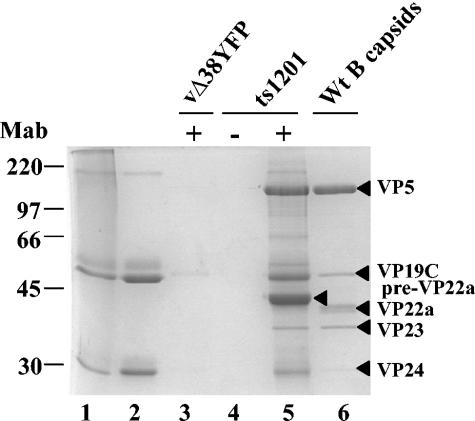
To determine whether UL17 was assembled into capsids at an early stage in their formation, procapsids were isolated using an immunoprecipitation method developed by Newcomb et al. (21) and refined by Sheaffer et al. (35). BHK cells were infected at the nonpermissive temperature with the HSV-1 strain 17 temperature-sensitive protease mutant ts1201, which has a block in virus assembly and is unable to cleave the scaffolding protein, preVP22a, and full-length protease efficiently at this temperature. As a consequence of this defect, the mutant-infected cells accumulate procapsids in the nuclei. Extracts of virus-infected cells were incubated with a mixture of VP5 monoclonal antibodies 6F10 and DM165, and the antibody-procapsid complexes were concentrated by centrifugation. As a control, the ts1201-infected cell extract was treated in the same way as that described above, except that phosphate-buffered saline was used instead of antibody. To ensure that capsids were selectively precipitated from the virus-infected cell extract by VP5 antibodies, BHK cells were infected with the VP19C null mutant under the same conditions as those used for ts1201, and the virus-infected cell extracts were incubated with VP5 antibodies. To assess the purity of the procapsid preparation, the procapsid protein profile was compared to that of B capsids purified from wt virus-infected cells. Figure 4 shows a Coomassie blue-stained SDS polyacrylamide gel of proteins from purified B capsids, immunoprecipitated procapsids, and the controls. In the absence of VP5 antibody, no procapsids were immunoprecipitated from the ts1201-infected cell extract (Fig. 4, lane 4). Similarly, no capsid proteins were detected in immunoprecipitates of the VP19C null mutant-infected cell extracts treated with VP5 antibody, as expected, because this mutant fails to assemble capsids (Fig. 4, lane 3). The two controls confirm that the VP5 antibody selectively immunoprecipitated procapsids from the ts1201-infected cell extract (Fig. 4, lane 5). In contrast to the B-capsid sample, the procapsid preparation had large amounts of preVP22a, the uncleaved UL26.5 protein (Fig. 4, lanes 5 and 6). Consistent with previous work, the level of scaffolding proteins in procapsids was greater than the amount observed in wt virus B capsids (25, 35).
FIG. 4.
Comparison of proteins from immunoprecipitated ts1201 procapsids with those of wt HSV-1 strain 17 B capsids. Proteins of immunoprecipitated procapsids from ts1201-infected cells (lane 5) and purified wt virus B capsids (lane 6) were analyzed by SDS-PAGE, and the separated proteins were detected by staining with Coomassie brilliant blue. As a control, an extract of ts1201-infected cells was treated in the same way as that described for the isolation of procapsids, except no VP5 antibody was added (lane 4). In addition, extracts of cells that had been infected with the VP19C deletion mutant, vΔ38YFP, under the same conditions as those for ts1201 were also treated with VP5 monoclonal antibody (Mab), using the procapsid isolation protocol (lane 3). Purified 6F10 and DM165 monoclonal antibodies were analyzed in lanes 1 and 2, respectively. Protein size markers (in kilodaltons), indicated by the short lines, are shown at the far left-hand side of the panel. The HSV-1 capsid proteins are indicated by the closed triangles. The presence (+) or absence (−) of monoclonal antibody in the immunoprecipitation reaction is indicated at the top of the image of the gel.
To determine the relative amount of UL17 present in these capsids, Western blot analysis was carried out using a monoclonal antibody to UL17. The amount of each capsid preparation was standardized by using a polyclonal antibody to the capsid shell protein VP23. Figure 5 shows that UL17 was present in similar amounts in procapsids and B capsids, suggesting that UL17 is incorporated into the capsid shell at an early stage in its formation. The blots were also probed with monoclonal antibody to UL6 and UL25 to confirm that procapsids had the expected pattern of proteins. As reported previously, the UL25 packaging protein was present at lower levels in procapsids than in B capsids (35). The amounts of each DNA packaging protein in capsids and procapsids were determined relative to the level of VP23 and are shown in Table 2 together with values from an independent experiment.
FIG. 5.
Comparison of the relative amounts of packaging proteins in procapsids and B capsids. Serial twofold dilutions of purified wt B capsids and immunoprecipitated ts1201 procapsids were prepared, and the proteins were separated by SDS-PAGE. Packaging and structural proteins were detected by Western blotting using antibodies specific for UL17, UL6, UL25, and VP23. Mab, monoclonal antibody.
TABLE 2.
Percentage of the protein relative to that of B capsidsa
| VP23 | UL17 | UL6 | UL25 |
|---|---|---|---|
| 100 | 103 | 110 | 25 |
| 100 | 114 | 101 | 28 |
The relative amount of protein was determined by densitometric analysis of digital images of exposed films of Western blots. The results are shown as the percentage of protein relative to the level in B capsids, which was set at 100%. The two sets of figures are derived from independent experiments.
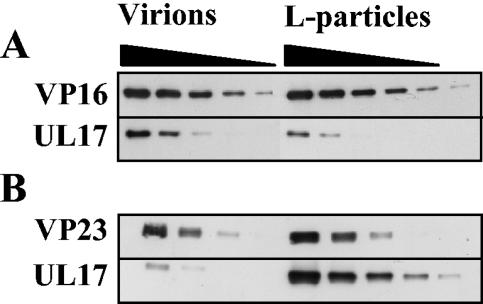
Analysis of UL17 in virions and L particles.
The above results and previous findings by Goshima et al. (8) show that UL17 is associated with the capsid. To reconcile our data with earlier work indicating that UL17 was a tegument protein (34), we compared the amount of UL17 in L particles with the levels in virions. Because the overall composition of the tegument in L particles and virions appears to be identical (39), the amounts of virions and L particles were standardized by using a monoclonal antibody to VP16, a major tegument protein, in Western blot analysis. Under these conditions UL17 was approximately twofold more abundant in virions than L particles (Fig. 6A). The amount of capsid contamination in the L-particle preparation was determined by Western blot analysis using a monoclonal antibody to the triplex protein VP23. The level of capsid contamination ranged from about 5 to 10% (data not shown). The L-particle preparation and virion sample were also standardized with respect to the capsid protein VP23 and were analyzed. Under these conditions the number of L particles greatly exceeded that of virions. If UL17 was located only in the capsid, it should be present in similar amounts in L-particle and virion preparations in this experiment. Considerably more UL17 was found in the L-particle preparation than could be accounted for by capsid contamination (Fig. 6B). There was approximately 10-fold more UL17 in the preparation of L particles than in virions, although both samples contained similar amounts of VP23 and therefore similar numbers of capsids. These results suggest that UL17 is likely to be a genuine component of L particles.
FIG. 6.
Analysis of UL17 in wt virions and L particles. Serial twofold dilutions of virions and L particles were prepared, and the proteins were separated by SDS-PAGE. Proteins were detected by Western blotting using antibodies against UL17, VP16, and VP23. For panel A, virions and L particles were equalized on the basis of their VP16 content. For panel B, virions and L particles were equalized on the basis of their VP23 content to determine whether capsid contamination of L particles accounted for the presence of UL17.
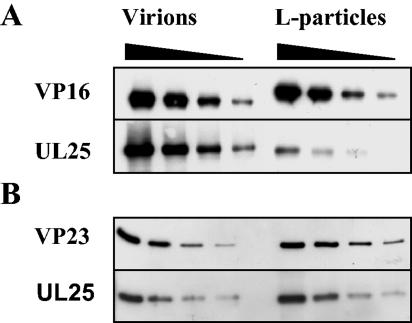
Analysis of UL25 and UL6 in virions and L particles.
Although UL17 had been reported to copurify with L particles, the two other virion-associated packaging proteins, UL6 and UL25, had not been examined (34). To determine whether UL25 and UL6 were present in significant amounts in L particles, the levels in virions and L particles standardized with VP16 were investigated. Virions contained approximately 10-fold more UL25 than L particles (Fig. 7A). When virions and L particles were standardized using VP23, there were similar amounts of UL25 present in both particles, suggesting that this protein was only present as a result of capsid contamination (Fig. 7B).
FIG. 7.
Analysis of UL25 in wt virions and L particles. Serial twofold dilutions of virions and L particles were prepared, and the proteins were separated by SDS-PAGE. Proteins were detected by Western blotting using antibodies against UL25, VP16, and VP23. For panel A, virions and L particles were equalized on the basis of their VP16 contents. For panel B, virions and L particles were equalized on the basis of their VP23 contents to determine whether capsid contamination of L particles accounted for the presence of UL25.
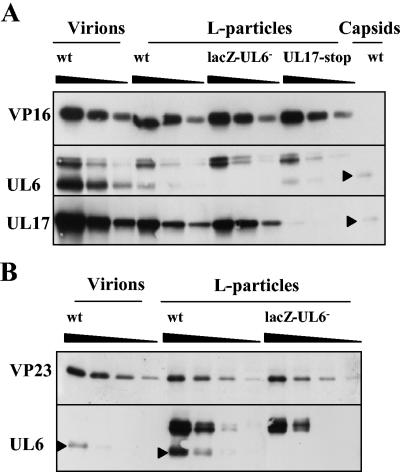
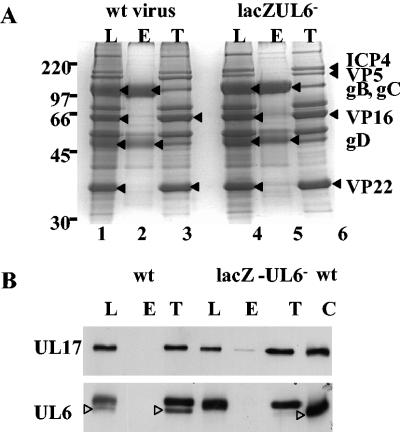
In Fig. 3, the relative amount of UL6 in virions compared to that of angularized capsids was not determined, because the UL6-specific band migrated close to a presumed cross-reacting protein. When, however, virions and L particles, standardized with VP16, were analyzed on a large 7.5% polyacrylamide gel under denaturing conditions and UL6 was identified by Western blotting, the UL6 present in capsids formed a single band as expected, whereas the protein in virions and L particles resolved into three bands (Fig. 8A). The species with the lowest apparent molecular weight comigrated with the UL6 present in capsids. To exclude the possibility that the proteins with higher molecular weight were modified forms of UL6, L particles were also purified from BHK cells infected with lacZ-UL6− virus and the proteins were screened for the presence of the higher-molecular-weight species reacting with UL6-specific monoclonal antibody. Because the mutant L particles, equalized with wt virus L particles using antibody to VP16, contained the higher-molecular-weight species, it was concluded that these proteins were cross-reacting with the primary antibody and were not modified forms of UL6 (Fig. 8A). The levels of UL17 in L particles derived from BHK cells infected with the UL17 stop mutant virus were considerably lower than those in wt virus L particles, confirming that the protein recognized by UL17-specific monoclonal antibody in wt virus L particles was UL17 and not a cross-reacting protein (Fig. 8A). Virions and L particles, standardized with VP23, were examined to determine whether the UL6 in wt L-particle preparation was derived from contaminating capsids or virions. The level of UL6 was at least twofold greater than the amount present in contaminating virions or capsids (Fig. 8B).
FIG. 8.
Comparison of UL6 in wt virions and wt and mutant L particles. Serial twofold dilutions of virions and L particles were prepared, and the proteins were separated by SDS-PAGE. Proteins were detected by Western blotting using antibodies against UL6, UL17, VP16, and VP23. Virions and L particles were equalized on the basis of their VP16 content (A). A sample of wt B capsids was also included to distinguish UL6 from cross-reacting proteins. For panel B, wt virions and L particles were equalized on the basis of their VP23 content to determine whether capsid contamination of L particles accounted for the presence of UL6. UL6 protein is indicated by a closed triangle.
To exclude the possibility that the association of UL17 with L particles was nonspecific, L particles were treated with NP-40 and the tegument fraction was concentrated by centrifugation. The proteins in the tegument fraction and the soluble fraction were separated and either stained with Coomassie blue (Fig. 9A) or blotted onto a nitrocellulose membrane. UL17 was present in the insoluble tegument fraction of L particles, confirming previous results of Salmon et al. (34) (Fig. 9B). Because UL6 was found in slightly greater amounts in L particles than could be accounted for by contaminating capsids, the location of UL6 in L particles was also examined. UL6, like UL17, localized in the tegument fraction of L particles (Fig. 9B).
FIG. 9.
Localization of UL17 and UL6 within L particles. Wt L particles (L) were treated with NP-40, and the insoluble proteins, referred to as the tegument fraction (T), were separated from the soluble products, mainly envelope proteins (E), by centrifugation. The proteins were analyzed by SDS-PAGE and were detected either by silver staining (A) or by Western blotting (B) using antibodies specific for UL17 and UL6.
DISCUSSION
UL17 was detected in capsids and in L particles, suggesting that it was present in both tegument and capsid compartments of the virion. This conclusion was reinforced by the finding that there was more UL17 in virions than in C capsids and that UL17 associated with the tegument component of L particles stripped of their envelopes. Because the tegument is not attached to DNA-containing capsids in the nucleus (2), it is likely that UL17 protein participating in DNA packaging is bound to capsids and is not in the tegument layer.
Our results confirm the study by Goshima et al. (8), which suggested that UL17 was present in B and C capsids. Previously, Salmon et al. (34) reported that they were unable to detect UL17 by Western blotting in a purified B-capsid preparation and concluded that the protein was not an integral component of capsids. To date we have been unable to purify the UL17 protein to homogeneity to determine its copy number in capsids. It is clear, however, from a comparison of the protein profile of UL17-stop mutant capsids with that of wt virus capsids that UL17 is a minor component and is not readily detectable when the proteins are stained with Coomassie blue (34 and our unpublished data). It is therefore probable that the failure of Salmon et al. (34) to identify UL17 in capsids was due to a lower sensitivity of their Western blot analysis.
The finding that UL17 is present in procapsids suggests that the protein is required at an early stage in DNA packaging, consistent with the studies on the UL17 null mutant showing that UL17 is essential for cleavage and packaging of the viral genome into capsids (34). In this respect UL17 differs from UL25, which is present in considerably smaller amounts in procapsids than in B capsids and appears to be required at a later stage (18, 35, 37). Both proteins, however, were repeatedly found in C capsids in greater amounts than in B capsids, whereas in the same experiments no increase in the amount of UL6 was observed. The elevated levels of UL17 in C capsids suggest that UL17 may have a second function after the DNA has been packaged. Because virions reproducibly contained higher levels of UL25 than C capsids and UL25 did not appear to be a tegument protein, it is likely that some UL25 protein was lost from C capsids during their purification. This possibility is supported by the finding that the relative amount of UL25 bound to C capsids compared to that of B capsids in our experiments was lower than previously published values (26, 35). Interestingly and unexpectedly, the levels of both UL17 and UL25 in A capsids were similar to those in B capsids. Because A capsids are thought to result from abortive packaging events, these results suggest either that some of the UL17 and UL25 is loosely attached to capsids and dependent on the presence of packaged DNA or that binding of a proportion of UL17 and UL25 requires a completed packaging event.
The observation that Vero cells infected with the UL17 null mutant contain large aggregates of B capsids suggests that UL17 may be required for capsid dispersal within the nucleus (33). The fact that UL17 is more abundant in C capsids than in procapsids indicates that UL17 may have a role in the initial envelopment of C capsids, a function previously proposed for UL25 (37). To date, no interactions have been reported between UL17 and UL31 or UL34, the two proteins identified so far that are involved in primary envelopment. Alternatively, UL17 and possibly UL25 may be important for stabilizing the DNA-containing capsid.
To date, it is not known whether the asymmetric portal vertex interacts with the tegument. The observation that UL6 was not present in large amounts in the tegument is consistent with the idea that the UL6 portal does not directly associate with the tegument layer in the virion. Experiments are in progress to determine whether UL17 is located at the portal vertex and is involved in attaching the tegument to the portal vertex or whether it is located elsewhere and has an indirect role in DNA packaging.
The finding that UL6 and UL25 were present in L particles in considerably reduced amounts compared to those of UL17 strengthens the idea that UL17 specifically interacts with the tegument. Because the amount of UL6 present in L-particle preparations was only about twofold greater than the amount in contaminating capsids or virions, it is likely that UL6 is not a genuine component of the tegument. Earlier findings by Patel and McLean (27) concluded that UL6 was not a tegument protein, because it was not present in immune precipitates from extracts of L particles incubated with UL6 polyclonal antibody 2C2. The UL6 antibody in their studies was not very potent, and in order to detect UL6 in virus particles or HSV-infected cells, UL6 was usually immunoprecipitated prior to Western blot analysis. It is likely that, in comparison to the amount in virions, the reduced level of UL6 (three- to fourfold reduction) found in L particles would not have been detected in their assay.
Present evidence favors the idea that tegument in the virion is added to the capsid in the cytoplasm, possibly at two different stages (reviewed by Mettenleiter [20]). The L particles analyzed in the present study were purified from the virus-infected cell medium and had a protein composition similar to that of virions apart from the capsid proteins and a few phosphorylated proteins, including ICP4 (38). It is therefore likely that the tegument in L particles is acquired in the same way as the tegument of the maturing virus. This assumption is supported by the finding that the levels of UL17 in L particles were similar to those in the tegument layer of virions, taking into account the amount of UL17 attached to the DNA-containing capsid. Although UL17 is predominately nuclear in HSV-1-infected cells (8), it is probably incorporated into the tegument in the cytoplasm, presumably shortly after it is synthesized. Three-dimensional reconstruction of herpes simplex virions examined by cryoelectron microscopy has revealed that the tegument is attached to the capsid via the pentons at the vertices (42), and this finding has been confirmed recently by using cryoelectron tomography to study the structure of HSV-1 virions (9). VP1-3, encoded by UL36, is a strong candidate for binding to the pentons, because it interacts with VP5 and is tightly attached to the capsid in virions. Also, the UL36 null mutant has a late block in virus assembly, accumulating capsids which lack tegument in the cytoplasm of nonpermissive cells (5, 7, 19). Interestingly, this protein is present in L particles, suggesting that although it is tightly associated with cytoplasmic capsids, it can, like UL17, bind to tegument in the absence of capsids (39). No information, however, is available as to whether VP1-3 interacts with UL17. The reason that UL17 is a constituent of both the capsid and the tegument layers of the virion is unclear. At this stage it is also not known whether UL17 is fortuitously incorporated into the tegument because the capsid-bound UL17 interacts with a tegument protein or because it has a specific role later in the virus growth cycle or at very early times.
Acknowledgments
We thank D. McGeoch and N. Stow for critically reading the manuscript. The rabbit skin cells were a generous gift from B. Roizman. We are especially grateful to S. Graham for the isolation of the monoclonal antibodies, to Joyce Mitchell for wt virus L particles, to Colleen White for her contribution of UL6 monoclonal antibodies, and S. Weller for the plasmid pAPV.
This work was supported by the UK Medical Research Council. J. Thurlow was funded by an MRC Research Training studentship.
REFERENCES
- 1.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the growth of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booy, F. P., W. W. Newcomb, B. L. Trus, J. C. Brown, T. S. Baker, and A. C. Steven. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S. M., D. A. Ritchie, and J. H. Subak-Sharpe. 1973. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J. Gen. Virol. 18:329-346. [DOI] [PubMed] [Google Scholar]
- 4.De la Luna, S., and J. Ortin. 1992. Pac gene as an efficient dominant marker and reporter gene in mammalian cells. Methods Enzymol. 216:376-387. [DOI] [PubMed] [Google Scholar]
- 5.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiIanni, C. L., J. T. Stevens, M. Bolgar, D. R. O'Boyle, Jr., S. P. Weinheimer, and R. J. Colonno. 1994. Identification of the serine residue at the active site of the herpes simplex virus type 1 protease. J. Biol. Chem. 269:12672-12676. [PubMed] [Google Scholar]
- 7.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goshima, F., D. Watanabe, H. Takakuwa, K. Wada, T. Daikoku, M. Yamada, and Y. Nishiyama. 2000. Herpes simplex virus UL17 protein is associated with B capsids and colocalizes with ICP35 and VP5 in infected cells. Arch. Virol. 145:417-426. [DOI] [PubMed] [Google Scholar]
- 9.Grünewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 10.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 12.Kirkitadze, M. D., P. N. Barlow, N. C. Price, S. M. Kelly, C. Boutell, F. J. Rixon, and D. M. McClelland. 1998. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J. Virol. 72:10066-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matz, B., J. H. Subak-Sharpe, and V. G. Preston. 1983. Physical mapping of temperature-sensitive mutations of herpes simplex virus type 1 using cloned restriction endonuclease fragments. J. Gen. Virol. 64:2261-2270. [DOI] [PubMed] [Google Scholar]
- 15.McClelland, D. A., J. D. Aitken, D. Bhella, D. McNab, J. Mitchell, S. M. Kelly, N. C. Price, and F. J. Rixon. 2002. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J. Virol. 76:7407-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 17.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269-276. [DOI] [PubMed] [Google Scholar]
- 18.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 20.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Cheng, A. Steven, F. Booy, and J. C. Brown. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb, W. W., D. R. Thomsen, F. L. Homa, and J. C. Brown. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal protein complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel, A. H., and J. B. McLean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465-478. [DOI] [PubMed] [Google Scholar]
- 28.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 29.Preston, V. G., M. F. Al-Kobaisi, I. M. McDougall, and F. J. Rixon. 1994. The herpes simplex virus gene UL26 proteinase in the presence of the UL26.5 gene product promotes the formation of scaffold-like structures. J. Gen. Virol. 75:2355-2366. [DOI] [PubMed] [Google Scholar]
- 30.Preston, V. G., J. A. V. Coates, and F. J. Rixon. 1983. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 45:1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rixon, F. J., C. Addison, A. McGregor, S. J. Macnab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 77:2251-2260. [DOI] [PubMed] [Google Scholar]
- 32.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 34.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, K. O. 1964. Relationships between the envelope and the infectivity of herpes simplex virus. Proc. Soc. Exp. Biol. Med. 115:814-816. [DOI] [PubMed] [Google Scholar]
- 37.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szilágyi, J. F., and J. Berriman. 1994. Herpes simplex virus L particles contain spherical membrane-enclosed inclusion vesicles. J. Gen. Virol. 75:1749-1753. [DOI] [PubMed] [Google Scholar]
- 39.Szilágyi, J. F., and C. Cunningham. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72:661-668. [DOI] [PubMed] [Google Scholar]
- 40.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 41.Valpuesta, J. M., and J. L. Carrascosa. 1994. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys. 27:107-155. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, Z. H., B. V. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]