Abstract
1,25-Dihydroxyvitamin D3 [1,25(OH)2D3] induces the synthesis of 25-hydroxyvitamin D3 24-hydroxylase [24(OH)ase], an enzyme involved in its catabolism, thereby regulating its own metabolism. Here we demonstrate that CCAAT enhancer binding protein β (C/EBPβ) is induced by 1,25(OH)2D3 in kidney and in osteoblastic cells and is a potent enhancer of vitamin D receptor (VDR)-mediated 24(OH)ase transcription. Transfection studies indicate that 1,25(OH)2D3 induction of 24(OH)ase transcription is enhanced a maximum of 10-fold by C/EBPβ. Suppression of 1,25(OH)2D3-induced 24(OH)ase transcription was observed with dominant negative C/EBP or osteoblastic cells from C/EBPβ−/− mice. A C/EBP site was identified at positions −395 to −388 (−395/−388) in the rat 24(OH)ase promoter. Mutation of this site inhibited C/EBPβ binding and markedly attenuated the transcriptional response to C/EBPβ. We also report the cooperation of CBP/p300 with C/EBPβ in regulating VDR-mediated 24(OH)ase transcription. We found that not only 1,25(OH)2D3 but also parathyroid hormone (PTH) can induce C/EBPβ expression in osteoblastic cells. PTH potentiated the induction of C/EBPβ and 24(OH)ase expression in response to 1,25(OH)2D3 in osteoblastic cells. Data with the human VDR promoter (which contains two putative C/EBP sites) indicate a role for C/EBPβ in the protein kinase A-mediated induction of VDR transcription. From this study a fundamental role has been established for the first time for cooperative effects and cross talk between the C/EBP family of transcription factors and VDR in 1,25(OH)2D3-induced transcription. These findings also indicate a novel role for C/EBPβ in the cross talk between PTH and 1,25(OH)2D3 that involves the regulation of VDR transcription.
Vitamin D is a principal factor that maintains calcium homeostasis (14). The hormonally active form of vitamin D, 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], is produced by two sequential hydroxylations of vitamin D by 25-hydroxylase in the liver and by 25-hydroxyvitamin D3 1α-hydroxylase in the kidney. 1,25(OH)2D3 regulates gene expression in target cells by binding to the vitamin D receptor (VDR), a ligand-activated transcription factor that belongs to the nuclear receptor superfamily (26, 54). Ligand-activated VDR heterodimerizes with the retinoid X receptor (RXR), and the VDR/RXR complex binds to vitamin D response elements (VDREs) in the promoters of target genes (26, 34, 54). Recent studies have led to the identification of coactivators that play a role in mediating VDR transcriptional activity. In addition to general transcription factors, ligand-activated VDR is known to interact with p160 coactivators that have histone acetylase activity (steroid receptor coactivator/NCoA1, glucocorticoid receptor-interacting protein 1 [GRIP-1/TIF-2], and the activator and thyroid and retinoic acid receptors [ACT]/pCIP), as well as with the coactivator complex VDR-interacting proteins (DRIP) that act through the recruitment of the RNA polymerase II holoenzyme (14, 26, 34, 54, 55). Although the identification of cofactors involved in VDR-mediated transcription has been a major focus of research, relatively few 1,25(OH)2D3-regulated genes are known to occur in target tissues that maintain calcium homeostasis.
One of the most pronounced effects of 1,25(OH)2D3 is increased synthesis of the 25-hydroxyvitamin D3 24-hydroxylase [24(OH)ase] enzyme (50). 24(OH)ase is expressed at high levels in kidney and can be induced by 1,25(OH)2D3 in kidney, intestine, and osteoblastic cells as well as in many other tissues (50). Hydroxylation of 1,25(OH)2D3 at carbon 24 by 24(OH)ase is the first step in the metabolic inactivation of 1,25(OH)2D3 (59). Thus, 1,25(OH)2D3 regulates its own metabolism by inducing the 24(OH)ase enzyme, protecting against hypercalcemia. Other genes regulated by 1,25(OH)2D3 in target tissues that maintain calcium homeostasis include the calcium-binding protein calbindin (in intestine and kidney), which has been proposed to act as a facilitator of calcium diffusion, and the bone calcium-binding proteins osteocalcin and osteopontin (14).
We used a gene chip array in order to examine what other genes are regulated in response to 1,25(OH)2D3. We found that a gene, in addition to 24(OH)ase and calbindin, that is activated by a factor greater than 50% by 1,25(OH)2D3 in mouse kidney is the transcriptional activator CCAAT enhancer-binding protein β (C/EBPβ). Induction of C/EBPβ by 1,25(OH)2D3 was observed in primary murine osteoblasts as well as in mouse kidney. C/EBPβ belongs to the CCAAT enhancer binding protein family of transcription factors, which have been reported to be involved in the regulation of cell growth, differentiation, inflammation, and the expression of cell type-specific genes (22, 56). The C/EBP family of transcription factors consists of six members (C/EBPα to -ζ). They all contain a highly conserved basic leucine zipper domain at the C terminus that is involved in DNA binding and in dimerization. Different C/EBP family members are capable of forming heterodimers, and C/EBPs can also interact with other transcription factors (22, 56). C/EBPβ is expressed at relatively high levels in liver, intestine, adipose tissue, kidney, and myelomonocytic cells (56). In addition, recent studies have indicated that C/EBPβ and -δ are present in osteoblasts and that they play a role in the regulation of specific genes expressed in osteoblasts (osteocalcin, prostaglandin G/H synthase, and IGF-I) (15, 21, 23, 38, 66).
In the present study we report that C/EBPβ is a 1,25(OH)2D3 target and that one role for C/EBPβ as a target of 1,25(OH)2D3 is as an enhancer of VDR-mediated 24(OH)ase transcription. Thus, this study establishes a novel mechanism of cross talk between the C/EBP family of transcription factors and VDR in 1,25(OH)2D3-induced transcription. We found that not only 1,25(OH)2D3 but also PTH can induce C/EBPβ in osteoblastic cells. Since C/EBPβ was found to enhance PKA-mediated induction of VDR transcription, our findings also indicate for the first time a role for C/EBPβ in the cross talk between PTH and 1,25(OH)2D3 that involves enhancement of PKA-induced VDR transcription.
MATERIALS AND METHODS
Materials.
[14C]chloramphenicol (50 mCi/mmol), [γ-32P]ATP (3,000 Ci [111 TBq]/mmol), Easytag l-[35S] methionine (>1,000 Ci [37.0 TBq]/mmol), nylon membranes, and the enhanced-chemiluminescence (ECL) detection system were purchased from NEN Life Science Products (Boston, Mass.). Prestained molecular weight markers were obtained from Bio-Rad Laboratories, Inc. (Hercules, Calif). The TNT coupled reticulocyte lysate system was from Promega Corp. (Madison, Wis.). Acetyl coenzyme A, formamide, and the PTH fragment (residues 1 to 34) were obtained from Sigma (St. Louis, Mo.). Dulbecco's modified Eagle's medium (DMEM), DMEM plus Ham's F-12 nutrient mixture (DMEM-F-12), fetal bovine serum (FBS), charcoal-stripped FBS, a random-primer DNA labeling kit, T4 polynucleotide kinase, and oligo(dT) cellulose were purchased from Life Technologies, Inc. (Gaithersburg, Md.). H89 and Go-6983 were from Calbiochem (San Diego, Calif.). C/EBPβ antiserum was obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). 1,25(OH)2D3 was a generous gift from Milan Uskokovic (Hoffmann-LaRoche, Nutley, N.J.).
Cell cultures and animals.
COS-7 African green monkey kidney cells and UMR-106 rat osteoblastic cells were obtained from the American Type Culture Collection (Manassas, Va.) and were cultured in DMEM supplemented with 10% heat-inactivated FBS from Gemini Biological Products (Calabasas, Calif.) and DMEM-F-12 supplemented with 5% FBS, respectively. JEG-3 human choriocarcinoma cells were cultured in MEM supplemented with 10% FBS and 1 mM sodium pyruvate and 0.1 mM nonessential amino acids. Studies using the human VDR (hVDR) promoter were performed with JEG-3 cells since these cells express VDR and are strongly responsive to stimulation of the cyclic AMP (cAMP) signal transduction pathway (24). The hVDR promoter construct showed no activity or showed minimal response in rat UMR-106 osteoblastic cells as well as in murine and rat primary osteoblastic cells, perhaps due to species specificity. A 1% antibiotic mixture (penicillin, streptomycin, and neomycin) was added to all of the growth media. All cells were grown in a humidified atmosphere of 95% air-5% CO2 at 37°C. Cells were grown to 60 to 70% confluence and changed to medium supplemented with 2% charcoal-dextran-treated FBS before treatment. Osteoblast-enriched bone cells were isolated from 22-day-old fetal rat parietal bones or from neonatal murine calvaria by serial collagenase digestion as previously described (16, 37) and were cultured in DMEM supplemented with 10% FBS. Osteoblast-enriched bone cells were also obtained by collagenase digestion of calvaria from 19-day-old fetal or neonatal mice with a targeted deletion in the C/EBPβ locus (C/EBPβ−/− mice) and control wild-type mice (62). C/EBPβ−/− and wild-type mice were obtained for this study by breeding female heterozygous mice with a targeted deletion in the gene for C/EBPβ with heterozygous male mice. Mice were genotyped by PCR. C/EBPβ−/− mice have hypoglycemia, and 50% die within 1 to 2 h after birth (62). Osteoblastic cultures from these mice were established in MEM with lipoic acid supplemented with 15% fetal bovine serum. Cultures were combined based on genotype. For studies related to 1,25(OH)2D3 administration, C/EBP−/− mice or wild-type mice (3 to 4 months old) were injected with a single dose of 1,25(OH)2D3 (45 ng) and killed at 16 h after injection. Control mice received an intraperitoneal injection of 0.1 ml of vehicle (9:1 mix of propylene glycol and ethanol) and were killed 16 h later. Experiments involving animals for the preparation of osteoblastic cells were approved by the Institutional Animal Care and Use Committees at University of Medicine and Dentistry of New Jersey-New Jersey Medical School and Yale University (for studies using osteoblastic cells for Northern and Western blot analyses) and at Case Western Reserve University (for studies using osteoblastic cells from C/EBPβ−/− mice and wild-type controls). Cells were treated with vehicle or the compounds noted in the figures at the indicated times and concentrations.
Transfection constructs and transfections and assay of CAT or luciferase activity.
For transfection studies, constructs of a chimeric gene in which the rat 24(OH)ase promoter (positions −1367 to +74 [−1367/+74], which contain both VDREs at −258/−244 and −151/−137) was linked to the chloramphenicol acetyltransferase (CAT) gene and the deletion mutant construct (−671/+74 and −291/+74), previously described (27), were used for transfection in COS-7 cells. Within −671/+74 of the 24(OH)ase promoter is a putative C/EBP site at −395/−388. The putative C/EBP site, −395/−388 (TGGCAAG), was mutated to GACTAGT with the Quick Change site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The oligonucleotides used to generate the C/EBPβ-mutated site (shown in lowercase letters) were as follows: 5′-CTCCACTTGCgactagtGCGCGGGCGGCC-3′ (top strand) and 5′-GGCCGCCCGCGCactagtcGCAAGTGGAG-3′ (bottom strand). The mutated construct was generated by amplifying the original plasmid by PCR using Pfu DNA polymerase and the primers containing the desired mutation. Following digestion of the original plasmid by DpnI, Epicurian Coli XL 10-Gold ultracompetent cells were transformed with the nicked plasmid that contains the desired mutation. Plasmid DNA was isolated from the positive clones, and the mutation was confirmed by DNA sequencing. The following C/EBP expression plasmids were also used: pMEX-C/EBPβ and pMEX-C/EBPα (gifts of Simon Williams, Texas Tech University, Lubbock, Tex.). The C/EBPδ expression vector and a dominant negative C/EBP construct (C/EBP DN) were created as previously described (38). COS-7 cells were also transfected with the hVDR expression plasmid pAVhVDR (from J. W. Pike, University of Wisconsin, Madison). In some studies COS-7 cells were transfected with the AF-2-defective (L417S) VDR mutant (7), prepared in the laboratory of L. P. Freedman (Merck and Co., Inc., West Point, Pa.). A-CREB (a dominant negative CREB) was from C. Vinson (National Institutes of Health, Bethesda, Md.). The expression vector containing the inducible cAMP early repressor II-γ coding sequence (pSV2 ICER II-γ) was from C. Molina (UMDNJ—New Jersey Medical School, Newark) (24). COS-7 cells were used for transfection studies since they contained lower levels of endogenous C/EBP. The expression plasmid encoding the catalytic subunit of PKA was obtained from G. S. McKnight (University of Washington, Seattle). pRSV-CBP was described previously (45). pCMVp300 (53) was obtained from R. Stein (Vanderbilt Medical Center, Nashville, Tenn.). The E1A expression plasmid (pCMV E1A) containing the E1A 12S cDNA was a gift from Michael B. Matthews (UMDNJ—New Jersey Medical School, Newark). PCMV-YY1 and the pCMV vector were provided by T. Shenk, Princeton University, Princeton, N.J. In addition, cells were transfected with the β-galactosidase expression vector pCH110 (Pharmacia Biotech, Piscataway, N.J.) as an internal control for transfection efficiency. Empty vectors were used to keep the total DNA concentration the same. Cells were transfected using the calcium phosphate DNA precipitation method (6) or using the Lipofectamine 2000 reagent (Life Technologies, Inc.) according to the manufacturer's protocol. Twelve to sixteen hours after transfection, cells were shocked for 1 min with phosphate-buffered saline (PBS) containing 10% dimethyl sulfoxide, washed with PBS, and treated as described in Results in the appropriate medium supplemented with 2% charcoal-dextran-treated FBS. After treatment with vehicle or the compounds noted at the concentrations and times indicated in Results, cells were harvested and cell extracts were prepared by freezing and thawing five times. The CAT assay was performed using equivalent amounts of protein and/or with constant β-galactosidase activity (6, 19). Autoradiograms were analyzed by densitometric scanning using a model CS9000U dual-wavelength flying spot scanner (Shimadzu Scientific Instruments, Princeton, N.J.). For some experiments, different autoradiographic exposure times were needed for the densitometric analysis. CAT activity was also quantitated by scanning thin-layer chromatography plates with the Packard Constant Imager system (Packard Instrument Co., Meriden, Conn.). In some studies cells were transfected with the hVDR promoter (−1500/+60) luciferase construct, generated as previously described (24), or the C/EBPβ promoter luciferase constructs (−1400/+16, −426/+16, −168/+16, −121/+16, −121/+16MUT1, −121/+16MUT2, and −121/+16 MUT1 and -2) previously described (46). These constructs were named LAPPRO1, LAPPRO2, LAPPRO7, LAPPRO8, LAPPRO8 MUT I, LAPPRO8 MUT II, LAPPRO8 MUT I, and LAPPRO8 MUT II, respectively, in the original publication (46). Luciferase results were quantitated in relative light units by using a luminometer.
Microarray analysis.
Gene expression analysis was done essentially as described in the Affymetrix GeneChip expression analysis technical manual (1). C57BL6 mice were made 1,25(OH)2D3 deficient by placing them on a 0.8% strontium diet [which has been reported to produce functional vitamin D deficiency by inhibiting the conversion of 25(OH)D3 to 1,25(OH)2D3 (4)] for 6 days as described previously (32, 63). Deficient mice were injected with 1,25(OH)2D3 [48, 24, and 6 h prior to sacrifice; 30 ng of 1,25(OH)2D3 per injection]. Total RNA was prepared from the kidneys of deficient and 1,25(OH)2D3-treated mice with RNA-Bee RNA extraction solution (Tel-Test, Friendswood, Tex.) by following the manufacturer's instructions. Polyadenylated [poly(A)+] mRNA was isolated from the total RNA by oligonucleotide (deoxythymidine)-cellulose column chromatography. cDNA synthesis was carried out at 37°C for 60 min with the T7-(dT)24 primer (Operon) and Superscript II (200 U/μg of RNA sample; Invitrogen, Carlsbad, Calif.). This cDNA was used as a template for the synthesis of cRNA with T7 RNA polymerase (Ambion, Inc., Austin, Tex.) (37°C, 4 to 5 h). The cRNA was purified on an affinity resin (QIAGEN RNeasy) minicolumn, biotin labeled, and hybridized to MG-U74 GeneChip mouse oligonucleotide array (Affymetrix) overnight at 45°C with the GeneChip model 640 hybridization oven (Affymetrix). Washing and staining was done using the Affymetrix GeneChip fluidics station 400 and standard Affymetrix protocols (1). Fluorescence intensities were captured with a GeneArray scanner II (Hewlett-Packard). Affymetrix Microarray suite 4.0 software was used to analyze the results. Hybridization of cRNA with the array chip was performed three times with different control and 1,25(OH)2D3-treated samples. A comparative analysis between treatment and control samples was performed with the Affymetrix statistical algorithm using default parameters.
Northern blot analysis, reverse transcription-PCR (RT-PCR), and Western blot analysis.
Total RNA was extracted from cultured cells and mouse kidney with the RNA-Bee reagent. Poly(A)+ RNA was isolated by oligo(deoxythymidine)-cellulose chromatography. Northern blot analysis was performed as previously described (68). 32P-labeled cDNA probes were prepared using random primer DNA labeling systems (Life Technologies, Inc.) according to the random primer method (6). The 400-bp C/EBPβ cDNA (encoding the 133 N-terminal amino acids to avoid possible cross-reaction with other family members [the C-terminal region is highly conserved]), used for Northern analysis, was obtained by NcoI and HindIII digestion of pMEX C/EBPβ (from Simon Williams, Texas Tech University, Lubbock, Tex.). The 3.2-kb rat 24(OH)ase cDNA was obtained by EcoRI digestion and was a gift from K. Okuda (Hiroshima University School of Dentistry, Hiroshima, Japan [48]). The β-actin cDNA was purchased from Clontech Laboratories, Inc. (Palo Alto, Calif.). The blots were hybridized to the 32P-labeled cDNA probes for 16 h at 42°C, washed, air dried, and exposed to Kodak BIOMAX MR film at −80°C in the presence of intensifying screens. Autoradiograms were analyzed by densitometric scanning using the dual-wavelength flying spot scanner. The relative optical density obtained using the C/EBPβ or the 24(OH)ase probe was divided by the relative optical density obtained after probing with β-actin to normalize for sample variation.
For RT-PCR analysis of 24(OH)ase mRNA in primary osteoblastic cells, RT-PCR was performed using 1 μg of total RNA and the Superscript one-step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen). Primers used were 24(OH)ase (5′-GCCGAGCCTGCTGGAA3′ and 5′-CCCCATAAAATCAGCCAAGAC-3′) and actin (5′-CCTGTGGATCTGACAGCTGAA-3′ and 5′-TCCCAAATCGGTTGGAGATA-3′). PCR cycle numbers used were as follows: with 24(OH)ase, 32 cycles, and with actin, 30 cycles. The cycles were chosen so that the amplification was conducted in the linear range of amplification efficiency. The resulting PCR products were subjected to electrophoresis on a 1% agarose gel containing ethidium bromide, and bands were visualized under UV light. Gel data were recorded using the Gene Genius bioImaging system (Syngene, Frederick, Md.), and relative densities of the bands were determined using Gene Tool software (Syngene). Data were normalized for the expression of β actin within the sample.
For Western blot analysis, total cellular protein was prepared using a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 1.0% NP-40, and protease inhibitors. Fifty micrograms of protein was separated by SDS-10% polyacrylamide gel electrophoresis (PAGE) and analyzed by Western blot analysis as previously described (7) by the enhanced-chemiluminescence Western blotting detection system (NEN Life Sciences). Protein concentration was assayed by the method of Bradford (11).
Nuclear extracts.
C/EBPβ was expressed in transiently transfected COS-7 cells. Cells were transfected by calcium phosphate precipitation using 5 μg of the expression plasmid pMEX-C/EBPβ. For nuclear extract preparation from COS-7 cells as well as from osteoblastic cells, cells were rinsed twice with PBS at 4°C, harvested by scraping, gently pelleted, washed, and lysed in hypotonic buffer containing 10 mM HEPES (pH 7.4), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, phosphatase inhibitors (1 mM sodium orthovanadate, 10 mM sodium fluoride), protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 1 mg of pepstatin A per ml, 2 mg of leupeptin per ml, 2 mg of aprotinin per ml), and 1% Triton X-100. Nuclei were pelleted at 3,500 × g for 5 min, and cytoplasmic supernatants were separated. Nuclei were resuspended in hypertonic buffer containing 0.42 M NaCl, 0.2 mM EDTA, 25% glycerol, and the phosphatase and protease inhibitors indicated above. Soluble nuclear proteins were released by 60 min of incubation at 4°C, and insoluble material was separated by centrifugation at 12,000 × g for 5 min. The protein concentration of the supernatant was measured by Bradford's method (11), and aliquots were stored at −80°C.
EMSA.
29-mer complementary oligonucleotides spanning the C/EBP motif of the 24(OH)ase promoter containing the wild-type (−395/−388) or the mutated (prepared by the UMD Molecular Resource Facility, Newark, N.J.) C/EBP site were used for electrophoretic mobility shift assays (EMSA). The sequences of the oligonucleotides used were 5′-CTC CAC TTG CTT GGC AAG CGC GGG CGG CC-3′ and 5′-GGC CGC CCG CGC TTG CCA AGC AAG TGG AG-3′ for the wild type and 5′-CTC CAC TTG Cga cta gtG CGC GGG CGG CC-3′ and 5′-GGC CGC CCG CGC act agt cGC AAG TGG AG-3′ for the mutant construct. The C/EBP motif is shown in boldface type, and the corresponding mutated sequences are in lowercase type. Overlapping oligonucleotide strands were heat denatured and annealed overnight. Fifty nanograms of duplex oligonucleotides were 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase (Invitrogen) and purified using a Micro Bio-Spin P-30 column (Bio-Rad). The eluted probe was used for EMSA as described previously (6). Briefly, aliquots of the nuclear preparations from C/EBPβ-transfected COS cells (2 to 8 μg of protein) were incubated for 20 min at 27°C with 2 μg of poly(dI-dC) with or without unlabeled specific or nonspecific DNA competitor or antibodies in binding buffer (4 mM Tris-HCl [pH 7.9], 1 mM EDTA [pH 8.0], 60 mM KCl, 12% glycerol, 12 mM HEPES, 1 mM dithiothreitol), followed by the addition of 0.3 to 0.5 ng of the labeled oligonucleotide probe (∼100,000 cpm) and incubation for 30 min at 27°C. For EMSA done using primary osteoblastic cells (38) treated with 1,25(OH)2D3 (10−8 M for 24 h), immunoblots were used to measure the C/EBPβ content of the nuclear extracts after 24 h of treatment. When the nuclear C/EBPβ content was changed due to the treatment, corrections were made so that equal amounts of C/EBPβ were used in the EMSA. The samples were separated by electrophoresis on a 6% nondenaturing polyacrylamide gel that had been preelectrophoresed for 30 min at 100 V/cm at 4°C in 45 mM Tris-45 mM boric acid-1 mM EDTA. Electrophoresis was conducted for 2.5 h under identical conditions. The dried gels were exposed to X-ray film at −80°C with an intensifying screen.
Protein-protein interaction assay.
Glutathione S-transferase (GST)-C/EBPβ [pGEX-C/EΒPβ22-297, created by S. C. Williams using PCR-generated fragments encoding amino acids 22 to 297 of C/EBP β inserted into pGEX-4T3 (Pharmacia), was grown in E. coli DH5α cells for 3 to 4 h and induced by isopropyl-β-d-thiogalactopyranoside (IPTG) for an additional 2 h. Cells were harvested by centrifugation and resuspended in 1/50 volume of PBS containing 0.1% Triton X-100, protease inhibitors (leupeptin, aprotinin, trypsin inhibitor, and phenylmethylsulfonyl fluoride). The bacteria were lysed by pulse sonication on ice. The lysate was clarified by centrifugation at 12,000 × g for 10 min at 4°C. Glutathione-Sepharose beads (500 μl) were washed with PBS containing 0.1% Triton X-100, and the supernatant obtained from the lysate (5 ml) was gently mixed with the glutathione-Sepharose beads for 2 to 3 h in binding buffer (50 mM Tris [pH 7.2], 1 mM EDTA, 1 mM dithiothreitol, 150 mM NaCl, and 0.1% Triton X-100). Beads containing the GST protein were collected by low-speed centrifugation, followed by three successive washes in the same buffer. pcDNA3-p3001-855, pcDNA3-p300806-1234, pcDNA3-p3001195-1673, and pcDNA3-p3001661-2414 were gifts from D. M. Livingston (18). p300 fragments were translated in vitro with the TNT translation system (TNT quick coupled transcription-translation system; Promega Corp., Madison, Wis.) in the presence of [35S]methionine. The labeled protein (4 μl of crude translated protein) was incubated with equal amounts of GST fusion protein coupled to glutathione-Sepharose, washed, eluted, and fractionated by SDS-PAGE and autoradiography.
For coimmunoprecipitation assays, nuclear extracts (NE) were isolated using the NE-PER nuclear extraction reagent kit from Pierce (Woburn, Mass.). NE were incubated with 1,25(OH)2D3 (10−8 M) or vehicle for 1 h at 4°C. After the incubation, 1 μg of primary antibody (CBP/p300, rabbit polyclonal from Santa Cruz Biotechnology) was added to the NE in 500 μl of immunoprecipitation buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) and incubated for 4 h with rotation at 4°C. Twenty-five microliters of protein A-Sepharose 4 fast flow beads (Amersham Biosciences) was added to the NE-antibody mix, and it was further incubated with rotation at 4°C for 1 h. After centrifugation at 1,500 × g at 4°C for 5 min, the supernatant was discarded and the immunoprecipitated complex was separated by SDS-10% PAGE and probed with C/EBPβ antibody.
Statistical analysis.
Results are expressed as means ± standard errors (SE), and significance was determined by analysis by Student's t test for two-group comparison or by analysis of variance for multiple-group comparison.
RESULTS
C/EBPβ mRNA induction by 1,25(OH)2D3 precedes the induction of 24(OH)ase mRNA.
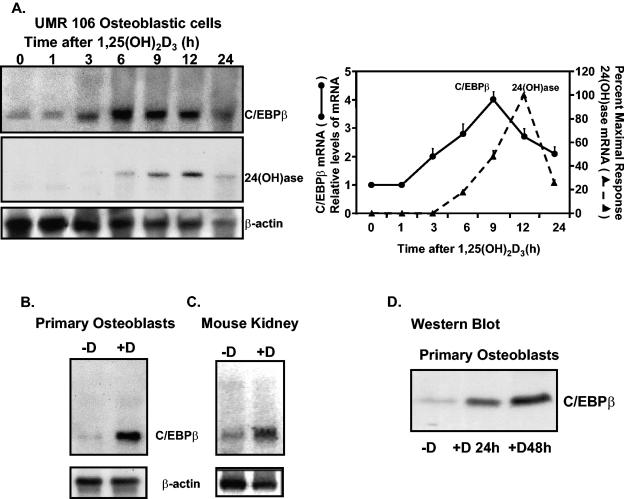
The effect of 1,25(OH)2D3 administered by intraperitoneal injection upon mRNA levels in vitamin D-deficient mouse kidney was examined using the GeneChip array. We observed that most array hybridization signals did not change significantly upon 1,25(OH)2D3 treatment. Genes that did alter their hybridization signals included genes that have previously been identified as 1,25(OH)2D3 regulated [24(OH)ase] (57.7- ± 0.4-fold induction; P < 0.05) and the calbindin-D28k gene (2.5- ± 0.2-fold induction; P < 0.05). A further 17 genes whose hybridization signals were activated by a factor greater than 50% by 1,25(OH)2D3 included the C/EBPβ gene, a transcriptional activator (4.4- ± 0.2-fold induction; P < 0.05), and the ceruloplasmin gene (4.9- ± 0.3-fold induction; P < 0.05). Because C/EBP family members have previously been reported to be regulated by other steroids and because of a possible interrelationship between C/EBPβ and 24(OH)ase [suggested by putative C/EBP sites in the 24(OH)ase promoter], we focused on C/EBPβ as a target of 1,25(OH)2D3. The induction of C/EBPβ by 1,25(OH)2D3 was verified by Northern and Western blot analyses. Induction of C/EBPβ mRNA was observed not only in mouse kidney (Fig. 1C) but also in UMR osteoblastic cells (Fig. 1A) and in primary osteoblasts (Fig. 1B). Induction of C/EBPβ protein in primary osteoblasts is shown in Fig. 1D. Results of Northern analyses indicate that the first significant induction of C/EBPβ by 1,25(OH)2D3 in UMR osteoblastic cells precedes the induction of 24(OH)ase mRNA (3 h compared to 6 h) (Fig. 1A), consistent with a possible role for C/EBPβ in the 1,25(OH)2D3 induction of 24(OH)ase transcription.
FIG. 1.
C/EBPβ is induced by 1,25(OH)2D3 in osteoblastic cells and in mouse kidney. (A, left panels) Representative autoradiogram. Northern analysis was performed using 5 μg of poly(A)+ RNA per lane from UMR osteoblastic cells that had been treated with 10−8 M 1,25(OH)2D3 and harvested at various times after treatment. The filter was hybridized with 32P-labeled C/EBPβ cDNA. The blots were then rehybridized with 32P-labeled rat 24(OH)ase and β-actin cDNAs. (Right panel) Graphic representation of Northern blot analyses of time-dependent effects of 1,25(OH)2D3 on the expression of C/EBPβ and 24(OH)ase mRNAs in UMR osteoblastic cells. Data represent the means ± SE of results of three to five independent experiments. The first significant induction in C/EBPβ mRNA occurred at 3 h (P < 0.05 compared to levels at zero time), while the first significant induction in 24(OH)ase mRNA was observed at 6 h (P < 0.05 compared to levels at zero time). (B) Northern blot analysis of poly(A)+ RNA from primary osteoblasts treated with vehicle (−D) or 1,25(OH)2D3 (10−8 M) (9 h). (C) Northern blot analysis of kidney from vitamin D-deficient mice treated with vehicle or 1,25(OH)2D3 as described in Materials and Methods. (B and C) Northern blots were hybridized with C/EBPβ cDNA followed by β-actin cDNA. Autoradiograms are representative of results observed in at least three different experiments. (D) Western blot analysis using NE from osteoblasts treated with vehicle (−D) or with 1,25(OH)2D3 (10−8 M) for 24 or 48 h.
C/EBPβ enhances the inductive action of 1,25(OH)2D3 on 24(OH)ase transcription.
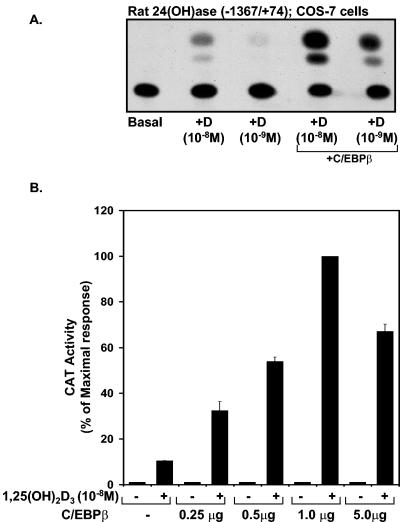
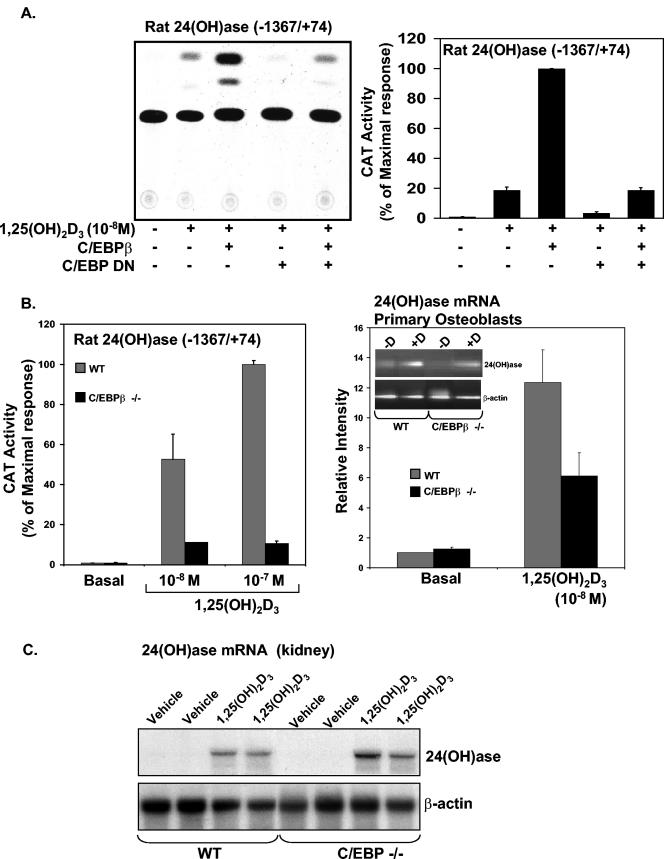
The rat 24(OH)ase promoter contains two functional VDREs (at −258/−244 and −150/−136) (27, 49). Since examination of the rat 24(OH)ase promoter suggests, by sequence homology, two putative C/EBP sites (at −395/−388 and at −964/−955), the possibility that C/EBPβ may be involved in the 1,25(OH)2D3 induction of 24(OH)ase transcription was tested in experiments in which COS-7 cells were transfected with the rat 24(OH)ase promoter (−1,367/+74) and the hVDR in the presence or absence of an expression plasmid driving the expression of C/EBPβ. In dose-response studies, we found that suboptimal 1,25(OH)2D3 induction of 24(OH)ase transcription is enhanced a maximum of 10-fold by C/EBPβ (Fig. 2). In the absence of 1,25(OH)2D3 and with C/EBPβ at 0.25 to 5.0 μg (Fig. 2, lower panel) or in the presence of 1,25(OH)2D3, the 24(OH)ase promoter failed to respond to C/EBPβ in COS-7 cells transfected with an AF-2-defective (L417S) VDR mutant (data not shown), indicating that C/EBPβ requires ligand-activated VDR in a transcriptionally active state to enhance 24(OH)ase transcription. A dose-dependent enhancement by C/EBPδ of the inductive action of 1,25(OH)2D3 was also observed (a maximum enhancement of 6.9- ± 0.6-fold was observed at a concentration of 0.5 μg of the C/EBPδ expression vector [data not shown]). Compared to C/EBPβ and -δ, although C/EBPα enhanced 1,25(OH)2D3-induced 24(OH)ase transcription, it had the least pronounced effect, resulting in a maximum enhancement of 2.2- ± 0.2-fold (at a concentration of 5.0 μg of the C/EBPα expression vector). To further confirm the role of C/EBPβ in the inductive action of 1,25(OH)2D3 on 24(OH)ase transcription, studies were done with COS-7 cells transfected with hVDR using C/EBP DN. This construct lacks the trans-activation domain sequence. However, the dimerization and DNA binding domains are intact (38). C/EBP DN has previously been shown to suppress both C/EBPβ- and C/EBPδ-dependent gene expression (38). C/EBP DN inhibited the enhancement of VDR-mediated 24(OH)ase transcription observed in the presence of C/EBPβ [Fig. 3A, compare the lane with 1,25(OH)2D3 and C/EBPβ and the lane with 1,25(OH)2D3, C/EBPβ, and C/EBP DN (third lane versus fifth lane; P < 0.05)]. The induction by 1,25(OH)2D3 of 24(OH)ase transcription was also, in part, inhibited by C/EBP DN [Fig. 3A, compare the lane with 1,25(OH)2D3 and the lane with 1,25(OH)2D3] and C/EBP DN (second lane versus fourth lane; P < 0.05). The involvement of C/EBPβ in VDR-mediated 24(OH)ase transcription was also examined using primary osteoblasts isolated from mice with a targeted deletion in the C/EBPβ locus (C/EBPβ−/− mice) (62). Using osteoblastic cells from C/EBPβ−/− mice, 1,25(OH)2D3-induced 24(OH)ase transcription and 24(OH)ase mRNA were significantly decreased compared to 1,25(OH)2D3-induced 24(OH)ase transcription and mRNA observed with osteoblastic cells from control (wild-type) littermates (P < 0.05) (Fig. 3B, left and right panels, respectively). Significant levels (compared to the basal level) of 1,25(OH)2D3-induced transcription and mRNA were maintained in the C/EBPβ−/− mice (Fig. 3B), indicating that other factors are involved in 1,25(OH)2D3 induction of 24(OH)ase. When wild-type and C/EBPβ−/− mice were injected with 45 ng of 1,25(OH)2D3 and then killed after 24 h, 24(OH)ase mRNA was markedly induced in the kidneys of both wild-type and C/EBPβ−/− mice (Fig. 3C). No difference in the levels of induction of 24(OH)ase mRNA in the kidney was observed (P > 0.5), suggesting the possibility of compensatory mechanisms involving other C/EBP isoforms
FIG. 2.
Enhancement 1,25(OH)2D3-induced 24(OH)ase transcription by C/EBPβ. (A) COS-7 cells were cotransfected with 4 μg of the rat 24(OH)ase promoter CAT construct −1367/+74 and hVDR expression plasmid in the absence or presence (last two lanes) of pMEX C/EBPβ. Empty vectors were used to keep the total DNA concentration the same. A representative autoradiogram is shown. (B) CAT activity is represented graphically as percentages of the maximal response (mean ± SE; three to eight observations per group). COS-7 cells were transfected with the 24(OH)ase CAT reporter plasmid −1367/+74, hVDR, and increasing concentrations of the C/EBPβ expression vector (0.25 to 5.0 μg).
FIG.3.
Suppression of 1,25(OH)2D3-induced 24(OH)ase transcription using C/EBP DN or osteoblastic cells from C/EBPβ−/− mice. (A) C/EBP DN inhibits C/EBPβ enhancement and suppresses 1,25(OH)2D3-induced 24(OH)ase transcription. (Left panel) Representative autoradiogram. COS-7 cells were cotransfected with the CAT reporter plasmid [4 μg of the rat 24(OH)ase promoter construct −1367/+74 and the hVDR expression plasmid in the presence or absence of pMEX C/EBPβ and/or C/EBP DN]. Empty vectors were used to keep the total DNA concentration the same. Cells were treated with vehicle or 1,25(OH)2D3 (10−8 M). (Right panel) Graphic representation of the results of three separate experiments are expressed as mean percentages of the maximal response ± SE. C/EBPβ-mediated enhancement of transcription (third bar) and 1,25(OH)2D3-induced 24(OH)ase transcription (second bar) are significantly inhibited by C/EBP DN (second bar versus fourth bar and third bar versus fifth bar; P < 0.05). (B) Reduced VDR-mediated 24(OH)ase transcription and mRNA in osteoblastic cells isolated from C/EBPβ−/− mice. (Left and right panels) Primary osteoblastic cells were prepared from C/EBPβ−/− mice and control littermates (WT). (Left panel) Osteoblastic cells were transfected with the rat 24(OH)ase promoter (−1367/+74) and treated with 1,25(OH)2D3 (10−8 or 10−7 M). A significant inhibition of 1,25(OH)2D3-induced 24(OH)ase transcription was observed using cells from C/EBPβ−/− mice (P < 0.05 compared to values for the wild type). (Right panel) RT-PCR analysis of 24(OH)ase mRNA. A significant inhibition of 1,25(OH)2D3-induced 24(OH)ase mRNA was observed using osteoblastic cells from C/EBPβ−/− mice (P < 0.05 compared to values for the wild type). (C) Northern blot analysis of 24(OH)ase mRNA in the kidneys of C/EBPβ−/− and wild-type mice. C/EBPβ−/− or wild-type mice (3 to 4 months old) received either vehicle or 1,25(OH)2D3 (45 ng) 16 h before termination. A representative Northern blot is shown. There was no significant difference in 24(OH)ase mRNA levels induced by 1,25(OH)2D3 in the kidneys of the C/EBPβ−/− mice and in wild-type mice (P > 0.5).
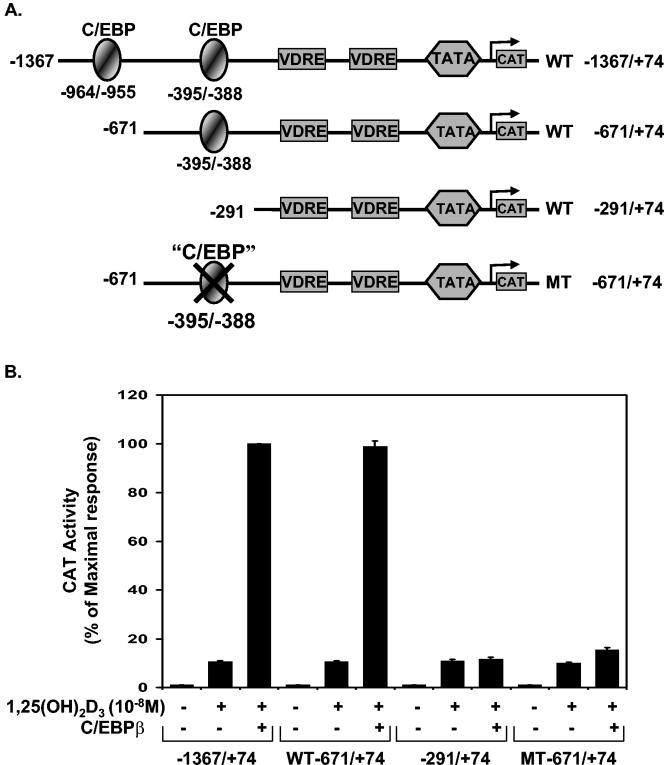
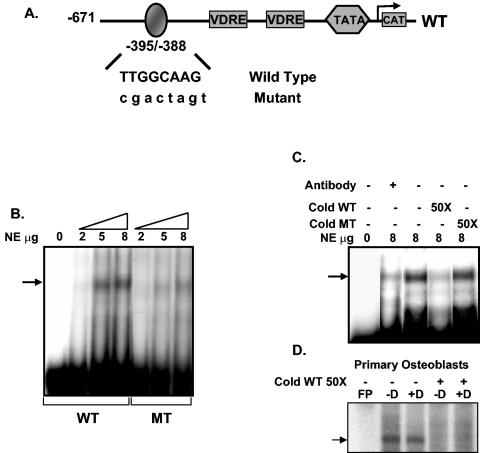
Identification of the C/EBPβ binding site in the rat 24(OH)ase promoter.
We next examined which sequences in the 24(OH)ase promoter were involved in this transactivation by C/EBPβ. The −1367/+74 24(OH)ase promoter construct contained both putative C/EBP sites (at −964/−955 and at −395/−388). Deletion of the 24(OH)ase promoter upstream of position −395 (−671/+74 phCAT) did not affect the enhancement of transcription by C/EBPβ (Fig. 4). Deletion of the 24(OH)ase promoter to −291, which did not include the −395/−388 C/EBP binding site, eliminated the ability of C/EBPβ to enhance transcription from the 24(OH)ase promoter. The C/EBP binding site at −395/−388 (TTGGCAA) differs from the reported optimal C/EBP binding site (TTGCGCAA) by a single base (51). To establish that this element directly mediates the enhancement of VDR-mediated 24(OH)ase promoter activity by C/EBPβ, site-directed mutagenesis was performed. Mutation of this site within the −671/+74 24(OH)ase promoter construct (MT −671/+74) blocked the response to C/EBPβ, suggesting that C/EBPβ acts through this site to enhance transcription from the 24(OH)ase promoter.
FIG. 4.
Identification of the C/EBPβ activation domain in the rat 24(OH)ase promoter. (A) Schematic of CAT constructs of the 24(OH)ase promoter and specific mutations and deletions introduced in the promoter as described in Materials and Methods. Regulatory sequences in the rat 24(OH)ase promoter constructs −1367/+74 and −671/+74, including two putative C/EBP motifs and the two VDREs, are shown. The −671/+74 rat 24(OH)ase promoter with a mutated C/EBP motif (GACTAGT) is also represented here. The sequence of the rat 24(OH)ase C/EBP motif at −395/−388 (TTGGCAA) differs from the reported optimal C/EBP binding site (TTGCGCAA) by a single base (51). (B) CAT activities determined in extracts of COS-7 cells transfected with hVDR and the CAT constructs of the 24(OH)ase promoter shown in A with or without cotransfection of 1 μg of the C/EBPβ expression vector. CAT activity is represented as a percentage of the maximal response (mean ± SE; three to six observations per group). Cells were treated with vehicle or 1,25(OH)2D3 (10−8 M).
The ability of C/EBPβ to bind this element was tested by gel mobility shift assays using synthetic oligonucleotides corresponding to the wild-type (−395/−388) or mutated C/EBP binding sequences (Fig. 5A) and nuclear extracts from C/EBPβ-transfected COS-7 cells. C/EBPβ protein interacted with the wild-type oligonucleotide in a dose-dependent manner, while the oligonucleotide containing the mutated C/EBP sequences failed to show efficient binding (Fig. 5B). Preincubation with C/EBP antibody (Fig. 5C, lane 2) or cold wild-type oligonucleotide (Fig. 5C, lane 4) but not mutant oligonucleotide (Fig. 5C, lane 5) depleted the binding of C/EBPβ to the labeled probe, indicating the specificity of this interaction. No increase in the binding to the C/EBP site was observed using nuclear extracts from primary osteoblastic cells treated with 1,25(OH)2D3 (Fig. 5D). Similar results were obtained using nuclear extracts from 1,25(OH)2D3-treated UMR osteoblastic cells (not shown).
FIG. 5.
C/EBPβ binding motif in the 24(OH)ase promoter detected by EMSA. (A) Schematic of the regulatory sequences in the −671/+74 rat 24(OH)ase promoter, including the C/EBP site at −395/−388, and the two VDREs. Oligonucleotides corresponding to the wild-type (TTGGCAAG) or mutated (CGACTAGT) C/EBP site were used for EMSA. (B) 32P-labeled oligonucleotide probes containing wild-type or mutated sequences were incubated with increasing concentrations (2 to 8 μg) of nuclear protein from COS-7 cells transfected with the C/EBPβ expression vector. (C) 32P-labeled oligonucleotide containing the wild-type C/EBP site from the 24(OH)ase gene (−395/−388) was incubated with 8 μg of nuclear protein alone (lane 3) or in the presence of C/EBPβ antibody (lane 2) or a 50-fold molar excess of wild-type (WT) or mutant (MT) cold competitor oligonucleotide. (D) The 32P-labeled oligonucleotide containing the wild-type C/EBP site (−395/−388) was incubated with nuclear protein prepared from primary osteoblastic cells treated with vehicle (−D) or 10−8 M 1,25(OH)2D3 (+D) for 24 h. Preincubation with cold C/EBP site (cold WT 50×) depleted the binding to the labeled probe. FP refers to free probe. Gel mobility shift data are representative of at least three experiments.
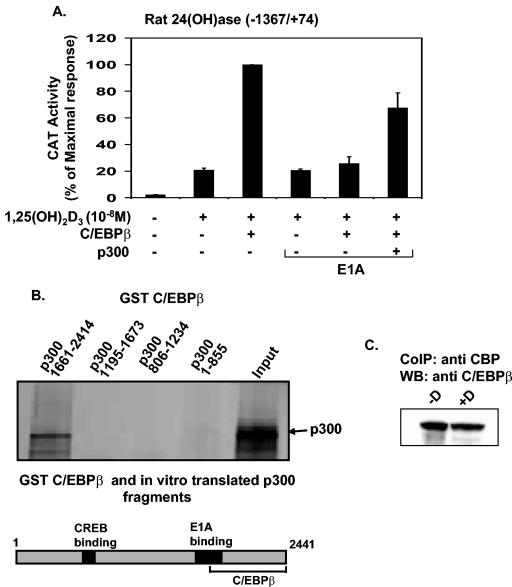
Functional association between CBP/p300 and C/EBPβ in the regulation of VDR-mediated 24(OH)ase transcription.
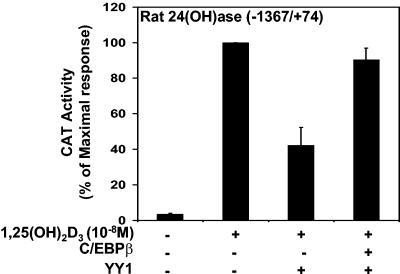
To examine the possibility that CBP/p300 may play a role in the enhancement of 24(OH)ase transcription by C/EBPβ, we studied the effect of the adenovirus E1A protein, which blocks the association of CBP/p300 with coactivators, on C/EBPβ-mediated transactivation (30). C/EBPβ was cotransfected in COS-7 cells with VDR and the rat 24(OH)ase promoter construct −1367/+74 in the presence or absence of the E1A expression vector. As shown in Fig. 6A, E1A inhibited the stimulatory effect of C/EBPβ on VDR-mediated 24(OH)ase transcription [compare transcription with 1,25(OH)2D3 and C/EBPβ in the absence and presence of E1A; P < 0.05]. Transfection and overexpression of p300 relieved the inhibitory effect of E1A (Fig. 6A, last lane). Similar results were observed using the CBP expression vector (not shown). Although relief of the inhibitory effect of E1A was observed in the presence of p300, transfection of the p300 expression vector (at concentrations from 1 to 5 μg) did not enhance 1,25(OH)2D3-induced 24(OH)ase transcription (data not shown), consistent with our previously reported findings (57). GST pull-down assays indicated that C/EBPβ interacts with the C terminus of p300 (positions 1661 to 2414). The N-terminal region of p300, containing the CREB binding domain, did not bind to C/EBPβ (Fig. 6B). Using UMR osteoblastic cells and coimmunoprecipitation, 1,25(OH)2D3 was not found to affect the interaction between C/EBPβ and CBP (Fig. 6C). We previously showed that the C terminus of p300/CBP can bind to YY1, which inhibits VDR-mediated 24(OH)ase transcription (57). Thus, YY1 represses 24(OH)ase transcription in part by sequestering coactivator proteins. Since both C/EBPβ and YY1 can bind to the C terminus of p300/CBP, we examined the effect of C/EBPβ on YY1-mediated repression of 24(OH)ase transcription. C/EBPβ was found to relieve YY1-mediated repression [Fig. 7; compare data obtained with 1,25(OH)2D3 plus YY1 versus data obtained with 1,25(OH)2D3 plus YY1 plus C/EBPβ; P < 0.05]. In the presence of YY1, C/EBPβ relieved the repression but was unable to enhance the response over 1,25(OH)2D3-induced levels observed in the absence of YY1 (Fig. 7). Since YY1 binds TFIIB, which is required for VDR-mediated transcription, as well as CBP/p300 (57), in the presence of 2 μg YY1, 1 μg of C/EBPβ may not be sufficient to completely overcome the inhibitory effects of YY1 and results not only in a reversal of the inhibition but also in an enhancement. These findings suggest that a role for CBP/p300 may be to provide a means of cross talk between key transcriptional regulators of the 24(OH)ase gene resulting in either enhancement or inhibition of responsiveness to 1,25(OH)2D3.
FIG. 6.
Functional cooperation between CBP/p300 and C/EBPβ. (A) COS-7 cells were cotransfected with the CAT reporter plasmid [4 μg of the rat 24(OH)ase promoter construct −1367/+74] and hVDR expression plasmid in the presence or absence of pMEX C/EBPβ, pCMV p300, and pCMV E1A 12S as indicated. Empty vectors were used to keep the total DNA concentration the same. The results are expressed as percentages of the maximum response (means ± SE) and are representative of three or more separate experiments. E1A significantly inhibited the C/EBPβ enhancement of 1,25(OH)2D3-induced transcription [1,25(OH)2D3 plus C/EBPβ plus E1A versus 1,25(OH)2D3 plus C/EBPβ; P < 0.05). At the concentration of pCMV E1A 12S used (4 ng), there was no effect of E1A on 1,25(OH)2D3-induced 24(OH)ase transcription. However at a higher concentration (200 ng of the E1A expression vector), suppression of the 1,25(OH)2D3 response was observed (data not shown). (B) Cooperation between CBP/p300 and C/EBPβ by direct protein-protein interaction to regulate VDR-mediated 24(OH)ase transcription. GST or the GST fusion proteins p300-(1-855), p300-(806-1234), p300-(1195-1673), and p300-(1661-2414) were bound to glutathione-Sepharose beads, and equal amounts of fusion proteins were incubated with in vitro-translated 35S-labeled C/EBPβ. After extensive washing, the bound proteins were eluted in SDS loading buffer and analyzed by SDS-10% PAGE followed by autoradiography. (Lower panel) Schematic representation of p300 showing the region that binds C/EBPβ. (C) Interaction of CBP/p300 in mammalian cells after incubation of UMR cell nuclear extracts in the presence of vehicle or 10 to 8 M 1,25(OH)2D3. Complexes were coprecipitated (CoIP) using CBP/p300 antibody and protein A beads. The immunoprecipitated complex was separated by SDS-PAGE and probed with C/EBPβ antibody. 1,25(OH)2D3 did not alter the interaction between CBP/p300 and C/EBPβ. Data are representative of three experiments. WB, Western blot.
FIG. 7.
C/EBPβ relieves YY1-mediated inhibition of 24(OH)ase transcription. COS-7 cells were cotransfected with the CAT reporter plasmid [4 μg of the rat 24(OH)ase promoter construct −1367/+74] and the hVDR expression plasmid in the presence or absence of pCMV-YY1 alone (2 μg) or pCMV-YY1 (2 μg) and pMEX C/EBPβ (1 μg). Empty vectors were used to keep the total DNA concentration the same. The results are expressed as percentages of the maximum response (means ± SE; three observations per group).
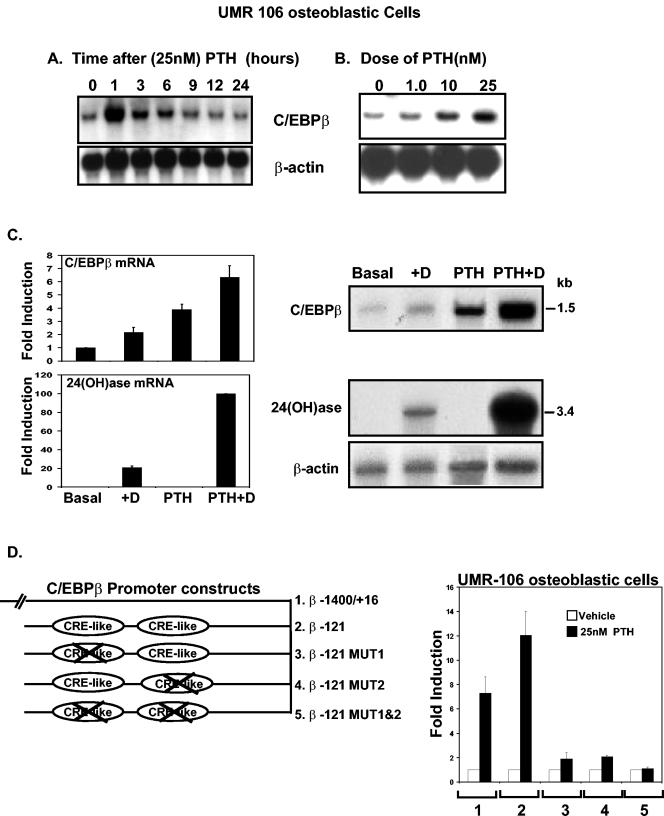
C/EBPβ mRNA is induced by PTH as well as by 1,25(OH)2D3.
We found that not only 1,25(OH)2D3 but also PTH can induce C/EBPβ expression in osteoblastic cells. Results of Northern analysis are shown in Fig. 8. When UMR-106-01 osteoblastic cells were treated with 25 nM PTH, the maximal induction of C/EBPβ mRNA was observed at 1 h (19- ± 3-fold; P < 0.05 compared to values at zero time) (Fig. 8A). By 24 h after PTH treatment, levels of C/EBPβ mRNA were similar to those observed in control cells (P > 0.5, values at zero time versus those at 24 h) (Fig. 8A). Induction of C/EBPβ mRNA by PTH was dose dependent. The first significant induction was observed at 1 nM PTH (1.4- ± 0.1-fold; P < 0.05 versus the value with the vehicle control [lane 0]), and the highest level of induction was observed at 25 nM PTH (3.8- ± 0.5-fold at 4 h after treatment) (Fig. 8B). Treatment of UMR-106-01 cells with 1,25(OH)2D3 (10−8 M) for 16 h followed treatment with PTH for 4 h (25 nM) [PTH plus 1,25(OH)2D3] resulted in a 3.2-fold enhancement of C/EBPβ mRNA and a 5-fold enhancement of 24(OH)ase mRNA (Fig. 8C) over the levels observed with 1,25(OH)2D3 alone. When mRNA was prepared from osteoblastic cells from wild-type or C/EBPβ−/− mice, although a decrease was observed in the 1,25(OH)2D3 response in osteoblasts from C/EBPβ−/− mice (Fig. 3B, right panel), after treatment in the presence of 25 nM PTH for 4 h, the 1,25(OH)2D3 response was enhanced similarly in the osteoblastic cells from both wild-type and C/EBPβ−/− mice (not shown). Since we observed that C/EBPδ as well as C/EBPβ can be induced by PTH in osteoblastic cells, it is possible that C/EBPδ can compensate for the lack of C/EBPβ.
FIG. 8.
C/EBPβ mRNA and transcription are induced by PTH in UMR osteoblastic cells. (A) Time-dependent effect of PTH (25 nM) on C/EBPβ mRNA in UMR-106 osteoblastic cells. Northern analysis was performed using total RNA isolated from UMR-106 osteoblastic cells that had been treated with 25 nM PTH or a vehicle control and harvested at various times after PTH treatment. The filter was hybridized with 32P-labeled C/EBPβ cDNA and rehybridized with β-actin cDNA. A representative autoradiogram is shown. Similar results were observed in threeseparate experiments. (B) Dose-dependent effect of PTH on C/EBPβ mRNA in UMR-106 osteoblastic cells. Northern analysis was performed using total RNA isolated from UMR cells treated for 4 h with the indicated concentrations of PTH. The filter was hybridized with 32P-labeled C/EBPβ cDNA and rehybridized with β-actin cDNA. A representative autoradiogram is shown. Similar results were observed in three separate experiments. (C) PTH enhances 1,25(OH)2D3-induced C/EBPβ and 24(OH)ase mRNA expression in UMR osteoblastic cells. (Left panels) Graphic representation of Northern analyses. Northern blot analysis was performed using total RNA isolated from UMR cells that had been treated with PTH (25 nM) for 4 h followed by treatment with 1,25(OH)2D3 (10−8 M) for 16 h. The filters were hybridized with 32P C/EBPβ and then stripped and rehybridized sequentially with 32P-24(OH)ase cDNA and β-actin cDNA. (Right panels) Representative autoradiogram. Data are from three independent experiments (means ± SE). (D) PTH-dependent activation of functional CREs in the proximal region of the C/EBPβ gene promoter. (Left panel) Schematic of deletion and mutation luciferase reporter constructs containing the 5′ upstream regulatory sequences of the C/EBPβ promoter. Mutations introduced into the C/EBPβ promoter fragment −121/+16 (diagram 2) disrupt either the CRE response element at −111/−107 (diagram 3) or the CRE response element at −65/−61 (diagram 4) or both (diagram 5) CRE response elements. (Right panel) UMR osteoblastic cells were transfected with 1.4 kb of the C/EBPβ promoter construct (bars 1) or with deletion and mutation luciferase reporter constructs (bars 2 to 5) (the numbers represent the specific constructs used shown in the left panel). Cells were treated with vehicle (white bars) or PTH (25 nM) (black bars) for 24 h. The relative luciferase activity of the respective transfection without stimulation was set to 1, and the changes after PTH stimulation are shown as increases in induction (n-fold). Increases in the induction (n-fold) of luciferase activity are given as means of results of three independent experiments ± SE.
To examine the mechanism of activation of C/EBPβ by PTH and 1,25(OH)2D3, UMR-106 osteoblastic cells were transfected with different deletion constructs of the C/EBPβ promoter. These constructs (−1400/+16, −426/+16, −168/+16, and −121/+16) were found to be unresponsive to 1,25(OH)2D3 (10−9 to 10−7 M) (data not shown). The C/EBPβ promoter constructs were also unresponsive to 1,25(OH)2D3 when VDR-transfected COS-7 cells or primary osteoblasts were used, suggesting possible posttranscriptional regulation of C/EBPβ by 1,25(OH)2D3. However, a region located 121 bp upstream of the start site of transcription was found to be required for activation of the C/EBPβ promoter by PTH (Fig. 8D). We found that the first significant induction is observed at 1 nM PTH and that maximum induction is at 25 nM PTH (12- ± 2.0-fold). The PKA inhibitor H89 (5 μM), a dominant negative CREB (1 μg), and the ICER (1 μg) blocked the PTH-dependent effect on C/EBPβ promoter activity (data not shown), suggesting that PTH activation occurs via a PKA-dependent pathway. The PKC inhibitor Go-6983 (5 μM) did not affect the PTH-dependent induction of C/EBPβ transcription. Mutation constructs of the C/EBPβ promoter demonstrated that the activation of the C/EBPβ promoter by PTH is mediated through two CREB binding sites at −111/−107 and at −65/−61 (Fig. 8D).
We previously reported that PTH can enhance VDR-mediated 24(OH)ase transcription in osteoblastic cells (24). Since the enhancement by PTH and/or activation by PKA of 1,25(OH)2D3 action in osteoblastic cells has been suggested to be due, at least in part, to the upregulation of VDR (24, 72), we tested the possibility that the induction of C/EBPβ may have an effect on the transcription of the VDR. The hVDR promoter (−1500/+60) luciferase construct, which contains two putative C/EBP sites (Fig. 9A), was cotransfected with an expression vector for the catalytic subunit of PKA (41) in the presence or absence of the C/EBPβ expression vector. Expression of PKA resulted in a 3.5-fold induction of hVDR promoter activity that was further enhanced in the presence of C/EBPβ (PKA versus PKA with C/EBPβ; P < 0.05) (Fig. 9B, bar 2 versus bar 3). C/EBPβ alone did not affect hVDR transcription (Fig. 9B, bar 4). C/EBP DN (100 ng of the expression vector) (Fig. 9B, bars 5 to 7) inhibited C/EBPβ-mediated enhancement (PKA and C/EBPβ [bar 3]) versus results with PKA, C/EBPβ, and C/EBP DN [bar 7]; P < 0.05). At higher doses of C/EBP DN, C/EBPβ enhancement was inhibited and PKA-induced VDR transcription was significantly suppressed as well (Fig. 9B, bar 2 [with PKA] versus bar 9 [with PKA and C/EBP DN] and bar 2 versus bar 12; P < 0.05). At the highest concentration of C/EBP DN, the basal level of hVDR promoter activity was also significantly affected (Fig. 9B, bar 1 versus bar 11; P < 0.05). These finding suggest a role for C/EBPβ in PKA-induced VDR transcription.
FIG. 9.
C/EBPβ enhances PKA-mediated transcription of hVDR. (A) Schematic of the luciferase construct of the hVDR promoter including two putative C/EBP motifs and two putative CRE sites. (B) JEG cells were cotransfected with 2 μg of the hVDR promoter construct −1500/+60 in the presence or absence of 1 μg of the PKA catalytic subunit expression vector, 5 μg of pMEX C/EBPβ, and increasing concentrations of C/EBP DN (100, 250, and 500 ng; bars 5 to 7, 8 to 10, and 11 to 13, respectively). Empty vectors were used to keep the total DNA concentration the same. Results of three or more separate experiments are presented (means ± SE). C/EBPβ enhancement of PKA-induced VDR transcription is significantly inhibited by 100, 250, or 500 ng of C/EBP DN (with PKA and C/EBPβ versus with PKA and C/EBP DN; bar 3 versus bar 7, bar 3 versus bar 10 or bar 3 versus bar 13; P < 0.05). The PKA induction of VDR transcription is significantly inhibited by 250 and 500 ng of C/EBPβ DN (PKA versus PKA plus 250 or 500 ng of C/EBP DN; bar 2 versus bar 9 or bar 2 versus bar 12; P < 0.05). Basal levels of hVDR transcription are inhibited at 500 ng of C/EBP DN (basal level plus 500 ng of C/EBPβ DN versus basal level; bar 11 versus bar 1; P < 0.05).
DISCUSSION
The results of this study establish C/EBPβ as a 1,25(OH)2D3 target gene that enhances the 1,25(OH)2D3 response of the 24(OH)ase gene. We describe for the first time the functional cooperation between the C/EBP family of transcription factors and VDR in 1,25(OH)2D3-induced transcription. Regulation of C/EBP members by other steroid hormones has previously been reported. Glucocorticoids enhance the expression of C/EBPβ and C/EBPδ in osteoblasts as well as in other cell types (35, 38, 73) and thyroid hormone (T3) regulates C/EBPα in adipocytes (42). An interrelationship between other steroid receptors and C/EBP proteins in the regulation of target genes has also been noted. Similar to the effect of C/EBPβ on VDR-mediated 24(OH)ase transcription, the glucocorticoid receptor (GR) and C/EBPβ act cooperatively in the regulation of a number of genes, including those for phosphoenolpyruvate carboxykinase (PEPCK), α-1 acid glycoprotein, herpes simplex virus thymidine kinase, and β casein (2, 10, 47, 70, 71). C/EBPβ has also been reported to cooperate with the thyroid receptor in mediating T3 induction of PEPCK transcription (52). However steroid receptors and C/EBP proteins do not always cooperate to enhance the regulation of gene expression. Ligand-activated retinoic acid receptor inhibits the transcriptional activity of C/EBP proteins, and the regulatory effect of C/EBPs on interleukin-6 (IL-6) and cAMP-induced IGF-I expression in osteoblastic cells is blocked by the hormone-activated estrogen receptor (ER) (40, 61, 65). It has been reported that C/EBPβ can bind to GR, ER, and retinoic acid receptor (10). It was suggested that the binding of GR to C/EBPβ enhances the ability of C/EBPβ to activate transcription and that the ability of C/EBPβ to bind to ER and RAR may be important for the repression of C/EBPβ activation (10). In vitro-translated VDR is able to bind to C/EBPβ (P. Dhawan and S. Christakos, unpublished data). It will be of interest in future studies to determine the functional significance in the cell of the potential for direct binding between VDR and C/EBPβ.
In our study we found that there was no effect of C/EBPβ on basal 24(OH)ase transcription. C/EBPβ could not enhance 24(OH)ase transcription either in the absence of 1,25(OH)2D3 and VDR or in the presence of an AF-2-defective VDR mutant, indicating that C/EBPβ requires the presence of ligand-bound VDR in a transcriptionally active state to enhance 24(OH)ase transcription. Similarly, C/EBPβ alone does not activate casein gene transcription. Cooperative effects of STAT5 (signal transducer and activator of transcription 5) and C/EBPβ on casein gene transcription were observed only in the presence of GR (70). Wyszomierski and Rosen suggested that the GR dependence of C/EBPβ activation of β-casein may be due to the recruitment of coactivators in the presence of GR needed to relieve an inhibitory conformation of C/EBPβ (70). It is of interest that we found that the viral oncoprotein E1A, which blocks the association of CBP/p300 with coactivators, inhibited the stimulatory effect of C/EBPβ on VDR-mediated transcription. CBP or p300 relieved the inhibition of E1A, and GST pull-down assays indicate that C/EBPβ interacts with the C terminus of p300 (positions 1661 to 2414) (Fig. 6) (28, 43). Previous studies indicated similar functional interactions between C/EBPδ and CBP (25, 28). YY1, which also binds the C terminus of CBP (5, 57) and has been reported to bind C/EBPβ (8), represses 1,25(OH)2D3-induced 24(OH)ase transcription (57). We found that CBP (57) or C/EBPβ (Fig. 7) can relieve YY1-mediated repression of 24(OH)ase transcription. Thus, YY1 may inhibit 1,25(OH)2D3-induced 24(OH)ase transcription by binding not only to CBP but also to C/EBPβ. These findings suggest that the 1,25(OH)2D3 dependence of C/EBPβ activation of the 24(OH)ase gene, similar to the GR dependence of C/EBPβ activation of the casein gene, may require the ligand-activated VDR/RXR complex that allows recruitment of coactivators such as CBP/p300 in order to relieve the inhibitory conformation of C/EBPβ. It is of interest that the Ras-activated Ets transcription factors were recently shown to cooperate with the VDR in the regulation of 24(OH)ase (17). Similar to our results, Ets was unable to function to affect 24(OH)ase transcription in the absence of 1,25(OH)2D3. Those authors suggested that this inability to function may be due to repression by the unliganded VDR/RXR complex. It is possible that repression by non-ligand-activated VDR/RXR may also be one factor contributing to the lack of effect of C/EBPβ on basal 24(OH)ase transcription.
Cooperative relationships between C/EBP proteins and transcription factors in addition to steroid receptors have been reported. C/EBPβ was found to regulate another cytochrome P450 gene, cyp2D5, present in liver (31). Similar to our the results of our study, C/EBPβ was unable to affect basal levels. Transactivation of the cyp2D5 promoter by C/EBPβ requires Sp1 (31). C/EBPβ and NF-κB have also been shown to cooperate to regulate the transcription of the IL-6 and IL-8 genes (36). Thus, similarities exist between the cooperative effects of C/EBPβ and other transcription factors and those we have observed for C/EBPβ and VDR.
Our findings suggest that C/EBP proteins may enhance VDR-mediated 24(OH)ase transcription in both osteoblastic cells and kidney cells. In osteoblastic cells C/EBPα is unaffected by 1,25(OH)2D3, while C/EBPβ and C/EBPδ are induced by 1,25(OH)2D3 (21). C/EBP factors have been shown to regulate a number of other genes expressed in osteoblasts. C/EBPδ activates IGF-I gene transcription in osteoblasts, and Runx2, the nuclear factor essential for osteogenesis, binds C/EBPδ in addition to regulating its synthesis, suggesting the importance of the interaction between these two factors in regulating C/EBPδ-sensitive genes in osteoblasts (39, 66). C/EBPβ and -δ synergize with Runx2 to regulate bone-specific osteocalcin expression (21), and both C/EBPβ and C/EBPδ enhance basal and IL-1-stimulated prostaglandin G/H synthase 2 (PGHS-2) transcription in osteoblastic cells (23). These findings suggest the importance of C/EBPβ and -δ in the regulation of osteoblast gene expression. C/EBPβ has also been reported to contribute to the transcriptional activation of a number of genes in the kidney. C/EBPβ contributes to the cAMP activation of transcription of PEPCK, a gluconeogenic enzyme present in kidney as well as liver, as well as to the activation of the nitric oxide synthase 2 gene in the medullary thick ascending limb (20, 33). Basal levels of C/EBPα and C/EBPβ mRNAs and high levels of inducible C/EBPβ and C/EBPδ mRNAs have been reported to occur in kidney (3). In our studies using null mice, although decreased, significant levels of 1,25(OH)2D3-induced 24(OH)ase mRNA were maintained in the osteoblastic cells from the C/EBP−/− mice, indicating the involvement of other factors in the induction of 24(OH)ase by 1,25(OH)2D3. In addition, in the studies in which the mice were injected with 1,25(OH)2D3 (45 ng), a difference between wild-type and C/EBPβ−/− mice in their levels of induction of 24(OH)ase mRNA in kidney was not observed at this dose of 1,25(OH)2D3, suggesting the possibility of compensation by other C/EBP isoforms. Thus, it is possible that in kidney and in osteoblastic cells functional cooperation with VDR in the regulation of 24(OH)ase can occur through C/EBPβ as well as other C/EBP isoforms such as C/EBPδ.
Although we found, as with most genes regulated by C/EBPs, that functional cooperation is not specific for C/EBPβ, a few promoters have been reported to have specificity for a C/EBP family member. Cooperation with GR in the regulation of the β casein gene as well as in the regulation of the herpes simplex virus thymidine kinase gene promoter has been reported to be specific for C/EBPβ (10, 70). In addition, synergy with Sp1 for the regulation of the cyp2D5 gene in HepG2 hepatocarcinoma cells is specific for C/EBPβ (31). It has been suggested that specificity may be conferred by an obligate interaction of the specific C/EBP protein with other proteins or transcription factors (70). Although all three C/EBP proteins, C/EBPα, -β, and -δ, can activate the macrophage colony-stimulating factor receptor promoter in macrophages and the a1 acid glycoprotein promoter in hepatoma cells, it has been suggested that the C/EBP proteins do not act simultaneously (2, 74). For these genes it has been suggested that the presence of interacting proteins may affect which C/EBP is involved in activation. In future studies it will be of interest to determine whether not only cell type specificity but also the presence of interacting proteins determine which C/EBP protein is involved in cooperating with VDR in regulating 24(OH)ase in different tissues.
We found that not only 1,25(OH)2D3 but also PTH can induce C/EBPβ expression in osteoblastic cells. PTH potentiated the induction of C/EBPβ and 24(OH)ase expression by 1,25(OH)2D3 (Fig. 8). It has previously been suggested that PTH and/or PKA activation of vitamin D hormone action in osteoblastic cells occurs, in part, by the upregulation of VDR (24, 29, 67, 72). A novel finding of our study is that C/EBPβ is involved in the PKA activation of VDR transcription, thus establishing a role for C/EBPβ in the cross talk between PTH and 1,25(OH)2D3 that involves enhancement of PKA-induced VDR transcription. A role for C/EBP proteins in the cross talk between hormone signaling pathways has previously been suggested. C/EBP proteins (α and β) have been reported to participate in the T3 and cAMP induction of PEPCK gene transcription in HepG2 cells (52). Furthermore, preexposure of osteoblastic cells to glucocorticoids increases the stimulatory effect of PGE2 on IGF-I synthesis through an increase in new C/EBPδ and C/EBPβ expression (38). In addition, an induction of C/EBPδ by both cAMP and glucocorticoid in fetal lung has been reported (12). We found that the mechanism of the PTH induction of C/EBPβ expression involves the activation of C/EBPβ gene transcription, that the PTH activation occurs via a PKA-dependent pathway, and that two CREB binding sites are important for the activation (Fig. 8D). A PKA-dependent accumulation of C/EBPδ and C/EBPβ in the nucleus has been suggested as an additional mechanism involved in the second messenger-mediated enhancement of gene expression via activation of C/EBPs (9, 13, 38). C/EBPβ has also been reported to be a substrate for PKA, and its phosphorylation has been shown to be required for its nuclear translocation (13). In preliminary studies using immunocytochemistry and fluorescence microscopy, we found that PTH (10 nM) can stimulate the nuclear accumulation of C/EBPβ in osteoblastic cells (Y. Liu and S. Christakos, unpublished data). Phosphorylation has also been suggested to be required for DNA binding and transactivation by C/EBPδ (25, 58). It will be of interest in future studies to further examine the multiple mechanisms involved in the regulation of C/EBP proteins by 1,25(OH)2D3 and PTH.
Our findings suggest that in osteoblastic cells, PTH can induce C/EBP expression, which can enhance not only 1,25(OH)2D3-induced 24(OH)ase transcription but also PKA-mediated VDR transcription. Studies in our lab using AOK-B50 cells (a proximal tubule cell line with stably transfected PTH receptors) did not indicate that PTH (25 nM) can induce C/EBPβ (after 2, 4, 8, or 16 h of treatment) (F. A. Weyts, Z. Bowen, and S. Christakos, unpublished data). Also, in these cells PTH does not affect VDR levels, and 1,25(OH)2D3-induced 24(OH)ase activity is downregulated by PTH (75), indicating cell type specificity. Although we previously reported that upregulation of VDR in osteoblastic cells by PKA may be one factor involved in the PTH enhancement of 1,25(OH)2D3-induced 24(OH)ase transcription (72), little is known about the mechanisms involved in the regulation of VDR by PKA. Our study represents the first demonstration of the regulation of VDR transcription by C/EBP proteins. C/EBP DN inhibited the C/EBPβ enhancement of PKA-induced VDR transcription as well as PKA-induced VDR transcription. In the VDR promoter, there are two CRE-like sequences (−369/−347 and −579/−558) as well as two putative C/EBP sites (−1490/−1482 and −920/−911) (44). C/EBP family members have been reported to bind both CRE and C/EBP promoter sites (69). CREB and C/EBP family members are involved in the cAMP response of the PEPCK gene by a mechanism that includes binding to the PEPCK gene CRE (60, 71). Thus, it is possible that the inhibition of the PKA activation of the VDR promoter by C/EBP DN may be due to disruption of the binding of endogenous C/EBP (which is activated by PKA) to a VDR promoter CRE or C/EBP site. We previously reported that ICER can inhibit the PTH enhancement of 1,25(OH)2D3-induced 24(OH)ase transcription in osteoblastic cells (24). The mechanism involved inhibition of the PKA induction of VDR transcription, and it was suggested that this inhibition is mediated in part by the binding of ICER to the proximal CRE in the VDR promoter. Whether this CRE is involved in C/EBP activation remains to be determined. It will be of interest in future studies to determine the C/EBP binding sites in the VDR promoter as well as to determine whether different regions of C/EBP proteins are involved in enhancing 1,25(OH)2D3-induced 24(OH)ase transcription and in regulating the PKA activation of VDR transcription.
In this study C/EBP family members were found to be important enhancers of VDR-mediated 24(OH)ase transactivation in kidney and osteoblasts. It is possible that the C/EBP family members enhance 24(OH)ase transcription in other cell types as well. The physiological significance of our findings with kidney and bone cells is that enhanced 24(OH)ase expression would be important for preventing elevated 1,25(OH)2D3 levels in serum and subsequent hypercalcemia as well as for preventing elevated 1,25(OH)2D3 levels in bone cells that would result in impaired bone formation (64).
In summary, this study provides new insight into the mechanisms involved in the regulation of 24(OH)ase. We have described for the first time functional cooperation between the C/EBP family of transcription factors and VDR in 1,25(OH)2D3-induced 24(OH)ase transcription. In addition, our findings indicate a novel role for C/EBPβ in the cross talk between PTH and 1,25(OH)2D3 that involves the regulation of VDR transcription. It will be of interest in future studies to examine in more detail the kinetics of the molecular events involved in the functional cross talk in the regulation of the 24(OH)ase gene and to examine C/EBP proteins as mediators of other PTH actions that affect osteoblast function and skeletal integrity.
Acknowledgments
We are indebted to the investigators who contributed reagents to this study (see Materials and Methods). We thank Michael Huening for his help and advice.
This work was supported by grants from the National Institutes of Health (grant DK38961 to S.C., grant DK53980 to P.N.M., and grant DK56310 to T.L.M.). A.L.M.S. was supported by a predoctoral fellowship from the Pharmaceutical Research Manufacturer's Association.
REFERENCES
- 1.Affymetrix Inc. 2002. GeneChip expression analysis. Technical manual. Affymetrix Inc., Santa Clara, Calif.
- 2.Alam, T., M. R. An, R. C. Mifflin, C. C. Hsieh, X. Ge, and J. Papaconstantinou. 1993. trans-activation of the alpha 1-acid glycoprotein gene acute phase responsive element by multiple isoforms of C/EBP and glucocorticoid receptor. J. Biol. Chem. 268:15681-15688. [PubMed] [Google Scholar]
- 3.Alam, T., M. R. An, and J. Papaconstantinou. 1992. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 267:5021-5024. [PubMed] [Google Scholar]
- 4.Armbrecht, H. J., R. H. Wasserman, and M. E. Bruns. 1979. Effect of 1,25-dihydroxyvitamin D3 on intestinal calcium absorption in strontium-fed rats. Arch. Biochem. Biophys. 192:466-473. [DOI] [PubMed] [Google Scholar]
- 5.Austen, M., B. Luscher, and J. M. Luscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272:1709-1717. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Bent, R. E. Kingston, D. D. Moore, J. G. Steidman, J. A. Smith, and K. Struhl. 2002. Current protocols in molecular biology. Wiley, New York, N.Y.
- 7.Barletta, F., L. P. Freedman, and S. Christakos. 2002. Enhancement of VDR-mediated transcription by phosphorylation: correlation with increased interaction between the VDR and DRIP205, a subunit of the VDR-interacting protein coactivator complex. Mol. Endocrinol. 16:301-314. [DOI] [PubMed] [Google Scholar]
- 8.Bauknecht, T., R. H. See, and Y. Shi. 1996. A novel C/EBP beta-YY1 complex controls the cell-type-specific activity of the human papillomavirus type 18 upstream regulatory region. J. Virol. 70:7695-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billiard, J., Y. Umayahara, K. Wiren, M. Centrella, T. L. McCarthy, and P. Rotwein. 2001. Regulated nuclear-cytoplasmic localization of CCAAT/enhancer-binding protein delta in osteoblasts. J. Biol. Chem. 276:15354-15361. [DOI] [PubMed] [Google Scholar]
- 10.Boruk, M., J. G. Savory, and R. J. Hache. 1998. AF-2-dependent potentiation of CCAAT enhancer binding protein beta-mediated transcriptional activation by glucocorticoid receptor. Mol. Endocrinol. 12:1749-1763. [DOI] [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Breed, D. R., L. R. Margraf, J. L. Alcorn, and C. R. Mendelson. 1997. Transcription factor C/EBPdelta in fetal lung: developmental regulation and effects of cyclic adenosine 3′,5′-monophosphate and glucocorticoids. Endocrinology 138:5527-5534. [DOI] [PubMed] [Google Scholar]
- 13.Chinery, R., J. A. Brockman, D. T. Dransfield, and R. J Coffey. 1997. Antioxidant-induced nuclear translocation of CCAAT/enhancer-binding protein beta. A critical role for protein kinase A-mediated phosphorylation of Ser299. J. Biol. Chem. 272:30356-30361. [DOI] [PubMed] [Google Scholar]
- 14.Christakos, S., P. Dhawan, Y. Liu, X. Peng, and A. Porta. 2003. New insights into the mechanisms of vitamin D action. J. Cell Biochem. 88:695-705. [DOI] [PubMed] [Google Scholar]
- 15.Delany, A. M., D. Durant, and E. Canalis. 2001. Glucocorticoid suppression of IGF I transcription in osteoblasts. Mol. Endocrinol. 15:1781-1789. [DOI] [PubMed] [Google Scholar]
- 16.Divieti, P., B. Lanske, H. M. Kronenberg, and F. R. Bringhurst. 1998. Conditionally immortalized murine osteoblasts lacking the type 1 PTH/PTHrP receptor. J. Bone Miner. Res. 13:1835-1845. [DOI] [PubMed] [Google Scholar]
- 17.Dwivedi, P. P., J. L. Omdahl, I. Kola, D. A. Hume, and B. K. May. 2000. Regulation of rat cytochrome P450C24 (CYP24) gene expression. Evidence for functional cooperation of Ras-activated Ets transcription factors with the vitamin D receptor in 1,25-dihydroxyvitamin D(3)-mediated induction. J. Biol. Chem. 275:47-55. [DOI] [PubMed] [Google Scholar]
- 18.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 19.Gorman, C. M., L. F. Moffat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta, A. K., and B. C. Kone. 1999. CCAAT/enhancer binding protein-beta trans-activates murine nitric oxide synthase 2 gene in an MTAL cell line. Am. J. Physiol. 276:F599-F605. [DOI] [PubMed] [Google Scholar]
- 21.Gutierrez, S., A. Javed, D. K. Tennant, M. van Rees, M. Montecino, G. S. Stein, J. L. Stein, and J. B. Lian. 2002. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277:1316-1323. [DOI] [PubMed] [Google Scholar]
- 22.Hanson, R. W. 1998. Biological role of the isoforms of C/EBP minireview series. J. Biol. Chem. 273:28543. [DOI] [PubMed] [Google Scholar]
- 23.Harrison, J. R., P. L. Kelly, and C. C. Pilbeam. 2000. Involvement of CCAAT enhancer binding protein transcription factors in the regulation of prostaglandin G/H synthase 2 expression by interleukin-1 in osteoblastic MC3T3-E1 cells. J. Bone Miner. Res. 15:1138-1146. [DOI] [PubMed] [Google Scholar]
- 24.Huening, M., G. Yehia, C. A. Molina, and S. Christakos. 2002. Evidence for a regulatory role of inducible cAMP early repressor in protein kinase A-mediated enhancement of vitamin D receptor expression and modulation of hormone action. Mol. Endocrinol. 16:2052-2064. [DOI] [PubMed] [Google Scholar]
- 25.Ji, C., W. Chang, M. Centrella, and T. L. McCarthy. 2003. Activation domains of CCAAT enhancer binding protein delta: regions required for native activity and prostaglandin E2-dependent transactivation of insulin-like growth factor I gene expression in rat osteoblasts. Mol. Endocrinol. 17:1834-1843. [DOI] [PubMed] [Google Scholar]
- 26.Jurutka, P. W., G. K. Whitfield, J. C. Hsieh, P. D. Thompson, C. A. Haussler, and M. R. Haussler. 2001. Molecular nature of the vitamin D receptor and its role in regulation of gene expression. Rev. Endocr. Metab. Disord. 2:203-216. [DOI] [PubMed] [Google Scholar]
- 27.Kerry, D. M., P. P. Dwivedi, C. N. Hahn, H. A. Morris, J. L. Omdahl, and B. K. May. 1996. Transcriptional synergism between vitamin D-responsive elements in the rat 25-hydroxyvitamin D3 24-hydroxylase (CYP24) promoter. J. Biol. Chem. 271:29715-29721. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs, K. A., M. Steinmann, P. J. Magistretti, O. Halfon, and J. R. Cardinaux. 2003. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J. Biol. Chem. 278:36959-36965. [DOI] [PubMed] [Google Scholar]
- 29.Krishnan, A. V., S. D. Cramer, F. R. Bringhurst, and D. Feldman. 1995. Regulation of 1,25-dihydroxyvitamin D3 receptors by parathyroid hormone in osteoblastic cells: role of second messenger pathways. Endocrinology 136:705-712. [DOI] [PubMed] [Google Scholar]
- 30.Kurokawa, R., D. Kalafus, M. H. Ogliastro, C. Kioussi, L. Xu, J. Torchia, M. G. Rosentfeld, and C. K. Glass. 1998. Differential use of CREB binding protein-coactivator complexes. Science 279:700-703. [DOI] [PubMed] [Google Scholar]
- 31.Lee, Y. H., S. C. Williams, M. Baer, E. Sterneck, F. J. Gonzalez, and P. F. Johnson. 1997. The ability of C/EBP beta but not C/EBP alpha to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol. Cell. Biol. 17:2038-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, H., and S. Christakos. 1991. Differential regulation by 1,25-dihydroxyvitamin D3 of calbindin-D9k and calbindin-D28k gene expression in mouse kidney. Endocrinology 128:2844-2852. [DOI] [PubMed] [Google Scholar]
- 33.Liu, X., Q. T. Wall, L. Taylor, and N. P. Curthoys. 2001. C/EBPbeta contributes to cAMP-activated transcription of phosphoenolpyruvate carboxykinase in LLC-PK(1)-F+ cells. Am. J. Physiol. Renal Physiol. 281:F649-F657. [DOI] [PubMed] [Google Scholar]
- 34.MacDonald, P. N., T. A. Baudino, H. Tokumaru, D. R. Dowd, and C. Zhang. 2001. Vitamin D receptor and nuclear receptor coactivators: crucial interactions in vitamin D-mediated transcription. Steroids 66:171-176. [DOI] [PubMed] [Google Scholar]
- 35.Matsuno, F., S. Chowdhury, T. Gotoh, K. Iwase, H. Matsuzaki, K. Takatsuki, M. Mori, and M. Takiguchi. 1996. Induction of the C/EBP beta gene by dexamethasone and glucagon in primary-cultured rat hepatocytes. J. Biochem. (Tokyo) 119:524-532. [DOI] [PubMed] [Google Scholar]
- 36.Matsusaka, T., K. Fujikawa, Y. Nishio, N. Mukaida, K. Matsushima, T. Kishimoto, and S. Akira. 1993. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. USA 90:10193-10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy, T. L., M. Centrella, and E. Canalis. 1988. Further biochemical and molecular characterization of primary rat parietal bone cell cultures. J. Bone Miner. Res. 3:401-408. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy, T. L., C. Ji, Y. Chen, K. Kim, and M. Centrella. 2000. Time- and dose-related interactions between glucocorticoid and cyclic adenosine 3′,5′-monophosphate on CCAAT/enhancer-binding protein-dependent insulin-like growth factor I expression by osteoblasts. Endocrinology 141:127-137. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy, T. L., C. Ji, Y. Chen, K. K. Kim, M. Imagawa, Y. Ito, and M. Centrella. 2000. Runt domain factor (Runx)-dependent effect on CCAAT/enhancer-binding protein δ expression and activity in osteoblasts. J. Biol. Chem. 275:21746-21753. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy, T. L., C. Ji, and M. Centrella. 2000. Links among growth factors, hormones and nuclear factors with essential roles in bone formation. Crit. Rev. Oral Biol. Med. 11:409-422. [DOI] [PubMed] [Google Scholar]
- 41.Mellon, P. L., C. H. Clegg, L. A. Correll, and G. S. McKnight. 1989. Regulation of transcription by cyclic AMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 86:4887-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menendez-Hurtado, A., A. Santos, and A. Perez-Castillo. 2000. Characterization of the promoter region of the rat CCAAT/enhancer-binding protein [alpha] gene and regulation by thyroid hormone in rat immortalized brown adipocytes. Endocrinology 141:4164-4170. [DOI] [PubMed] [Google Scholar]
- 43.Mink, S., B. Haenig, and K. H. Klempnauer. 1997. Interaction and functional collaboration of p300 and C/EBPβ. Mol. Cell. Biol. 17:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamoto, K., R. A. Kesterson, H. Yamamoto, Y. Taketani, E. Nishiwaki, S., Tatsumi, Y. Inoue, K. Morita, E. Takeda, and J. W. Pike. 1997. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol. Endocrinol. 11:1165-1179. [DOI] [PubMed] [Google Scholar]
- 45.Nakajima, T., C. Uchida, S. F. Anderson, C. G. Lee, J. Hurwitz, J. D. Parvin, and M. Montminy. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107-1112. [DOI] [PubMed] [Google Scholar]
- 46.Niehof, M., M. P. Manns, and C. Trautwein. 1997. CREB controls LAP/C/EBP beta transcription. Mol. Cell. Biol. 17:3600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishio, Y., H. Isshiki, T. Kishimoto, and S. Akira. 1993. A nuclear factor for interleukin-6 expression (NF-IL6) and the glucocorticoid receptor synergistically activate transcription of the rat alpha 1-acid glycoprotein gene via direct protein-protein interaction. Mol. Cell. Biol. 13:1854-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohyama, Y., M. Noshiro, and K. Okuda. 1991. Cloning and expression of cDNA encoding 25-hydroxyvitamin D3 24-hydroxylase. FEBS Lett. 278:195-198. [DOI] [PubMed] [Google Scholar]
- 49.Ohyama, Y., K. Ozono, M. Uchida, M. Yoshimura, T. Shinki, T. Suda, and O. Yamamoto. 1996. Functional assessment of two vitamin D-responsive elements in the rat 25-hydroxyvitamin D3 24-hydroxylase gene. J. Biol. Chem. 271:30381-30385. [DOI] [PubMed] [Google Scholar]
- 50.Omdahl, J. L., H. A Morris, and B. K. May. 2002. Hydroxylase enzymes of the vitamin D pathway: expression, function, and regulation. Annu. Rev. Nutr. 22:139-166. [DOI] [PubMed] [Google Scholar]
- 51.Osada, S., H. Yamamoto, T. Nishihara, and M. Imagwa. 1997. DNA binding specificity of the CCAAT/ enhancer-binding protein transcription factor family. J. Biol. Chem. 271:3891-3896. [DOI] [PubMed] [Google Scholar]
- 52.Park, E. A., S. Song, C. Vinson, and W. J. Roesler. 1999. Role of CCAAT enhancer-binding protein beta in the thyroid hormone and cAMP induction of phosphoenolpyruvate carboxykinase gene transcription. J. Biol. Chem. 274:211-217. [DOI] [PubMed] [Google Scholar]
- 53.Qiu, Y., A. Sharma, and R. Stein. 1998. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol. Cell. Biol. 18:2957-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rachez, C., and L. P. Freedman. 2000. Mechanisms of gene regulation by vitamin D(3) receptor: a network of coactivator interactions. Gene 246:9-21. [DOI] [PubMed] [Google Scholar]
- 55.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Naar, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 56.Ramji, D. P., and P. Foka. 2002. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365:561-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raval-Pandya, M., P. Dhawan, F. Barletta, and S. Christakos. 2001. YY1 represses vitamin D receptor-mediated 25-hydroxyvitamin D(3)24-hydroxylase transcription: relief of repression by CREB-binding protein. Mol. Endocrinol. 15:1035-1046. [DOI] [PubMed] [Google Scholar]
- 58.Ray, B. K., and A. Ray. 1994. Expression of the gene encoding alpha 1-acid glycoprotein in rabbit liver under acute-phase conditions involves induction and activation of beta and delta CCAAT-enhancer-binding proteins. Eur. J. Biochem. 222:891-900. [DOI] [PubMed] [Google Scholar]
- 59.Reddy, G. S., and K. Y. Tserng. 1989. Calcitroic acid, end product of renal metabolism of 1,25-dihydroxyvitamin D3 through C-24 oxidation pathway. Biochemistry 28:1763-1769. [DOI] [PubMed] [Google Scholar]
- 60.Roesler, W. J., E. A. Park, and P. J. McFie. 1998. Characterization of CCAAT/enhancer-binding protein alpha as a cyclic AMP-responsive nuclear regulator. J. Biol. Chem. 273:14950-14957. [DOI] [PubMed] [Google Scholar]
- 61.Schwarz, E. J., M. J. Reginato, D. Shao, S. L. Krakow, and M. A. Lazar. 1997. Retinoic acid blocks adipogenesis by inhibiting C/EBP beta-mediated transcription. Mol. Cell. Biol. 17:1552-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Screpanti, I., L. Romani, P. Musiani, A. Modesti, E. Fattori, D. Lazzaro, C. Sellitto, S. Scarpa, D. Bellavia, and G. Lattanzio. 1995. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J. 14:1932-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song, Y., X. Peng, A. Porta, H. Takanaga, J. B. Peng, M. A. Hediger, J. C. Fleet, and S. Christakos. 2003. Calcium transporter 1 and epithelial calcium channel messenger ribonucleic acid are differentially regulated by 1,25 dihydroxyvitamin D3 in the intestine and kidney of mice. Endocrinology 144:3885-3894. [DOI] [PubMed] [Google Scholar]
- 64.St.-Arnaud, R., A. Arabian, R. Travers, F. Barletta, M. Raval-Pandya, K. Chapin, J. Depovere, C. Mathieu, S. Christakos, M. B. Demay, and F. H. Glorieux. 2000. Deficient mineralization of intramembranous bone in vitamin D-24-hydroxylase-ablated mice is due to elevated 1,25-dihydroxyvitamin D and not to the absence of 24,25-dihydroxyvitamin D. Endocrinology 141:2658-2666. [DOI] [PubMed] [Google Scholar]
- 65.Stein, B., and M. X. Yang. 1995. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-κB and C/EBPβ. Mol. Cell. Biol. 15:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Umayahara, Y., J. Billiard, C. Ji, M. Centrella, T. L. McCarthy, and P. Rotwein. 1999. CCAAT/enhancer-binding protein delta is a critical regulator of insulin-like growth factor-I gene transcription in osteoblasts. J. Biol. Chem. 274:10609-10617. [DOI] [PubMed] [Google Scholar]
- 67.van Leeuwen, J. P., J. C. Birkenhager, G. C. van den Bemd, and H. A. Pols. 1996. Evidence for coordinated regulation of osteoblast function by 1,25-dihydroxyvitamin D3 and parathyroid hormone. Biochim. Biophys. Acta 1312:54-62. [PubMed] [Google Scholar]
- 68.Varghese, S., S. Lee, Y. C. Huang, and S. Christakos. 1988. Analysis of rat vitamin D-dependent calbindin-D28k gene expression. J. Biol. Chem. 263:9776-9784. [PubMed] [Google Scholar]
- 69.Wilson, H. L., and W. J. Roesler. 2002. CCAAT/enhancer binding proteins: do they possess intrinsic cAMP-inducible activity? Mol. Cell Endocrinol. 188:15-20. [DOI] [PubMed] [Google Scholar]
- 70.Wyszomierski, S. L., and J. M. Rosen. 2001. Cooperative effects of STAT5 (signal transducer and activator of transcription 5) and C/EBP beta (CCAAT/enhancer-binding protein-beta) on beta-casein gene transcription are mediated by the glucocorticoid receptor. Mol. Endocrinol. 15:228-240. [DOI] [PubMed] [Google Scholar]
- 71.Yamada, K., D. T. Duong, D. K. Scott, J. C. Wang, and D. K. Granner. 1999. CCAAT/enhancer-binding protein beta is an accessory factor for the glucocorticoid response from the cAMP response element in the rat phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 274:5880-5887. [DOI] [PubMed] [Google Scholar]
- 72.Yang, W., S. J. Hyllner, and S. Christakos. 2001. Interrelationship between signal transduction pathways and 1,25(OH)2D3 in UMR106 osteoblastic cells. Am. J. Physiol. Endocrinol. Metab. 281:E162-E170. [DOI] [PubMed] [Google Scholar]
- 73.Yeh, W. C., Z. Cao, M. Classon, and S. L. McKnight. 1994. Cascade regulation of terminal adipocyte differentiation by three members of C/EBP family of leucine zipper proteins. Genes Dev. 9:168-181. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, D. E., C. J. Hetherington, S. Meyers, K. L. Rhoades, C. J. Larson, H. M. Chen, S. W. Hiebert, and D. G. Tenen. 1996. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF α2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol. Cell. Biol. 16:1231-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zierold, C., G. Reinholz, J. A. Mings, J. M. Prahl, and H. F. DeLuca. 2000. Regulation of porcine 1,25-dihydroxyvitamin D3-24-hydroxylase (Cyp24) by 1,25-dihydroxyvitamin D3 and parathyroid hormone in AOK-B50 cells. Arch. Biochem. Biophys. 381:323-327. [DOI] [PubMed] [Google Scholar]