Abstract
Osteoclast differentiation factor (ODF)/receptor activator of NF-κB ligand is essential for inducing the differentiation of mature osteoclasts. We find that nuclear factor Y (NF-Y) binds to the CCAAT box on the ODF promoter and regulates its basal transcriptional activity. The CCAAT box on the ODF gene is required for its transcriptional induction by vitamin D3, suggesting that NF-Y coregulates this promoter along with VDR. Chromatin immunoprecipitation analysis reveals that NF-Y is required for the recruitment of RNA polymerase II (RNAPII) and TATA box binding protein on the ODF promoter. Stimulation with vitamin D3 facilitates the recruitment of VDR and p300 onto the ODF promoter, resulting in acetylation of histone H4 in an NF-Y-independent manner. ODF gene induction by parathyroid hormone or prostaglandin E is also dependent on NF-Y. Furthermore, NF-Y is essential for the recruitment of RNAPII onto other CCAAT box-containing promoters, such as those of osteopontin, CYP24, and E2F1. These results suggest that NF-Y recruits RNAPII and general transcription factors onto various CCAAT box-containing promoters in response to various inductions to permit strong transcriptional activation independently of histone modifications.
Eukaryotic transcription is regulated by multiple DNA elements, such as promoter-proximal elements, and is often combined with distal enhancer elements, which are recognized by several transcription factors. Core promoter regions often contain TATA, GC, and CCAAT boxes. The CCAAT box is one of the most typical elements for transcriptional activation. Several proteins, such as transcription factors of the C/EBP family and NF-I, have been shown to bind to CCAAT sequences. Nuclear factor Y (NF-Y) also binds to CCAAT boxes on many promoter regions (8). NF-Y is a well-conserved, ubiquitous transcription factor that consists of three subunits, NF-YA, NF-YB, and NF-YC, all of which are necessary for CCAAT box binding. NF-YB and NF-YC contain histone-folding motifs similar to H2A and H2B, respectively, and a tight complex of NF-YB and NF-YC associates with NF-YA (25). It has been reported that NF-Y associates with TATA box binding protein (TBP) (5) and several TBP-associated factors (TAFs) (11). NF-Y also interacts with p300/CBP, PCAF, and GCN5 and mediates histone acetylation on several promoters through these interactions (6, 15, 24, 33).
Osteoclast differentiation factor (ODF) is an essential factor for differentiation and activation of osteoclasts that is expressed on the osteoblast membrane (22, 43). ODF is also known as TRANCE (tumor necrosis factor [TNF]-related activation-induced cytokine) and RANKL (receptor activator of NF-κB ligand) (3, 39). ODF knockout mice exhibit typical osteoporosis with total occlusion of bone marrow space within endosteal bone (21) and also fail to develop mammary glands (10). In ODF−/− mice, mature osteoclasts are not found, whereas osteoclast progenitor cells are present. These observations indicate that ODF is an essential factor for developing mature osteoclasts and inducing bone resorption.
ODF gene expression is induced by vitamin D3, parathyroid hormone (PTH), interleukin-1 (IL-1), TNF-α, and PGE in primary osteoblasts or stromal cells (12, 27, 35, 43). The mouse ODF promoter region contains a vitamin D3 response element (VDRE) (19, 20). A heterodimer of vitamin D3 receptor (VDR) and retinoid X receptor (RXR) mediates vitamin D3 response (30). VDR-RXR transactivates target genes by recruiting transcriptional coactivators and mediators, such as the DRIP complex, which mediates transcriptional initiation, and the SRC-1/p300 complexes, which induce histone acetylation on target genes (13, 31, 32). The mouse ODF promoter also contains a CCAAT sequence near the transcriptional start site, but its significance is not known.
In this report, we show that NF-Y is essential for transcriptional regulation of the ODF promoter. NF-Y exhibits a synergistic effect with vitamin D3 in activating this promoter. Knockdown analysis for an NF-Y subunit reveals that NF-Y is essential for the recruitment of RNAPII prior to induction but not for histone acetylation after vitamin D3 induction on the ODF promoter. Furthermore, NF-Y is also required for RNAPII recruitment onto other CCAAT box-containing promoters, such as those of osteopontin (OPN), CYP24, and E2F1.
MATERIALS AND METHODS
Plasmids.
The mouse ODF gene fragment spanning the region −961 to +142 was amplified by PCR from C3N/HeJ liver genomic DNA. ODF gene fragments in pODF961, pODF500, pODF205, pODF105, pODF80, and pODF55 were generated through PCR amplification and were ligated into NheI and HindIII sites of pGV-B (TOYO Ink Co.). The CCAAT box mutation in ODF gene fragments was introduced with the CCAAT box mutated primer (underlined) 5′-TTTTGCTAGCACCCTCCTGGAAGCTGAAAAACTCTGGAGG-3′ and was cloned as described above. The VDRE sequence was introduced in ODF reporter plasmids by oligonucleotides containing VDRE sequence (underlined): sense strand, 5′-TTTTGAGCTCTGGTGACTCACCGGGTGAACGGGGGCAGCTAGCTTT-3′; antisense strand, 5′-AAAGCTAGCTGCCCCCGTTCACCCGGTGAGTCACCAGAGCTCAAA-3′. The oligonucleotides were annealed, digested with SacI and NheI, and inserted into ODF gene reporter plasmids.
The mouse OPN gene was a kind gift of T. Yamamoto (Ohita Medical University). pOPN, which includes the region −2977 to +13, was constructed as follows. Fragments of the OPN gene were amplified by PCR and modified by enzymatic reactions to create a protruding NheI site and an end-filled EcoRI site. These fragments were ligated into the NheI site and the end-filled HindIII site of pGV-B. pOPN72 and pOPN72CCCAATmt were constructed as follows. OPN gene fragments −72 to +13, with or without mutation in the CCAAT box, were generated by PCR, digested with EcoRI and HindIII, and end filled. These fragments were ligated into the end-filled HindIII site of pOPN.
Cell culture and reporter gene assay.
ST-2 cells were maintained in MEMα (Gibco BRL) supplemented with 10% fetal bovine serum. For the reporter gene assays, 0.2 μg of luciferase expression vector containing various promoters and 0.2 μg of pSV-β-galactosidase (Promega) were cotransfected into ST-2 cells (5 × 104 cells/35-mm-diameter dish) using Effectene reagent (QIAGEN). The cells were incubated for 12 h before the transfection mixture was replaced with fresh medium, followed by treatment with or without 30 nM calcitriol (ROUSSEL UCLAF). After 36 h, the cells were harvested and luciferase and β-galactosidase activities were analyzed.
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts (NE) were prepared from ST-2 cells using the Dignam method (7). The following oligonucleotides were used as probes: mouse ODF promoter, region −80 to −50, sense strand (5′-gggACCCTCCTGGAAGCTGATTGGCTCTGGAGG-3′) and antisense strand (5′-gggCCTCCAGAGCCAATCAGCTTCCAGGAGGGT-3′) for the CCAAT box; and region +33 to approximately +47, sense strand (5′-gggGGACCTCTGTGAACC-3′) and antisense strand (5′-gggGGTTCACAGAGGTCC-3′) for VDRE. The annealed oligonucleotides were labeled with Klenow fragments in the presence of [α-32P]dCTP. The labeled probes were incubated with ST-2 nuclear extracts in a reaction mixture containing 20 mM HEPES (pH 7.9), 100 mM KCl, 12% glycerol, 0.5 mM EDTA, 0.6 mM dithiothreitol, and 50 ng of poly(dI-dC)/μl at 30°C for 30 min. A mixture of recombinant VDR and RXRα was generated with an in vitro translation system (Promega), and 2 μl of the product was used for the assay. The samples were loaded on 5% acrylamide gels and electrophoresed at 200 V for 2 h and were autoradiographed.
Reverse transcription-PCR (RT-PCR).
ST-2 cells (106 cells/100-mm-diameter dish) were grown for 24 h and treated with 100 nM calcitriol for various time periods from 0 to 48 h. After incubation, the cells were washed twice with phosphate-buffered saline(−) [PBS(−)], and total RNA was extracted with Sepasol-RNA I (Nacalai tesque). The first-strand cDNA was synthesized from total RNA by SuperScript II (Life Technologies). PCRs were performed with KOD DNA polymerase (TOYOBO) using GeneAmp 9600 and the following primers: ODF sense, 5′-CGCAGATGGATCCTAACAGAATATCA-3′; ODF antisense, 5′-CTTGGGATTTTGATGCTGGTTTTAAC-3′; OPN sense, 5′-AGTTTCCAGGTTTCTGATGAACAG-3′; OPN antisense, 5′-TTAGTTGACCTCAGAAGATGAACT-3′; GAPDH sense, 5′-ACTTTGTCAAGCTCATTTCC-3′; GAPDH antisense, 5′-TGCAGCGAACTTTATTGATG-3′; epidermal growth factor receptor (EGFR) sense, 5′-CACTACATTGATGGCCCACACTGTG-3′; EGFR antisense, 5′-TTAGATCCAAAAGGTCTTTAAGATC-3′; CYP24 sense, 5′-CCAAGTGCTGGGCTCTAGCGAAG-3′; CYP24 antisense, 5′-CTACCGTGGACAGAACGCAATGGG-3′; E2F1 sense, 5′-ATCGGAGCCTCCGTCGTCACA-3′; E2F1 antisense, 5′-AGGCCGCGGCGAGGGCTCGAT-3′; OPN sense, 5′-AGTTTCCAGGTTTCTGATGAACAG-3′; OPN antisense, 5′-TTAGTTGACCTCAGAAGATGAACT-3′. A thermal cycle of 98°C, 15 s; 68°C (ODF), 63°C (OPN), or 60°C, 10 s; and 74°C, 30 s was repeated for 35 cycles for ODF, 30 cycles for OPN, CYP24, p21, EGFR, and E2F1, or 25 cycles for GAPDH. PCR products were electrophoresed on 1.5% agarose gels and were stained with ethidium bromide (EtBr).
Chromatin immunoprecipitation assay (ChIP).
ST-2 cells (3 × 105 cells/100-mm-diameter dish) were incubated for 3 days with or without vitamin D3 and for an additional 24 h with 5 mM sodium butyrate, a histone deacetylase inhibitor. Formaldehyde was added to the culture medium at a final concentration of 1% for 10 min at 25°C, and then fixation was stopped by the addition of 0.125 M glycine. The fixed cells were washed twice with PBS(−) and harvested. Cells were suspended in a buffer containing 20 mM HEPES (pH 7.9), 85 mM KCl, 0.5% NP-40, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF), incubated for 20 min on ice, and then vortexed. Nuclei were collected by centrifugation at 5,000 rpm (TOMMY XL-160) at 4°C for 5 min, suspended in a solution of 1% sodium dodecyl sulfate (SDS), 50 mM Tris (pH 8.0), and 0.5 mM PMSF, and incubated for 10 min on ice. The samples were sonicated for 10 min to cleave genomic DNA to an average length of 1,000 bp and were centrifuged at 15,000 rpm at 4°C for 10 min. Following a fivefold dilution of the nuclear lysates using NP-40 lysis buffer (20 mM Tris, 100 mM NaCl, 1 mM EDTA, and 0.1% NP-40), samples were incubated overnight at 4°C with 5 μg of antibody/ml against NF-YB (Santa Cruz), VDR (ARB Co.), RNA polymerase II (8WG16 or H5), TBP (3G3; a kind gift from L. Tora), p300 (Santa Cruz), or acetylated histone H4 (Upstate Biotechnology). A 60-μl aliquot of protein A- or G-conjugated Sepharose beads (Upstate Biotechnology) containing 1 μg of salmon sperm DNA fragments/μl and 1 μg of bovine serum albumin/μl was added and incubated at 4°C for 1 h. Sepharose beads were washed four times with NP-40 lysis buffer. The beads were incubated with 0.3 M NaCl for 6 h at 65°C. The eluates were incubated with proteinase K for 1 h at 45°C and extracted with phenol-CHCl3, and DNA was recovered by ethanol precipitation. PCR was performed using the following primers: ODF forward primer, 5′-TTTGCTAGCGGGCAGATGTGGGAGTGA-3′; ODF reverse primer, 5′-AAGCTTCCCGCCCCGACCGTTCACA-3′; OPN forward primer, 5′-TTTGAATTCAAACCAGAGGAGGAGTGTA-3′; OPN reverse primer, 5′-AAAAGGCTTTTGGTGGTGGGGTCAGACCTCCCAGA-3′; ODF −2000 forward primer, 5′-ACTTTGTCAAGCTCATTTCC-3′; ODF −1500 reverse primer, 5′-TGCAGCGAACTTTATTGATG-3′; CYP24 forward primer, 5′-AGATGAAAATACAAGGGCTGGC-3′; CYP24 reverse primer, 5′-TATTCACCTGTATCGTCAGATC-3′; p21 forward primer, 5′-AACCCAGGGCTTCACTTCCAGCAAG-3′; p21 reverse primer, 5′-TGAACCAACTGTGGGAAGGACTAACTCT-3′; EGFR forward primer, 5′-AGCCCCAGCGCAACGCGCAGCAGCC-3′; EGFR reverse primer, 5′-CGCGTCCAGGTGACCTGTCGTCTGTC-3′; E2F forward primer, 5′-ATCGGAGCCTCCGTCGTCACA-3′; E2F reverse primer, 5′-AGGCCGCGGCGAGGGCTCGAT-3′. A thermal cycle of 98°C, 15 s; 60°C, 10 s; and 74°C, 30 s was repeated for 30 cycles. PCR products were separated on 1.5% agarose gels and were stained with EtBr or quantified with a GeneAmp 5700 (Applied Biotechnologies).
Silencer RNA for NF-YA mRNA.
A silencer RNA (siRNA) for NF-YA was designed in the region 142 to 164 of the mouse NF-YA mRNA as follows: sense strand, 5′-GUCCAGACCCUCCAGGUAGUU-3′; antisense strand, 5′-CUACCUGGAGGGUCUGGACUU-3′. cRNA strands were transcribed in vitro using the Silencer small interfering RNA (siRNA) construction kit (Ambion), and double-stranded RNA (dsRNA) was generated. siRNA products were transfected into ST-2 cells (104 cells/24-well dish) using DMRIE-C reagents (Invitrogen). Six hours later, the transfection mixtures were replaced with fresh medium and cells were treated with or without 30 nM vitamin D3 for 48 h. Cells were washed with PBS(−) twice and were lysed with lysis buffer (50 mM Tris [pH 7.6], 400 mM NaCl, and 1% Nonidet P-40). Cell lysates (10 μg) were separated on a SDS-12.5% polyacrylamide gel and transferred to polyvinylidene difluoride membrane (Millipore). Proteins of interest were visualized using anti-NF-YA (Santa Cruz) or RNAPII (8WG16) antibody and ECL (Amersham). RT-PCR analysis and ChIP assays were performed as described above.
RESULTS
NF-Y binds to the CCAAT box of the mouse ODF promoter.
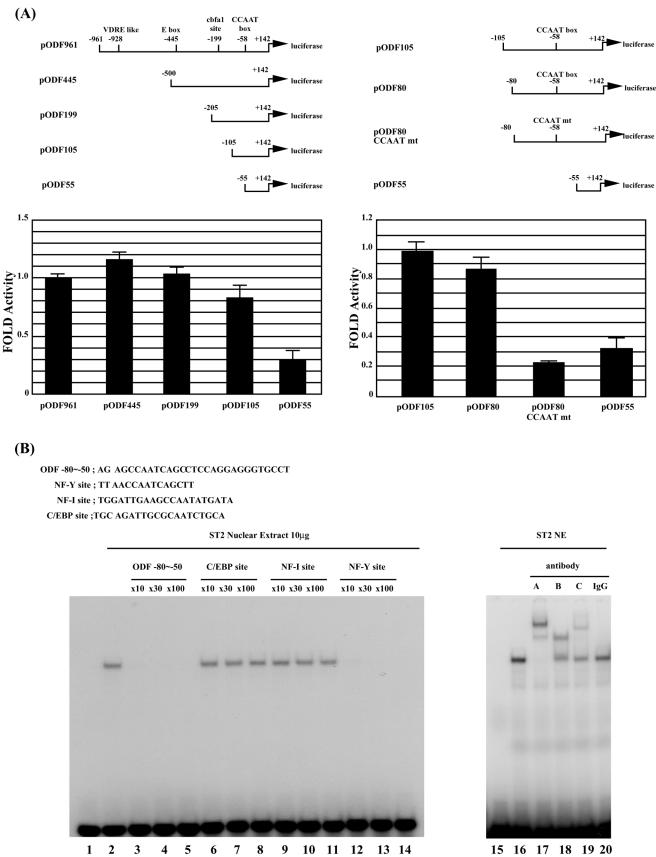
The 5′ region of the mouse ODF gene up to −961 bp upstream of the putative transcription start site has previously been reported (19). This region contains putative sites for an inverted TATA box (at position −28), an inverted CCAAT box (at position −58), a cbfa1 site (at position −199), and a vitamin D3-responsive element (VDRE) (at position −928) (Fig. 1A). To determine sequences important for transcriptional regulation of mouse ODF, we performed a reporter gene assay with the ST-2 mouse stromal cell line using several plasmids with truncated versions of the ODF promoter, as indicated in Fig. 1A. Deletions up to 105 bp upstream of the transcription start site resulted in a less than 20% reduction of reporter gene activity. However, when the ODF promoter was truncated to 55 bp upstream of the transcription start site, a significant decrease in reporter gene activity was observed. To test the significance of the CCAAT box, we used reporter plasmids with a mutated CCAAT box. pODF80CCAATmt and pODF55 showed significantly reduced activities compared to that of pODF80 (25 and 38%, respectively), whereas little difference was observed between pODF105 and pODF80. These results demonstrate the importance of the CCAAT sequence in regulating the basal transcriptional activity of the ODF gene.
FIG.1.
NF-Y is required for basal transcriptional activity of the mouse ODF gene. (A) ST-2 cells were transiently cotransfected with 0.2 μg of a series of reporter plasmids containing truncated versions of the mouse ODF gene, as indicated, and 0.2 μg of pSV-β-galactosidase. The constructs contain a putative TATA box, CCAAT box, and an cbfa1 site, respectively. Luciferase activity was normalized to β-galactosidase activity. Results shown are the averages (means ± standard errors of the means) of three independent experiments. (B) Electrophoretic mobility shift assay of binding to the CCAAT box of the ODF promoter. A 32P-labeled double-stranded oligonucleotide containing the −80 to −50 region of the ODF promoter was incubated with 10 μg of ST-2 cell nuclear extract (lanes 2 to 14 and 16 to 20). Lanes 1 and 15 were used as controls. A 10- to 100-fold molar excess of unlabeled competitor oligonucleotides (lanes 3 to 14), 1 μg of antibodies against NF-YA, NF-YB, and NF-YC (lanes 17 to 19), or rabbit preimmune immunoglobulin G (IgG) (lane 20) was added to the mixture. The sequences of the probe and the competitor oligonucleotides are shown in the upper panel.
To identify the protein that binds to the CCAAT box, we performed an electrophoretic mobility shift assay (EMSA) using the −80 to −50 region of the ODF gene, which contains the CCAAT box, as a probe (Fig. 1B). When ST-2 cell nuclear extracts were incubated with the DNA probe, a single protein-DNA complex was observed (lane 2). Excess amounts of the nonlabeled DNA probe completely outcompeted the shifted band (lanes 3 to 5). To characterize the binding factor, we used different competitor oligonucleotides containing consensus sequences for known CCAAT binding transcriptional factors (lanes 6 to 14). Band intensity was not affected by addition of excessive competitors containing either a C/EBP site (lanes 6 to 8) or an NF-I site (lanes 9 to 11). However, binding was abolished by an excess of a double-stranded oligonucleotide containing an NF-Y binding site (lanes 12 to 14). Furthermore, addition of antibodies against the NF-Y subunits, NF-YA, NF-YB, and NF-YC, caused supershift of the band (lanes 17 to 19). These results indicate that NF-Y binds to the CCAAT box of the ODF gene.
NF-Y is required for transcription of the ODF gene and recruitment of RNAPII to the promoter.
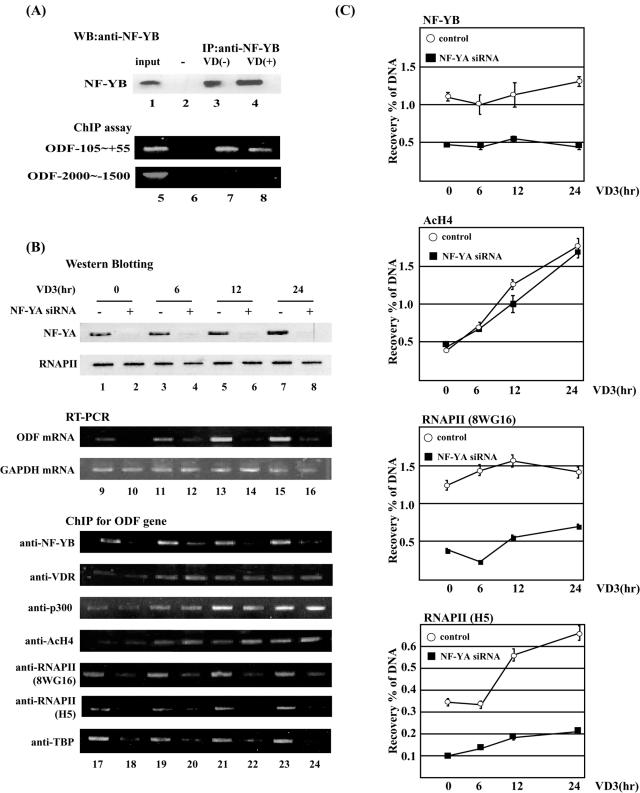
To examine the interaction between NF-Y and the CCAAT box of the ODF gene in vivo, we employed a chromatin immunoprecipitation assay (Fig. 2A). Western blot analysis revealed that similar amounts of endogenous NF-YB subunit were specifically immunoprecipitated from the formaldehyde cross-linked ST-2 cell extracts with anti-NF-YB antibody, irrespective of vitamin D3 stimulation (Fig. 2A, upper panel). Under these conditions, immunoprecipitated DNA fragments were analyzed by PCR. Amplified DNA fragments were detected in anti-NF-YB-immunoprecipitated samples irrespective of vitamin D3 stimulation when primers designed for the ODF (−105 to +55) promoter were used but not when primers designed for the distal region (−2000 to −1500) were used (Fig. 2A, lanes 7 and 8). These results suggest that NF-Y specifically recognizes the CCAAT box under both induced and uninduced conditions in ST-2 cells.
FIG. 2.
NF-Y is required for transcription of the ODF gene and recruitment of RNAPII to the promoter. (A) Chromatin immunoprecipitation analysis of NF-Y binding to the mouse ODF gene in vivo. ST-2 cells were exposed to 30 nM vitamin D3 (lane 3) for 24 h and were cross-linked with 1% formaldehyde. Nuclear extracts were immunoprecipitated with anti-NF-YB antibody (lanes 2 and 3) or preimmune goat immunoglobulin G (IgG) (lane 1). Eluates were subjected to Western blot (WB) analysis with anti-NF-YB antibody or PCR analysis with appropriate primers. PCR products were separated on 1.5% agarose gels and were stained with EtBr. (B) ST-2 cells were transfected with 2 nM siRNA for the NF-YA mRNA for 48 h. Transfectants were treated for 0, 12, 24, or 48 h with 30 nM vitamin D3. The upper panel (lanes 1 to 8) shows Western blot analysis for NF-YA or RNAPII using 10 μg of total cell lysates. The middle panel (lanes 9 to 16) shows RT-PCR analysis for ODF and GAPDH expression. The bottom panel (lanes 17 to 24) shows ChIP assays for the ODF core promoter region (−105 to +55) using indicated antibodies. (C) Quantitative PCR analysis for ChIP materials used in panel B. Results shown are the averages (means ± standard errors of the means) of three independent experiments. IP, immunoprecipitation. AcH4, acetylated histone H4.
To further analyze the role of NF-Y in ODF expression, we generated an siRNA targeted to the mouse NF-YA mRNA sequence. Treatment of ST-2 cells with 2 nM siRNA for 48 h reduced the level of endogenously expressed NF-YA protein to less than one-fifth but had no effect on the level of the largest RNAPII subunit (Fig. 2B, lanes 1 to 8). Judging by green fluorescent protein (GFP) expression, transfection efficiency was greater than 80%, but transfected cells of siRNA for NF-YA were significantly decreased over 96 h, suggesting that the knockdown of NF-YA subunit might inhibit cell proliferation (data not shown).
In ST-2 cells, the level of ODF mRNA was clearly increased by vitamin D3 treatment (Fig. 2B, lanes 9, 11, 13, and 15). Under induced and uninduced conditions, NF-YA siRNA treatment led to a significant decrease in the expression level of ODF with little effect on GAPDH expression (Fig. 2B, lanes 10, 12, 14, and 16). This indicates that NF-Y is essential for ODF gene expression in ST-2 cells.
We next examined the molecular function of NF-Y on the ODF promoter (lanes 17 to 24). NF-YB constitutively bound to the ODF promoter throughout the 24-h period of vitamin D3 treatment, and this binding was decreased by siRNA for NF-YA. These results are consistent with the idea that formation of the ternary complex of NF-YA, NF-YB, and NF-YC is necessary for DNA binding. VDR bound to the ODF promoter region after vitamin D3 stimulation. The coactivator p300 was also recruited to the ODF promoter after treatment with vitamin D3. Similarly, histone H4 on the ODF promoter was acetylated after vitamin D3 treatment. VDR binding, p300 recruitment, and histone H4 acetylation were not affected by treatment with siRNA for NF-YA. On the contrary, RNAPII and TBP constitutively bound to the ODF promoter, and knockdown of NF-YA inhibited the recruitment of these factors regardless of vitamin D3 stimulation. Quantitative PCR analysis (Fig. 2C) showed that NF-YA siRNA reduced binding of NF-YB and RNAPII to the ODF promoter by 60% but had no effect on acetylation of histone H4. Interestingly, association of the IIo form of RNAPII (H5) with the ODF promoter was increased 1.5- to 2-fold by vitamin D3 treatment, and NF-YA knockdown suppressed this enhancement. These results indicate that NF-Y presets RNAPII and general transcription factors (GTF) (TBP) on the ODF promoter and that vitamin D3 induces transcriptional activation through phosphorylation of the RNAPII C-terminal domain (CTD) and acetylation of histones on the ODF promoter.
The CCAAT box is required for transcriptional induction by vitamin D3 on the ODF and OPN genes.
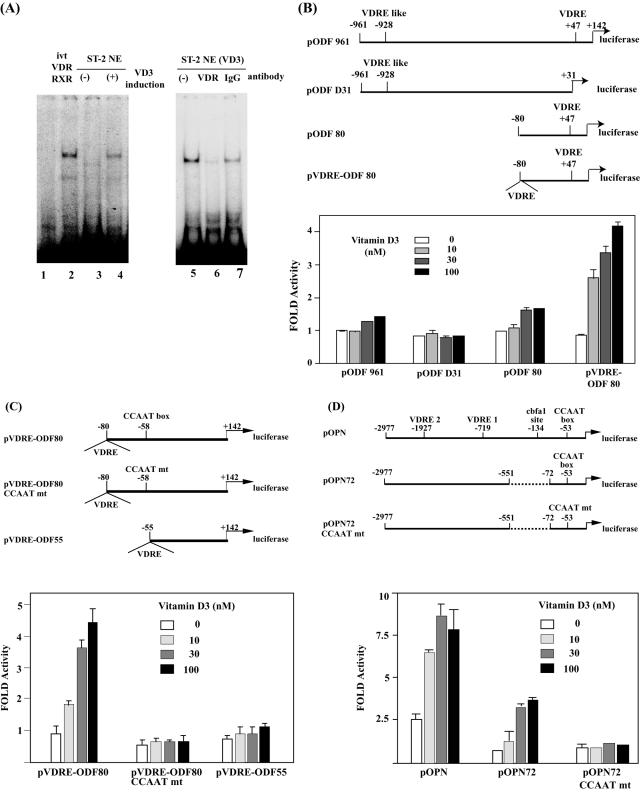
Because the importance of NF-Y in both basal and inducible transcription of the endogenous ODF gene was established, we next examined cis-acting elements on the ODF gene in greater detail (Fig. 3). Previously, Kitazawa et al. reported that a putative VDRE is located at −928 bp on the mouse ODF promoter (19). As shown above, VDR is assembled onto the ODF core promoter region (Fig. 2B). Using EMSA, we identified another VDRE sequence at the region +33 to +47 (GGTTCAcagAGGTCC) in the 5′ untranslated region (5′UTR) (Fig. 3A). When this region was used as a DNA probe, a shifted band was detected by incubation with in vitro-translated VDR and RXRα (Fig. 3A, lane 2). A shifted band was also detected at a similar position by incubation with vitamin D3-treated ST-2 NE (lane 4) but not with untreated NE (lane 3). Anti-VDR antibody inhibited formation of the protein-DNA complex (lane 6). These results suggest that the VDR-RXR complex binds to the region +33 to +47 of the mouse ODF gene.
FIG. 3.
The CCAAT box is required for transcriptional induction by vitamin D3 on the ODF and OPN genes. (A) Electrophoretic mobility shift assay of binding to the region +33 to +47 of the ODF gene. A 32P-labeled double-stranded oligonucleotide was incubated with 10 μg of nuclear extracts of ST-2 cells with or without prior treatment of 30 nM VD3 for 12 h (lanes 3 to 7) or in vitro-translated (ivt) VDR and RXRα (lane 2). Antibodies (1 μg) against VDR and rabbit preimmune immunoglobulin G (IgG) (lanes 6 and 7) were added to the mixture. ST-2 cells were transiently cotransfected with 0.2 μg of a series of reporter plasmids containing either the mouse ODF promoter (B and C) or the OPN promoter (D) and 0.2 μg of pSV-β-galactosidase. After transfection, cells were exposed to increasing concentrations (0 to 100 nM) of vitamin D3 and were incubated for 36 h. Luciferase activity was normalized to β-galactosidase activity. Results shown are the averages (means ± standard errors of the means) of three independent experiments.
To test the activity of the 5′UTR VDRE of the ODF gene, we performed reporter gene assays (Fig. 3B). When the reporter plasmid pODF961 containing the region −961 to +142 was used, we observed 1.5-fold transcriptional induction by vitamin D3. When the truncated construct pODF80 containing the region −80 to +142, including the 5′UTR VDRE, was used, 1.7-fold transcriptional induction by vitamin D3 was observed. In contrast, pODFΔ31 containing −961 to +31, but not including the 5′UTR VDRE, exhibited no transcriptional induction by vitamin D3. These results suggest that the 5′UTR VDRE is required for vitamin D3 induction. Because we observed only a low level of induction by vitamin D3 on these constructs, we next prepared the reporter plasmid pVDRE-ODF80, which is pODF80 with an extra VDRE derived from the human osteocalcin gene. Using this construct, 4.2-fold transcriptional induction was observed. Next we examined the role of the CCAAT box on the ODF promoter in vitamin D3 induction using the reporter plasmids shown in Fig. 3C. While pVDRE-ODF80 showed more than threefold transcriptional induction by vitamin D3, pVDRE-ODF80CCAATmt and ODF55, whose CCAAT box was either mutated or deleted, showed little induction. These results indicate that not only the VDREs but also the CCAAT box are required for transcriptional induction of the ODF gene by vitamin D3.
Osteopontin (OPN) is also expressed from osteoblasts following vitamin D3 stimulation. As shown in Fig. 3D, the mouse OPN promoter region includes two VDRE sites and a CCAAT box. Previously, Kim and Sodek reported that the NF-Y complex binds to the CCAAT box of the OPN gene in nuclear extracts of mouse fibroblast NIH 3T3 cells (18). We also found that NF-Y binds to the CCAAT box with ST-2 NE on EMSA (data not shown). We examined the role of the CCAAT box in vitamin D3 induction of the OPN gene. When the reporter plasmid pOPN, containing −2977 to +13 of the OPN promoter, was used, eightfold transcriptional induction by vitamin D3 was observed. When pOPN72, lacking the cbfa1 site, was used, a 75% decrease in basal transcriptional activity was observed, whereas induction by vitamin D3 was unaffected. In contrast, induction by vitamin D3 was abolished by mutation of the CCAAT box. These results indicate that the CCAAT box is necessary for vitamin D3-induced expression of the OPN gene.
NF-Y is essential for ODF gene expression following PTH or PGE stimulation.
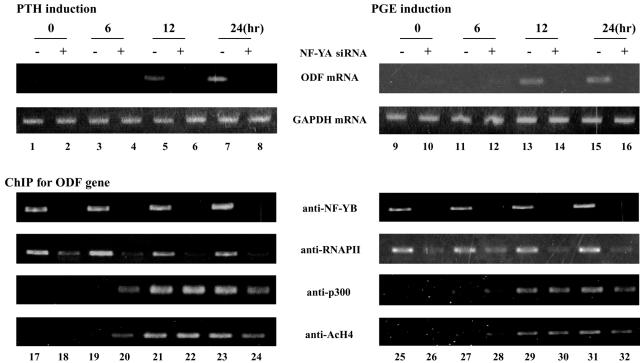
ODF gene expression is also induced by PTH or PGE stimulation (42). We further analyzed transcriptional regulation on the ODF promoter with these stimulations (Fig. 4). PTH or PGE stimulation is known to induce activation of the CREB family of transcription factors through protein kinase A phosphorylation in osteoblasts, but the mechanism of transcriptional regulation of ODF is not clear. The level of the ODF mRNA increased 12 or 24 h after treatment of ST-2 cells with either PTH or PGE (lanes 5, 7, 13, and 15). The NF-YA siRNA treatment significantly reduced these inductions (lanes 6, 8, 14, and 16). These results indicate that NF-Y is essential for ODF expression following PTH or PGE stimulation. Under these conditions, ChIP assay was performed. NF-YB and RNAPII were assembled onto the ODF promoter independent of PTH or PGE stimulation, and p300 recruitment and acetylation of histone H4 was gradually induced in response to these stimulations. Knockdown of NF-YA inhibited the assembly of NF-YB and RNAPII on the ODF promoter but did not affect p300 recruitment and acetylation of histone H4. These results underscore the importance of NF-Y on both basal and inducible transcription of the ODF gene.
FIG. 4.
NF-Y is required for ODF gene expression following PTH or PGE stimulation. ST-2 cells were transfected with 2 nM siRNA for the NF-YA mRNA for 48 h. Transfectants were treated with 100 ng of PTH/ml or 300 nM PGE for 0, 6, 12, or 24 h. The upper panel shows RT-PCR analysis for ODF and GAPDH expression. The bottom panel shows ChIP assays for the ODF core promoter region (−105 to +55) using indicated antibodies.
NF-Y presets RNAPII on CCAAT box-containing promoters independently of p300 recruitment and histone modification.
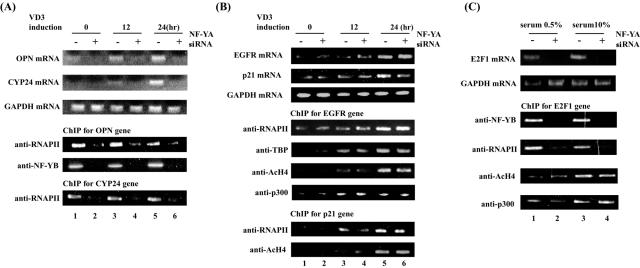
We further analyzed transcriptional regulation of the OPN and CYP24 genes, both of which contain a CCAAT box on the core promoter regions (28) (Fig. 5A). CYP24 expression was induced by vitamin D3, similar to OPN expression. Expression of these genes was abolished by treatment of NF-YA siRNA. NF-YB and RNAPII constitutively bound to the OPN and CYP24 promoters, and their binding was inhibited by NF-YA siRNA. We next analyzed vitamin D3-inducible genes with no CCAAT box on their promoters, namely the EGFR and p21 genes (Fig. 5B). Expression of these genes was induced by vitamin D3 and was not affected by the treatment of NF-YA siRNA. Interestingly, RNAPII and TBP recruitment to the EGFR and p21 promoters was strongly induced by vitamin D3 and was not affected by NF-YA siRNA. These results indicate that vitamin D3-inducible transcription of target genes with and without CCAAT boxes is achieved through activation of different steps of transcription.
FIG. 5.
NF-Y is required for RNAPII recruitment on several CCAAT box-containing promoters. ST-2 cells were transfected with 2 nM siRNA for the NF-YA mRNA for 48 h. Transfectants were treated with 30 nM vitamin D3 for 0, 6, 12, or 24 h. Upper panels show RT-PCR analysis for OPN, CYP24, and GAPDH expression (A) and EGFR, p21, and GAPDH expression (B). Lower panels show ChIP assays performed using indicated antibodies and promoter-specific primers for the OPN and CYP24 genes (A) and the p21 and EGFR genes (B). (C) NF-YA siRNA-transfected cells were incubated with 0.5 or 10% serum for 12 h. The upper panel shows RT-PCR analysis for E2F1 and GAPDH expression. The lower panel shows ChIP assays performed using indicated antibodies and promoter-specific primers for the E2F1 gene.
The E2F1 gene contains a CCAAT box but not a TATA box on the core promoter region (14, 38). Finally, we examined possible regulation of E2F1 expression by NF-Y (Fig. 5C). Basal and serum-stimulated expression of E2F1 was abolished by NF-YA siRNA. NY-B and RNAPII constitutively bound to the E2F1 promoter, and recruitment of these factors was abolished by NF-YA knockdown. These results indicate that NF-Y facilitates E2F1 expression by recruiting RNAPII to the promoter independently of the TATA box.
DISCUSSION
In this study, we have shown the mechanism of the transcriptional regulation of CCAAT box-containing promoters in vivo. We showed clearly that NF-Y is essential for ODF gene expression, recognizing the CCAAT box on the ODF core promoter region. NF-Y is essential for the recruitment of RNAPII and GTFs to the core promoter.
NF-Y is a ubiquitous transcription factor that activates the basal transcriptional activity of various promoters through CCAAT boxes. Over 25% of eukaryotic promoters contain CCAAT boxes, in most cases within 100 bp of transcription start sites (26). We showed that NF-Y constitutively binds to all the CCAAT box-containing promoters examined. RNAPII and TBP also constitutively bind to these promoters in an NF-Y-dependent manner. Because NF-Y has been shown to associate with TBP and several TAFs (5, 11), NF-Y may recruit preinitiation complexes through binding to TFIID. NF-Y is also necessary for recruiting RNAPII to the TATA-less E2F1 gene. This indicates that a CCAAT box, when recognized by NF-Y, efficiently facilitates preinitiation complex formation in the absence of a TATA box. DNA binding subunits NF-YB and NF-YC have a histone folding motif and, together with NF-YA, tightly bind to a CCAAT sequence (17). NF-Y may create a core promoter architecture that is suitable for assembly of a preinitiation complex. In a recent study, Duan et al. (9) have shown that a CCAAT box is required for the formation of the transcription initiation complex containing RNAPII, TBP, and TFIIB. The authors did so by introducing DNA fragments of the γ-globin gene promoter, containing a wild-type or mutated CCAAT box, into mouse MEL cells exogenously (9). On the other hand, we clearly showed that NF-Y is necessary for recruitment of RNAPII in vivo by using siRNA for NF-YA.
In this study, we found that the CCAAT boxes of the ODF and OPN genes are required for transcriptional induction by vitamin D3. Furthermore, knockdown of NF-YA expression significantly decreased vitamin D3-induced ODF and OPN gene expression. However, physical interaction between VDR and NF-Y could not be detected in vivo or in vitro (data not shown). It has been reported that NF-Y cooperates with several transcription factors, such as SP-1, ATF-2, RFX, and SREBP1, and that it synergistically enhances transcriptional activity of several promoters (2, 16, 36, 40). However, there are few reports on physical interactions of these transactivators with NF-Y. NF-Y may therefore exhibit a synergistic effect through interacting with some coactivators. Contrary to this view, however, knockdown of NF-YA did not affect recruitment of p300. These points suggest that VDR and NF-Y may act independently of each other through binding to their respective cis elements.
It is known that transcription factors of the nuclear receptor family, including VDR, transactivate through recruiting several coactivators (SRC-1/p300) or mediators (DRIP/TRAP complex) in a ligand-dependent manner (30). The estrogen receptor recruits the preinitiation complex through recruiting coactivators upon estrogen induction (34). It was also reported that the mediator DRIP complex interacts with several TAFs and mediates the preinitiation complex upon induction (4, 41). We consistently observed that the recruitment of RNAPII is dependent on vitamin D3 induction on the EGFR and p21 promoters, which do not contain the CCAAT box. However, RNAPII and TBP (GTFs) are preset by NF-Y on the CCAAT box-containing promoters of the ODF, OPN, and CYP24 genes. Interestingly, phosphorylation of the CTD of RNAPII that is preset on the ODF promoter is induced by vitamin D3 (Fig. 4B and C). Phosphorylation of the RNAPII CTD is thought to enhance the rate of elongation reaction (41). Thus, mediators and coactivators might transactivate a subset of vitamin D3-inducible genes in part through phosphorylation of the RNAPII CTD.
Recruitment of VDR and p300 to the ODF gene was only observed following vitamin D3 induction. This effect was time-dependent and synchronized with increased acetylation of histone H4. NF-YA knockdown showed no effect on the progression of acetylation of histone H4. This suggests that the acetylation of histone H4 is independent of NF-Y. NF-Y is known to associate with p300/PCAF and GCN5, coactivators with histone acetyltransferase activity. It has been reported on the basis of results of DNase I protection assays and ChIP analysis that NF-Y potentiates histone acetylation on the Xenopus hsp70 promoter (23, 24) and the cyclin B2 promoter (33). These promoters have two or three CCAAT boxes, and NF-Y binds to each CCAAT box independently. It is thought that binding of multiple NF-Ys recruits p300 (33). On promoters containing a CCAAT box, such as the ODF promoter, histone modification might be directed by transactivators, like VDR, other than NF-Y.
According to a recent report, SP1 plays a role in presetting RNAPII. Before TNF-α stimulation, RNAPII is preset on the A20 core promoter region, which is dependent on the GC box of the A20 gene (1). The OPN, E2F1, and p21 genes also have GC boxes that are recognized by SP1 (29, 36, 37). Because RNAPII recruitment was significantly decreased by NF-Y knockdown on the OPN and E2F1 genes, RNAPII recruitment is dependent on NF-Y on these promoters. Furthermore, RNAPII recruitment on the p21 gene was at a very low level prior to induction, and stimulation with vitamin D3 significantly induced the RNAPII recruitment. Thus, the role for SP1 in presetting RNAPII is very small, if any, on these promoters.
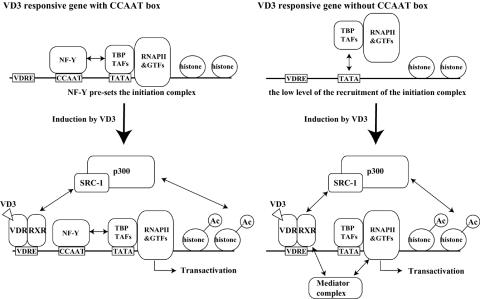
On the basis of our results, we present a model of transcriptional regulation of vitamin D3-inducible genes, which is illustrated schematically in Fig. 6. On vitamin D3-inducible genes containing a CCAAT box, such as ODF, OPN, and CYP24, NF-Y constitutively binds to the CCAAT box and presets the basal transcriptional state by recruiting RNAPII and GTFs to the promoter. Following vitamin D3 stimulation, VDR-RXR and p300 are assembled onto the promoter, increase histone acetylation, and activate transcription at a postpreinitiation complex assembly step. Thus, recruitment of RNAPII by NF-Y permits rapid transcriptional induction and synthesis of a greater amount of transcripts. On the other hand, on vitamin D3-inducible genes lacking a CCAAT box, such as EGFR and p21, promoters are not occupied by RNAPII and GTFs prior to induction. Following vitamin D3 stimulation, VDR-RXR, p300, and probably the mediator complex are assembled onto the promoter and activate transcription at a preinitiation complex assembly step and the subsequent steps.
FIG. 6.
Models for transcriptional regulation on the vitamin D3-responsive gene with or without the CCAAT box. On CCAAT box-containing genes, in the absence of vitamin D3 induction the NF-Y complex is located on the CCAAT box and presets GTFs and RNAPII. When induced by vitamin D3, transcriptional coactivators such as SRC-1/p300 are recruited by VDR-RXR independently of NF-Y and stimulate histone acetylation (indicated as Ac) on the promoter, leading to transcriptional activation and gene expression. On genes lacking a CCAAT box, upon vitamin D3 induction VDR-RXR recruits coactivators (SRC-1/p300) and mediators (DRIP complex) and thereby causes recruitment of RNAPII and GTFs and acetylation of histones.
In this report, we show directly that NF-Y is necessary for expression of CCAAT box-containing genes through presetting RNAPII on core promoter regions. By contrast, other transactivators, such as VDR, recruit the preinitiation complex on promoters lacking a CCAAT box. Further studies will be required to address the difference of transcriptional regulation for CCAAT-containing genes and others.
Acknowledgments
This work was supported in part by a Grant for Research and Development Projects in Cooperation with Academic Institutions from the New Energy and Industrial Technology Development Organization to H.H. This work was also supported by a grant from the 21st Century COE Program, Ministry of Education, Culture, Sports, Science, and Technology.
REFERENCES
- 1.Ainbinder, E., M. Revach, O. Wolstein, S. Moshonov, N. Diamant, and R. Dikstein. 2002. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-κB. Mol. Cell. Biol. 22:6354-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, C. R., C. G. Pesce, and A. R. Kornblihtt. 1996. The CCAAT-binding proteins CP1 and NF-I cooperate with ATF-2 in the transcription of the fibronectin gene. J. Biol. Chem. 271:22271-22279. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. M., E. Maraskovsky, W. L. Billingsley, W. C. Dougall, M. E. Tometsko, E. R. Roux, M. C. Teepe, R. F. DuBose, D. Cosman, and L. Galibert. 1997. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 390:175-179. [DOI] [PubMed] [Google Scholar]
- 4.Baek, H. J., S. Malik, J. Qin, and R. G. Roeder. 2002. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell. Biol. 22:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellorini, M., D. K. Lee, J. C. Dantonel, K. Zemzoumi, R. G. Roeder, L. Tora, and R. Mantovani. 1997. CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Res. 25:2174-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Currie, R. A. 1998. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J. Biol. Chem. 273:1430-1434. [DOI] [PubMed] [Google Scholar]
- 7.Dignam, J. D. 1990. Preparation of extracts from higher eukaryotes. Methods Enzymol. 182:194-203. [DOI] [PubMed] [Google Scholar]
- 8.Dorn, A., J. Bollekens, A. Staub, C. Benoist, and D. Mathis. 1987. A multiplicity of CCAAT box-binding proteins. Cell 50:863-872. [DOI] [PubMed] [Google Scholar]
- 9.Duan, Z., G. Stamatoyannopoulos, and Q. Li. 2001. Role of NF-Y in in vivo regulation of the gamma-globin gene. Mol. Cell. Biol. 21:3083-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fata, J. E., Y. Y. Kong, J. Li, T. Sasaki, J. Irie-Sasaki, R. A. Moorehead, R. Elliott, S. Scully, E. B. Voura, D. L. Lacey, W. J. Boyle, R. Khokha, and J. M. Penninger. 2000. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103:41-50. [DOI] [PubMed] [Google Scholar]
- 11.Frontini, M., C. Imbriano, A. diSilvio, B. Bell, A. Bogni, C. Romier, D. Moras, L. Tora, I. Davidson, and R. Mantovani. 2002. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J. Biol. Chem. 277:5841-5848. [DOI] [PubMed] [Google Scholar]
- 12.Hofbauer, L. C., D. L. Lacey, C. R. Dunstan, T. C. Spelsberg, B. L. Riggs, and S. Khosla. 1999. Interleukin-1β and tumor necrosis factor-α, but not interleukin-6, stimulate osteoprotegerin ligand gene expression in human osteoblastic cells. Bone 25:255-259. [DOI] [PubMed] [Google Scholar]
- 13.Hong, H., K. Kohli, M. J. Garabedian, and M. R. Stallcup. 1997. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 17:2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsiao, K. M., S. L. McMahon, and P. J. Farnham. 1994. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 8:1526-1537. [DOI] [PubMed] [Google Scholar]
- 15.Jin, S., and K. W. Scotto. 1998. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol. Cell. Biol. 18:4377-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jump, D. B., A. P. Thelen, and M. K. Mater. 2001. Functional interaction between sterol regulatory element-binding protein-1c, nuclear factor Y, and 3,5,3′-triiodothyronine nuclear receptors. J. Biol. Chem. 276:34419-34427. [DOI] [PubMed] [Google Scholar]
- 17.Kim, I. S., S. Sinha, B. de Crombrugghe, and S. N. Maity. 1996. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex. Mol. Cell. Biol. 16:4003-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, R. H., and J. Sodek. 1999. Transcription of the bone sialoprotein gene is stimulated by v-Src acting through an inverted CCAAT box. Cancer Res. 59:565-571. [PubMed] [Google Scholar]
- 19.Kitazawa, R., S. Kitazawa, and S. Maeda. 1999. Promoter structure of mouse RANKL/TRANCE/OPGL/ODF gene. Biochim. Biophys. Acta 1445:134-141. [DOI] [PubMed] [Google Scholar]
- 20.Kitazawa, R., and S. Kitazawa. 2002. Vitamin D(3) augments osteoclastogenesis via vitamin D-responsive element of mouse RANKL gene promoter. Biochem. Biophys. Res. Commun. 290:650-655. [DOI] [PubMed] [Google Scholar]
- 21.Kong, Y. Y., H. Yoshida, I. Sarosi, H. L. Tan, E. Timms, C. Capparelli, S. Morony, A. J. Oliveira-dos-Santos, G. Van, A. Itie, W. Khoo, A. Wakeham, C. R. Dunstan, D. L. Lacey, T. W. Mak, W. J. Boyle, and J. M. Penninger. 1999. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397:315-323. [DOI] [PubMed] [Google Scholar]
- 22.Lacey, D. L., E. Timms, H. L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y. X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:65-176. [DOI] [PubMed] [Google Scholar]
- 23.Landsberger, N., and A. P. Wolffe. 1995. Role of chromatin and Xenopus laevis heat shock transcription factor in regulation of transcription from the X. laevis hsp70 promoter in vivo. Mol. Cell. Biol. 15:6013-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Q., M. Herrler, N. Landsberger, N. Kaludov, V. V. Ogryzko, Y. Nakatani, and A. P. Wolffe. 1998. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 17:6300-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maity, S. N., and B. de Crombrugghe. 1998. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 23:174-178. [DOI] [PubMed] [Google Scholar]
- 26.Mantovani, R. 1998. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagai, M., and N. Sato. 1999. Reciprocal gene expression of osteoclastogenesis inhibitory factor and osteoclast differentiation factor regulates osteoclast formation. Biochem. Biophys. Res. Commun. 257:719-723. [DOI] [PubMed] [Google Scholar]
- 28.Ohyama, Y., M. Noshiro, G. Eggertsen, O. Gotoh, Y. Kato, I. Bjorkhem, and K. Okuda. 1993. Structural characterization of the gene encoding rat 25-hydroxyvitamin D3 24-hydroxylase. Biochemistry 32:76-82. [DOI] [PubMed] [Google Scholar]
- 29.Prowse, D. M., L. Bolgan, A. Molnar, and G. P. Dotto. 1997. Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J. Biol. Chem. 272:1308-1314. [DOI] [PubMed] [Google Scholar]
- 30.Rachez, C., and L. P. Freedman. 2000. Mechanisms of gene regulation by vitamin D(3) receptor: a network of coactivator interactions. Gene 246:9-21. [DOI] [PubMed] [Google Scholar]
- 31.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Naar, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 32.Rachez, C., Z. Suldan, J. Ward, C. P. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salsi, V., G. Caretti, M. Wasner, W. Reinhard, U. Haugwitz, K. Engeland, and R. Mantovani. 2003. Interactions between p300 and multiple NF-Y trimers govern cyclin B2 promoter function. J. Biol. Chem. 278:6642-6650. [DOI] [PubMed] [Google Scholar]
- 34.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 35.Tsukii, K., N. Shima, S. Mochizuki, K. Yamaguchi, M. Kinosaki, K. Yano, O. Shibata, N. Udagawa, H. Yasuda, T. Suda, and K. Higashio. 1998. Osteoclast differentiation factor mediates an essential signal for bone resorption induced by 1 alpha,25-dihydroxyvitamin D3, prostaglandin E2, or parathyroid hormone in the microenvironment of bone. Biochem. Biophys. Res. Commun. 246:337-341. [DOI] [PubMed] [Google Scholar]
- 36.Villard, J., M. Peretti, K. Masternak, E. Barras, G. Caretti, R. Mantovani, and W. Reith. 2000. A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y. Mol. Cell. Biol. 20:3364-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, D., S. Yamamoto, N. Hijiya, E. N. Benveniste, and C. L. Gladson. 2000. Transcriptional regulation of the human osteopontin promoter: functional analysis and DNA-protein interactions. Oncogene 23:5801-5809. [DOI] [PubMed] [Google Scholar]
- 38.Wang, W., L. Dong, B. Saville, and S. Safe. 1999. Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Sp1/estrogen receptor interactions. Mol. Endocrinol. 13:1373-1387. [DOI] [PubMed] [Google Scholar]
- 39.Wong, B. R., J. Rho, J. Arron, E. Robinson, J. Orlinick, M. Chao, S. Kalachikov, E. Cayani, F. S. Bartlett III, W. N. Frankel, S. Y. Lee, and Y. Choi. 1997. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J. Biol. Chem. 272:25190-25194. [DOI] [PubMed] [Google Scholar]
- 40.Xiong, S., S. S. Chirala, and S. J. Wakil. 2000. Sterol regulation of human fatty acid synthase promoter I requires nuclear factor-Y- and Sp-1-binding sites. Proc. Natl. Acad. Sci. USA 97:3948-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi, Y., T. Narita, N. Inukai, T. Wada, and H. Handa. 2001. SPT genes: key players in the regulation of transcription, chromatin structure and other cellular processes. J. Biochem. (Tokyo) 129:185-191. [DOI] [PubMed] [Google Scholar]
- 42.Yanagisawa, J., H. Kitagawa, M. Yanagida, O. Wada, S. Ogawa, M. Nakagomi, H. Oishi, Y. Yamamoto, H. Nagasawa, S. B. McMahon, M. D. Cole, L. Tora, N. Takahashi, and S. Kato. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell 9:553-562. [DOI] [PubMed] [Google Scholar]
- 43.Yasuda, H., N. Shima, N. Nakagawa, K. Yamaguchi, M. Kinosaki, S. Mochizuki, A. Tomoyasu, K. Yano, M. Goto, A. Murakami, E. Tsuda, T. Morinaga, K. Higashio, N. Udagawa, N. Takahashi, and T. Suda. 1998. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA 95:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]