Abstract
Recent studies show overlap between Fanconi anemia (FA) proteins and those involved in DNA repair mediated by homologous recombination (HR). However, the mechanism by which FA proteins affect HR is unclear. FA proteins (FancA/C/E/F/G/L) form a multiprotein complex, which is responsible for DNA damage-induced FancD2 monoubiquitination, a key event for cellular resistance to DNA damage. Here, we show that FANCD2-disrupted DT40 chicken B-cell line is defective in HR-mediated DNA double-strand break (DSB) repair, as well as gene conversion at the immunoglobulin light-chain locus, an event also mediated by HR. Gene conversions occurring in mutant cells were associated with decreased nontemplated mutations. In contrast to these defects, we also found increased spontaneous sister chromatid exchange (SCE) and intact Rad51 foci formation after DNA damage. Thus, we propose that FancD2 promotes a subpathway of HR that normally mediates gene conversion by a mechanism that avoids crossing over and hence SCEs.
Fanconi anemia (FA) is a rare autosomal-recessive disorder in childhood that is characterized by progressive bone marrow failure, developmental abnormalities, and prominent predisposition to leukemia and squamous carcinoma (reviewed in references 12, 16, and 25). Cells from FA patients are highly susceptible to chromosomal breakage and killing from DNA interstrand cross-links (ICLs) produced by agents such as diepoxybutane or mitomycin C (MMC) (12, 16, 25, 45). This property led to the early proposal that the basic defect in FA involves ICL repair (44, 45).
Eleven genes have been identified by complementation analysis (FancA/B/C/D1/D2/E/F/G/I/J/L) (12, 16, 25, 30, 32). Many of the FA proteins interact with each other in the nucleus, forming a multiprotein complex (the core complex) containing FancA/C/E/F/G/L, BLM helicase, and other unidentified components (12, 16, 25, 32, 33). During S phase or after DNA damage, FancD2 protein is monoubiquitinated on a specific lysine residue (K561 in the human protein) (15, 54). This event is apparently mediated by FancL/PHF9 E3 ligase but also requires the integrity of the core complex (12, 32, 58). This monoubiquitination appears to be essential for FancD2 function, since FancD2 K561R mutant protein cannot complement the MMC sensitivity of human FANCD2-deficient cells (15). Interestingly, FancD2 is phosphorylated at serine 222 by ATM protein kinase after ionizing irradiation (IR) (55). This phosphorylation is required for the IR-induced S-phase checkpoint but not for MMC tolerance, whereas the monoubiquitination is dispensable for the S-phase checkpoint (54).
Several lines of evidence indicate that the FA pathway influences homologous recombination (HR)-mediated DNA repair. The monoubiquitinated FancD2 (FancD2-L form) is targeted to nuclear foci that colocalize with HR proteins Rad51 and BRCA1 (15, 54). BRCA1, a tumor suppressor for familial breast cancer that is important for HR (reviewed in references 46 and 59), colocalizes with FancD2 after DNA damage (15, 58). Other FA proteins are reported to overlap or interact with HR proteins (24, 36, 50). The discovery that FANCD1 gene is identical to BRCA2, another tumor suppressor gene involved in familial breast cancer, provides a strong link between HR and the FA “pathway” (22). BRCA2 promotes nuclear localization and nucleoprotein filament formation of Rad51 (13, 46, 59) and physically interacts with FancD2 (23, 61). Finally, we have recently reported mild HR defects in FANCG-deficient cells derived from DT40 (66), a chicken B-cell line that is highly proficient in gene targeting (6).
FancD2 and its putative activator FancL are found even in Caenorhabditis elegans and Drosophila (32, 57), whereas other FA proteins exist only in vertebrates, with the exception of FancD1/BRCA2, which is conserved in Ustilago maydis (28). Notably, The Fancd2-null mouse has a more severe phenotype (21) than knockouts of Fanca (63), Fancc (9), and Fancg (29, 67). However, FancD2's precise biochemical function remains enigmatic.
Recently, three mechanisms of immunoglobulin (Ig) diversification (Ig gene conversion, class switch recombination, and somatic hypermutation) were shown to be dependent on activation-induced deaminase (AID) (2, 18, 20, 35). Avian species utilize gene conversion (templated substitutions by pseudogene donor sequences located upstream of the Ig gene), and DT40 cells recapitulate this process during growth in culture (1, 2, 5). Since Ig gene conversion is mediated by HR proteins, including Rad54 (4) and Rad51 paralogs (43, 51), DT40 provides a system to define a role for FancD2 in this process.
We report here the generation and phenotypic characterization of FANCD2-deficient DT40 cells (6). Compared to FANCG mutant (66), FANCD2 deficiency was associated with a severe phenotype. We find that FancD2 is required for efficient gene targeting, HR-directed repair of chromosomal double-strand breaks (DSBs), and gene conversion at the Ig light-chain locus without increased frequency of nontemplated point mutations. In contrast, spontaneous sister chromatid exchange (SCE) is elevated in FANCD2-deficient cells. Since SCE is thought to be mediated by HR (49), this phenotype indicates that not all HR processes are affected similarly. We propose that there are subpathways of HR in mitotic cells, which are dependent or independent of FancD2 protein.
MATERIALS AND METHODS
Cell culture, transfection, and cell cycle analysis.
DT40 wild type and derivatives were cultured at 39.5°C with 5% CO2 by using RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 1% chicken serum, 2-mercaptoethanol (50 μM), and l-glutamine (2 mM). Transfection by electroporation and subsequent selection were done as described previously (66). Cell cycle profiles were analyzed as described previously (52). Measurement of cell cycle length was carried out by chasing cell cycle profile after pulse-labeling with bromodeoxyuridine (BrdU) as described in an earlier study (52).
Chicken FANCD2 cloning and gene targeting.
An expressed sequence tag clone containing only C-terminal ∼200-bp chicken FANCD2 cDNA was obtained by searching the chicken expressed sequence tag database and then used for cDNA library (Clontech, Palo Alto, Calif.) screening. The library was also screened by using human FANCD2 probe that was obtained by reverse transcription-PCR from human cDNA. Full-length chicken FANCD2 was PCR amplified from DT40 cDNA and then inserted into pcDNA3 vector (Invitrogen, Carlsbad, Calif.). The sequence of the cloned cDNA was compared to that from direct sequencing of reverse transcription-PCR products; three PCR-associated errors were corrected by site-directed mutagenesis (Stratagene, La Jolla, Calif.). The human Rad51 expression vector has been described (34).
Genomic clones of FANCD2 were isolated by library screening (Stratagene). Targeting vectors were designed to replace a genomic segment containing exons with a resistance marker gene cassette. As a result, chicken FancD2 amino acids 554 to 653, which contain the conserved monoubiquitination site (K563 in chicken protein), were deleted.
FANCD2 disruption was achieved in three lines of DT40: wild type, DT40 carrying the recombination substrate SCneo (14, 27), and DT40Cre1. DT40Cre1 cells express transfected v-myb and MerCreMer (mutated estrogen receptor fused to Cre recombinase, which is translocated from cytoplasm to nucleus upon addition of 4-hydroxy tamoxifen) (2, 68). We disrupted FANCD2 loci in this cell line by sequential targeting by using single targeting vector (FANCD2-bsr) and MerCreMer-mediated recycling of the drug resistance marker. FANCC disruption will be described elsewhere (S. Hirano et al., unpublished data).
Sensitivity of cells to X-rays, UV, MMC, and cisplatin.
Colony formation was assayed in medium containing 1.4% methylcellulose. Serially diluted cells were irradiated with 4-MV X-rays (Linear Accelerator; Mitsubishi Electric, Inc., Tokyo, Japan) as described previously (66). Irradiation of synchronized cells was done after nocodazole arrest (52). UV irradiation was performed on cells suspended in phosphate-buffered saline. Cells were exposed for 1 h to MMC (Kyowa-Hakkou, Tokyo, Japan) or continuously to cisplatin (Nihon-Kayaku, Tokyo, Japan).
Western blot analysis and Rad51 focus assay.
Anti-chicken FancD2 serum was raised by injecting recombinant chicken FancD2 protein (amino acids 3 to 250) into rabbits. Anti-Rad51 serum was kindly provided by Akira Shinohara (Osaka University). Whole-cell extracts were separated with sodium dodecyl sulfate-6% polyacrylamide gel electrophoresis and then transferred to a membrane. Detection was done with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences, Piscataway, N.J.) by using ECL Western blotting detection reagents (Amersham Biosciences). To visualize subnuclear focus formation of Rad51, Cytospin slides were fixed and stained with the antiserum. Images were captured by using TCS-NT laser scanning confocal microscopy (Leica Microsystems, Bannockburn, Ill.) and processed with Adobe Photoshop software. Cells with more than four brightly fluorescing foci were scored as positive. At least 100 morphologically intact cells were examined at each time point.
Analysis of chromosomal aberrations and SCE.
Chromosome analysis was done as previously described (51, 52). For SCE analysis, Colcemid (0.1 μg/ml) was added for the last 2 h of this incubation before harvest, and scoring was performed as previously described on coded slides (51).
Measurement of HR-mediated repair of DSB induced by I-SceI expression.
I-SceI expression vector was transiently transfected by electroporation, and cells were selected in 96-well culture plates containing 2.0 mg of G418/ml. Surviving colonies were counted after 14 days. To determine the nature of repair events, genomic DNA was extracted from expanded colonies, restricted with KpnI and SacI double digestion, and examined by Southern blotting with a neo fragment as a probe (66). To determine cell survival after I-SceI expression, cells were transiently transfected with enhanced green fluorescent protein (EGFP)-I-SceI expression vector. After 16 h, EGFP-positive cells were clone sorted into 96-well culture plates by using FACSAria cell sorter (BD Biosciences, San Jose, Calif.) and cultured without G418. The EGFP-I-SceI expression vector was made by subcloning I-SceI coding sequence to pEGFP-C1 (Clontech).
Measurement of gene conversion events at Ig locus.
The rate of generation of surface IgM (sIgM) loss or gain variants was monitored by flow cytometric analysis of cells that had been kept in culture for the indicated time after limiting dilution and then stained with fluorescein isothiocyanate (FITC)-conjugated anti-chicken IgM (Bethyl, Inc., Montgomery, Tex.). The percent sIgM-positive or -negative cells was calculated for live cells based on propidium iodide (PI) staining and light scatter. Cells were sorted after staining with anti-chicken IgM-FITC and anti-FITC magnetic beads (Miltenyi Biotech, Auburn, Calif.). PCR amplification and sequencing of the rearranged Vλ gene was done as described previously (43). Nucleotide changes were aligned with the pseudo Vλ database (kindly provided by Julian Sale) and then classified into a gene conversion, nontemplated point mutation, or ambiguous category according to the criteria described (43). The tract length of gene conversion was defined as number of the nucleotides that were changed from the original sequence. Statistical analysis was done by using StatView software (SAS Institute, Inc., Cary, N.C.).
RESULTS
Targeted disruption of FANCD2 in DT40 cells.
To isolate the chicken version of FANCD2, we searched the BursalEST database (http://swallow.gsf.de/DT40/dt40Est.html) and found one clone that apparently contained part of the chicken FANCD2 gene. Library screening was done with this clone, and the C-terminal part of FANCD2 cDNA was isolated. To obtain the N-terminal region, human FANCD2 was amplified and used as a probe to screen a cDNA library. PCR primers were designed from sequences of the resulting cDNA clones and used to amplify full-length chicken FANCD2 from DT40 cDNA.
The full-length cDNA (DDBJ accession number AB162932) encodes a 1,439-amino-acid protein, which is very similar in size to the 1,451 amino acids in human FancD2 protein. Chicken and human FancD2 are 55% identical and 70% similar. Both the monoubiquitination (15) residue (K561) and the ATM phosphorylation (55) residue (S222) are conserved in chicken FancD2.
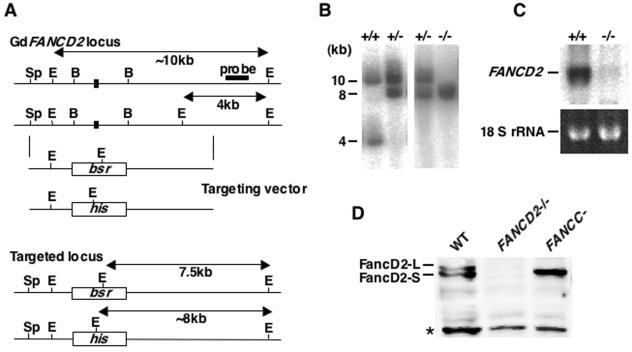
We also obtained chicken FANCD2 genomic clones by library screening and designed FANCD2 disruption vectors that should abolish the exon containing the putative monoubiquitination site (K563 in chicken sequence) (Fig. 1A). Gene targeting events were verified by Southern, Northern, and Western blotting analysis (Fig. 1B, C, and D). Of note, our anti-FancD2 sera detected two forms of FancD2, FancD2-S and FancD2-L (Fig. 1D). FancD2-L, presumed to be a monoubiquitinated form, was absent in DT40 fancc mutant as in human cells (Fig. 1D).
FIG. 1.
Targeted disruption of chicken FANCD2 loci in DT40 cells. (A) Schematic representation of partial chicken FANCD2 locus, the gene disruption constructs, and the configuration of targeted alleles. There is a polymorphism indicated by the EcoRI site between two alleles. E, EcoRI; B, BamHI; Sp, SpeI. Only the exon that contains lysine (K563) corresponding to the monoubiquitination site in the human FancD2 protein is indicated (solid box). (B) Southern blot analysis of EcoRI-digested genomic DNA from cells with indicated genotypes by using flanking probe as shown in panel A. (C) Northern blot analysis of total RNA from wild-type and fancd2 cells with chicken FANCD2 cDNA probe. (D) Western blot analysis of whole-cell lysate from wild type, fancc and fancd2 cells. The asterisk indicates a nonspecific band. The blot was probed with anti-chicken FancD2 antisera and detected with horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence reagents.
FANCD2-deficient cells show sensitivity to cross-linking agents and IR.
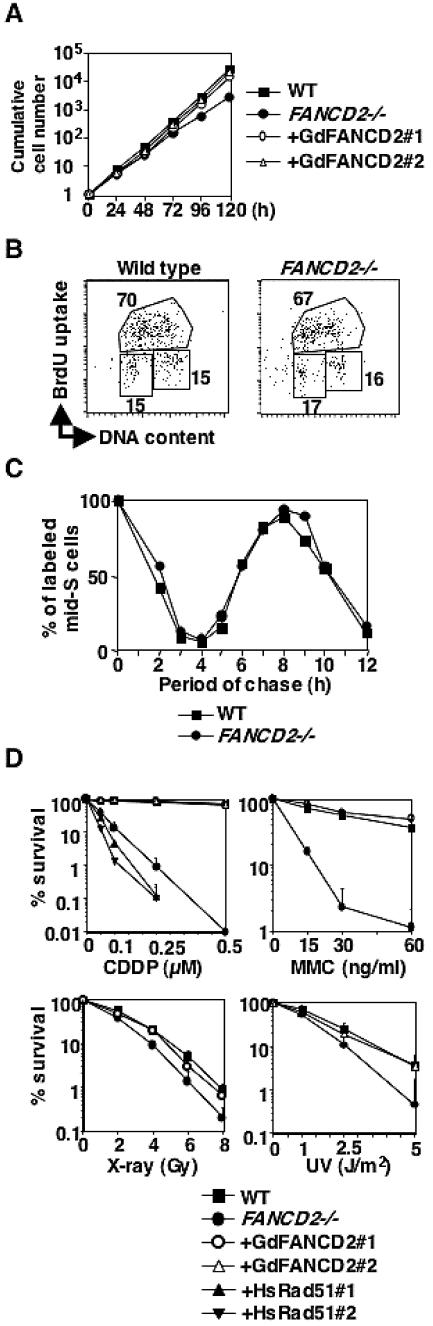
To characterize FANCD2-targeted cells (hereafter referred to as fancd2 cells), we measured the proliferation rate and obtained profiles of cell cycle progression. These cells grew slower than wild type, and two clones of complemented fancd2 cells (Fig. 2A) but did not accumulate in any particular cell-cycle phase (Fig. 2B). Fancd2 cells appeared to die spontaneously with an increased number of subdiploid cells (1.4% in wild type versus 10.5% in the fancd2 mutant), providing an explanation for the slower cell proliferation rate. Indeed, there was no significant difference in the length of the cell cycle between wild-type and fancd2 cells (Fig. 2C).
FIG. 2.
Characterization of fancd2 cells. (A) Growth curves of cells with indicated genotypes. Cell numbers were monitored by flow cytometry using fixed numbers of plastic beads as standards. (B) Cell cycle distributions. Cells were pulse-labeled with BrdU and stained with anti-BrdU antibody and PI (DNA content). Numbers in lower left, upper, and lower right regions indicate the G1, S, and G2/M phases, respectively. (C) The calculated relative percentage of cells in the mid-S phase is plotted over time. Cells were pulse-labeled with BrdU, and the labeled cells (i.e., S phase) were chased by using flow cytometer. The length of single cell cycle is ∼8 h in both wild-type and fancd2 cells. (D) Sensitivity curves of cells to various DNA-damaging agents. The fraction of surviving colonies in methylcellulose plates is shown for each agent.
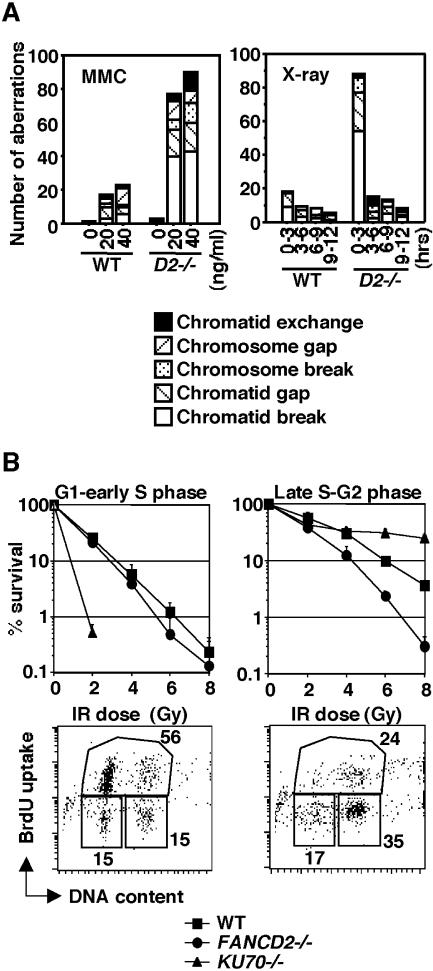
Based on colony survival assays, fancd2 cells are slightly more sensitive to X-rays and UV (Fig. 2D) than the wild-type controls but extremely sensitive to the DNA cross-linkers cisplatin and MMC (Fig. 2D). These cells also showed high sensitivity to MMC-induced chromosomal aberrations (Fig. 3A), a finding in common with cells derived from FA patients. Levels of spontaneous chromosomal aberrations were slightly elevated (3.0 versus 1.3 for wild-type total aberrations per 100 cells) (Fig. 3A). We transfected fancd2 cells with a chicken FANCD2 cDNA expression plasmid and identified FANCD2-complemented cells by Western blotting. The hypersensitivity of fancd2 cells to DNA damage was reverted to wild-type levels in these clones (Fig. 2D), indicating that these defects were indeed caused by FANCD2 disruption.
FIG. 3.
Analysis of HR repair capacity in fancd2 cells. (A) Number of chromosomal aberrations in wild-type and fancd2 cells after treatment with MMC or X-ray. In the right panel, cells were sampled at 3-h intervals after irradiation. (B) Cell cycle phase-specific X-ray sensitivity. Cells were arrested at M phase in the presence of nocodazole, released into fresh medium, and irradiated at 4 h (G1 to early S) or 9 h (late S to G2). Representative data of cell cycle profiles at both time points after synchronization are shown.
It has been reported that human Rad51 expression can rescue cisplatin sensitivity in Rad51 paralog mutants (51, 53) but not in the fancg mutant (66). Human Rad51 expression in fancd2 cells caused increased cisplatin toxicity compared to nonexpressing cells (Fig. 2D), similar to fancg cells (66). This result suggests that increased Rad51 in fancd2 cells disturbs their ability to handle DNA damage.
Gene targeting is defective in fancd2 cells.
Mechanisms of targeted integration of homologous DNA are poorly understood, but it is known that this process requires HR factors. To examine whether FancD2 is involved in targeted integration, we measured the gene targeting efficiency at several loci. After transfection, cells were selected with appropriate drugs, and individual colonies were examined by Southern blot analysis. All four targeting vectors showed a clearly decreased ratio of targeted to random integration events in fancd2 cells compared to the wild type (Table 1). Furthermore, chicken FANCD2 expression clearly increased the targeting efficiency using three of the four targeting vectors (Table 1). The lack of increase at the FANCG locus may be a statistical anomaly.
TABLE 1.
Targeted integration efficiencies in fancd2 cells
| Genotype | No. of targeted clones/total no. of clones (%)a with targeting vector:
|
|||
|---|---|---|---|---|
| FANCG-puro | Xrcc2-puro | Xrcc3-puro | Ovalbumin-puro | |
| Wild type | 3/14 (21) | 12/24 (50) | 4/28 (14) | 10/24 (42) |
| FANCD2−/− | 0/17 (0) | 3/52 (5.8) | 0/40 (0) | 0/24 (0) |
| FANCD2−/− + GdFANCD2 | 0/25 (0) | 14/27 (52) | 2/63 (3.2) | 16/26 (62) |
Data are the numbers of targeted clones per the total number of clones analyzed by Southern blotting. The percentage of targeted integration events is given in parentheses.
Repair of IR-induced DSBs is defective in late S to G2 phase.
In DT40 cells, DSB repair during the late S and G2 phases requires Rad54-mediated HR, whereas Ku70-dependent nonhomologous end joining (NHEJ) is highly active in the G1 and early S phases (52). Since fancd2 cells are mildly IR sensitive, we wanted to examine whether their IR sensitivity is cell cycle dependent. We synchronized wild-type, ku70, and fancd2 cells in mitosis by ∼8 h incubation with nocodazole, followed by release into drug-free medium (14, 52). In G1 to early S phase (4 h after the release) or in late S to G2 phase (9 h), cells were X-irradiated and seeded onto plates containing medium supplemented with methylcellulose. Ku70 cells showed extreme IR sensitivity in G1 to early S but were more resistant than wild type in late S to G2 (Fig. 3B) as previously described (14, 52). In contrast, fancd2 cells were more sensitive than wild type in late S to G2, whereas they were only marginally sensitive in G1 to early S phase (Fig. 3B). Because breaks and gaps in metaphase chromosomes probably represent unrepaired DSBs, we also counted these aberrations in four consecutive 3-h time intervals starting immediately after the IR (2-Gy) exposure. Cells that entered M phase 0 to 3 h after IR would have been irradiated in S or G2 phase. Only in this first interval did fancd2 cells exhibit highly increased aberrations compared to the wild type (Fig. 3A). These results, along with the survival data of Fig. 3B, suggest that FancD2 contributes to HR repair of DSBs produced directly by X-rays.
HR-directed repair of I-SceI-induced DSBs is reduced in fancd2 cells.
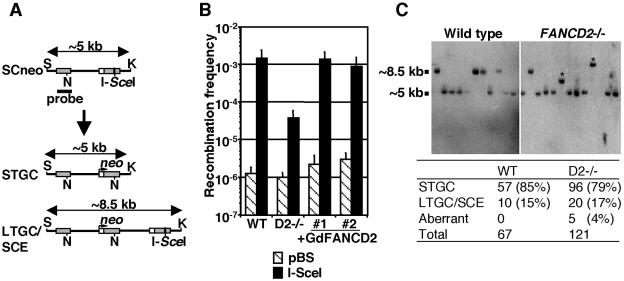
To measure HR capacity by using the artificial recombination substrate SCneo (27), we disrupted FANCD2 alleles in DT40 cells carrying SCneo at OVALBUMIN locus (14). In addition, mutant cells were transfected with chicken FANCD2 cDNA, and two complemented clones were established. After DSB induction in a nonfunctional neo segment in SCneo by transient expression of I-SceI, G418 selection was applied for ∼2 weeks until the colonies were clearly visible. Cells should be G418 resistant only if the DSB was repaired correctly by HR with the upstream neo fragment as a template (Fig. 4A). Significantly, the number of G418-resistant colonies was reduced ∼40-fold in fancd2 cells compared to the wild type and FANCD2-complemented fancd2, whereas transfection with control DNA (pBluescript) yielded a similar number of colonies in all genotypes (Fig. 4B). We isolated neo-resistant clones from wild-type or fancd2 cells after I-SceI expression and determined the plating efficiency in the presence of G418. The mean plating efficiencies of wild-type (three clones) and fancd2 (three clones) cells were 52 and 34%, respectively. Correcting for the reduced plating efficiency of fancd2 cells compared to that of wild type, the HR deficiency determined by this assay was calculated to be ∼26-fold.
FIG. 4.
Analysis of HR-mediated repair of I-SceI-induced DSBs. (A) Schematic representation of HR-directed repair of chromosomal DSBs induced by I-SceI expression. A DSB is induced at the I-SceI site in the downstream neo fragment. Structure of the expected repair products, STGC and LTGC/unequal SCE (LTGC/SCE), and position of neo probe for Southern blot analysis are shown. S, SacI; K, KpnI; N, NcoI. (B) Recombination frequency as measured by number of G418-resistant colonies. Cells were transiently transfected with either control plasmid (pBS) or I-SceI expression vector and then selected in G418-containing medium. (C) Southern blot analysis of SacI/KpnI double-digested genomic DNA from G418-resistant colonies. Positions of STGC or LTGC/SCE repair products are shown. In right panel, two aberrant sized bands from fancd2 cells are indicated with asterisks. The total number of each repair events is summarized in the table.
We also examined colony formation of the cells after DSB induction by I-SceI. To this end, we utilized the EGFP-I-SceI expression vector, which is fully functional in inducing neo resistance after transient transfection (data not shown). GFP-I-SceI-positive cells were clone sorted into 96-well plates and cultured without G418. Cell growth was observed in 84 of 384 wells (∼22%) containing wild type, and 36 of 384 wells (∼9%) containing fancd2 cells. Given the difference in the plating efficiency, this result indicates that the overall DSB repair efficiency (including NHEJ activity) in fancd2 cells was not drastically compromised.
Next, we examined the structure of the repaired SCneo substrate by Southern blot analysis of genomic DNA from randomly picked G418-resistant colonies. Most (79%) HR repair events in fancd2 cells were short-tract gene conversions (STGC) as in the wild type (85%, as described in reference 66) (Fig. 4C). The remaining colonies underwent either long-tract gene conversion (LTGC) or unequal SCE events with expansion of two to three neo repeats in wild-type (15%) and fancd2 (17%) cells. These two types of events could not be distinguished, since daughter cells having a short neo fragment originating from SCE would be killed by G418. Although the DSB repair with unequal SCE has been reported to be very rare in the SCneo substrate in hamster cells (26), we could not exclude the possibility that there were some changes in LTGC/SCE ratio in fancd2 cells. Taken together, these results indicate that overall gene conversion frequencies were reduced in fancd2 cells.
In addition, it seems noteworthy that, in five clones (4%) from fancd2 cells, aberrant repair products were found (Fig. 4C and data not shown). The sizes of these products were variable and larger than that of STGC but incorrect for LTGC events. No such products were observed in wild-type colonies, suggesting a qualitative HR defect in fancd2 cells. We isolated genomic DNA from three of such clones and tried to determine the structure of repair products by genomic Southern blotting and PCR, followed by sequencing. In two of the clones, the repair products were compatible with premature termination of repair DNA synthesis, followed by ligation with the other broken ends without homology (HR/NHEJ mechanisms) (26). In another clone, the product was associated with an extensive expansion of neo segments, which might be created by repeated cycles of repair (LTGC) and DSB induction (see data in the supplemental material).
Ig gene conversion is defective in fancd2 cells.
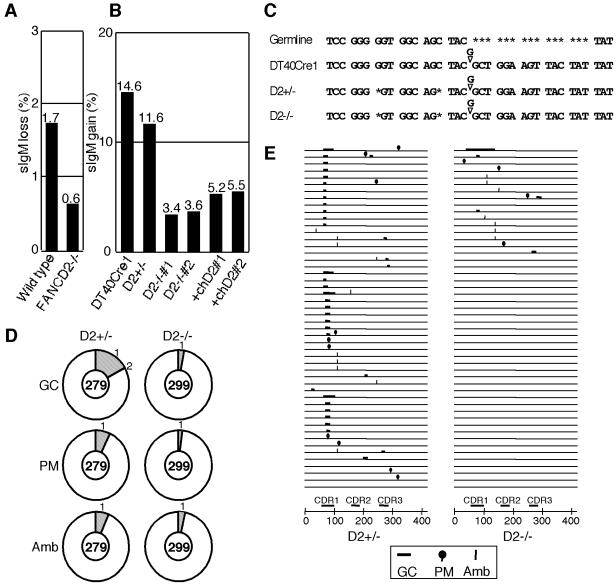
To examine the role of FancD2 in a more physiological context, we measured Ig gene conversion in fancd2 cells. DT40 cells continue to diversify the Ig V locus through gene conversion that depends on the HR machinery using upstream pseudogene segments as a donor template (1, 4, 5). Since our fancd2 cells were derived from an sIgM-positive subline of DT40, we first measured the sIgM conversion rate from positive to negative. Subclones were expanded for 3 weeks and then analyzed. The subclones derived from fancd2 cells contained fewer (∼3-fold reduction) sIgM-negative revertants than subclones from wild type (Fig. 5A). Since the loss of sIgM in wild type was shown to be mostly due to gene conversion (43), these data suggest that FANCD2 disruption likely reduces gene conversion events at Ig loci.
FIG. 5.
Analysis of gene conversion frequencies in fancd2 cells. (A) Gene conversion frequencies quantified by loss of sIgM expression. Cultures (sIgM positive) were subcloned into sets of 48 cultures, kept for 3 weeks, and analyzed for sIgM expression by flow cytometer. Each black bar and the number on top of it represent mean of the percent sIgM-negative cells in 48 subcultures. (B) Gene conversion frequencies quantified by gain of sIgM expression in DT40Cre1 and its derivatives. Subcultures from each genotype were analyzed after expanding for 4 weeks; the rest of the analysis was done as in panel A. Each black bar and the number on top of it represent mean percent sIgM-positive cells in 48 subcultures. The experiment was repeated twice with the same results. (C) Part of the Ig Vλ nucleotide sequence in cells with indicated genotypes. DT40Cre1 carries a single guanine insertion (as reported in reference 2), creating a frameshift and a truncated protein. In all our derivatives of DT40Cre1, two additional nucleotides were deleted separately (see asterisks). (D) Analysis of nucleotide changes in unsorted pooled subclones after 4 weeks culture. Vλ gene was PCR amplified from genomic DNA, subcloned, and sequenced. The number in the center of the pie chart is the number of the Vλ sequences analyzed, and the outer numbers are the number of the indicated events in the single sequence. GC, gene conversion; PM, point mutation; Amb, ambiguous category. (E) Distribution of the sequence changes in each Vλ sequences. Fifty representative sequences from each genotype are shown. CDR, complementarity-determining region.
Additional experiments were designed to examine in more detail the role of FancD2 in Ig gene conversion by using the previously established sIgM-negative DT40Cre1 subline, in which Ig conversion events occur at a very high rate (2). Gene conversion in DT40Cre1 reverts the cells to sIgM positive by correcting a frameshift mutation present in the light chain locus (Vλ) (Fig. 5C) (2). Thus, we prepared FANCD2 disruptants in DT40Cre1 cells. These mutants are cisplatin sensitive like mutants derived from wild-type DT40 (data not shown). Similarly to the data shown in Fig. 2C, the length of the cell cycle measured by BrdU pulse-labeling did not differ significantly between the wild-type and the fancd2 DT40Cre1 (∼8 h in both cases). Subclones of the mutant and wild-type DT40Cre1 were expanded for 4 weeks and assayed for sIgM expression by flow cytometry. Strikingly, the percentage of sIgM-positive cells in subclones from fancd2 cells was remarkably reduced (∼3.5-fold) compared to wild-type and heterozygous FANCD2+/− cells (Fig. 5B). FANCD2-transfected fancd2 cells showed only very partial complementation (Fig. 5B), suggesting that fine regulation of expression levels by endogenous cis elements might be necessary for normal activity in this assay. Consistent with this idea, gene conversion defects in rad54 cells also show partial correction by chicken Rad54 cDNA expression (4).
To verify that the configuration of the Vλ gene was the same in DT40Cre1 and its derivatives, we carried out nucleotide sequencing. As shown in Fig. 5C, we found two single-nucleotide deletions (of ambiguous origin) in each derivative of DT40Cre1. Thus, the drastic reduction of gene conversion events in fancd2 cells is not due to these mutations, since the FANCD2+/− cells carrying the same mutations showed nearly normal levels of gene conversion.
Ig V gene conversion is associated with a decrease in nontemplated mutations in fancd2 cells.
The subclones of FANCD2+/− and fancd2 cells derived from DT40Cre1 that were kept in culture for 4 weeks were pooled (unsorted populations), and the Vλ nucleotide sequences were determined. In fancd2 cells, only 8 Vλ sequences among 299 sequences had gene conversion events (2.7%), whereas FANCD2+/− cells had 48 conversion events among 279 sequences (17.1%) (Fig. 5D and E). Thus, our estimate for the degree of reduction of Ig gene conversion events conferred by FANCD2 disruption is ∼6.3-fold.
Nontemplated single nucleotide substitution in the Ig V gene is another important mechanism for diversification, which is analogous to somatic hypermutation in germinal center B cells (20, 43). RAD51 paralog mutants (e.g., XRCC2, XRCC3, and RAD51B) exhibit a striking shift from gene conversion to nontemplated substitutions (43). Thus, we looked at the nontemplated changes in the sequence data from DT40Cre1 FANCD2+/− and fancd2 cells. Relatively few of these events were noted in cells with both genotypes, but they appeared to be present at a reduced level in fancd2 cells (2%; 6 events in 299 sequences) compared to FANCD2+/− cells (7.2%; 20 events in 279 sequences) (PM class in Fig. 5D and E). This difference was statistically significant (P = 0.0028 [Mann-Whitney U test]).
We next looked at the nature of gene conversion events in the fancd2 background. Sequencing analysis of the Vλ in sorted sIgM-positive subpopulations revealed that the tract length of gene conversion was not different in the absence of FancD2 (mean ± the standard deviation [SD]; 11.8 ± 13.7 bp in FANCD2+/−, n = 43 versus 15.6 ± 23.9 bp in FANCD2−/−, n = 38). In addition, we could not detect any aberrant events, such as nucleotide changes surrounding gene conversion tracts that may suggest compromised fidelity of the gene conversion events in fancd2 cells.
Spontaneous SCEs are elevated, and Rad51 focus formation is normal in fancd2 cells.
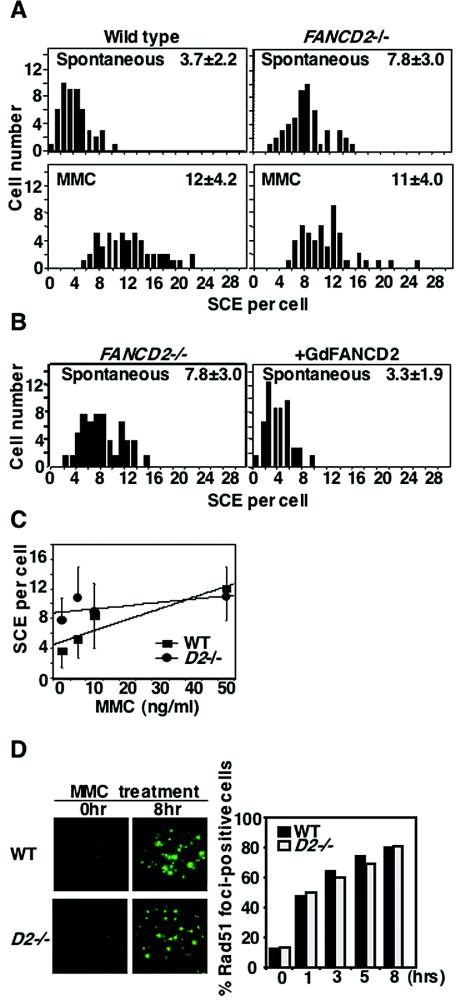
Unexpectedly, we found that spontaneous SCEs were ∼2-fold higher in fancd2 cells than in wild-type and in complemented fancd2 cells (Fig. 6A and B). In contrast, after MMC treatment (50 ng/ml), the levels of SCEs were similar in fancd2 cells compared to the wild type. With increasing concentrations of MMC, fancd2 cells showed only a minor increase compared to the more pronounced increase in the wild type (Fig. 6C). Inefficient S-phase checkpoints in response to ICL in human FA cells (7, 42) might be relevant to this phenotype.
FIG. 6.
Levels of SCE and Rad51 foci formation in fancd2 cells. (A) Distribution of spontaneous and MMC-induced SCEs with the means and SDs for 50 metaphases. (B) Spontaneous SCE levels in fancd2 cells with or without complementation with chicken FANCD2. Means and SDs from 50 metaphases are shown. (C) SCE dose response for 8 h MMC exposure scored as in panel A. Error bars indicate SD. Linear fitting was done by DeltaGraph software. (D) Rad51 focus formation after MMC treatment. Cells were treated for 1 h with MMC (500 ng/ml), washed, and sampled at each time point. At least 200 cells were scored for each preparation.
Another measure of HR capacity is subnuclear Rad51 focus formation, which is cytologically detectable in S phase and enhanced by DNA damage. Rad51 focus formation, which may reflect Rad51's polymer formation on single-stranded DNA, depends on several proteins including the Rad51 paralogs and BRCA2 in higher eukaryotes (62). Rad51 focus formation was normal in fancd2 cells after MMC (Fig. 6D) or IR treatment (data not shown).
DISCUSSION
FANCD2 plays a crucial role in a variety of HR processes.
Replication forks often stall because of endogenous DNA damage during normal replication (10). Connections have been proposed between repair/processing of the stalled or collapsed forks and the role of FA proteins (60). The ICL sensitivity characteristic of FA cells could be related to ICL's obvious ability to prevent fork progression. It is now widely accepted that restart of the stalled forks is mediated by at least two processes: HR and translesion synthesis (TLS). Emerging evidence has indicated a link between FA proteins and HR (15, 22-24, 36, 61, 66), further suggesting the FA proteins may help stabilize chromosomes against collapse during replication. Interestingly, FANCD1/BRCA2 was recently shown to prevent the breakdown of stalled replication forks, possibly via its important function in HR (31).
In the present study, we provide genetic evidence for the involvement of vertebrate FancD2 in a variety of processes that require HR. First, fancd2 cells are more IR sensitive than wild type to killing in late S to G2 phase when DSB repair predominantly takes place through HR. In contrast, fancd2 cells have only slight sensitivity in G1 to early S phase when NHEJ predominates (52). Consistent with this result are our data showing that fancd2 cells display extensive chromosomal aberrations in the 0-to-3-h interval after IR, which corresponds to irradiation in late S or G2 phase. Second, HR repair efficiency of chromosomal DSBs created by I-SceI endonuclease is reduced in fancd2 cells. Physical analysis revealed some aberrant repair products that were not observed in wild-type cells. Third, Ig gene conversion events are greatly reduced in fancd2 cells. Ig gene conversion is known to depend on AID (2, 18) and the HR machinery (4, 43, 51), although the AID-mediated initiating DNA lesion in V gene is not necessarily a DSB (1, 38). Fourth, the efficiency of targeted integration events at multiple loci is drastically reduced in fancd2 cells. Previous studies showed that mutants of many HR genes display this phenotype (4, 49, 51, 53, 56, 65). Interestingly, we found that DNA damage-induced focus formation of Rad51, a central molecule of HR, is not impaired in the absence of FancD2, suggesting that FancD2 acts either independently of Rad51 or in later steps after the focus formation.
FANCD2 is dispensable for spontaneous SCEs.
A portion of the HR-mediated restart events at stalled forks results in crossing over when two Holliday junctions are resolved, leading to the cytologically detectable SCE. We found that spontaneous SCE was clearly elevated in fancd2 cells. A similar elevation of SCEs was noted in fancc DT40 cells as well (Hirano et al., unpublished). Although SCEs are known to depend on HR proteins, including Rad51 (49), Rad54 (49), the five Rad51 paralogs (51, 53), and Nbs1 (56), FancD2 and FancC are clearly dispensable for spontaneous SCE in DT40 cells. Taken together, we suggest a role of FancD2 might be restricted to a certain subpathway of HR. For example, mitotic HR events in general are thought to proceed via processes that avoid crossing over such as through synthesis-dependent strand annealing (11, 17, 26, 41, 62). It is possible that FancD2 might be required for synthesis-dependent strand annealing but not for HR that accompanies double Holliday junction formation such as SCE.
Of note, we recently found that the elevated spontaneous SCE in fancc or fancd2 cells could be explained by partially impaired relocalization of BLM helicase (Hirano et al., submitted). Bloom syndrome is characterized by chromosomal instability and greatly elevated (10-fold) spontaneous SCE (8). Notably, the BLM helicase, which suppresses crossing over via the topoisomerase III-dependent “dissolution” of double Holliday junctions (19, 64), is associated with the core complex of FANCA/C/E/F/G (33).
Roles of FANCD2 in Ig gene conversion and hypermutation.
AID initiates gene conversion or hypermutation (nontemplated nucleotide substitutions) in chicken Ig V gene probably by creating a common DNA intermediate (3, 20, 37). The processed initiating events eventually trigger either HR (leading to gene conversion) or TLS (leading to hypermutation) (4, 43, 51). Interestingly, defective gene conversion in DT40 mutants lacking XRCC2, XRCC3, and RAD51B is associated with drastically increased hypermutation (43). On the other hand, the decrease in gene conversion in rad54 cells does not accompany a significant change in hypermutation, probably because the lesions in those cells may be committed to HR and are no longer available for the TLS pathway (43, 47).
We found that defective gene conversion in fancd2 cells is associated with decreased nontemplated mutations (Fig. 5D). Thus, it seems likely that FancD2 promotes TLS and spontaneous mutagenesis. The well-documented hypomutability of human FA cells at the hprt locus is quite consistent with this hypothesis (39, 40). Fancd2 cells share extremely high sensitivity to MMC or cisplatin with TLS mutants such as rev1 or rev3 cells (47, 48). Elucidation of the precise biochemical function of FancD2 will be required to clarify this issue.
In summary, the current study has established a role for FancD2 in a variety of HR-mediated DNA repair processes. However, the overall phenotype of fancd2 cells clearly differs significantly from that of mutants of Brca2, Rad51, Rad51 paralogs, Rad54, and Nbs1, which consistently show reduced spontaneous SCEs. Given that spontaneous SCE is not decreased in fancd2 cells, we propose that FancD2 promotes a subpathway of HR that normally mediates gene conversion by a mechanism that avoids crossing over.
Supplementary Material
Acknowledgments
We thank Yoko Moriya for expert technical assistance in nucleotide sequencing; Ian Hickson, Peter McHugh, and Hitoshi Kurumizaka for critically reading the manuscript and for suggestions; Ketan J. Patel for discussions; Julian Sale for the pseudo-Vλ sequence database and suggestions for analyzing gene conversion and hypermutation events; Akira Shinohara for anti-Rad51 antibody; and Maria Jasin for providing the SCneo and I-SceI expression vector.
This study was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan (M.T. and N.M.). Financial support was also provided by Kawasaki Medical School (Project Research Grants No. 13-211, 14-203, and 15-201A), the Naito Foundation, the Takeda Foundation, and the Sagawa Foundation for Promotion of Cancer Research. A portion of the study was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory under contract no. W-7405-Eng-48, with funding from NIH/NCI grant CA89405 (L.H.T.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arakawa, H., and J. M. Buerstedde. 2004. Immunoglobulin gene conversion: insights from bursal B cells and the DT40 cell line. Dev. Dyn. 229:458-464. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, H., J. Hauschild, and J. M. Buerstedde. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295:1301-1306. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa, H., H. Saribasak, and J. M. Buerstedde. 2004. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2:967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezzubova, O., A. Silbergleit, Y. Yamaguchi-Iwai, S. Takeda, and J. M. Buerstedde. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell 89:185-193. [DOI] [PubMed] [Google Scholar]
- 5.Buerstedde, J. M., C. A. Reynaud, E. H. Humphries, W. Olson, D. L. Ewert, and J. C. Weill. 1990. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 9:921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buerstedde, J. M., and S. Takeda. 1991. Increased ratio of targeted to random integration after transfection of chicken B-cell lines. Cell 67:179-188. [DOI] [PubMed] [Google Scholar]
- 7.Centurion, S. A., H. R. Kuo, and W. C. Lambert. 2000. Damage-resistant DNA synthesis in Fanconi anemia cells treated with a DNA cross-linking agent. Exp. Cell Res. 260:216-221. [DOI] [PubMed] [Google Scholar]
- 8.Chaganti, R. S., S. Schonberg, and J. German. 1974. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71:4508-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M., D. J. Tomkins, W. Auerbach, C. McKerlie, H. Youssoufian, L. Liu, O. Gan, M. Carreau, A. Auerbach, T. Groves, C. J. Guidos, M. H. Freedman, J. Cross, D. H. Percy, J. E. Dick, A. L. Joyner, and M. Buchwald. 1996. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat. Genet. 12:448-451. [DOI] [PubMed] [Google Scholar]
- 10.Cox, M. M. 2001. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 35:53-82. [DOI] [PubMed] [Google Scholar]
- 11.Cromie, G. A., J. C. Connelly, and D. R. Leach. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8:1163-1174. [DOI] [PubMed] [Google Scholar]
- 12.D'Andrea, A. D., and M. Grompe. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3:23-34. [DOI] [PubMed] [Google Scholar]
- 13.Davies, A. A., J. Y. Masson, M. J. McIlwraith, A. Z. Stasiak, A. Stasiak, A. R. Venkitaraman, and S. C. West. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7:273-282. [DOI] [PubMed] [Google Scholar]
- 14.Fukushima, T., M. Takata, C. Morrison, R. Araki, A. Fujimori, M. Abe, K. Tatsumi, M. Jasin, P. K. Dhar, E. Sonoda, T. Chiba, and S. Takeda. 2001. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 276:44413-44418. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 16.Grompe, M., and A. D'Andrea. 2001. Fanconi anemia and DNA repair. Hum. Mol. Genet. 10:2253-2259. [DOI] [PubMed] [Google Scholar]
- 17.Haber, J. E. 1999. DNA recombination: the replication connection. Trends Biochem. Sci. 24:271-275. [DOI] [PubMed] [Google Scholar]
- 18.Harris, R. S., J. E. Sale, S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. AID is essential for immunoglobulin V gene conversion in a cultured B-cell line. Curr. Biol. 12:435-438. [DOI] [PubMed] [Google Scholar]
- 19.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 20.Honjo, T., K. Kinoshita, and M. Muramatsu. 2002. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 20:165-196. [DOI] [PubMed] [Google Scholar]
- 21.Houghtaling, S., C. Timmers, M. Noll, M. J. Finegold, S. N. Jones, M. S. Meyn, and M. Grompe. 2003. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17:2021-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howlett, N. G., T. Taniguchi, S. Olson, B. Cox, Q. Waisfisz, C. De Die-Smulders, N. Persky, M. Grompe, H. Joenje, G. Pals, H. Ikeda, E. A. Fox, and A. D. D'Andrea. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297:606-609. [DOI] [PubMed] [Google Scholar]
- 23.Hussain, S., J. B. Wilson, A. L. Medhurst, J. Hejna, E. Witt, S. Ananth, A. Davies, J. Y. Masson, R. Moses, S. C. West, J. P. De Winter, A. Ashworth, N. J. Jones, and C. G. Mathew. 2004. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 13:1241-1248. [DOI] [PubMed] [Google Scholar]
- 24.Hussain, S., E. Witt, P. A. Huber, A. L. Medhurst, A. Ashworth, and C. G. Mathew. 2003. Direct interaction of the Fanconi anaemia protein FANCG with BRCA2/FANCD1. Hum. Mol. Genet. 12:2503-2510. [DOI] [PubMed] [Google Scholar]
- 25.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2:446-457. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, R. D., N. Liu, and M. Jasin. 1999. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401:397-399. [DOI] [PubMed] [Google Scholar]
- 28.Kojic, M., C. F. Kostrub, A. R. Buchman, and W. K. Holloman. 2002. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell 10:683-691. [DOI] [PubMed] [Google Scholar]
- 29.Koomen, M., N. C. Cheng, H. J. van de Vrugt, B. C. Godthelp, M. A. van der Valk, A. B. Oostra, M. Z. Zdzienicka, H. Joenje, and F. Arwert. 2002. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum. Mol. Genet. 11:273-281. [DOI] [PubMed] [Google Scholar]
- 30.Levitus, M., M. A. Rooimans, J. Steltenpool, N. F. Cool, A. B. Oostra, C. G. Mathew, M. E. Hoatlin, Q. Waisfisz, F. Arwert, J. P. De Winter, and H. Joenje. 2003. Heterogeneity in Fanconi anemia: evidence for two new genetic subtypes. Blood 103:2498-2503. [DOI] [PubMed] [Google Scholar]
- 31.Lomonosov, M., S. Anand, M. Sangrithi, R. Davies, and A. R. Venkitaraman. 2003. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 17:3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meetei, A. R., J. P. de Winter, A. L. Medhurst, M. Wallisch, Q. Waisfisz, H. J. van de Vrugt, A. B. Oostra, Z. Yan, C. Ling, C. E. Bishop, M. E. Hoatlin, H. Joenje, and W. Wang. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35:165-170. [DOI] [PubMed] [Google Scholar]
- 33.Meetei, A. R., S. Sechi, M. Wallisch, D. Yang, M. K. Young, H. Joenje, M. E. Hoatlin, and W. Wang. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23:3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison, C., A. Shinohara, E. Sonoda, Y. Yamaguchi-Iwai, M. Takata, R. R. Weichselbaum, and S. Takeda. 1999. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell. Biol. 19:6891-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553-563. [DOI] [PubMed] [Google Scholar]
- 36.Nakanishi, K., T. Taniguchi, V. Ranganathan, H. V. New, L. A. Moreau, M. Stotsky, C. G. Mathew, M. B. Kastan, D. T. Weaver, and A. D. D'Andrea. 2002. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell. Biol. 4:913-920. [DOI] [PubMed] [Google Scholar]
- 37.Nambu, Y., M. Sugai, H. Gonda, C. G. Lee, T. Katakai, Y. Agata, Y. Yokota, and A. Shimizu. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science 302:2137-2140. [DOI] [PubMed] [Google Scholar]
- 38.Neuberger, M. S., R. S. Harris, J. Di Noia, and S. K. Petersen-Mahrt. 2003. Immunity through DNA deamination. Trends Biochem. Sci. 28:305-312. [DOI] [PubMed] [Google Scholar]
- 39.Papadopoulo, D., C. Guillouf, H. Mohrenweiser, and E. Moustacchi. 1990. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc. Natl. Acad. Sci. USA 87:8383-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papadopoulo, D., B. Porfirio, and E. Moustacchi. 1990. Mutagenic response of Fanconi's anemia cells from a defined complementation group after treatment with photoactivated bifunctional psoralens. Cancer Res. 50:3289-3294. [PubMed] [Google Scholar]
- 41.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sala-Trepat, M., D. Rouillard, M. Escarceller, A. Laquerbe, E. Moustacchi, and D. Papadopoulo. 2000. Arrest of S-phase progression is impaired in Fanconi anemia cells. Exp. Cell Res. 260:208-215. [DOI] [PubMed] [Google Scholar]
- 43.Sale, J. E., D. M. Calandrini, M. Takata, S. Takeda, and M. S. Neuberger. 2001. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature 412:921-926. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki, M. S. 1975. Is Fanconi's anaemia defective in a process essential to the repair of DNA cross links? Nature 257:501-503. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki, M. S., and A. Tonomura. 1973. A high susceptibility of Fanconi's anemia to chromosome breakage by DNA cross-linking agents. Cancer Res. 33:1829-1836. [PubMed] [Google Scholar]
- 46.Scully, R., and D. M. Livingston. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simpson, L. J., and J. E. Sale. 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 22:1654-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonoda, E., T. Okada, G. Y. Zhao, S. Tateishi, K. Araki, M. Yamaizumi, T. Yagi, N. S. Verkaik, D. C. van Gent, M. Takata, and S. Takeda. 2003. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 22:3188-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19:5166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart, G. S., B. Wang, C. R. Bignell, A. M. Taylor, and S. J. Elledge. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961-966. [DOI] [PubMed] [Google Scholar]
- 51.Takata, M., M. S. Sasaki, E. Sonoda, T. Fukushima, C. Morrison, J. S. Albala, S. M. Swagemakers, R. Kanaar, L. H. Thompson, and S. Takeda. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 20:6476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21:2858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taniguchi, T., I. Garcia-Higuera, P. R. Andreassen, R. C. Gregory, M. Grompe, and A. D. D'Andrea. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100:2414-2420. [DOI] [PubMed] [Google Scholar]
- 55.Taniguchi, T., I. Garcia-Higuera, B. Xu, P. R. Andreassen, R. C. Gregory, S. T. Kim, W. S. Lane, M. B. Kastan, and A. D. D'Andrea. 2002. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109:459-472. [DOI] [PubMed] [Google Scholar]
- 56.Tauchi, H., J. Kobayashi, K. Morishima, D. C. van Gent, T. Shiraishi, N. S. Verkaik, D. vanHeems, E. Ito, A. Nakamura, E. Sonoda, M. Takata, S. Takeda, S. Matsuura, and K. Komatsu. 2002. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420:93-98. [DOI] [PubMed] [Google Scholar]
- 57.Timmers, C., T. Taniguchi, J. Hejna, C. Reifsteck, L. Lucas, D. Bruun, M. Thayer, B. Cox, S. Olson, A. D. D'Andrea, R. Moses, and M. Grompe. 2001. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell 7:241-248. [DOI] [PubMed] [Google Scholar]
- 58.Vandenberg, C. J., F. Gergely, C. Y. Ong, P. Pace, D. L. Mallery, K. Hiom, and K. J. Patel. 2003. BRCA1-independent ubiquitination of FANCD2. Mol. Cell 12:247-254. [DOI] [PubMed] [Google Scholar]
- 59.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 60.Venkitaraman, A. R. 2004. Tracing the network connecting brca and Fanconi anaemia proteins. Nat. Rev. Cancer 4:266-276. [DOI] [PubMed] [Google Scholar]
- 61.Wang, X., P. R. Andreassen, and A. D. D'Andrea. 2004. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24:5850-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell. Biol. 4:435-445. [DOI] [PubMed] [Google Scholar]
- 63.Wong, J. C., N. Alon, C. McKerlie, J. R. Huang, M. S. Meyn, and M. Buchwald. 2003. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum. Mol. Genet. 12:2063-2076. [DOI] [PubMed] [Google Scholar]
- 64.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi-Iwai, Y., E. Sonoda, J. M. Buerstedde, O. Bezzubova, C. Morrison, M. Takata, A. Shinohara, and S. Takeda. 1998. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol. 18:6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamoto, K., M. Ishiai, N. Matsushita, H. Arakawa, J. E. Lamerdin, J. M. Buerstedde, M. Tanimoto, M. Harada, L. H. Thompson, and M. Takata. 2003. Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol. Cell. Biol. 23:5421-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, Y., Y. Kuang, R. M. De Oca, T. Hays, L. Moreau, N. Lu, B. Seed, and A. D. D'Andrea. 2001. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood 98:3435-3440. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, Y., J. Wienands, C. Zurn, and M. Reth. 1998. Induction of the antigen receptor expression on B lymphocytes results in rapid competence for signaling of SLP-65 and Syk. EMBO J. 17:7304-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.