Abstract
The rad18 gene of Schizosaccharomyces pombe is an essential gene that is involved in several different DNA repair processes. Rad18 (Smc6) is a member of the structural maintenance of chromosomes (SMC) family and, together with its SMC partner Spr18 (Smc5), forms the core of a high-molecular-weight complex. We show here that both S. pombe and human Smc5 and -6 interact through their hinge domains and that four independent temperature-sensitive mutants of Rad18 (Smc6) are all mutated at the same glycine residue in the hinge region. This mutation abolishes the interactions between the hinge regions of Rad18 (Smc6) and Spr18 (Smc5), as does mutation of a conserved glycine in the hinge region of Spr18 (Smc5). We purified the Smc5-6 complex from S. pombe and identified four non-SMC components, Nse1, Nse2, Nse3, and Rad62. Nse3 is a novel protein which is related to the mammalian MAGE protein family, many members of which are specifically expressed in cancer tissue. In initial steps to understand the architecture of the complex, we identified two subcomplexes containing Rad18-Spr18-Nse2 and Nse1-Nse3-Rad62. The subcomplexes are probably bridged by a weaker interaction between Nse2 and Nse3.
Structural maintenance of chromosomes (SMC) proteins are found in eubacteria, archaea, and eukaryotes and appear to be present in all eukaryotes examined so far (5). In eukaryotes, six classes of SMC proteins have been identified, and they form the heterodimeric cores of three conserved protein complexes (15). Smc1 and -3 comprise the core of the cohesins, which prevent sister chromatid separation following DNA replication by holding the sister chromatids together. Smc2 and -4 form the core of condensins, which play an important role in condensing chromosomes during mitosis. Smc5 and -6 are the core of a complex involved in DNA repair (7).
SMC proteins have a common structure. The N- and C-terminal domains are globular and are separated by two long coiled coils interrupted by a flexible hinge. The N- and C-terminal domains contain Walker A and B motifs, respectively, which are brought together by the molecule folding back on itself via the hinge such that the two coiled coils interact in an antiparallel structure. For cohesin and condensin, it has been shown that the Smc1-Smc3 and Smc2-Smc4 heterodimers are formed by intermolecular interactions of the hinge domains (10, 14). In addition, it has been proposed that the globular domains of the two SMC monomers of cohesin can also interact to form a ring around the chromosomal DNA, encompassing the two sister chromatids (10). The mean evolutionary distance of the Smc5 and -6 families from the root of the evolutionary tree is about twice that of other eukaryotic SMC proteins, indicating a higher degree of sequence divergence (5). This is particularly true of the hinge region. While the hinges of Smc1 to -4 share considerable sequence similarity, which also extends to the prokaryotic SMCs, the Smc5 and Smc6 hinges are markedly different.
The prototype member of the Smc5-6 complex is the Rad18 (Smc6) protein of Schizosaccharomyces pombe. (Note that S. pombe rad18 and Saccharomyces cerevisiae RAD18 are completely unrelated genes.) rad18.X (encoding an R706C mutation in the second coiled coil region close to the hinge) and rad18-74 (encoding an A151T mutation in the N-terminal globular domain) cells are sensitive to both UV and γ-irradiation and to methyl methanesulfonate (MMS) treatment (19, 26) and are deficient in the repair of double strand breaks in DNA (26). Epistasis analysis suggests that Rad18 (Smc6) is involved in several recombinational repair processes, including a pathway for removing UV damage that is distinct from nucleotide excision repair, a UV DNA damage tolerance pathway, and a double-strand-break repair pathway (19, 23). After the treatment of cells with γ-irradiation, rad18.X and rad18-74 cells arrest the cell cycle and then, after a delay, return to the cell cycle with the same kinetics as wild-type cells, even though the DNA damage is not repaired in the mutant cells (1, 26). It has therefore been suggested that Rad18 (Smc6) is not required to establish a checkpoint but is needed to maintain it (11, 26). Rad18 (Smc6) also has an essential function (19). The lethality of a rad18 deletion strain may result either from an accumulation of unrepaired DNA damage or from a separate function that is unrelated to the protein's involvement in repair. Several viable hypomorphic mutations have been reported which confer sensitivity to DNA damage while retaining viability. These include the mutation of serine 1045 to alanine (7). By extrapolation from the SMC-like structure of the Rad50 and Smc1 proteins, Ser1045 is likely to be involved in binding of the γ-phosphate of ATP and required for ATP hydrolysis (2, 17, 27).
Purification of Rad18 (Smc6) from S. pombe cells showed it to be part of a high-molecular-weight complex with several components, one of which is its SMC protein partner, Spr18 (Smc5). Two other proteins that interact with Rad18 (Smc6) were recently identified by mass spectroscopy and were designated Nse1 and Nse2 (20). Like Rad18 (Smc6), Spr18 (Smc5), Nse1, and Nse2 are essential genes (7, 20).
In the present work, we show that four randomly isolated temperature-sensitive mutants of Rad18 (Smc6) have mutations at an identical glycine residue in the hinge region, that Rad18 (Smc6) and Spr18 (Smc5) interact via their hinge regions, and that this interaction is destroyed in the temperature-sensitive rad18 mutants or if a conserved glycine in the hinge region of Spr18 (Smc5) is mutated. Aside from the Rad18 (Smc6) and Spr18 (Smc5) core proteins, we have identified four other polypeptides in the affinity-purified Rad18 (Smc6) complex. These include Nse1, Nse2, and two new components. We have begun to delineate the architecture of the complex by determining which of the individual subunits interact in vitro.
MATERIALS AND METHODS
Isolation of temperature-sensitive rad18 mutants.
A rad18 deletion construct in which the 3.7-kb AccI fragment spanning the entire open reading frame (ORF) was replaced with the arg3 gene was generated essentially as described by Lehmann et al. (19). A diploid strain was constructed with the genotype ura4-D18/ura4-D18 leu1-32/leu1-32 arg3-D4/arg3-D4 h+/h+, and the deletion plasmid was used to transform this strain to arginine prototrophy. Colonies were screened for the replacement event by Southern blotting, and an h+/h90 derivative was isolated. This strain was transformed with the MH42-rad18+ plasmid (ura4+) and sporulated, and haploid progeny with the genotype rad18::arg3+ ura4-D18 leu1-32 arg3-D4 h+ (MH42-rad18+) were selected. A randomly mutagenized pool of rad18 genes was created by treating the MH41-rad18 plasmid (leu+) with hydroxylamine (18). Mutagenized plasmids were transformed into the rad18::arg3 (MH42-rad18+) strain, and transformants were selected on medium lacking leucine and arginine and then replica plated onto medium containing fluoroorotic acid (FOA) to select for the loss of the MH42-rad18+ plasmid. For the identification of temperature-sensitive alleles of rad18, the resulting 15,000 colonies were screened by replica plating onto plates that were incubated at 27 and 36°C. Four clones that could grow at 27°C but not at 36°C were isolated. Plasmids were recovered from these temperature-sensitive strains, and the mutation sites were identified by sequencing. For genomic integration of temperature-sensitive alleles (T1 to T3), the mutations were incorporated into the 7.8-kb genomic clone in plasmid SpF (19), and rad18::arg3 haploid strains harboring each of these plasmids (SpF.T1 to SpF.T3) were generated. These strains were each transformed with the appropriate 7.8-kb linear fragment of rad18 DNA containing the T1, T2, or T3 mutation, and integration events were screened by the use of FOA to select for the loss of the SpF plasmid. Southern blotting was used to confirm the correct replacement of the arg3 deletion with the rad18 temperature-sensitive mutant alleles.
DNA damage sensitivity tests.
For UV irradiation, cells were grown to mid-log phase at the appropriate temperature (25 or 30°C), plated on yeast extract (YE) plates, and irradiated at 254 nm by use of a Stratalinker chamber (Stratagene). For γ-irradiation, exponentially growing cells were irradiated in suspension by a 137Cs source (dose rate, 8.5 Gy min−1) prior to plating on YE plates. All plates were incubated at 25 or 30°C as appropriate, and colonies were counted after 4 to 5 days. Survival was expressed as a percentage of the number of colonies that were formed in the absence of irradiation.
Cell cycle synchronization.
Cells were synchronized by lactose gradient centrifugation as described by Barbet and Carr (3). G2 cells were resuspended in fresh prewarmed YE and incubated at 36°C for the duration of the experiment. Aliquots were removed at 20-min intervals, plated on YE plates, and incubated at 25°C for 4 to 5 days to determine cell viability. At the same time, aliquots were fixed in methanol and stained with DAPI (4′,6′-diamidino-2-phenylindole) and calcofluor in order to determine the percentage of cells that passed mitosis.
Yeast two-hybrid analysis.
The ProQuest two-hybrid system (Invitrogen) was used to analyze interactions between human Smc5 (hSmc5) and hSmc6 hinge regions. Transformation of the plasmids pPC86-hSmc5 hinge, pDBLeu-hSMC6 hinge, and pDBLeu-hSmc6gly506 into S. cerevisiae MaV203, growth on selective media, and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) assays were performed according to the manufacturer's instructions.
The Matchmaker Gal4 two-hybrid system 3 (Clontech) was used to analyze interactions between the individual components of the S. pombe complex. Transformation of the constructs cloned in pGBKT7, pGADT7, and/or pACT2 (details available on request) into S. cerevisiae Y190, growth on selective media, and X-Gal assays were performed according to the manufacturer's instructions.
Antibody generation.
A cDNA fragment corresponding to Rad18 (Smc6) amino acids (aa) 152 to 684 was cloned into the vector pQE-30 (Qiagen) and expressed in Escherichia coli as an N-terminal hexahistidine-tagged fusion protein. The protein was purified to near homogeneity under denaturing conditions by Ni2+-nitrilotriacetic acid affinity chromatography and then was used to inoculate two rabbits. Antibodies were affinity purified by use of an antigen immobilized with Aminolink Plus coupling gel (Pierce) according to the manufacturer's instructions.
Polyclonal antibodies to Spr18 (Smc5), raised in sheep, and to Nse2, raised in rabbits, are described elsewhere (7) and in the accompanying paper (1a). Antibodies to the Nse1 and Nse3 proteins were raised in rabbits against a mix of two peptides each (for Nse1, EKERQDGLSDKHKFIL and DFKIKRVQDQLDGRL; for Nse3, LRRPATSNANSSNL and TEYRQEYQNQSSSSAA). Nse3 antibodies were purified against the same peptides linked to N-hydroxysuccinimide-activated Sepharose (Pharmacia).
These antibodies identified the corresponding in vitro-translated proteins. They also detected proteins of the appropriate sizes in cell extracts which were not detected by the corresponding preimmune sera.
Purification of the Rad18 (Smc6) complex.
Forty liters of S. pombe Myc-Rad18 cells in which the genomic rad18 gene was N-terminally tagged with the c-myc epitope were grown to mid-log phase, harvested, and washed, and the cell pellet was resuspended in an equal volume of lysis buffer A (45 mM HEPES [pH 7.8], 300 mM KCl, 10% glycerol, 5 mM EDTA, 5 mM EGTA, 80 mM β-glycerophosphate, 12 mM NaF, 0.1 mM sodium orthovanadate, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], and a protease inhibitor cocktail consisting of 5 μg each of trypsin inhibitor, pepstatin, leupeptin, and aprotinin/ml, 10 μg each of bestatin and E-64/ml, and 50 μg of chymostatin/ml). The cells were snap-frozen as droplets in liquid nitrogen and lysed with a Retsch grinder, with frequent additions of liquid nitrogen. The lysate was clarified by centrifugation and subjected to ammonium sulfate precipitation. The 13-to-50%-cut precipitate containing Rad18 (Smc6) was resuspended in 1 volume of binding buffer B (25 mM HEPES [pH 7.8], 300 mM KCl, 10% glycerol, 2 mM EDTA, 3 mM EGTA, 3 mM MgCl2, 0.2 mM DTT, 0.5 mM PMSF, and the protease inhibitor cocktail described above) and extensively dialyzed against the same buffer. The dialyzed lysate was diluted onefold with binding buffer B and incubated overnight at 4°C with 5 ml of protein G-Sepharose beads that had been previously cross-linked to a monoclonal anti-Myc antibody (9E10). The beads were washed once with 10 volumes of binding buffer B, four times with 10 volumes of wash buffer 1 (25 mM HEPES [pH 7.8], 500 mM KCl, 8% glycerol, 2 mM EDTA, 2 mM MgCl2, 0.5 mM PMSF, 0.1% NP-40), and twice with wash buffer 2 (25 mM HEPES [pH 7.8], 100 mM KCl, 8% glycerol, 2 mM EDTA, 2 mM MgCl2, 0.5 mM PMSF, 0.01% NP-40). The beads were transferred to a column, incubated with 2 volumes of wash buffer 2 containing 5 mM ATP for 10 min at 4°C, and then washed once more with 10 volumes of wash buffer 2. The Rad18 (Smc6) complex was eluted at 30°C for 1 h, with 3 mg of peptide/ml corresponding to the Myc epitope (AEEQKLISEEDL).
Mass spectrometric analysis of the Rad18 (Smc6) complex.
Purified Myc-Rad18 (Smc6) complex components were precipitated with 4 volumes of acetone, separated by sodium dodecyl sulfate-4 to 20% polyacrylamide gradient gel electrophoresis (SDS-PAGE), and stained with colloidal Coomassie blue after fixation with 40% methanol-10% acetic acid for 1 h. Individual bands were excised and identified by trypsin digestion and mass fingerprinting by mass spectrometry at the COGEME Proteome Service Facility 1, University of Aberdeen. Proteins were identified by searching databases with the MASCOT search engine (Matrix Science).
In vitro transcription-translation.
ORFs corresponding to proteins or protein fragments were amplified from S. pombe cDNAs and singly or coexpressed in vitro as Myc-, hemagglutinin (HA)-, S- or untagged proteins from the pEPEX, pET-Duet, pTriEx, or pGem-T-Easy vector by use of the TNT Quick coupled transcription-translation system (Promega) in 50-μl reactions according to the manufacturer's instructions.
The following constructs were used: for Rad18, pEPEX-myc-Rad18, pEPEX-myc-Rad18-hinge (aa 504 to 719), pEPEX-HA-Rad18-hinge (aa 504 to 719), and pEPEX-myc-Rad18-gly551-hinge; for Spr18, pEPEX-Spr18, pEPEX-Spr18-gly529, pEPEX-myc-Spr18-hinge (aa 393 to 620), pEPEX-HA-Spr18-hinge (aa 393 to 620), pEPEX-myc-Spr18-gly529-hinge, pEPEX-HA-Spr18-gly529-hinge, pET-Duet-Spr18 “headless” (aa 171 to 910), pGem-Spr18 (aa 226 to 836), pGem-Spr18 (aa 324 to 731), and pET-Duet-Spr18 “heads” (aa 2 to 174 plus aa 897 to 1065); for Nse1, pEPEX-Nse1 and pGem-T-Easy-Nse1(aa 1 to 116); for Nse2, pET-Duet-Nse2 (aa 1 to 178) and pET-Duet-Nse2 (aa 114 to 250); for Nse3, pEPEX-myc-Nse3 and pEPEX-HA-Nse3; and for Rad62, pTriEx-4-Rad62. Details of the construction of the plasmids are available on request.
Preparation of recombinant proteins.
The Nse1, Nse2, and Nse3 proteins were expressed as glutathione S-transferase (GST) fusions. The expression of pGEX-Nse1 and pGEX-Nse2 constructs was carried out in BL21 cells, while pGEX-Nse3 and pTriEx-Rad62 constructs were expressed from Rosetta-gami B cells (Novagen). The cells were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and harvested after 4 h of induction. The bacteria were lysed with HEPES buffer C (25 mM HEPES [pH 7.5], 200 mM KCl, 3 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM PMSF, 0.2% NP-40, and protease inhibitors).
GST pull-down experiments.
For GST pull-down experiments, 50 μl of GST fusion proteins immobilized on glutathione-Sepharose beads (Amersham Pharmacia Biotech) was incubated with 5 to 10 μl of in vitro-expressed proteins in a total volume of 200 μl of HEPES buffer C containing 1 mM ATP, 50 mM creatine phosphate, and 1 U of creatine phosphokinase. After 2 h at 4°C, the beads were washed twice with HEPES buffer C containing 400 mM KCl and 0.4% NP-40 and once with HEPES buffer C containing 100 mM KCl and 0.1% NP-40. Input, unbound, and bound fractions were separated by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by phosphorimaging and immunoblotting with anti-GST (Amersham Pharmacia Biotech).
Myc pull-down experiments.
For Myc pull-down experiments, 5 to 10 μl of in vitro-expressed proteins in a total volume of 200 μl of HEPES buffer C containing 1 mM ATP, 50 mM creatine phosphate, and 1 U of creatine phosphokinase was mixed with 10 μl of an anti-Myc monoclonal antibody and incubated overnight at 4°C. Fifty microliters of protein G-Sepharose beads (Amersham Pharmacia Biotech) was then added, and the mixture was incubated for 1 h. The beads were washed twice with HEPES buffer C containing 400 mM KCl and 0.4% NP-40 and once with HEPES buffer C containing 100 mM KCl and 0.1% NP-40. Alternatively, 20 μl of the reaction was added to 180 μl of binding buffer B (described above) and incubated overnight at 4°C with 100 μl of protein G-Sepharose beads that had been previously cross-linked with a monoclonal anti-Myc antibody (9E10). The beads were washed twice with 10 volumes of wash buffer 1 (described above) and twice with 10 volumes of wash buffer 2. Input, unbound, and bound fractions were separated by SDS-PAGE and analyzed by phosphorimaging and/or immunoblotting with respective antibodies.
S tag pull-down experiments.
For S tag pull-down experiments, 50 μl of S tag fusion proteins immobilized on protein S-agarose beads (Novagen) was incubated with 5 to 10 μl of in vitro-expressed proteins in a total volume of 200 μl of HEPES buffer MB (25 mM HEPES [pH 7.2], 100 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, 1 mM PMSF, 0.1% NP-40, and protease inhibitors). After 2 h at 4°C, the beads were washed three times with HEPES buffer MB. Input, unbound, and bound fractions were separated by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by phosphorimaging and immunoblotting with protein S conjugated with horseradish peroxidase (Novagen).
RESULTS
Temperature-sensitive mutants of rad18 (smc6) with mutations in the hinge region.
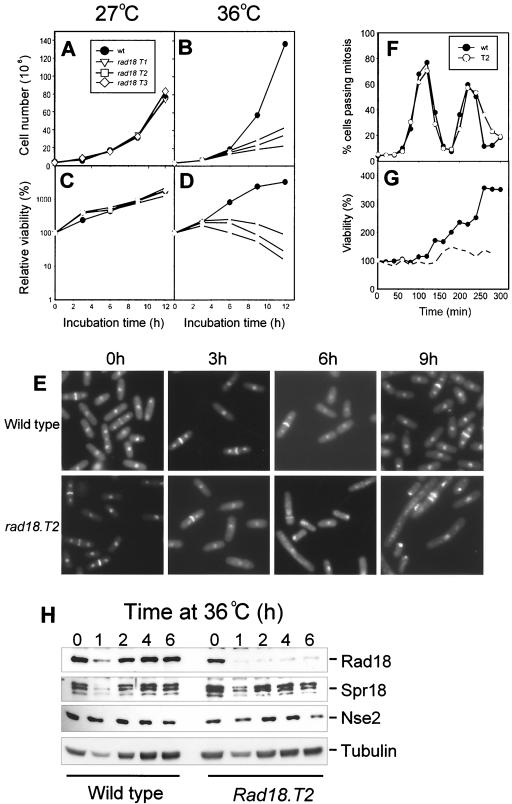
Since rad18 (smc6) is an essential gene in S. pombe, we were interested in isolating conditionally lethal mutants. To this end, we randomly mutagenized a rad18 (smc6)-containing plasmid by using hydroxylamine and then used the mutagenized population to isolate plasmids that were temperature sensitive in their ability to rescue the lethality of rad18 (smc6) deletion mutants. Four independent temperature-sensitive plasmid-containing clones (T1 to T4) were obtained, from which the plasmids were recovered and sequenced. Strikingly, all of the plasmids were mutated at Gly551 in the hinge region of the protein. In plasmids T2 and T4, Gly551 was mutated to Arg, whereas in T1 and T3 it was mutated to Glu and Lys, respectively. Each of these three mutations at Gly551 (T2 was used for the Gly-to-Arg mutation) was then introduced into the genome of wild-type S. pombe cells. The resulting strains were analyzed for growth and viability at 27 and 36°C. All three strains grew normally and were viable at 27°C (Fig. 1A and C). At 36°C, growth was severely impaired and cells began to lose viability after 6 h (Fig. 1B and D). At 27°C, the morphology of the rad18 (smc6) mutant cells was indistinguishable from that of wild-type cells, whereas within 6 h of shifting the temperature to 36°C, the cells showed a variety of abnormal phenotypes, including aberrant nuclear segregation, cut phenotypes, abnormal septa, and elongated cells (Fig. 1E). These phenotypes are characteristic of terminal phenotypes that were observed previously for rad18 (smc6) deletion mutants (19). Using synchronized cells, we found that at 36°C, the cells were able to pass through at least two rounds of mitosis (Fig. 1F), and viability was gradually lost after each round of mitosis (Fig. 1G). To determine if this ability to maintain viability following the shift to the restrictive temperature reflected a relatively long half-life of the Rad18 (Smc6) protein, we incubated the rad18.T2 mutant at 36°C for different times and analyzed the amount of Rad18 (Smc6) protein by immunoblotting. Figure 1H shows that there was a substantial reduction in the amount of Rad18 (Smc6) protein 1 h after the temperature shift and that this reduced level remained fairly constant for several hours. Interestingly, the amounts of Spr18 (Smc5) and Nse2 remained unchanged despite the loss of the Rad18 (Smc6) protein.
FIG. 1.
Temperature-sensitive mutants of rad18 (smc6). The wild type and three temperature-sensitive mutants of rad18 (smc6) were incubated at 27°C (A and C) or 36°C (B and D), and the growth (A and B) and viability (C and D) of each strain were measured at different times. (E) Wild-type and rad18.T2 cells were grown at 36°C, and at different times the cells were fixed in methanol and stained with DAPI and calcofluor. Cells that were synchronized by lactose gradient centrifugation were incubated at 36°C, and at various times the numbers of cells passing mitosis (F) and the cell viability (G) were measured. (H) Cell lysates from wild-type and rad18.T2 cells grown at 36°C for 0 to 6 h were analyzed by SDS-PAGE and immunoblotting with antibodies to Rad18 (Smc6), Spr18 (Smc5), Nse2, and tubulin.
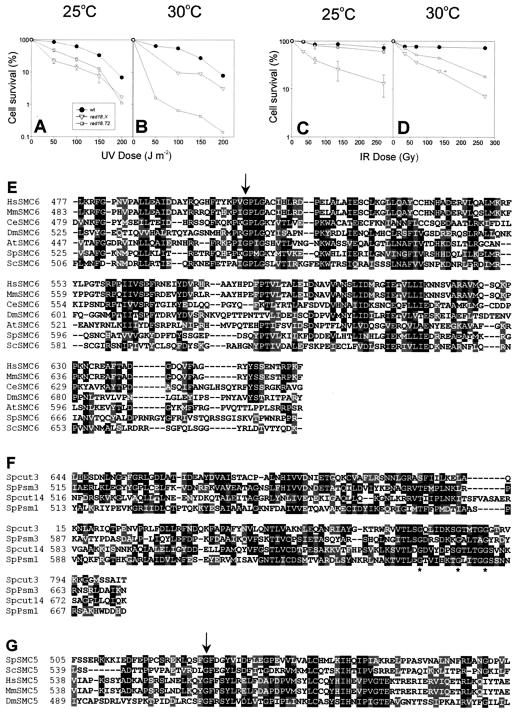
We also examined the sensitivity of the rad18.T2 cells to DNA damage at the permissive and semipermissive temperatures. Figure 2A shows that the mutant cells were mildly sensitive to UV irradiation at 25°C but that this sensitivity was dramatically increased at 30°C (Fig. 2B) and far exceeded the sensitivity of the rad18.X mutant. Sensitivity to γ-irradiation was barely affected at 25°C (Fig. 2C) but was more marked at 30°C, although the cells were less sensitive than the rad18.X mutant (Fig. 2D).
FIG. 2.
Mutations in the hinge region cause sensitivity to DNA damage. Wild-type, rad18.X, and rad18-T2 cells grown at 25°C (A and C) or 30°C (B and D) were exposed to different doses of UV (A and B) or γ-rays (C and D), and their viabilities were determined at 25°C (A and C) or 30°C (B and D). (E) Alignment of the hinge regions of SMC6 proteins from humans, mice, Caenorhabditis elegans, D. melanogaster, A. thaliana, S. pombe, and S. cerevisiae. The conserved residue corresponding to Gly551 in Rad18 (Smc6) is indicated with an arrow. (F) Alignment of the hinge regions of the Smc1 (Psm1), Smc2 (Cut3), Smc3 (Psm3), and Smc4 (Cut14) proteins of S. pombe. Conserved glycine residues that were shown to be important for hinge-mediated interactions (13) are indicated with asterisks. (G) Alignment of part of the hinge regions of SMC5 proteins corresponding to aa 505 to 585 of the S. pombe protein (the hinge region spans aa 400 to 620). The conserved Gly529 residue is indicated.
To determine if the temperature sensitivity of rad18.T2 cells could be suppressed by overexpression of other genes involved in either the DNA damage checkpoint, DNA repair, or DNA replication, we transformed cells with plasmids expressing a variety of genes involved in these processes under the control of the thiamine-repressible nmt1 promoter. In no case did the overexpression of any of these genes suppress the temperature sensitivity of rad18.T2 (data not shown).
Interaction between the hinge regions of Smc5 and Smc6.
Our finding that four independent temperature-sensitive clones were all mutated at the same glycine residue in the hinge region of Rad18 (Smc6) points to the importance of this region in maintaining the function of the Smc5-6 complex. The hinge region is highly conserved within the Smc6 family (Fig. 2E), and Gly551 is conserved in all species. Although the hinge region mediates the heterodimerization of Smc1 with Smc3 in cohesin and of Smc2 with Smc4 in condensin, the hinge regions of Smc6 and Smc5 have minimal sequence similarity to the hinges of Smc1 to -4 (compare Fig. 2E with Fig. 2F). In particular, there is a series of glycine residues that have been shown to be vital for hinge-mediated dimerization and that are conserved not only in the Smc1 to -4 families (Fig. 2F) but also in bacterial SMC proteins (13, 14). These glycine residues are not found in the Smc6 or Smc5 family.
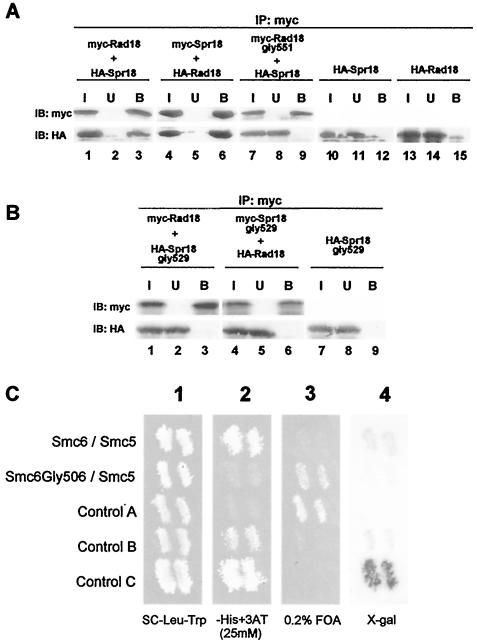
In order to determine if the hinge region also mediates the interaction between Rad18 (Smc6) and Spr18 (Smc5), we generated different tagged fragments of the proteins by in vitro transcription-translation and analyzed the interactions between them. We were not able to detect any interaction of His-tagged N-terminal globular domains of Rad18 (Smc6) or Spr18 (Smc5) with the C-terminal globular domain of Rad18 (Smc6) (data not shown). However when the hinge region of Rad18 (Smc6) tagged with the c-Myc epitope was mixed with the HA-tagged Spr18 (Smc5) hinge region and immunoprecipitated with an anti-Myc antibody, both Myc-Rad18 (Smc6) and HA-Spr18 (Smc5) were quantitatively recovered in the immune precipitate (Fig. 3A, lane 3). In a converse experiment with Myc-Spr18 (Smc5) mixed with HA-Rad18 (Smc6) and immunoprecipitated with anti-Myc, both proteins were again found in the immune complex (Fig. 3A, lane 6). When comparable experiments were performed with the Rad18 (Smc6)-Gly551 mutant construct, the interaction between the Myc-Spr18 (Smc6) and HA-Spr18 (Smc5) hinge fragments was abolished (Fig. 3A, lane 9, bottom panel). Singly expressed HA-tagged hinge constructs were not precipitated by the anti-Myc beads alone (Fig. 3A, lanes 12 and 15). These data provide strong evidence that Rad18 (Smc6) and Spr18 (Smc5), like the other SMC proteins, interact through their hinge regions and that the mutation of glycine 551 confers temperature sensitivity by weakening this interaction.
FIG. 3.
SMC6 hinge domain interacts with SMC5 hinge domain, and mutations at conserved glycine residues abolish this interaction. (A) Tagged S. pombe hinge constructs (Myc-Rad18 [Smc6], Myc-Spr18 [Smc5], HA-Rad18 [Smc6], HA-Spr18 [Smc5], and Myc-Rad18gly551) were expressed in vitro as indicated and were incubated overnight at 4°C with an anti-Myc antibody that was previously cross-linked to protein G-Sepharose beads to precipitate Myc-tagged constructs. The beads were washed extensively, and the input (I), unbound (UB), and bound (B) proteins were analyzed by SDS-PAGE and immunoblotting with either anti-Myc or anti-HA antibodies. Lanes 10 to 15 are negative controls without any Myc-tagged protein. (B) Immunoprecipitations were performed as described for panel A, but with constructs containing the Spr18 Gly529 mutation. (C) Yeast two-hybrid analysis of hSmc5 and hSmc6. Yeast strains harboring the plasmids GAL4-DB hSmc6 hinge/GAL4-AD hSmc5 hinge and GAl4-DB hSmc6 Gly506 hinge/GAL4-AD hSmc5 hinge along with the control strains A (no interaction), B (weak interaction), and C (strong interaction) were patched onto an SC-Leu-Trp master plate and incubated at 30°C for 18 h (panel 1). Cells from this plate were replica plated onto SC-Leu-Trp-His plus 25 mM 3-amino-2,4,6-triazole (panel 2), SC-Leu-Trp plus 0.2% 5-FOA (panel 3), and enriched yeast extract medium containing a nylon membrane (for X-Gal assay) (panel 4) before growth at 30°C for an additional 2 days.
The hinge regions of Spr18 (Smc5) orthologs are well conserved between species but differ from those of other SMC proteins. As shown in Fig. 2G, there is a conserved glycine at aa 529 in the hinge region of the S. pombe Spr18 (Smc5) protein. We mutated this glycine to arginine and examined the effect of this mutation on the interaction with the Rad18 (Smc6) hinge. Figure 3B shows that, as with the Gly551 mutation in Rad18 (Smc6), a Gly529Arg mutation in Spr18 (Smc5) abolishes the interaction between the hinges (compare lanes 3 and 6 of Fig. 3B with the corresponding lanes of Fig. 3A).
Spr18 (Smc5) and Rad18 (Smc6) have human orthologs (25). We used a yeast two-hybrid system to analyze interactions between the hinge regions of hSmc5 and hSmc6. Figure 3C shows the results of this analysis. As with S. pombe, our results show that the hinge regions of the two proteins interact. Interactions were measured between the hinge regions of hSmc5 and either wild-type hSmc6 or hSmc6 with a mutation to Arg at Gly506 (corresponding to Gly551 of the S. pombe protein) by the use of three different reporter genes (HIS3, URA3, and lacZ). Interaction-dependent transcriptional activation of the HIS3 and URA3 genes allowed growth on plates lacking histidine (Fig. 3C, panel 2) but inhibited growth on medium containing 5-FOA (Fig. 3C, panel 3). Induction of the lacZ gene resulted in a blue color when assayed with X-Gal (Fig. 3C, panel 4). Positive controls in the assay were the weakly interacting proteins Rb and E2F1 (control B) and the strongly interacting Drosophila proteins DP and E2F1 (control C). The negative control was empty vectors (control A). Our results clearly demonstrate that the hinge regions of hSmc5 and hSmc6 interact (Fig. 3C, top row). The strength of the interaction was intermediate between the weakly and strongly interacting controls. The mutation of Gly506 in hSmc6 completely abolished its interaction with hSmc5 (Fig. 3C, second row). Thus, the interaction between the hinge regions and the importance of Gly551 in this interaction are conserved in yeast and humans.
Composition of the SMC5-6 complex.
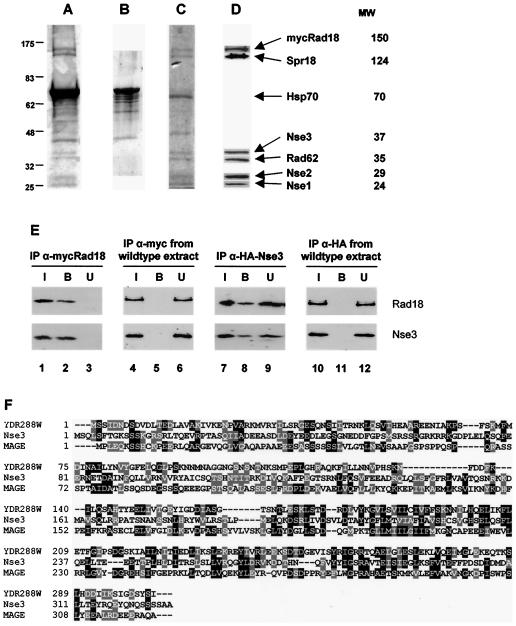
In an earlier study, it was reported that Rad18 (Smc6) and Spr18 (Smc5) are part of a high-molecular-weight complex which contains several other components (7). Using cells in which the genomic rad18 gene was N-terminally tagged with the c-Myc epitope, we were able to purify the complex partially by immuno-affinity chromatography with anti-Myc beads but were hindered from analyzing the components because of a strong contaminating 70-kDa protein in all of our purifications (see Fig. 3C of reference 7) (Fig. 4A). Even though it was shown previously that this protein bound nonspecifically to anti-Myc beads (7), we were unable to remove it from the complex by use of a variety of different chromatographic procedures. Using mass spectroscopy, we identified the contaminating protein as Hsp70 and ultimately were able to remove it by washing the anti-Myc beads with ATP (Fig. 4B). The complex, which remained attached to the beads, was eluted with the c-Myc peptide, electrophoresed in an SDS-PAGE gel, and stained with colloidal Coomassie blue (Fig. 4C). The bands were excised and analyzed by mass spectroscopy. Apart from Rad18 (Smc6) and Spr18 (Smc5), four other proteins were identified (Table 1). Two of them corresponded to the previously identified Nse1 (8, 11, 20) and Nse2 (20) proteins. One of the other two we designated Nse3, and the other is orthologous to S. cerevisiae Qri2. The ortholog of Qri2 was recently shown to be identical to the S. pombe Rad62 protein (21). We raised antibodies to Nse1, Nse2, Nse3, and Rad62 and used these to probe immunoblots of the complex after electrophoresis in SDS-PAGE gels. Figure 4D confirms that these four proteins are indeed present in the complex. The presence of Nse1, Nse2, and Nse3 as members of the complex and as free proteins was also analyzed by gel filtration (see the accompanying paper [1a]).
FIG. 4.
Purification and identification of Rad18 (Smc6)/Spr18 (Smc5) complex components. A lysate prepared from a Myc-tagged Rad18 (Smc6) strain was incubated overnight at 4°C with an anti-Myc antibody that was previously cross-linked to protein G-Sepharose beads. The beads were washed extensively, and bound proteins were eluted by incubation with an excess peptide corresponding to the Myc epitope, separated by SDS-PAGE, and stained with colloidal Coomassie blue (A). (B) The intense contaminating band at 70 kDa was identified as Hsp70 by mass spectrometry and was effectively removed by washing the beads with 5 mM ATP. (C) The remaining proteins were eluted as before and separated by SDS-PAGE, and individual bands were excised and identified by trypsin digestion and mass fingerprinting. (D) Antibodies were raised against Nse1, Nse2, Nse3, and Rad62 and were used, along with Rad18 (Smc6) and Spr18 (Smc5) antibodies, to confirm the presence of the identified proteins in an independent preparation of the complex by immunoblotting. (E) Lysates prepared from Myc-tagged Rad18 (Smc6) (lanes 1 to 3), HA-tagged Nse3 (lanes 7 to 9), or untagged (lanes 4 to 6 and 10 to 12) strains were incubated overnight at 4°C with either anti-Myc or anti-HA antibodies that were previously cross-linked to protein G-Sepharose beads. The beads were washed extensively, and the input (I), bound (B), and unbound (U) proteins were analyzed by SDS-PAGE and immunoblotting with anti-Rad18 and anti-Nse3 antibodies. (F) Alignment of Nse3, S. cerevisiae YDR288W, and the MAGE consensus sequence KOG4562.
TABLE 1.
Identification of Rad18 (Smc5-6) complex components by mass spectrometry
| Protein | Protein size (aa) | No. of peptides matched/total no. searched | % Sequence coverage |
|---|---|---|---|
| Rad18 | 1,140 | 10/13 | 9 |
| Spr18 | 1,065 | 15/24 | 16 |
| Nse3 (spcc645.04) | 328 | 9/19 | 23 |
| Rad62 (spbc20F10.04c) | 300 | 6/25 | 26 |
| Nse2 | 250 | 5/14 | 27 |
| Nse1 | 232 | 8/13 | 26 |
Nse3.
To further confirm that Nse3 is part of the complex, we immunoprecipitated Rad18 (Smc6) from cells in which it was tagged with Myc in the genome and showed by immunoblotting that Nse3 was present in the immunoprecipitates (Fig. 4E, lane 2). No Nse3 was present in immunoprecipitates from untagged cells (lane 5). In converse experiments, we HA-tagged Nse3 in the genome and showed that Rad18 (Smc6) was present in anti-HA immunoprecipitates (Fig. 4E, lane 8; note that the immunoprecipitation of HA-Nse3 with anti-HA [lane 8] was less efficient than that of Myc-Rad18 (Smc6) with anti-Myc [lane 2]). Again, no Rad18 was precipitated from extracts of untagged cells (lane 11). These data confirm that Nse3 is in the same protein complex as Rad18 (Smc6). Deletion of most of the nse3 ORF showed that it is an essential gene, like all of the other components of the complex. The terminal morphology of the deletion mutant was a heterogeneous mixture of abnormal cells, as has been found previously with rad18, spr18, and rad62 mutants (H. Morikawa, I. Miyabe, T. Morishita, and H. Shinagawa, unpublished results).
Nse3 is a 328-aa protein and is designated SPCC645.04 in the S. pombe sequencing project. BLAST searching of the protein databases identified a putative ortholog in the S. cerevisiae database, an ORF encoding a 303-aa protein of unknown function designated YDR288W. An alignment of the two proteins showed a relatively low level of identity of 15.5% (Fig. 4F). Further iterative BLAST searching revealed sequence similarity to a melanoma antigen-encoding gene (MAGE) domain. The MAGE family is a large family of proteins, most of which are expressed in a wide variety of tumors but not in normal cells, with the exception of the male germ cells, the placenta, and possibly cells of the developing embryo. The cellular function of this family is unknown. Within the conserved domain database, both Nse3 and its S. cerevisiae homolog could be aligned with a MAGE family consensus sequence, KOG4562 (Fig. 4F), with 17 to 20% sequence identity. The sequence identity between Nse3 and both YDR288W and the MAGE consensus is highest for the C-terminal half of the protein, which is also the region with the most sequence similarity between different members of the MAGE family.
Interactions between components.
In order to dissect the interactions between the individual components of the complex, we expressed them as GST-tagged proteins in E. coli. GST-tagged Nse1, Nse2, and Nse3 were soluble. The proteins were also synthesized singly or together by in vitro transcription-translation (IVT), and their interactions were analyzed in several different ways.
Interaction of Nse2 with Spr18 (SMC5).
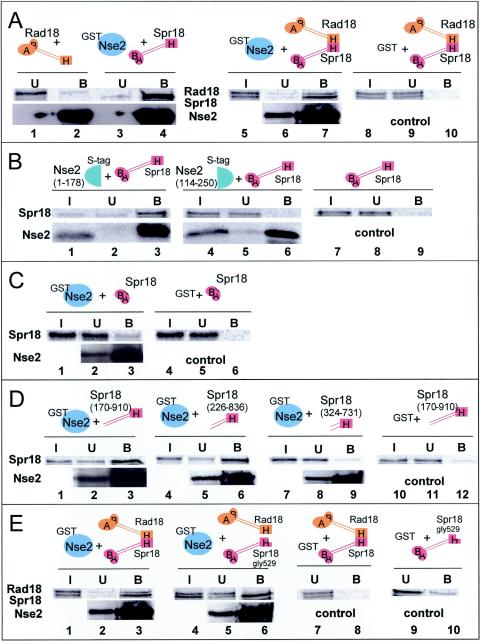
GST-Nse2 pull-down assays were performed with 35S-labeled Rad18 (Smc6) or Spr18 (Smc5). As shown in Fig. 5A (bottom panels), Nse2 was bound to the beads. Rad18 (Smc6) did not bind to the GST-Nse2 beads (lane 2, top panel), whereas Spr18 (Smc5) bound strongly (lane 4, top panel). When Rad18 (Smc6) and Spr18 (Smc5) were either synthesized together in vitro or synthesized separately and then mixed together, significant proportions of both Rad18 (Smc6) and Spr18 (Smc5) were bound (Fig. 5A, lane 7, top panel) compared to binding to GST beads alone (i.e., without GST-Nse2) (lane 10). Similar results were obtained irrespective of whether the Rad18 (Smc6) and Spr18 (Smc5) proteins were coexpressed or not (not shown). We conclude that Nse2 binds to the Rad18 (Smc6)-Spr18 (Smc5) heterodimer via the Spr18 (Smc5) component.
FIG. 5.
Interaction between Nse2 and Spr18 (Smc5). (A) A soluble protein extract prepared from E. coli strain BL21 expressing GST-Nse2 was preincubated with glutathione beads, and either IVT Rad18 (Smc6) (lanes 1 and 2), Spr18 (Smc5) (lanes 3 and 4), or a mixture of IVT Rad18 (Smc6) and Spr18 (Smc5) protein extracts (lanes 5 to 7) was added. The control experiment (lanes 8 to 10) was identical to that of lanes 5 to 7 except that GST was used instead of GST-Nse2. Input (I), unbound (U), and bound (B) fractions were collected and run in SDS-10% PAGE gels. The in vitro-expressed proteins were visualized with a phosphorimager. Bacterially expressed proteins were analyzed by immunoblotting with an anti-Nse2 rabbit antiserum. (B) IVT Spr18 (Smc5) was mixed with an IVT N-terminal (aa 1 to 178; lanes 1 to 3) and/or C-terminal fragment (aa 114 to 250; lanes 4 to 6) of Nse2-S tag and incubated with protein S beads. In the control experiment, the Spr18 (Smc5) protein was incubated with protein S beads in the absence of Nse2-S tag (lanes 7 to 9). (C) The IVT Spr18 (Smc5) head construct (aa 2 to 174 plus aa 897 to 1065) was mixed with either the GST-Nse2 (full-length) protein (lanes 1 to 3) or GST alone (lanes 4 to 6) and incubated with glutathione beads. (D) IVT Spr18 (Smc5) headless fragments containing aa 170 to 910 (lanes 1 to 3 and 10 to 12), aa 226 to 836 (lanes 4 to 6), or aa 324 to 731 (lanes 7 to 9) were mixed with either GST-Nse2 (full-length) prebound beads (lanes 1 to 9) or GST-only beads (lanes 10 to 12). (E) GST-Nse2 beads (lanes 1 to 6) or GST beads alone (lanes 7 to 10) were mixed with IVT Rad18 and Spr18 (lanes 1 to 3, 7, and 8) or Rad18-Spr18-gly529 (lanes 4 to 6). Note that in Fig. 5 to 7, the unbound and input fractions can be compared directly and represent 1/20 of the total reaction mix, whereas the bound fraction represents approximately 1/3 of the reaction.
Nse2 contains a RING finger motif that is characteristic of SUMO ligases in its C-terminal half. We have shown, in the accompanying paper, that Nse2 has SUMO ligase activity that is dependent on this RING finger motif (1a). In order to see if this motif was involved in binding to Spr18 (Smc5), we generated two fragments of Nse2, from aa 1 to 178 and from aa 114 to 250. Each of these fragments was C-terminally tagged with S protein and synthesized in vitro, incubated with IVT Spr18 (Smc5), and then bound to protein S beads. Figure 5B (bottom panels) shows that both S-tagged Nse2 fragments bound to the beads (lanes 3 and 6) but that Spr18 (Smc5) bound only to the N-terminal fragment (lane 3, top panel), not the C-terminal fragment (lane 6, top panel) or protein beads alone (lane 9). Similar results were obtained with bacterially expressed Nse2 fragments (not shown). Thus, the RING finger motif of Nse2 is not required for binding to Spr18 (Smc5).
To investigate which part of Spr18 (Smc5) is involved in the interaction with Nse2, we made several different constructs of Spr18 (Smc5). The first, Spr18 (Smc5) heads, contained the globular N- and C-terminal domains joined by a short linker. The other, Spr18 (Smc5) headless constructs, lacked the globular heads and contained different lengths of the central coiled coil together with the hinge domain. In GST pull-down assays with GST-Nse2 and IVT Spr18 (Smc5) heads, only a very small amount of the Spr18 heads bound to the beads (Fig. 5C, lane 3, top panel) compared to the control (lane 6). The Spr18 (Smc5) headless construct with about 250 aa of coiled coil bound strongly to the beads (Fig. 5D, lane 3, top panel) compared to the control (lane 12). Strong binding remained when we reduced the size of the coiled coil by a further 50 aa (Fig. 5D, lane 6) but was abolished when we deleted a further 100 aa from each arm (Fig. 5D, lane 9). We were not able to detect binding between Nse2 and Nse1, Nse3, or Rad62 under the conditions used for our experiments (not shown). We conclude that there is an interaction between Nse2 and Spr18 (Smc5) which is mediated by the N-terminal half of Nse2 and the coiled-coil region of Spr18 (Smc5). The regions of Spr18 (Smc5) between aa 226 and 324 of the N-terminal arm and aa 731 and 836 of the C-terminal arm are essential for this interaction.
The experiments depicted in Fig. 3B showed that the G529R mutation in the hinge region of Spr18 (Smc5) destroys the interaction between Rad18 (Smc6) and Spr18 (Smc5). Figure 5E shows that this mutation does not affect the interaction between Spr18 (Smc5) and Nse2. As described above, GST-Nse2 pulled down both Spr18 and Rad18 when the two Smc proteins were mixed together, as indicated by the doublet in Fig. 5E, lane 3. However when Rad18 (Smc6) and Spr18-G529R were incubated with GST-Nse2, Spr18-G529R was still pulled down, as indicated by the strong single band in lane 6, whereas the upper band corresponding to Rad18 (Smc6) was no longer bound to the beads. These data are consistent with binding of Spr18 (Smc5) and Rad18 (Smc6) via their hinges, whereas binding of Spr18 (Smc5) to Nse2 is mediated via the coiled coil region and does not involve the hinge.
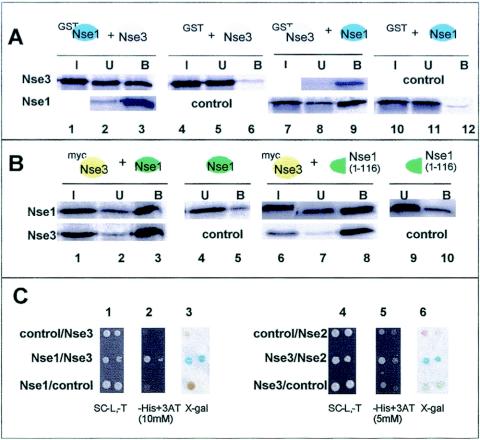
Interaction between Nse1 and Nse3.
In GST pull-down assays using GST-Nse1 and Rad18 (Smc6), Spr18 (Smc5), Nse2, Rad62, or mixtures of these proteins that were synthesized by IVT, we were unable to detect any interactions (data not shown). In contrast, we were able to detect an interaction between GST-Nse1 and Nse3 (Fig. 6A, lane 3) and, conversely, between GST-Nse3 and Nse1 (Fig. 6A, lane 9). In order to confirm our pull-down data, we used the yeast two-hybrid system. All Nse genes were fused to either Gal4(BD) or Gal4(AD) and cotransformed into the Y190 budding yeast strain. Figure 6C shows a strong Nse1-Nse3 interaction-dependent transcriptional activation of the HIS3 gene (panel 2). Induction of the lacZ gene resulted in a clear blue color when assayed with X-Gal (Fig. 6C, panel 3). Nse1 has a RING finger motif that is typical of E3 ubiquitin ligases close to its C terminus (8, 20). In further experiments, we mixed IVT Myc-tagged Nse3 with either the full length (232 aa) or the N-terminal half (aa 1 to 116) of Nse1 lacking the RING finger motif, followed by immunoprecipitation with anti-Myc antibodies. Although a small amount of Nse1 bound to the beads in the absence of Myc-Nse3 (Fig. 6B, lanes 5 and 10), there was much stronger binding in its presence (Fig. 6B, lanes 3 and 8, top panels). In contrast, the C-terminal half of Nse1 did not interact with Nse3 (data not shown). Thus, as with Nse2 and Spr18 (Smc5), the interaction of Nse1 with Nse3 is mediated by the N-terminal half of the protein and does not require the RING finger motif.
FIG. 6.
Interaction between Nse3 and Nse1 or Nse2. (A) Soluble protein extracts prepared from E. coli expressing either GST-Nse1 (lanes 1 to 3) or GST-Nse3 (lanes 7 to 9) were preincubated with glutathione beads, and in vitro-expressed Nse3 (lanes 1 to 3) or Nse1 (lanes 7 to 9), respectively, was added (I). In the control experiments (lanes 4 to 6 and 10 to 12), GST was used instead of GST-Nse1 and GST-Nse3. GST, GST-Nse1, and GST-Nse3 were detected by immunoblotting with anti-GST. (B) IVT Myc-Nse3 (lanes 1 to 3 and 6 to 8) was mixed with either IVT full-length Nse1 (lanes 1 to 3) or the N-terminal part of Nse1 (aa 1 to 116; lanes 6 to 8). Myc-Nse3 was immunoprecipitated with an anti-Myc monoclonal antibody and protein G beads. In the control experiments, Myc-Nse3 was not added (lanes 4, 5, 9, and 10). The proteins were visualized with a phosphorimager. (C) Yeast two-hybrid analysis of interactions between Nse1 and Nse3 (panels 1 to 3) and Nse3 and Nse2 (panels 4 to 6). In each set of three panels, the first panel is a positive control of cells growing in nonselective medium, the second panel shows growth in a selective medium, and the third panel shows β-galactosidase activity. Results for two concentrations of cells differing by a factor of 10 are shown. In each case, controls with appropriate empty vectors are provided above and below the test pairs.
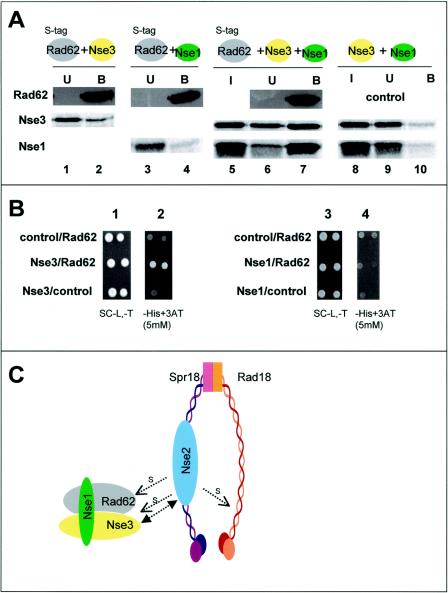
Interaction between Rad62 and Nse3.
In similar experiments to those described above, we examined the interaction of Rad62 with all of the other components of the complex, which were synthesized in vitro either together or separately. The only interaction we detected in this system was between S-tagged Rad62 and Nse3 (Fig. 7A, lane 2, middle panel). This interaction was rather weak compared to those described above. Only a small proportion of the Nse3 protein bound to the beads. We obtained further evidence for this interaction by yeast two-hybrid analysis (Fig. 7B, panel 2). Rad62 did not bind detectably to Nse1 either by pull-down assays (Fig. 7A, lane 4, bottom panel) or by yeast two-hybrid analysis (Fig. 7B, panel 4). However, when we mixed S-tagged Rad62 with both Nse3 and Nse1, all three proteins bound strongly to the beads (Fig. 7A, lane 7). This suggests that the interaction between Nse1 and Nse3 strengthens the binding of Nse3 to Rad62.
FIG. 7.
Interaction between Rad62 and Nse3. (A) A soluble protein extract prepared from E. coli strain Rosetta-gami B expressing S tag-Rad62 was preincubated with protein S beads and mixed with IVT Nse3 (lanes 1 and 2), Nse1 (lanes 3 and 4), or both Nse1 and Nse3 (lanes 5 to 7). In the control experiment, Nse3 and Nse1 were incubated with protein S beads in the absence of S tag-Rad62 (lanes 8 to 10). (B) Yeast two-hybrid analysis of interactions between Rad62 and Nse3 or Nse1. The details are the same as those for Fig. 6C. (C) Model for interactions between components. The two subcomplexes are indicated. The single arrows indicate components that are sumoylated by Nse2 in vitro (1a). The double-headed arrow indicates a tentative interaction between the subcomplexes, based on the data shown in Fig. 6C.
Interaction between Nse3 and Nse2.
The analysis described so far delineated two subcomplexes, made up of Rad18-Spr18-Nse2 and Nse1-Nse3-Rad62. Using pull-down assays, we were not able to identify any interactions between the components of the subcomplexes. However, by using yeast two-hybrid analysis, we detected a weak interaction between Nse3 and Nse2 (Fig. 6C, panels 5 and 6). Based on this observation, we tentatively propose that this may represent the link between the two subcomplexes.
DISCUSSION
In previous work, it was shown that rad18 (smc6) is an important DNA repair gene that is involved in several different repair pathways (7, 19, 23). Other genes, such as rad60 (22) and brc1 (26), have phenotypes which are very similar to those of rad18 (smc6) as well as being synthetically lethal with rad18 (smc6), suggesting the presence of a complex multiprotein network, whose mechanism of action remains to be elucidated. In this paper, we have begun to define the molecular architecture of the core Smc5-6 protein complex.
Our results show that the Smc5-6 complex is similar to the previously characterized cohesin and condensin complexes in that it has a SMC heterodimeric protein core associated with a series of non-SMC proteins. Although there is little sequence identity between the hinge regions of Smc6 and those of the other SMC proteins (Figs. 2E and F), as with the other complexes, Smc5 and -6 interact through their hinge regions. This suggests that SMC and SMC-like proteins may interact through their hinges in three different ways, firstly by using the structures determined for the Smc1-3 hinge (10), secondly via a presumably different structure in the Smc5-6 complex, and thirdly via a zinc hook, as found for the SMC-like protein Rad50 (16).
We generated four independent temperature-sensitive mutants of rad18 (smc6). Remarkably, they all carried a mutation at glycine 551 in the hinge region. Mutation of this conserved residue destabilized the interaction with the Smc5 hinge for both the S. pombe and human proteins, as did mutation of the conserved glycine 529 residue in Spr18 (Smc5). Temperature-sensitive mutations in S. cerevisiae SMC6 have also been isolated (24), but none of these mutations was in the hinge region. The cause of the temperature sensitivity of these S. cerevisiae mutants was not established. The phenotype of our temperature-sensitive mutants, namely, continued growth and proliferation for several hours despite a substantial decrease in the amount of Rad18 (Smc6) protein within 2 h at the restrictive temperature, suggests that cells can survive and remain viable for a significant period with low levels of Rad18 (Smc6) protein. This is consistent with previous observations of the phenotype of a dominant-negative mutation and the terminal phenotype of a rad18 (smc6) deletion mutant (7, 19) as well as with recent findings by Harvey et al. (11). These authors used an S. pombe strain, rad18-so, in which Rad18 (Smc6) could be conditionally depleted. When thiamine was added to this strain to deplete Rad18 (Smc6), the protein became undetectable after 12 to 18 h, but cells continued to grow at normal rates for 24 h.
We identified a total of six components in the Smc5-6 complex, namely, Rad18 (Smc6), Spr18 (Smc5), Nse1, Nse2, Nse3, and Rad62. In a recent paper, Hazbun et al. analyzed a series of anonymous protein complexes from S. cerevisiae by tagging essential genes and analyzing copurifying proteins by mass spectrometry and multidimensional protein identification technology (12). One complex was associated with ORF YDR288W and contained Rhc18, Smc5, Nse1, Mms21, and Qri2. This appears to be an analogous complex to the one that we have characterized from S. pombe, since these proteins are orthologs of Rad18 (Smc6), Spr18 (Smc5), Nse1, Nse2, and Rad62, respectively. As described above, Nse3 is related to and is probably an ortholog of YDR288W. These authors also suggested that Smc5 and -6 are associated with a second complex containing two different proteins, YML023C (64 kDa) and Kre29 (54 kDa). We have not detected this putative second complex in our experiments with S. pombe. No proteins corresponding to these molecular masses were evident in our affinity-purified Rad18 (Smc6) complex. However, in studies using gel filtration described in the accompanying paper (1a), we obtained evidence for a possible lower-molecular-weight complex containing Rad18 (Smc6) and Spr18 (Smc5) but lacking Nse1 and Nse2. The nature of this complex remains to be determined.
Nse3 shows sequence similarity to the MAGE family, many members of which are expressed in cancer tissues but not in normal cells and thus provide potential targets for tumor immunotherapy. There are 55 human MAGE genes or pseudogenes, and they have a conserved MAGE homology domain (4). No MAGE genes have been identified previously in yeasts. However, our analysis using iterative BLAST searches clearly demonstrated that both Nse3 and the S. cerevisiae ORF YDR288W are related to the MAGE family. There are also single related genes in Aspergillus nidulans, Neurospora crassa, Drosophila melanogaster, and Arabidopsis thaliana. Despite many publications about the MAGE family, little is known about their function. The implications of the relationship of Nse3 to the MAGE family are not clear at present. Many of the conserved residues in the MAGE family are not found in Nse3, so it is distantly related to the family. There is no obvious connection between what little is known at present of the functions of the MAGE family proteins and the role of Nse3 in the Smc5-6 complex. Our results raise the possibility that MAGE proteins are involved in DNA damage responses and/or the maintenance of chromatin structure.
We have initiated a study to understand the molecular architecture of the Smc5-6 complex. Our in vitro interaction studies have highlighted the following interactions between different components of the complex. (i) Rad18 (Smc6) and Spr18 (Smc5) interact via their hinge regions, and this interaction is dependent on conserved glycine residues in both Rad18 (Smc6) and Spr18 (Smc5). (ii) Nse2 interacts strongly with Spr18 (Smc5) via the N-terminal half of Nse2 and a central section of the coiled coil region of Spr18 (Smc5). (iii) Nse1 interacts with Nse3 via the N-terminal half of Nse1. (iv) Nse3 interacts with Rad62, and this interaction is strengthened substantially upon binding of Nse1. Furthermore, our preliminary data suggest that Rad62 also binds directly to Nse1 when Nse1 is bound to Nse3 (unpublished observations). Further interactions may not have been detected in our in vitro studies for several reasons, such as an interference of interactions by the epitope tags or a requirement for posttranslational modifications. Inside the cell, the Nse1-Nse3-Rad62 subcomplex must bind to the Rad18 (Smc6)-Spr18 (Smc5)-Nse2 subcomplex. Our yeast two-hybrid analysis suggests that the two subcomplexes may be bridged by an interaction between Nse2 and Nse3. This enabled us to propose a model for the complex, as indicated in Fig. 7C. Further work is required to understand the nature of the interactions between the two subcomplexes in more detail. Our work has so far not revealed any proteins that bridge the head domains of Rad18 (Smc6) and Spr18 (Smc5), as is the case for S. cerevisiae cohesin, in which Scc1p bridges the head domains of Smc1p and Smc3p (9). It may well be that the Smc5-6 complex is not, like cohesin, capable of forming a ring structure. Indeed, a model for the function of the complex that was proposed earlier, in which broken DNA ends are held together to enable the repair of double strand breaks to take place (7), would not be readily compatible with a ring structure. Different SMC complexes may have different overall structures, as discussed for the SMC-like Rad50 proteins (6, 16). These may depend on the nature of the hinge interaction, the flexibility of the coiled coils, the non-SMC proteins, and posttranslational modifications.
The sequences of Nse1 and Nse2 suggest that they are E3 ubiquitin and SUMO ligases, respectively. In a companion paper (1a), we show that Nse2 does indeed have SUMO ligase activity, which is capable of sumoylating Rad18 (Smc6), Nse3, and Rad62 in vitro (as indicated in Fig. 7C), and that within a Smc5-6 complex that was purified from cells, Rad18 (Smc6) was sumoylated. Furthermore, this sumoylating activity is involved in the response to DNA damage and the inhibition of DNA replication. Our future work will be directed toward uncovering further interactions within the complex and the effect of posttranslational modifications on these interactions and on the functions of the complex.
Acknowledgments
We are grateful to S. Smerdon (NIMR, London, United Kingdom) and D. Stead (COGEME-PSF1, Aberdeen, United Kingdom) for mass spectrometry analysis, to Elephtheria Papadeli and Emily Richardson for assistance with hinge interaction studies, to Jon Wing for cloning assistance, to M. Sehorn for suggesting the use of ATP to remove the heat shock protein, to J. Murray for the cDNA library, and to A. M. Carr and J. Murray for helpful comments on the manuscript.
This work was supported by an MRC programme grant and EU contract FIGH-CT-1999-00010 to A.R.L. and by BBSRC grant G/07462 to E.A.A. and F.Z.W.
REFERENCES
- 1.Al-Khodairy, F., and A. M. Carr. 1992. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Andrews, E. A., J. Palecek, J. Sergeant, E. Taylor, A. R. Lehmann, and F. Z. Watts. 2005. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25:194-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arumugam, P., S. Gruber, K. Tanaka, C. H. Haering, K. Mechtler, and K. Nasmyth. 2003. ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 13:1941-1953. [DOI] [PubMed] [Google Scholar]
- 3.Barbet, N. C., and A. M. Carr. 1993. Fission yeast wee1 protein kinase is not required for DNA damage-dependent mitotic arrest. Nature 364:824-827. [DOI] [PubMed] [Google Scholar]
- 4.Barker, P. A., and A. Salehi. 2002. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res. 67:705-712. [DOI] [PubMed] [Google Scholar]
- 5.Cobbe, N., and M. M. Heck. 2004. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol. Biol. Evol. 21:332-347. [DOI] [PubMed] [Google Scholar]
- 6.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8:1129-1135. [DOI] [PubMed] [Google Scholar]
- 7.Fousteri, M. I., and A. R. Lehmann. 2000. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujioka, Y., Y. Kimata, K. Nomaguchi, K. Watanabe, and K. Kohno. 2002. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J. Biol. Chem. 277:21585-21591. [DOI] [PubMed] [Google Scholar]
- 9.Gruber, S., C. H. Haering, and K. Nasmyth. 2003. Chromosomal cohesin forms a ring. Cell 112:765-777. [DOI] [PubMed] [Google Scholar]
- 10.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 11.Harvey, S. H., D. M. Sheedy, A. R. Cuddihy, and M. J. O'Connell. 2004. Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol. Cell. Biol. 24:662-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates III, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 13.Hirano, M., D. E. Anderson, H. P. Erickson, and T. Hirano. 2001. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 20:3238-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano, M., and T. Hirano. 2002. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 21:5733-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16:399-414. [DOI] [PubMed] [Google Scholar]
- 16.Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui, B. A. Owen, A. Karcher, B. Henderson, J. L. Bodmer, C. T. McMurray, J. P. Carney, J. H. Petrini, and J. A. Tainer. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418:562-566. [DOI] [PubMed] [Google Scholar]
- 17.Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101:789-800. [DOI] [PubMed] [Google Scholar]
- 18.Humphreys, G. O., G. A. Willshaw, H. R. Smith, and E. S. Anderson. 1976. Mutagenesis of plasmid DNA with hydroxylamine: isolation of mutants of multi-copy plasmids. Mol. Gen. Genet. 145:101-108. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann, A. R., M. Walicka, D. J. F. Griffiths, J. M. Murray, F. Z. Watts, S. McCready, and A. M. Carr. 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15:7067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald, W. H., Y. Pavlova, J. R. Yates III, and M. N. Boddy. 2003. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 278:45460-45467. [DOI] [PubMed] [Google Scholar]
- 21.Morikawa, H., T. Morishita, S. Kawane, H. Iwasaki, A. M. Carr, and H. Shinagawa. 2004. Rad62 protein functionally and physically associates with the Smc5/Smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell. Biol. 24:9401-9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morishita, T., Y. Tsutsui, H. Iwasaki, and H. Shinagawa. 2002. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22:3537-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onoda, F., M. Takeda, M. Seki, D. Maeda, J. Tajima, A. Ui, H. Yagi, and T. Enomoto. 2004. SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in Saccharomyces cerevisiae. DNA Repair (Amsterdam) 3:429-439. [DOI] [PubMed] [Google Scholar]
- 25.Taylor, E. M., J. S. Moghraby, J. H. Lees, B. Smit, P. B. Moens, and A. R. Lehmann. 2001. Characterisation of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell 12:1583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verkade, H. M., S. J. Bugg, H. D. Lindsay, A. M. Carr, and M. J. O'Connell. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10:2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weitzer, S., C. Lehane, and F. Uhlmann. 2003. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 13:1930-1940. [DOI] [PubMed] [Google Scholar]