Abstract
The Eastern African Afromontane forest is getting increased attention in conservation studies because of its high endemicity levels and shrinking geographic distribution. Phylogeographic studies have found evidence of high levels of genetic variation structured across the Great Rift System. Here, we use the epiphytic plant species Canarina eminii to explore causal explanations for this pattern. Phylogeographic analyses were undertaken using plastid regions and AFLP fragments. Population genetic analyses, Statistical Parsimony, and Bayesian methods were used to infer genetic diversity, genealogical relationships, structure, gene flow barriers, and the spatiotemporal evolution of populations. A strong phylogeographic structure was found, with two reciprocally monophyletic lineages on each side of the Great Rift System, high genetic exclusivity, and restricted gene flow among mountain ranges. We explain this pattern by topographic and ecological changes driven by geological rifting in Eastern Africa. Subsequent genetic structure is attributed to Pleistocene climatic changes, in which sky-islands acted as long-term refuges and cradles of genetic diversity. Our study highlights the importance of climate change and geographic barriers associated with the African Rift System in shaping population genetic patterns, as well as the need to preserve the high levels of exclusive and critically endangered biodiversity harboured by current patches of the Afromontane forest.
The Afromontane Floristic Region1,2 refers to a plant assemblage where a series of isolated highland forested areas are separated by lowland vegetation, and form an “archipelago-like” centre of endemism in the mountains of East and West Africa. The eastern part of this floristic region has been singled out as a biodiversity hotspot and is known as the Eastern Afromontane biodiversity hotspot3 (EABH) (Fig. 1a). The Eastern African “sky islands” (including both high plateaus and isolated mountain peaks), are sometimes referred to as the “Galapagos of Africa”4, and contain such flagship species as the mountain gorilla, the gelada, the Ethiopian wolf, or the coffee tree. These mountain ranges exhibit extraordinary levels of species endemism, for example, out of the > 10.000 plant species found in this region (7600 vascular plants), about one third are endemic5. This pattern of endemism has been attributed to long-term climatic stability, favouring lineage diversification and species accumulation over time6. The EABH is also one of the most threatened ecosystems in Africa5. Ethiopian montane forests, which two hundred years ago7,8 covered up to 35% of the Afromontane areas, are estimated to have had their range dramatically reduced by human activity in the last century9,10.
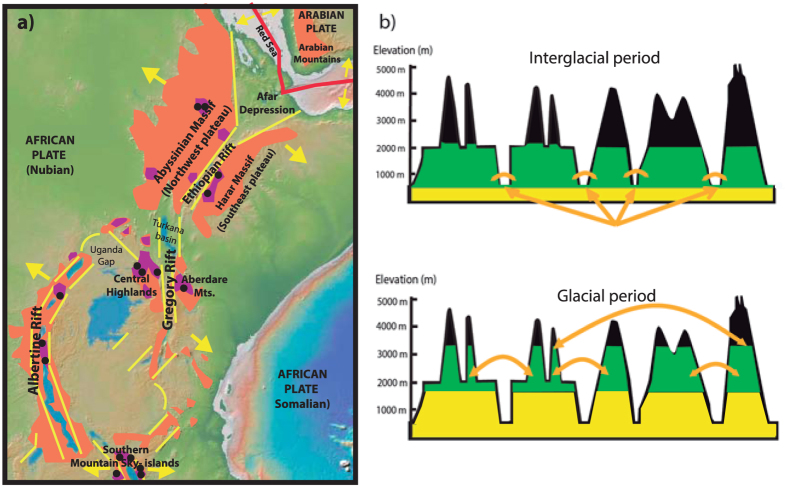
Figure 1. Geographic map of the Afromontane forests and main phylogeographic hypotheses.
(a) Geographic map of Eastern Africa showing the main geographic features named in the text. The Eastern Afromontane biodiversity hotspot is shaded in orange. The distribution of Canarina eminii is shaded in purple. The Rift System is shown in yellow; lines represent major tectonic faults and arrows indicate the direction of relative divergent Rift movement. The red line shows the tectonic plate boundaries. The map was generated using the software GeoMapApp (v. 2.3) (http://www.geomapapp.org/)85. Black dots represent the sampling for C. eminii. (b) Sky-islands housing Afromontane forests (in green) separated by savannahs and open forests (in yellow), showing the two main phylogeographic hypotheses postulated to explain patterns of genetic variation in Afromontane organisms. Upper) the “mountain forest bridge” hypothesis (arrows indicate forest reconnections); Lower) the “long-distance dispersal” hypothesis (arrows indicate migration events).
Geographically, the Eastern Afromontane region is divided longitudinally by the Great Rift System, a 4830 km north-south fissure whose terrestrial section extends from Djibouti to Mozambique. This orogenic fault marks the line along which the African Plate is splitting into two tectonic plates, the Somalian Plate and the Nubian Plate (Fig. 1a). The formation of the Rift commenced 31 Ma, as a result of the collision of the African and Arabian Plates with Eurasia; as extension continues, with ongoing volcanic activity11, complete breakup of the Somalian Plate is predicted to occur within 10 million years, with a new ocean basin originating where shallow lakes can now be found12. The northern part of the Rift runs south from the Afar Triangle and is surrounded by the Ethiopian Highland Plateaus (Fig. 1a): the Abyssinian massif (northwest Plateau) and the Harar massif (southeast Plateau). In the south tropical East Africa, the Rift System is divided into two arcs: the Eastern Gregory Rift and the Western Albertine Rift, which join at the northern end of Lake Malawi, where the Southern Mountain Sky-Islands (SMSI) lie (Fig. 1a). Due to its elevation and extent, this latter region is sometimes known as the “Roof of Africa”. Because of its geographic position, the Rift System is considered one of the most isolated mountain ranges in the world, compared to other tropical floras13.
The Afromontane vegetation is divided into three altitudinal belts, each with unique floristic composition14,15,16: the lowermost Afromontane forest (1300–3000 m), the ericaceous belt (3000–4100 m), and the uppermost Afroalpine zone ( > 3550 m). These vegetation belts are not continuous in their distribution but appear divided into forest patches isolated from one another by environmental barriers (Fig. 1b), including extensive savannah areas, semi-deserts, lowland forests (in the past), or (at present) an agricultural matrix of cultivated lands and forestry plantations1,17,18,19,20. When compared to other mountain systems, the Afromontane vegetation is considered to be rather uniform in composition and is formed by a large number of broad-range species13; though given the fragmented nature of the Afromontane region, with numerous forest patches, some of these widespread species are likely to include cryptic diversity21.
Interest on geographic patterns of genetic variance in Eastern African species has increased in recent years. Many of these phylogeographic studies (focusing on a diverse array of organisms, Table 1) have reported high levels of genetic variation structured with respect to the Great Rift Valley8,19,22, suggesting it has acted as a dispersal barrier for an extended period of time.
Table 1. Phylogeographic studies of organisms showing genetic variation structured around the Rift System.
| Organism | Phylogeographic disjunctions in the Ethiopian Rift | Phylogeographic disjunctions in the Volcanic arcs | Method (Reference) |
|---|---|---|---|
| Angiosperms(habitat) | |||
| Coffea arabica (Afromontane) | Two distinct groups across the Ethiopian Rift | — | ISSR; microsatellites50,86 |
| Lobelia giberroa (Afromontane) | Two distinct groups across the Ethiopian Rift: 1. Simien- Choke; 2. Chilallo-Bale-Gara Muleta | Two distinct groups across the Gregory Rift | AFLP8 |
| Hagenia abyssinica (Afromontane) | Structure both sides of the Rift with rare long-distance dispersal events crossing the Rift | — | microsatellites51 |
| Cordia africana (Afromontane) | Three different groups across the Ethiopian Rift: 1. Northwest plateau; 2. South-west Ethiopia; 3. Southeast plateau | — | AFLP and microsatellites87 |
| Juniperus procera (Afromontane) | Two relatively different groups across the Ethiopian Rift: 1.Goba-Yabelo; 2. Chilimo-Suba-Ziquala-Washa | — | AFLP88 |
| Prunus africana (Afromontane) | — | Two distinct groups across the Uganda Gap: 1. Albertine Rift and western Gregory Rift; 2. Eastern Gregory Rift | Plastid haplotypes, SSR52,89 |
| Warburgia ugandensis (Afro- montane transitional forest) | — | Two distinct groups across the Uganda Gap: 1. Albertine Rift and western Gregory Rift; 2. Eastern Gregory Rift | AFLP90 |
| Erica trimera (Ericaceous) | No difference for AFLP, but different haplotypes | A complex pattern in the Gregory Rift | AFLP and plastid haplotypes42 |
| Arabis alpina (Afroalpine) | Two distinct groups across the Ethiopian Rift: 1. Simien, Gara M.; 2. Choke, Elgon, Meru, Kilimanjaro | A complex pattern in the Gregory Rift | Plastid haplotypes19 |
| Cardus schimperi (Afroalpine) | Two distinct groups across the Ethiopian Rift: 1.Simien; 2. Elgon-Aberdare-Bale | Mt. Kenya population is a different subspecies; while populations from Mt. Elgon and Aberdare are closely related | AFLP26 |
| Trifolium cryptopodium (Afroalpine) | Two distinct groups across the Ethiopian Rift: 1. Simien Choke; 2. Bale-Aberdare–Elgon-Kilimanjaro | Populations related across the Rift Valley | AFLP26 |
| Deschampsia cespitosa (Afroalpine) | — | Two distinct groups across the Rift Valley: 1. Rwenzori; 2. Kilimanjaro-Bale | AFLP27 |
| Senegalia senegal (savannah, semi-desert) | — | Two distinct groups across the Gregory Rift with mixed population (Marigat): 1. Western side; 2. Eastern side. | microsatellite91 |
| Senegalia mellifera (savannah) | — | Two distinct groups across the Gregory Rift: 1. Western side; 2. Eastern side; 3. Central Gregory Rift | microsatellite49 |
| Vertebrates | |||
| Ostrich Struthio camelus | Two subspecies both sides of the Rift (molybdophanes/camelus) | Two subspecies both sides of the Rift (Molybdophanes/camelus) | mtDNA46 |
| Olive sunbird Nectarinia | — | Two distinct species across the Rift Valleys. 1. Western Rift Valley Nectarinia obscura. 2. Eastern Rift Valley Nectarinia olivacea | mtDNA44 |
| Springhare Pedetes capensis | — | Two distinct groups across the Rift Valleys: eastern populations/southern populations | mtDNA92 |
| Wildebeest Connochaetes taurinus | — | Two subspecies both sides of the Gregory Rift. 1. Western Gregory Rift: sbsp. mearnsi (Loliondo, Masai-Mara) 2. Eastern Gregory Rift: sbsp. albojubatus (Nairobi and Amboseli) | mtDNA93 |
| African wild dog Lycaon pictus | — | Two distinct groups across the Gregory Rift. 1. Eastern clade. 2. Southern clade | mtDNA and microsatellites94 |
| Sable antelope Hippotragus niger | — | Two distinct groups across the Gregory Rift: 1. West Tanzania and Kenya; 2. East Tanzania) | mitochondrial DNA and cyt. b95 |
| Ethiopian wolf Canis simensis | Three distinct groups across the Ethiopian Rift: Northwest plateau (1. Wollo/Shoa; 2. Simien/Mt. Guna) and southeast plateau (3. Arsi/Bale) | — | mtDNA22 |
| Lion Panthera leo | — | Two distinct groups across the Gregory Rift: 1. eastern (Tsavo-Transvaal); 2. western (Aberdare) | cytochrome b and NADH desh. subunit 596 |
| Baboon Theropithecus gelada | Two distinct groups across the Ethiopian Rift (northwest and southeast plateau) | — | RFLPs97 |
| Grass mouse Lemniscomys striatus | — | Two distinct groups across the Rift Valleys: Gregory Rift/Albertine Rift | cytochrome b98 |
| Rodent Mastomys natalensis | — | Two distinct groups across the Rift Valleys: eastern populations/southern populations | cytochrome b99 |
| African clawed frogs (Xenopus clivii and X. largeni) | Two distinct groups across the Ethiopian Rift (northwest and southeast plateau) | — | mtDNA, autosomal loci100 |
| Ethiopian anurans (Tomopterna, Amietia, Leptopelis, Ptychadena) | Two distinct groups across the Ethiopian Rift | — | Several mitochondrial and nuclear genes (101) |
| Insects | |||
| Mosquito Anopheles gambiae | — | Two distinct groups across the Rift Valleys: eastern populations (Kimili, Asembo bay, Kisian, Awendo)/western populations (Malindi, Jego) | microsatellites102,103,104,105 |
| Tsetse fly Glossina pallidipes | — | Two distinct groups across the Rift Valley: East Rift Valley (Dakabuko, Alangoshira, Shimba Hills, Kibwezi); West Rift Valley (Nguruman, Shompole, Marech) | Allozimes, microsatellites, mitochondrial loci106 |
| Mosquito Anopheles funestus | — | Two distinct groups across the Rift Valley: western Kenya pop. (Mbita, Udhoro)/Coastal populations (Majajani, Magaoni) | microsatellites107 |
| Termite Scledorhinotermes lamanianus | — | Two distinct groups across the Rift Valley: eastern populations/western populations | AFLPs57 |
The complex phylogeographic structure found in African highland species has also been explained by two non-mutually-exclusive dispersal-migration scenarios (Fig. 1b.): I) the “mountain-forest bridge” hypothesis postulates that current patterns might be explained by short-range or stepping-stone dispersal (SSD) between adjacent mountain ranges on each side of the Rift Valley, which would have been promoted by a more extensive forest coverage during Plio-Pleistocene warm and humid interglacial periods8,17,23,24,25; II) the “long-distance dispersal” hypothesis postulates that long-distance dispersal (LDD) events among isolated mountain populations during Pleistocene glacial periods are responsible for the observed phylogeographic patterns18,19,26. These two hypotheses are not mutually exclusive. For example, proponents of the SSD forest bridge hypothesis17 suggested LDD migration among some isolated mountain ranges18. A requisite to discriminate between these two scenarios is a temporal framework. Most Eastern African phylogeographic studies in plants do not include time estimates for colonization events and, therefore, do not allow discriminating between the mountain-forest bridge and long-distance dispersal hypotheses. This is further aggravated by the difficulties intrinsic to collecting material of widespread Afromontane species, due to the complex geographic landscape and the recent political instability27 (see below).
Here, we examine support for these two hypotheses, and the role of the Rift Valley as a long-term dispersal barrier, by reconstructing patterns of genetic variation in Canarina eminii Asch. & Schweinf, a strictly Afromontane species with a geographic distribution extending from the Ethiopian massifs to the SMSI in the northern end of Lake Malawi28, along the Rift Valley. This species grows mostly as a twig epiphyte on trees, mainly of genera Hagenia J.F.Gmel., Conopharyngia G. Don, and Afrocarpus (J. Buchholz & E.G. Gray) C.N. Page28, all characteristic elements of the Afromontane forest belt28. Little is known about the reproductive biology of C. eminii, except that the species is pollinated by sunbirds (unpublished field observations) and exhibits floral traits that have been associated with this type of ornitophilous pollination29 (e.g., large orange-red coloured flowers producing abundant diluted nectar). Fruits are fleshy, with the seeds enclosed in a sticky sweet jelly, suggesting endozoochorous dispersal (e.g., by monkeys30). Mairal et al.31 recently reconstructed the biogeographic history of Canarina L., a small genus of three species within tribe Platycodoneae (family Campanulaceae). This genus exhibits a wide disjunct distribution across North Africa and probably originated from a Central Asian ancestor, which arrived to Eastern Africa in the Mid Miocene (13.7 Ma) through the Arabian Plate31. The lineage leading to the Eastern African species C. abyssinica diverged first, followed by the split between C. eminii and the endemic Canarian species C. canariensis31 in the Late Miocene (c. 7 Ma). Divergence within C. eminii was traced back to 1.76 Ma31 (southern range of the Great Rift Valley and followed by subsequent colonisation of the northern range). However, the intraspecific sampling in the study by Mairal et al.31 was limited, which prevented analysis of patterns of genetic variation or examination of the processes population divergence in C. eminii.
In this study, we reconstruct the phylogeographic history of C. eminii among and within populations. Our aims were to: i) describe the geographic distribution of genetic variation within this species; ii) examine the role of the Rift Valley as a long-term dispersal barrier; iii) understand how Pleistocene climatic fluctuations might have affected population ranges, especially in relation to the “mountain-forest bridge” hypothesis and the role of isolated “sky islands”; and iv) provide insights into the phylogeography of an Afromontane epiphytic species. Since the epiphytic growth of this species is tied to dominant tree species of the Afromontane forests, it makes C. eminii a good case study for tracing the history of fragmentation and expansion of forest coverage in this region.
Results
Haplotype analyses
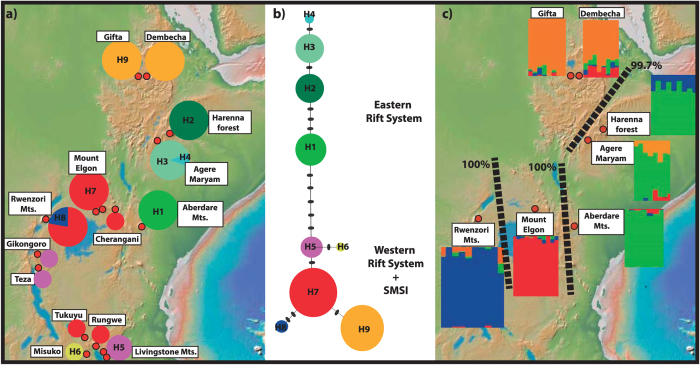
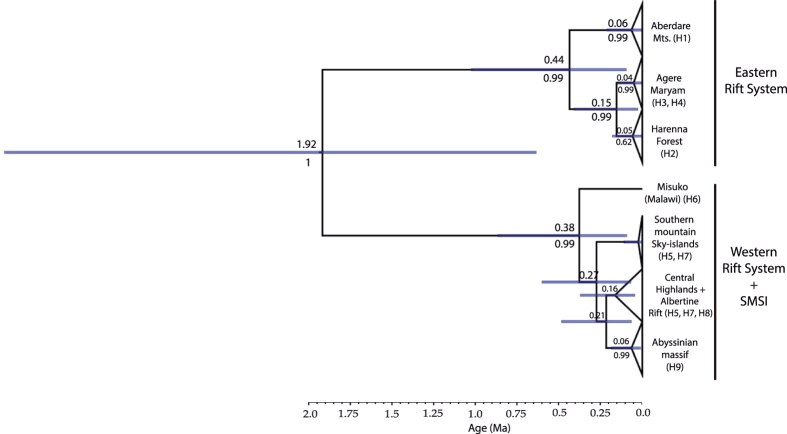
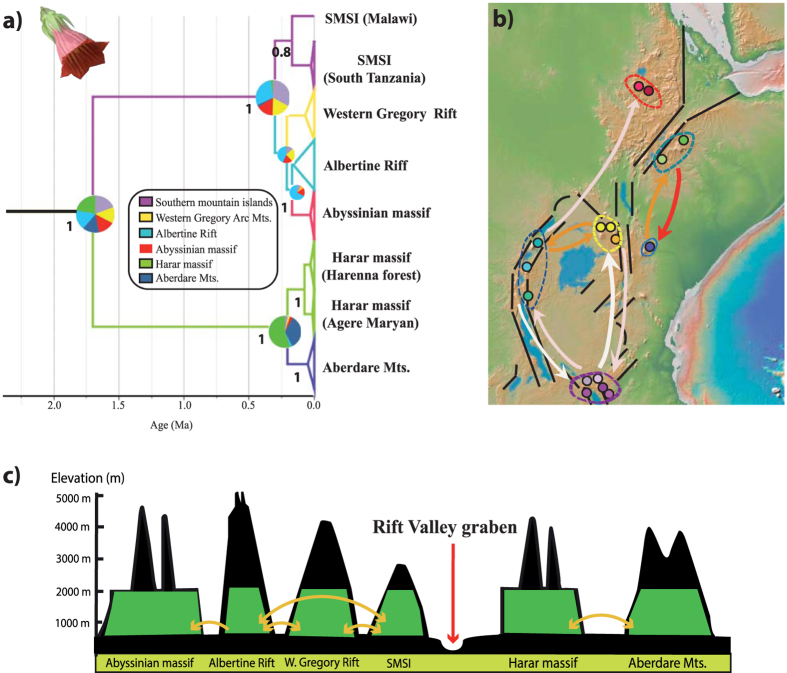
In order to examine patterns of genetic variation within C. eminii, we sequenced three highly variable plastid (pDNA) regions covering the entire distributional range of this species (see Materials and Methods). We generated 237 new sequences from 79 individuals: rpl32-trnLUAG (79 sequences), trnSGCU–trnGUCC (79 sequences), and petB1365–petD738 (79 sequences). The number of nucleotide sites ranged from 648 in trnSGCU–trnGUCC, 834 in pet B1365–petD738, to 963 in rpl32-trnLUAG. The concatenated matrix consisted of 79 sequences of C. eminii with 2445 nucleotide sites. The haplotype network recovered by TCS identified nine haplotypes (H1 to H9; Fig. 2a), divided into two main groups separated by five substitutions (Fig. 2b): one group east of the Rift Valley (Harenna Forest, Agere Maryam, and Aberdare Mts.), and a second group including populations of the Abyssinian massif, west of the Gregory Rifts, the Albertine Rift, and the SMSI. All haplotypes, save H5 and H7, were restricted to one population. Lineage divergence time estimation in the Bayesian software BEAST (Fig. 3, Supplementary Fig. S2.1) supported a similar geographic structure. The first population divergence event (1.92 Ma, 95% high posterior density (HPD) credibility interval (CI): 0.63–3.82 Ma) separated the populations situated at each side of the Rift Valley, with subsequent divergence events (starting 0.4 Ma) dividing populations located within each side of the Rift System (Fig. 3). We performed a phylogeographic analysis using Bayesian ancestral state reconstruction methods in BEAST (Fig. 4a,b); these were not conclusive, supporting a geographic origin of C. eminii in the Harar Massif or the SMSI, on each side of the main Rift System, with nearly equal probability. Nine migration routes were inferred by Bayesian Stochastic Search Variable Selection (BSSVS, see M&M) (Table S2.5); these seem to be arranged into parallel routes, following migration from north to south and vice versa on each side of the Rift System, and connecting also the intermediate mountain ranges (Fig. 4b).
Figure 2. Plastid and nuclear datasets analysed for Canarina eminii.
(a) Haplotype distribution. Red dots represent the geographic location of populations and pie charts show the frequency of occurrence of each haplotype. (b) Statistical Parsimony network inferred using the DNA plastid sequences by TCS. Black dashes on long connecting lines indicate nucleotide changes. Circle size is proportional to the frequency of haplotypes. Each haplotype is shown in a different colour, where codes (H1 to H9) correspond to the haplotypes shown in Fig. 2a. (c) Phylogroups using AFLP markers. Histograms showing the Bayesian clustering of individuals within populations (STRUCTURE); colours represent the proportion of individual membership to each inferred Bayesian group. Dashed lines indicate barriers to gene flow and their percentage, as inferred by BARRIER. The map was generated using the software GeoMapApp (v. 2.3) (http://www.geomapapp.org/)85.
Figure 3. Maximum clade credibility (MCC) tree obtained from the BEAST analysis of pDNA haplotypes of Canarina eminii.
Blue bars show 95% HPD credibility intervals. Numbers above branches show mean ages and numbers below branches indicate Bayesian posterior clade support values.
Figure 4. Phylogeographic analysis and reconstruction of the colonization of Canarina eminii.
(a) BEAST MCC tree showing the Bayesian ancestral range reconstruction analysis63. Coloured branches (see legend) represent the ancestral range with the highest posterior probability for each population; node pie charts show marginal probabilities for alternative ancestral ranges. Numbers below branches represent Bayesian posterior probabilities. (b) Map representing migration events that receive a BF support > 3, as recovered by BSSVS; colour tint is proportional to the support (dark red > orange > pale pink > white). The map was generated using the software GeoMapApp (v. 2.3) (http://www.geomapapp.org/)85 (c) A hypothetical reconstruction of the colonization of the Afromontane forest by Canarina eminii during interglacial periods based on our data and literature of other Afromontane groups. Orange arrows show possible short to medium distance dispersal events between isolated mountain ranges or across forest galleries. Expansion of the Afromontane forest (in green) during interglacial periods may have facilitated migration across Stepping Stone dispersal or forest bridges.
AFLP polymorphism, genetic diversity and structure
To complement the plastid signal (above), we carried out an analysis of fragment length polymorphism (AFLP) in the nuclear compartment. The final data set after scoring comprised 773 loci. The Bayesian software STRUCTURE assigned individuals to four geographic clusters based on patterns of genetic variance (K = 4, Fig. 2c; Fig. S2.2). These clusters were broadly coincident with those recovered by the haplotype network: i) a cluster of populations on the eastern side of the main Rift System (Harenna Forest, Agere Maryam and Aberdare Mts.), ii) the Abyssinian Plateau, iii) the Central Highlands, and iv) the Albertine Rift (Rwenzori Mts.). Neighbour-joining and neighbour-net diagrams (Fig. S2.3a,b) identified six well-supported groups coincident with main mountain ranges within the Rift, except for a group (Agere Maryam) that had a low bootstrap value (72.6%). The two populations of the Abyssinian Plateau were also genetically very close. Principal Component Analysis (PCO) of genetic variance also differentiated six groups: those of the eastern Rift occupied the centre of the plot, with the remaining located on the periphery (Fig. S2.4). Genetic differentiation among populations (Fst values) was higher among the western populations (Debre Markos, Mt. Elgon and Rwenzori, 0.28–0.38) than among those in the eastern side (Harenna Forest, Agere Maryam and Aberdare Mts., 0.17–0.21; Table S2.6), suggesting greater isolation among the populations west of the Rift than between those in the east. East of the Rift Valley System, a rarity index (frequency-down-weighted marker value, DW) was highest for the Harar Massif population, which also presented the highest number of polymorphic and private fingerprints (Table 2). Populations in the Central Highlands (Mt. Elgon and Cherangani Hills) had the highest DW values and a large number of polymorphic fragments, plus two fixed fragments. Populations west of the Rift Valley showed generally lower values of genetic diversity and no fixed fragments. No linear relationship was found between pairwise FST and geographic distance (Mantel analysis, r = 0.1884, p = 0.5012; Fig. S2.5). However, the correlation increased after excluding Mt. Elgon’s population from the analysis (r = 0.6198, p = 0.0560; see black stars in Supplementary Fig. S2.5). This population showed the largest genetic distance in pairwise-comparisons in relation to all other populations (Table 2.6, see Supplementary Fig. S2.5). In contrast, the population of the Abyssinian massif, which is separated by 500–1300 km from the southernmost populations (Mt. Elgon, Rwenzori), showed much lower pairwise FST values (Table S2.6). All intervals analysed with SPAGeDi (see Supplementary Fig. S2.6) gave a significant negative result, suggesting that populations are not more different with increasing distance. In agreement with this, BARRIER detected three major boundaries separating the four STRUCTURE clusters: i) Rwenzori Mountains from Mt. Elgon (100%), ii) Mt. Elgon from all areas in the east (100%), and iii) Abyssinian massif from the remaining areas (99.7%) (Fig. 2c). Hierarchical AMOVA showed the greatest genetic variance among the same four groups identified by STRUCTURE and BARRIER (Table S2.4). The relatively low among-group genetic variance (Table S2.4) might be explained by the limited number of populations sampled at each side of the Rift, and therefore should be taken with caution. Yet, the geographic structure recovered by AMOVA is fully congruent with the results from the Bayesian clustering (STRUCTURE and BARRIER) and with the Fst and DW estimates, lending support to this analysis.
Table 2. Descriptors of within-population genetic diversity in the cpDNA haplotypes and AFLP markers for each population studied of Canarina eminii.
| Haplotypes |
AFLPs |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | Mountain range | N° samples | Haplotypes | H (n) | H (d) | π | N° samples | N° of polymorphic fragments (% in brackets) | Hj (se) | N° of private fragments (% in brackets) | N° of fixed private fragments | DW index |
| West of the Rift | ||||||||||||
| Gifta, Debre Markos (Ethiopia) | Abyssinian massif (Northwest Plateau) | 8 | H9 | 1 | 0 | 0 | 8 | 477 (61.7) | 0.244 (0.0071) | 8 (1.68) | 0 | 93,187 |
| Dembecha, Debre Markos (Ethiopia) | Abyssinian massif (Northwest Plateau) | 7 | H9 | 1 | 0 | 0 | 8 | 443 (57.3) | 0.208 (0.0069) | 6 (1.35) | 0 | 99,213 |
| Rwenzori Mts. (Uganda) | Albertine Rift | 12 | H7, H8 | 2 | 0.303 | 0,00026 | 10 | 417 (53.9) | 0.193 (0.007) | 6 (1.439) | 0 | 111,177 |
| Gikongoro-Teza (Rwanda-Burundi) | Albertine Rift | 2 | H5 | 1 | 0 | 0 | – | — | — | — | — | — |
| Central highlands | ||||||||||||
| Mt. Elgon 1 & 2-Cherangani Hills (Kenya) | Central sky-islands | 19 | H7 | 1 | 0 | 0 | 11 | 477 (61.7) | 0.183 (0.0063) | 5 (1.048) | 2 | 140,694 |
| East of the Rift | ||||||||||||
| Agere Maryam, Yirga (Ethiopia) | Harar massif (Southeast Plateau) | 9 | H3, H4 | 2 | 0.222 | 0.0001 | 8 | 495 (64.0) | 0.241 (0.0067) | 13 (2.626) | 1 | 108,327 |
| Harenna Forest, (Ethiopia) | Harar massif (Southeast Plateau) | 8 | H2 | 1 | 0 | 0 | 9 | 491 (63.5) | 0.224 (0.0067) | 10 (2.037) | 2 | 128,342 |
| Aberdare Mts. (Kenya) | Aberdare Mts. (sky-island) | 9 | H1 | 1 | 0 | 0 | 7 | 435 (56.3) | 0.208 (0.0067) | 8 (1.839) | 2 | 91,057 |
| Southern Mountain Sky-Islands | ||||||||||||
| Misuku-Tukuyu-Rungwe-Livingstone Mts. (Tanzania-Malawi) | Southern Mountain Sky-islands | 5 | H5, H6, H7 | 3 | 0,4 | 0,00032 | — | — | — | — | — | — |
Abbreviations: H(n): number of haplotypes; H(d): haplotype diversity; π: nucleotide diversity; Hj (se): Nei’s gene diversity (standard error); DW: frequency-down-weighted value.
Discussion
The BEAST “nested dating” approach used here provided divergence time estimates similar to those in Mairal et al.31 for the origin of Canarina (8.98 My.) and the stem-age of C. eminii (7.14 My.) (Fig. S2.1). The first divergence event within C. eminii (ca. 1.92 Ma, 95% HPD 0.63–3.82 Ma, Fig. 3) postdates the age of establishment of the Afromontane forest16 (Early Pleistocene, ca. 2.4 Ma32), and is roughly coincidental with a period of aridification in East Africa, peaking at 1.7 Ma33,34. Our results also support that divergence in C. eminii followed the opening of the Rift Valley in the Early Pliocene, which divided the Rift System into two landstrips35.
A strong phylogeographic structure with the Great Rift Valley at its central axis was recovered for C. eminii populations. This agrees well with phylogeographic studies in other Afromontane groups (i.e, angiosperms, insects, and vertebrates, Table 1), all supporting the Rift System as an effective barrier to gene flow among populations. The depth of the phylogeographic split among STRUCTURE groups within C. eminii (Fig. 2c), the distribution of haplotypes (Fig. 2a,b), and the pattern of reciprocal monophyly and deep divergence times (1.92 Ma, HPD: 0.63–3.82 Ma) inferred with BEAST (Fig. 3), suggest a history of long-term isolation among populations east and west of the Rift Valley. In contrast, the large geographic distances and comparatively lower genetic distances between populations located on either side of the Rift Valley, suggest that geographic distance played a minor role in structuring patterns of genetic variation within C. eminii, confirmed by the lack of correlation the Mantel test establishes.
Patterns of colonization and biotic assemblage in the Eastern African Mountains have been argued to follow a northern (an “African pan-temperate element”28,36,37) and a southern (an “Afromontane track”38,39,40) component. Migrants coming from the north would have needed to overcome the widest point of the Rift Valley, the Afar Triangle (Fig. 1a), which acts as a major division before reaching the Ethiopian Plateaus, while southern migrants probably needed to split into two routes when reaching the two parallel volcanic arcs of the Albertine and the Gregory Rift39 (Fig. 1a). Thus, dispersal into the Eastern African Mountains from the north or from the south would have been forced by topography to follow two parallel routes, one on each side of the Rift Valley19,22. This scenario is supported by the Discrete Phylogeographic Approach (DPA) analysis of C. eminii, showing two colonization routes that split along the topography of the Rift System (Fig. 4b). Though this method has been recently criticized as being too decisive41, other studies in Afromontane taxa have found similar phylogeographic connections through the east26,27,42 and west8,43,44 (Table 1).
Which mechanisms are behind this pattern of geographic isolation between the two sides of the Rift Valley for C. eminii and other Afromontane taxa ? As mentioned above, the origin of C. eminii considerably postdates the formation of the Rift, so strict vicariance is not feasible (nor for any of the groups in Table 1). However, the major faults and flooding of graben associated to the Rift activity, may have acted as a corridor filtering migration, spatially restricting dispersal to two parallel paths on either side of the Rift Valley. Indeed, the volcanic desert of Afar in Ethiopia (the hottest place on Earth), and the drier floor and deep lakes at the bottom of Rift Valley, are ecologically very different from the surrounding higher elevation areas45,46, and constitute ecological barriers to dispersal (i.e., hostile environments that are beyond the tolerance limits of montane organisms). It would be interesting to test this hypothesis using genetic and ecological evidence in other lineages exhibiting a similar pattern of genetic isolation along the Rift Valley (Table 1).
Eastern Africa has experienced several climate fluctuations over the last 800,000 years, resulting in mountain forests extending and retracting successively, with further isolation and bottlenecking events for some species47,48. The Afromontane forest probably did not descend far enough to reach the bottom of the Rift Valley graben25 during the humid-climate expansion periods, and a widespread montane forest only seems likely on the Ethiopian plateaus, on either side of the Rift Valley25. However, some species might have had opportunities for gradual SSD migration through grasslands and open forest corridors between mountains25. This would have resulted in sister phylogroups occupying separate mountain ranges on either side of the Rift Valley, which could have been followed by in situ diversification during the arid glacial periods25,49. Patterns of genetic variation in C. eminii agree with this hypothesis, suggesting long-term isolation of populations in mountain enclaves on each side of the African Rift. High genetic differentiation and exclusiveness (Fig. 2a–c, Table S2.4, see Supplementary Fig. S2.3 and S2.4) was observed among populations on each sky-island, in agreement with other Afromontane phylogeographic studies8,50,51,52 and those in the Ericaceous42 and Afroalpine26,27,53 vegetation belts. In contrast to the glaciation-induced latitudinal range shifts in European taxa, which apparently resulted in a loss of genetic diversity (i.e., extinction and bottlenecks54), repeated events of fragmentation and reconnection during Pleistocene glacial/interglacial cycles may have contributed to the accumulation and maintenance of genetic diversity in the Eastern African mountains, and could explain their current status as centres of endemicity52,53. This agrees well with the hypothesis of a more moderate effect of glacial cycles at equatorial latitudes, in contrast with regions at higher latitudes54.
Another explanation for the east/west connectivity pattern and the observed low genetic variance between populations on each side of the Rift in C. eminii and other Afromontane endemics, is long-distance dispersal of seed and pollen through vectors such as wind, insects, or birds; this in turn could have been reinforced by wider forest coverage in the past, i.e., greater area availability for seedling establishment25. Long-range seed dispersal is likely the best explanation for connections among populations of C. eminii across the Turkana Basin and the Uganda Gap, which probably constitute ecological barriers (climatically unfavourable land) for gradual SSD migration through forest connections. Results from the plastid DNA suggest seed dispersal on each side of the Rift Valley, with more recent connectivity among the eastern side populations (Figs 2a and 3). On the other hand, the strong interpopulation structure found in the nuclear AFLP data suggests limited pollen dispersal (Fig. 2c). This could be explained by the strong territoriality and reduced mobility of Nectariniidae birds, the presumed pollinators of C. eminii. Some recent studies on these birds have revealed cryptic species within the same lineage occupying different mountain ranges, probably due to low connectivity between populations44,55. This pattern might have later been reinforced by a landscape strongly transformed by agriculture.
Both plastid and nuclear genetic data suggest a pattern of greater historical isolation among populations located in the western side of the Rift Valley than among those in the east (Fig. 2, Table 2). Whether this is associated to greater past connectivity through forest patches (mountain-bridge hypothesis) in the east, or whether it reflects a more recent historical pattern associated to agricultural activity, is not clear. Similar to other studies8,50,51, the highest level of genetic diversity was found in the Harar Massif, an area known for harbouring the best-preserved fragments of Afromontane forest (Table 2: Agere Maryam and Harenna Forest). This area may have acted as an important refuge for Afromontane forest organisms during glacial maxima. In the DPA analysis of C. eminii, the Harar Massif was inferred as the source of migration events on the eastern side of the Rift (Fig. 4b). For the western Rift, the SMSI was suggested as the ancestral area: these mountains harbour the highest plastid genetic diversity (Table 2), though these results could not be corroborated with the nuclear data (no AFLP sampling). The two arcs of the Rift Valley (Albertine and Gregory Rift) meet geographically in the north on the Central Highlands (Mt. Elgon and Cherangani Hills) and in the South in the SMSI (Fig. 1). These two regions (Mt. Elgon- Cherangani and the SMSI) apparently act as secondary contact points where the two parallel migration routes mentioned above interconnect19,44, and have been described as cradles of new genetic diversity53. This is congruent with our results in C. eminii, showing Mt. Elgon and the SMSI as a crossroad linking both sides of the west Rift System in the DPA analysis (Fig. 4), and both exhibiting high levels of genetic diversity (Table 2). Mount Elgon is also the oldest and most species-rich volcano of Eastern Africa13,56, with endemic lineages that are related to groups in either the eastern43 or the western side of the Rift Valley8,43. In contrast, the Afar Depression and the arid zone around the Turkana Basin seem to have acted as major barriers against gene flow (Fig. 1a and 4b). The Afar Depression separates the eastern and western Ethiopian populations, north of the two volcanic arcs, while the aridity around the Turkana Basin and the Uganda Gap, might have constituted an important obstacle to gene flow between the eastern and western populations57. The low haplotype diversity found in the Abyssinian massif population could reflect long-term isolation due to the influence of glaciations, though a low DW value suggests that recent dispersal could be a more likely explanation.
In summary, our study suggests that the topography and land formations of the Great Rift Valley underlie the phylogeographic pattern of east/west vicariance observed in C. eminii and other Afromontane endemic species. Though population sampling was limited to discriminate between the “mountain-forest” bridge and the LDD hypotheses, Pleistocene climatic oscillations and cyclical expansion and contraction of the Afromontane forest appear to have played a role in structuring levels of genetic variation in these species. Short-range dispersal among forest patches on each side of the Rift probably was complemented by LDD across some regions such as the Turkana Basin or the Uganda Gap, historically devoid of montane forests. Isolated sky-islands such as Mount Elgon or the Harar Massif acted as both refuges and cradles of genetic diversity. Given that the forested areas of Eastern Africa are currently in serious decline9 —especially the northern Ethiopian highlands subject to extensive logging— and that many of these harbour high levels of genetic variability (see Table 1, our study), designing new measures for the protection of these regions (e.g., new natural parks) may be crucial to preserve the endemic and highly-threatened Afromontane forest biota.
Methods
Sampling and DNA sequencing
The geographic distribution of C. eminii extends from the Ethiopian massifs to southern Tanganyika28. Despite our sampling effort (two field expeditions in 2009, 2015 to Ethiopia and one in 2010 to Kenya-Uganda), we failed to find C. eminii in many of the localities recorded by Hedberg in his classic monography of this species28. Much of the forest in these localities is highly degraded, threatened by the advancement of extensive agriculture, cattle rising, forestry, and other human activities (see Table S2.1 for a list of the visited localities for this study and a detailed description of the sampling). Despite this, we were able to sample fresh plant tissue from seven populations of C. eminii (Table S2.2): two populations in the Abyssinian massif (Gifta and Dembecha), two in the Harar Massif (Harenna forest and Agere Maryam), one in Rwenzori Mts. (Albertine Rift), one in Mt. Elgon (west of the Gregory Rift), and one in Aberdare Mts. (east of the Gregory Rift) (Fig. 2). Whenever possible leaves from 8–17 plants, 7–15 m apart were collected from each population. In addition, we added nine localities, obtained from old herbarium specimens (Royal Botanic Gardens, Kew): Cherangani (Kenya), Sasa trail in Mt. Elgon (Uganda), Gikongoro (Rwanda), Teza (Burundi), Tukuyu, Mwakelele, Livingstone Mountains, Mporoto Mountains (Tanzania) and Misuku (Malawi). Altogether, we sampled 16 localities of C. eminii (Table S2.2), representing all major geographic blocks in the distribution of the species (Fig. 1a). Three plastid (pDNA) regions were sequenced for a total of 79 individuals. For details on PCR amplification and sequence alignment see Appendix S1.1 and Table S2.3 in the Supplementary Material. Sources of the material examined, location of vouchers, GenBank accession numbers, and full references are listed in Table S2.2.
Phylogeographic analysis
Haplotype analyses were done on a concatenated dataset comprising all three sequenced plastid regions (see above). Genealogical relationships among haplotypes were inferred via the statistical parsimony algorithm58 implemented in TCS 1.2.159. The number of mutational steps resulting from single substitutions among haplotypes was calculated with 95% confidence limits, with gaps coded as missing data. Summary statistics for genetic diversity calculated for each population were: number of haplotypes H (n), haplotype diversity (Hd), and nucleotide diversity (π); using DnaSP 5.1 with the option “not consider gaps” selected60.
Haplotype divergence times were estimated using Bayesian relaxed clocks implemented in the software package BEAST v.1.7.561 and using the ‘nested dating approach’ described in Mairal et al.31; in this approach, a high phylogenetic-level dataset, including representatives of all three species of Canarina and nine outgroup taxa from Platycodoneae and Campanulaceae, was used to inform the clock rate of a linked population-level dataset of C. eminii, under a mixed Yule-coalescent model62. Congruence among results from the three plastid markers was tested by comparing clade support values for individual clades. See Appendix S1.2 for more details on these analyses.
The Discrete Phylogeographic Approach (DPA) of Lemey et al.63, implemented in BEAST, was used to infer ancestral ranges and to trace the history of migration events in C. eminii. It is based on a continuous-time Markov Chain process where the discrete states correspond to the geographic locations and the state transition rates to the migration rates between areas (Ronquist & Sanmartín, 2011). Bayesian Stochastic Search Variable Selection (BSSVS, Lemey et al.63) was used to identify the rates (colonization routes) that are best supported by the data, using a cut off value of three for the Bayes Factor comparison. We defined six discrete areas, assigning each plateau and Rift mountain ranges to a different area15,31: 1) Harar massif, 2) Abyssinian massif, 3) East Gregory Rift (Aberdare Mountains), 4) Central Highlands (Mount Elgon and Cherangani Hills), 5) Albertine Rift (Rwenzori, Rwanda and Burundi), and 6) Southern Mountain Sky-Islands (SMSI: South Tanzania and Malawi) (see Appendix S1.3 for more details).
AFLP fingerprinting
Laboratory molecular protocols for the AFLP analysis64 were implemented using the AFLP plant mapping kit (Applied Biosystems). We could not use the individuals sampled from herbarium collections for AFLP fingerprinting, as this approach is quite sensitive to the quality of the starting DNA and thus requires well-preserved material65. Therefore, sampling of populations in the nuclear dataset was more reduced than in the pDNA sequence dataset. Genomic DNA was digested with the enzymes EcoRI and MseI and linked to the adaptors EcoRI (5’-CTCGTAGACTGCGTACC-3’/5’-AATTGGTACGCAGTCTAC-3’) and MseI (5’- GACGATGAGTCCTGAC-3’/5’-ATCTCAGGACTCAT-3’). We tested 32 combinations of selective primers and chose the four pairs that produced the most polymorphic and clear profiles: 1-EcoRI6-FAM-ACT/MseI-CAA; 2-EcoRI6-FAM-ACT/MseI-CAT; 3-EcoRIVIC-AGG/MseI-CTA, and 4-EcoRIVIC-AGG/MseI-CTT.
Amplified fragments were analysed using GeneMapper 3.7 software (Applied Biosystems), and peaks ranging between 100 and 500 bp recorded. AFLP Scorer software66 was employed to run a reproducibility test, with the maximum acceptable error for each primer combination fixed at < 5%67. The AFLPdat R package68 was used to determine the numbers of private fragments per population. Only unambiguous fragments shared among duplicates were scored. Data reliability was assessed by comparison of duplicates, using one or two individuals per population (21 tests). The reproducibility value was 88–100%, with a mean of 97.5%.
AFLP data analysis
The resulting AFLP presence/absence matrix was analysed using AFLPSURV v.1.069 to estimate Nei’s gene diversity (Hj), pairwise differentiation among subpopulations (FST), the percentage of polymorphic fragments per population (P), and the bootstrapped Nei’s genetic distance matrix between individuals and populations70,71. The inbreeding coefficient (FIS) was set to 0.1 as suggested by Hardy72. The permutation test involved 20,000 permutations. In addition, a Bayesian method was employed to estimate allelic frequencies, using a non-uniform prior distribution73. Ten thousand permutations were performed to calculate FST values. Genetic distances between individuals, populations and geographic groups were also calculated. We used AFLPdat74 to calculate DW value per population, equivalent to the weighted endemism value75,76; this value is expected to be high in long-term isolated populations where rare markers should accumulate due to mutations, whereas newly established populations are expected to exhibit low values, thus helping to differentiate recent dispersal from more ancient isolation.
To distinguish genetic groups of individuals in the AFLP dataset, a comparison was made by constructing a pairwise similarity matrix for all individuals: Dice’s coefficient was calculated and the resulting matrix was transformed using principal coordinates analysis (PCO) with Ntsys v.2.177. Neighbour-nets of AFLP data were also calculated, both for individuals and populations, using the SplitsTree v.4.10 software78. To quantify the amount of genetic differentiation attributable to geographic and population subdivision, a hierarchical analysis of molecular variance was performed79 in ARLEQUIN v.3.0 (Table S2.4). For assessing the structure of populations, we used the Bayesian method implemented in STRUCTURE 2.280,81, assuming admixture conditions and uncorrelated allele frequencies between groups, 500,000 generations (plus a burn-in of 100,000) were run for K values of 1–10, with ten repetitions each. For each K value, only the run with the highest maximum likelihood value was considered. The LnP (D) for the successive decomposition of groups was used in all STRUCTURE analyses82. To test the effect of the spatial distance on the genetic structure of the C. eminii populations, correlations between genetic (measured as FST) and spatial distances between pairs of populations were determined using the Mantel permutation procedure in Ntsys. The genetic distance matrix used was based on the presence/absence matrix; the geographic distance matrix was based on absolute distances between the geographic coordinates for each collected population. In addition, the kinship multilocus coefficient (FIJ) was estimated using SPAGeDi83 to determine the spatial structure of the examined populations, taking spatial distances into account in the analyses. BARRIER v.2.284 was used to identify possible geographic locations acting as major genetic barriers among C. eminii populations, based on genetic distances. The significance of these was examined with 1000 bootstrapped distance matrices obtained using AFLPsurv.
Data Accessibility
DNA sequences: Genbank Accession nos KF028817–KM189329. GenBank accessions, sampling locations and/or online-only appendices uploaded as online Supplemental material.
Additional Information
How to cite this article: Mairal, M. et al. Geographic barriers and Pleistocene climate change shaped patterns of genetic variation in the Eastern Afromontane biodiversity hotspot. Sci. Rep. 7, 45749; doi: 10.1038/srep45749 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank E. Cano and M. I. Garcia (Parque Científico de Madrid) for technical assistance; J. M. Olesen, M.-X. Ren, E. Habiba and A. Sánchez-Meseguer for help in field collection and herbarium searches, and V. Culshaw for her useful comments on the language. We thank the authorities of the National Parks of the Bale Mountains and Harenna Forest (Ethiopia), the Aberdare Mountains Park (Kenya) and Rwenzori Mountains Park (Uganda) for help with fieldwork. Special thanks are given to P. Kamau from the National Museums of Kenya, Botany Department, for her help in herbarium and field searches. This study was supported by the Spanish government (grants CGL2006-09696 and CGL2010-18631/BOS to J.J.A., and CGL2012-40129-C02-01 and CGL2015-67849-P to I.S.), and from the Consejería de Educación de la Comunidad de Madrid, and EU FP7 Synthesys Programme award to L.P. (GB-TAF-5153). M.A. was funded by the Junta para la Ampliación de Estudios Doctorales programme (CSIC/FSE). M.M. was supported by a MINECO PhD fellowship (BES-2010-037261), under project CGL2009-13322-C03-01 to I.S.
Footnotes
The authors declare no competing financial interests.
Author Contributions M.M., M.A., I.S. and J.J.A. designed the study. J.J.A., M.A. and L.P. contributed samples. M.M., A.H., J.J.A., and M.A. performed the research. M.M., M.A., J.J.A. and I.S. wrote the manuscript. All authors commented and approved the final version.
References
- White F. The history of the Afromontane archipelago and the scientific need for its conservation. African Journal of Ecology. 19, 33–54 (1981). [Google Scholar]
- White F. The Vegetation of Africa. A Descriptive Memoir to Accompany The Unesco/AETFAT/UNSO Vegetation Map of Africa (3 Plates, Northwestern Africa, Northeastern Africa, and Southern Africa, 1: 5,000,000) (1983).
- Mittermeier R. A. et al. Hotspots revisited: earth’s biologically richest and most endangered ecoregions. CEMEX, Mexico City, Mexico (2004).
- Kingdon J. Island Africa: The Evolution Of Africa’s Rare Animals And Plants(ed. Collins, London, 1990). [Google Scholar]
- C.E.P.F., Eastern Afromontane Biodiversity Hotspot - Ecosystem Profile, http://www.cepf.net/Documents/Eastern_Afromontane_Ecosystem_Profile_FINAL.pdf, (2012) (11/10/16).
- Fjeldså J. & Lovett J. C. Geographical patterns of old and young species in African forest biota: the significance specific montane areas as evolutionary centres. Biodiversity & Conservation. 6, 325–346 (1997). [Google Scholar]
- EFAP. Ethiopian forestry action program: Final Report - Volume II: The Challenges For Development, Ministry of Natural Resources Development and Environmental Protection, Addis Ababa, Ethiopia (1994).
- Kebede M., Ehrich D., Taberlet P., Nemomissa S. & Brochmann C. Phylogeography and conservation genetics of a giant lobelia (Lobelia giberroa) in Ethiopian and Tropical East African mountains. Molecular Ecology. 16, 1233–1243 (2007). [DOI] [PubMed] [Google Scholar]
- F.A.O., Food and Agricultural Organization of the United Nations. Forest Resource Assessment, Page 4, http://www.fao.org/3/a-i4808e.pdf (2015) (11/10/16). [Google Scholar]
- Kloos H. & Legesse W. Water Resources Management In Ethiopia: Implications For The Nile Basin. Cambria Press (2010).
- Ring U. The east African Rift System. Austrian Journal of Earth Sciences. 107, 132–146 (2014). [Google Scholar]
- Fernandes R. M. S. et al. Angular velocities of Nubia and Somalia from continuous GPS data: implications on present-day relative kinematics. Earth and Planetary Science Letters. 222, 197–208 (2004). [Google Scholar]
- Sklenář P., Hedberg I. & Cleef A. M. Island biogeography of tropical alpine floras. Journal of Biogeography. 41, 287–297 (2014). [Google Scholar]
- Hedberg O. Vegetation belts of the East African mountains. Svensk Botanisk Tidskrift Utgifven af Svenska Botaniska Foreningen, Stockholm. 45, 140–202 (1951). [Google Scholar]
- Gehrke B. & Linder H. P. Species richness, endemism and species composition in the tropical Afroalpine flora. Alpine Botany. 124, 165–177 (2014). [Google Scholar]
- Linder H. P. The evolution of African plant diversity. Frontiers in Ecology and Evolution. 2, 1–14 (2014). [Google Scholar]
- Hedberg O. Evolution and speciation in a tropical high mountain flora. Biological Journal of the Linnean Society. 1, 135–148 (1969). [Google Scholar]
- Hedberg O. Evolution of the Afroalpine Flora. Biotropica. 2, 16–23 (1970). [Google Scholar]
- Assefa A., Ehrich D., Taberlet P., Nemomissa S. & Brochmann C. Pleistocene colonization of afro-alpine ‘sky islands’ by the arctic-alpine Arabis alpina. Heredity. 99, 133–142 (2007). [DOI] [PubMed] [Google Scholar]
- Popp M., Gizaw A., Nemomissa S., Suda J. & Brochmann C. Colonization and diversification in the African ‘sky islands’ by Eurasian Lychnis L. (Caryophyllaceae). Journal of Biogeography. 35, 1016–1029 (2008). [Google Scholar]
- Demos T. C., Peterhans J. C. K., Agwanda B. & Hickerson M. J. Uncovering cryptic diversity and refugial persistence among small mammal lineages across the Eastern Afromontane biodiversity hotspot. Molecular Phylogenetics and Evolution. 71, 41–54 (2014). [DOI] [PubMed] [Google Scholar]
- Gottelli D., Marino J., Sillero-Zubiri C. & Funk S. M. The effect of the last glacial age on speciation and population genetic structure of the endangered Ethiopian wolf (Canis simensis). Molecular Ecology. 13, 2275–2286 (2004). [DOI] [PubMed] [Google Scholar]
- Bonnefille R., Roeland J. & Guiot J. Temperature and rainfall estimates for the past 40,000 years in equatorial Africa. Nature. 346, 347–349 (1990). [Google Scholar]
- Mairal M., Sanmartín I. & Pellissier L. Lineage-specific climatic niche drives the tempo of vicariance in the Rand Flora. Journal of Biogeography, doi: 10.1111/jbi.12930 (2017). [DOI] [Google Scholar]
- Chala D., Zimmermann N. E., Brochmann C. & Vakkestuen V. Migration corridors for alpine plants among the ‘sky islands’ of eastern Africa: do they, or did they exist? Alpine Botany (in press).
- Wondimu T. et al. Crossing barriers in an extremely fragmented system: two case studies in the afro-alpine sky island flora. Plant Systematics and Evolution. 300, 415–430 (2014). [Google Scholar]
- Masao C. A. et al. Phylogeographic history and taxonomy of some afro-alpine grasses assessed based on AFLPs and morphometry: Deschampsia cespitosa, D. angusta and Koeleria capensis. Alpine Botany. 123, 107–122 (2013). [Google Scholar]
- Hedberg O. Monograph of the genus Canarina L. (Campanulaceae). Svensk Botanisk Tidskrift Utgifven af Svenska Botaniska Foreningen. Stockholm. 55, 17–62 (1961). [Google Scholar]
- Olesen J. M., Alarcon M., Ehlers B. K. & Aldasoro J. J. & Roquet, C. Pollination, biogeography and phylogeny of oceanic island bellflowers (Campanulaceae). Perspectives in Plant Ecology, Evolution and Systematics. 14, 169–182 (2012). [Google Scholar]
- Yamagiwa J. & Basabose A. K. Diet and seasonal changes in sympatric gorillas and chimpanzees at Kahuzi–Biega Nation. Primates. 47, 74–90. (2006). [DOI] [PubMed] [Google Scholar]
- Mairal M., Pokorny L., Aldasoro J. J., Alarcon M. & Sanmartin I. Ancient vicariance and climate-driven extinction explain continental-wide disjunctions in Africa: the case of the Rand Flora genus Canarina (Campanulaceae). Molecular Ecology. 24, 1335–1354 (2015). [DOI] [PubMed] [Google Scholar]
- Yalden D. & Largen. The endemic mammals of Ethiopia. Mammal Review. 22, 115–150 (1992). [Google Scholar]
- Bobe R. The evolution of arid ecosystems in eastern Africa. Journal of Arid Environments. 66, 564–584 (2006). [Google Scholar]
- Sepulchre P. et al. Tectonic uplift and eastern Africa aridification. Science. 313, 1419–1423 (2006). [DOI] [PubMed] [Google Scholar]
- Davidson A. & Rex D. Age of volcanism and rifting in southwestern Ethiopia. Nature. 283, 657–658 (1980). [Google Scholar]
- Engler A. Plants of the northern temperate zone in their transition to the high mountains of Tropical Africa. Annals of Botany. 4, 523–540 (1904). [Google Scholar]
- Gizaw A. et al. Colonization and diversification in the African ‘sky islands’: insights from fossil‐calibrated molecular dating of Lychnis (Caryophyllaceae). New Phytologist. 35, 1016–1029 (2016). [DOI] [PubMed] [Google Scholar]
- White F. The Afromontane Region. Biogeography And Ecology Of Southern Africa (ed. Springer) 463–513 (Netherlands, 1978).
- Galley C., Bytebier B., Bellstedt D. & Linder H. P. The Cape element in the Afrotemperate flora: from Cape to Cairo? Proceedings of the Royal Society B: Biological Sciences. 274, 535–543 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke B. & Linder H. P. The scramble for Africa: pan-temperate elements on the African high mountains. Proceedings of the Royal Society of London B: Biological Sciences. 276, 2657–2665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Maio N., Wu C. H., O’Reilly K. M. & Wilson D. New routes to phylogeography: A Bayesian structured coalescent approximation. PLoS Genetics. 11, e1005421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizaw A. et al. Phylogeography of the heathers Erica arborea and E. trimera in the afro-alpine ‘sky islands’ inferred from AFLPs and plastid DNA sequences. Flora. 208, 453–463 (2013). [Google Scholar]
- Knox E. & Palmer J. Chloroplast DNA evidence on the origin and radiation of the giant Lobelias in Eastern Africa. Systematic Botany. 23, 109–149 (1998). [Google Scholar]
- Bowie R. C., Fjeldsa J., Hackett S. J. & Crowe T. M. Molecular evolution in space and through time: mtDNA phylogeography of the Olive Sunbird (Nectarinia olivacea/obscura) throughout continental Africa. Molecular Phylogenetics and Evolution. 33, 56–74 (2004). [DOI] [PubMed] [Google Scholar]
- Benson C. W., Irwin M. S. & White C. M. N. The significance of valleys as avian zoogeographical barriers. Annals of Cape Province Museum of Natural History. 2, 155–189 (1962). [Google Scholar]
- Freitag S. & Robinson T. J. Phylogeographic Patterns in Mitochondrial DNA of the Ostrich (Struthio camelus). The Auk. 110, 614–622 (1993). [Google Scholar]
- DeMenocal P. B. Plio-Pleistocene African climate. Science. 270, 53–59 (1995). [DOI] [PubMed] [Google Scholar]
- Trauth M. H., Maslin M. A., Deino A. & Strecker M. R. Late Cenozoic moisture history of East Africa. Science. 309, 2051–2053 (2005). [DOI] [PubMed] [Google Scholar]
- Ruiz Guajardo J. C. et al. Landscape genetics of the key African acacia species Senegalia mellifera (Vahl)- the importance of the Kenyan Rift Valley. Molecular Ecology. 19, 5126–5139 (2010). [DOI] [PubMed] [Google Scholar]
- Aga E., Bekele E. & Bryngelsson T. Inter-simple sequence repeat (ISSR) variation in forest coffee trees (Coffea arabica L.) populations from Ethiopia. Genetica. 124, 213–221 (2005). [DOI] [PubMed] [Google Scholar]
- Ayele T. B., Gailing O., Umer M. & Finkeldey R. Chloroplast DNA haplotype diversity and postglacial recolonization of Hagenia abyssinica (Bruce) J.F. Gmel. in Ethiopia. Plant Systematics and Evolution. 280, 175–185 (2009). [Google Scholar]
- Kadu C. A. C. et al. Phylogeography of the Afromontane Prunus africana reveals a former migration corridor between East and West African highlands. Molecular Ecology. 20, 165–178 (2011). [DOI] [PubMed] [Google Scholar]
- Ehrich D. et al. Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Molecular Ecology. 16, 2542–2559 (2007). [DOI] [PubMed] [Google Scholar]
- Hewitt G. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 58, 247–276 (1996). [Google Scholar]
- Kahindo C., Bowie R. C. & Bates J. M. The relevance of data on genetic diversity for the conservation of Afromontane regions. Biological Conservation. 134, 262–270 (2007). [Google Scholar]
- Hedberg O. Afroalpine vegetation compared to paramo: convergent adaptations and divergent differentiation. Páramo: An Andean Ecosystem Under Human Influence(ed. Academic Press, London, 15–30, 1992). [Google Scholar]
- Wilfert L., Kaib M., Durka W. & Brandl R. Differentiation between populations of a termite in eastern Africa: implications for biogeography. Journal of Biogeography. 33, 1993–2000 (2006). [Google Scholar]
- Templeton A. R., Crandall K. A. & Sing C. F. A. Cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 132, 619–633 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M., Posada D. C. & Crandall K. A. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 9, 1657–1659 (2000). [DOI] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Drummond A. J. & Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 7, 214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorny L., Oliván G. & Shaw A. J. Phylogeographic patterns in two southern hemisphere species of Calyptrochaeta (Daltoniaceae, Bryophyta). Systematic Botany. 36, 542–553 (2011). [Google Scholar]
- Lemey P., Rambaut A., Drummond A. J. & Suchard M. A. Bayesian phylogeography finds its roots. PLoS Computational Biology. 5, e1000520 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P. et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 23, 4407–4414 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meudt H. M. & Clarke A. C. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends in plant science. 12, 106–117 (2007). [DOI] [PubMed] [Google Scholar]
- Whitlock R., Hipperson H., Mannarelli M., Butlin R. K. & Burke T. An objective, rapid and reproducible method for scoring AFLP peak-height data that minimizes genotyping error. Molecular Ecology Resources. 8, 725–735 (2008). [DOI] [PubMed] [Google Scholar]
- Bonin A. et al. How to track and assess genotyping errors in population genetics studies. Molecular Ecology. 13, 3261–3273 (2004). [DOI] [PubMed] [Google Scholar]
- Ehrich D. Aflpdat: a collection of R functions for convenient handling of AFLP data. Molecular Ecology Notes. 6, 603–604 (2006). [Google Scholar]
- Vekemans X., Beauwens T., Lemaire M. & Roldan-Ruiz I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology. 11, 139–151 (2002). [DOI] [PubMed] [Google Scholar]
- Nei M. & Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences. 76, 5269–5273 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. & Milligan B. G. Analysis of population genetic structure with RAPD markers. Molecular Ecology. 3, 91–99 (1994). [DOI] [PubMed] [Google Scholar]
- Hardy O. J. Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Molecular Ecology. 12, 1577–1588 (2003). [DOI] [PubMed] [Google Scholar]
- Zhivotovsky L. A. Estimating population structure in diploids with multilocus dominant DNA markers. Molecular Ecology. 8, 907–913. [DOI] [PubMed] [Google Scholar]
- Ehrich D. AFLPdat: a collection of R functions for convenient handling of AFLP data. Molecular Ecology Notes. 6, 603–604 (2006). [Google Scholar]
- Crisp M. D., Laffan S., Linder H. P. & Monro A. Endemism in the Australian flora. Journal of Biogeography. 28, 183–198 (2001). [Google Scholar]
- Schönswetter P. & Tribsch A. Vicariance and dispersal in the alpine perennial Bupleurum stellatum L.(Apiaceae). Taxon. 54, 725–732 (2005). [Google Scholar]
- Rohlf F. J. NTSYS-pc version 2.0. Numerical taxonomy and multivariate analysis system. Exeter software, Setauket, New York (1998).
- Huson D. H. & Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular biology and evolution. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G. & Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary bioinformatics online. 1, 47 (2005). [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M. & Pritchard J. K. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 7, 574–578 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S. & Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- Hardy O. J. & Vekemans X. Spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2, 618–620 (2002). [Google Scholar]
- Manni F., Guerard E. & Heyer E. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier’s algorithm. Human biology. 76, 173–190 (2004). [DOI] [PubMed] [Google Scholar]
- Ryan W. B. et al. Global multi-resolution topography synthesis. Geochemistry, Geophysics, Geosystems. 10, Q03014, doi: 10.1029/2008GC002332 (2009). [DOI] [Google Scholar]
- Silvestrini M. et al. Genetic diversity and structure of Ethiopian, Yemen and Brazilian Coffea arabica L. accessions using microsatellites markers. Genetic Resources and Crop Evolution. 54, 1367–1379 (2007). [Google Scholar]
- Derero A., Gailing O. & Finkeldey. R. Maintenance of genetic diversity in Cordia africana Lam., a declining forest tree species in Ethiopia. Tree Genetics & Genomes. 7, 1–9 (2010). [Google Scholar]
- Sertse D., Gailing O., Eliades N. G. & Finkeldey R. Anthropogenic and natural causes influencing population genetic structure of Juniperus procera Hochst. ex Endl. in the Ethiopian highlands. Genetic Resources and Crop Evolution. 58, 849–859 (2011). [Google Scholar]
- Kadu C. A. C. et al. Divergent pattern of nuclear genetic diversity across the range of the Afromontane Prunus africana mirrors variable climate of African highlands. Annals of Botany. 111, 47–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchugi A. et al. Genetic structuring of important medicinal species of genus Warburgia as revealed by AFLP analysis. Tree Genetics & Genomes. 4, 787–795 (2008). [Google Scholar]
- Omondi S. F. et al. Genetic Diversity and Population Structure of Acacia senegal (L) Willd. in Kenya. Tropical Plant Biology. 3, 59–70 (2010). [Google Scholar]
- Matthee C. A. & Robinson T. J. Molecular phylogeny of the springhare, Pedetes capensis, based on mitochondrial DNA sequences. Molecular Biology and Evolution 14, 20–29 (1997). [DOI] [PubMed] [Google Scholar]
- Arctander P., Johansen C. & Coutellec-Vreto M. A. Phylogeography of three closely related African bovids (tribe Alcelaphini). Molecular Biology and Evolution. 16, 1724–1739 (1999). [DOI] [PubMed] [Google Scholar]
- Girman D. J. et al. Patterns of population subdivision, gene flow and genetic variability in the African wild dog (Lycaon pictus). Molecular Ecology. 10, 1703–1723 (2001). [DOI] [PubMed] [Google Scholar]
- Pitra C., Hansen A. J., Lieckfeldt D. & Arctander P. An exceptional case of historical outbreeding in African sable antelope populations. Molecular Ecology. 11, 1197–1208 (2002). [DOI] [PubMed] [Google Scholar]
- Dubach J. et al. Molecular genetic variation across the southern and eastern geographic ranges of the African lion, Panthera leo. Conservation Genetics. 6, 15–24 (2005). [Google Scholar]
- Belay G. & Mori A. Intraspecific phylogeographic mitochondrial DNA (D-loop) variation of Gelada baboon, Theropithecus gelada, in Ethiopia. Biochemical Systematics and Ecology. 34, 554–561 (2006). [Google Scholar]
- Nicolas V. et al. Phylogeographic structure and regional history of Lemniscomys striatus (Rodentia: Muridae) in tropical Africa. Journal of Biogeography. 35, 2074–2089 (2008). [Google Scholar]
- Colangelo P. et al. A mitochondrial phylogeographic scenario for the most widespread African rodent, Mastomys natalensis. Biological Journal of the Linnean Society. 108, 901–916 (2013). [Google Scholar]
- Evans B. J., Bliss S. M., Mendel S. A. & Tinsley R. C. The Rift Valley is a major barrier to dispersal of African clawed frogs (Xenopus) in Ethiopia. Molecular Ecology. 20, 4216–4230 (2011). [DOI] [PubMed] [Google Scholar]
- Freilich X., Anadón J. D., Bukala J., Calderon O., Chakraborty R. & Boissinot S. Comparative Phylogeography of Ethiopian anurans: impact of the Great Rift Valley and Pleistocene climate change. BMC evolutionary biology 16, 206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field L. M. et al. Analysis of genetic variability in Anopheles arabiensis and Anopheles gambiae using microsatellite loci. Insect Molecular Biology. 8, 287–297 (1999). [DOI] [PubMed] [Google Scholar]
- Lehmann T. et al. The Rift Valley complex as a barrier to gene flow for Anopheles gambiae in Kenya. Journal of Heredity. 90, 613–621 (1999). [DOI] [PubMed] [Google Scholar]
- Lehmann T. et al. Brief communication. The Rift Valley complex as a barrier to gene flow for Anopheles gambiae in Kenya: the mtDNA perspective. Journal of Heredity. 91, 165–168 (2000). [DOI] [PubMed] [Google Scholar]
- Lehmann T. et al. Population Structure of Anopheles gambiae in Africa. Journal of Heredity. 94, 133–147 (2003). [DOI] [PubMed] [Google Scholar]
- Krafsur E. S. Population structure of the tsetse fly Glossina pallidipes estimated by allozyme, microsatellite and mitochondrial gene diversities. Insect Molecular Biology. 11, 37–45 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braginets O. P., Minakawa N., Mbogo C. M. & Ya G. Population genetic structure of the African malaria mosquito Anopheles funestus in Kenya. The American journal of tropical medicine and hygiene. 69, 303–308 (2003). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: Genbank Accession nos KF028817–KM189329. GenBank accessions, sampling locations and/or online-only appendices uploaded as online Supplemental material.