Abstract
To elucidate the physiological significance of MEK5 in vivo, we have examined the effect of mek5 gene elimination in mice. Heterozygous mice appear to be healthy and were fertile. However, mek5−/− embryos die at approximately embryonic day 10.5 (E10.5). The phenotype of the mek5−/− embryos includes abnormal cardiac development as well as a marked decrease in proliferation and an increase in apoptosis in the heart, head, and dorsal regions of the mutant embryos. The absence of MEK5 does not affect cell cycle progression but sensitizes mouse embryonic fibroblasts (MEFs) to the ability of sorbitol to enhance caspase 3 activity. Further studies with mek5−/− MEFs indicate that MEK5 is required for mediating extracellular signal-regulated kinase 5 (ERK5) activation and for the regulation of the transcriptional activity of myocyte enhancer factor 2. Overall, this is the first study to rigorously establish the role of MEK5 in vivo as an activator of ERK5 and as an essential regulator of cell survival that is required for normal embryonic development.
The mitogen-activated protein kinase (MAPK) cascades constitute a complex network of signaling pathways that are involved in the regulation of numerous cell functions (9). They consist of the sequential activation of protein kinases that include MAPKs, MAPK/extracellular signal-regulated kinase (ERK) kinases (MEKs or MKKs), and MEK kinases (MEKKs) (9). MAPKs are activated by dual phosphorylation on threonine (T) and tyrosine (Y) residues within a T-X-Y motif by MEKs. MEKs are activated by MEKKs. Two main mechanisms have been proposed to ensure specific transmission of the signals from upstream kinases to MAPKs (38, 40): (i) scaffold proteins that assemble the different components of a cascade; (ii) physical interactions between the components of a cascade. Both mechanisms may operate in parallel and allow different responses of the same MAPK signaling pathways to different stimuli.
At least four MAPK subfamilies have been identified: ERK1/2, ERK5, c-Jun NH2-terminal protein kinases (JNKs), and p38 MAPKs. MAPK activators include MEK1 and MEK2 for ERK1/2, MEK5 for ERK5, MKK4 and MKK7 for JNKs, and MKK3 and MKK6 for p38 MAPKs (9). Targeted deletion of the mapk and mek/mkk genes has contributed substantially to our increased understanding of the physiological role of these pathways in development and pathogenesis. In particular, the recent elimination of the erk5 gene in mice has provided genetic evidence that ERK5 is required for normal cardiac development (31, 34, 42).
ERK5, also known as big MAPK, is almost twice the size (815 amino acids) of the other MAPKs (45). Its unique COOH-terminal tail contains a myocyte enhancer factor 2 (MEF2)-interacting domain and a potent transcriptional activation domain (12). The ERK5 catalytic NH2-terminal domain is 50% identical to ERK2. The activity of a number of transcription factors has been shown to be regulated by ERK5, including MEF2, c-Fos and Fra-1, Sap1, c-Myc, and NF-κB (6, 11, 13, 15, 28, 37). In vitro, the ERK5 signaling pathway has been implicated in MEF2-dependent gene expression during muscle differentiation and neuronal survival (4, 20, 33). The signaling cascade that leads to ERK5 activation is stimulated in response to mitogens and a number of stresses (1, 11, 14, 39, 41).
In vitro protein kinase assays and transfection studies with constitutively activated MEK5 have demonstrated that MEK5 is a potent activator of ERK5 (5, 45). The MEK5 cDNA encodes a 444-amino-acid protein, which displays more than 50% homology with the other known MEKs. Two alternative splice variants encoding two isoforms, MEK5α (50 kDa) and MEK5β (40 kDa), have been identified (5). MEK5β is ubiquitously distributed and primarily cytosolic while MEK5α is expressed mostly in liver and brain and is in the particulate fraction. MEK5 activity is regulated by MEKK2 and MEKK3 (2, 35). Consistent with the abnormal phenotype displayed by the erk5−/− mice, transgenic mice overexpressing activated MEK5 in the heart display eccentric cardiac hypertrophy that progresses to dilated cardiomyopathy and sudden death (26).
The in vivo role of MEK5 has not been rigorously established, mainly because of the lack of available pharmacological and genetic reagents that specifically alter MEK5 activity (24). Therefore, we have engineered a novel genetically modified mouse model deficient in MEK5 expression. Our results show that the targeted deletion of the mek5 gene causes early embryonic death. Embryonic day 10.5 (E10.5) mek5−/− embryos display cardiovascular defects suggesting that, like ERK5, MEK5 is required for normal cardiac development. In addition to the heart, the head and dorsal regions of the mutant embryos exhibit a marked decrease in proliferation and an increase in apoptosis compared to wild-type littermates. Consistent with these results, mek5−/− fibroblasts are more sensitive than wild-type cells to sorbitol-induced caspase 3 activity. However, no marked difference was observed in the cell cycle progression of the wild-type and mutant cells. Further studies demonstrate that MEK5 is required for mediating ERK5 activation in response to both mitogenic and stress signals. Moreover, the absence of MEK5 expression prevents the MEK kinases MEKK2 and MEKK3 from increasing the transcriptional activity of MEF2A and MEF2D, two well-characterized downstream substrates of ERK5 (15). Altogether, these results provide clear genetic evidence that MEK5 is a nonredundant component of the ERK5/MEF2-dependent cell survival pathway that is required for mediating normal cardiac development.
MATERIALS AND METHODS
Generation of mek5−/− mice.
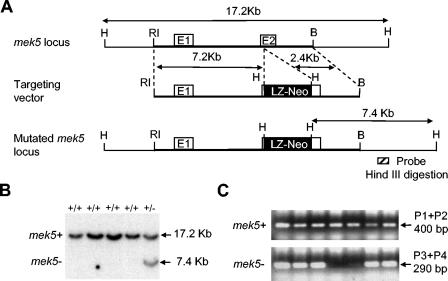
The sequence of the mek5 gene locus was obtained from GenBank (accession no. AC124753). Gene analysis revealed that the mek5 gene is encoded by at least 16 exons and 15 introns spanning ∼120 kb. MEK5 genomic DNA was cloned from a 129/Sv mouse strain-derived genomic RPCI-21 PAC library (United Kingdom HGMP Resource Centre) with a MEK5 cDNA probe. A 9.6-kb EcoRI-BamHI genomic fragment encompassing exons 1 and 2 of the mek5 gene was subcloned into pBluescript II KS vector (Stratagene). A β-galactosidase (β-Gal) and neomycin resistance (LZ-Neo) cassette containing a stop codon and a polyadenylation termination signal was inserted in frame into exon 2 by using an engineered HindIII restriction site. This gave rise to a targeting vector comprising 7.2-kb EcoRI-HindIII and 2.4-kb HindIII-BamHI fragments of MEK5 homologous sequences at its 5′ and 3′ extremities, respectively (Fig. 1). The resulting plasmid (50 μg) was linearized with NotI and electroporated into R1 embryonic stem (ES) cells (kindly provided by Andras Nagy, Samuel Lunenfeld Research Centre, Mount Sinai Hospital, Toronto, Canada). Neomycin-resistant clones selected with 500 μg of G418 (Invitrogen)/ml were screened by Southern blotting. Homologous recombined ES cell clones were injected into C56BL/6 blastocysts and transferred into pseudopregnant CD1 females to generate chimeras. Resulting high-grade agouti-marked male chimeras were mated with C57BL/6 females for germ line transmission of the mutation. All mice employed for this study were hosted in a pathogen-free facility at the University of Manchester. The animal studies were carried out according to Home Office and Institutional guidelines.
FIG. 1.
Strategy for the targeted disruption of the mek5 gene. (A) The genomic region at the mek5 locus, the mek5 targeting vector, and the predicted structure of the mutated mek5 gene are depicted. Restriction enzyme sites are indicated (B, BamH1; RI, EcoRI; H, HindIII). White boxes are mek5 exons. The black box is the β-Gal neomycin cassette (LZ-Neo). (B) Southern blotting analysis of HindIII-restricted genomic DNA prepared from ES cell clones indicates the presence of wild-type (+/+) and heterozygous (+/−) genotypes. The blot was probed with a random-primed 32P-labeled mouse MEK5 genomic probe (see hatched box in panel A). (C) Genomic DNAs isolated from mouse tails were amplified by two separate PCRs with primers specific for the mek5 gene (P1 + P2) and for the LZ-Neo cassette (P3 + P4).
Southern blot analysis.
Genomic DNA prepared from ES cells by standard protocols (36) was digested with HindIII, analyzed by electrophoresis, blotted onto a nylon membrane, and hybridized with a radiolabeled 600-bp genomic fragment located outside the 3′ region of the targeting vector (Fig. 1). This probe hybridizes to a 7.4-kb fragment (disrupted allele) and to a 17.2-kb fragment (endogenous mek5 gene).
Genotype determination of mice and embryos.
mek5+/− offspring were identified by two separate PCRs on tail DNA. The wild-type allele was amplified with forward (P1, 5′-GCTCATGTTTCTGTG-3′ ) and reverse (P2, 5′-TGTGCCGTATGATGATC-3′ ) primers. The mutant allele was amplified with a neomycin-specific primer set (P3, 5′-CTTGGGTGGAGAGGCTATTATTC-3′; P4, 5′-AGGTGAGATGACAGGAGATC-3′ ). Genotype determination of embryos was performed by PCR of genomic DNA isolated from the yolk sacs by using a three-primer set (forward primer 1, 5′-ATCAGAATGAGGCTCAGG-3′; forward primer 2, 5′-GCGCATCGCCTTCTATCG-3′; reverse primer, 5′-TGTGCCGTATGATGATC-3′ ). Fragments (600 and 700 bp) were amplified from the wild-type and disrupted alleles, respectively.
Tissue culture and preparation of lysates.
Mouse embryonic fibroblasts (MEFs) obtained from wild type and mek5−/− E9.5 embryos were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen), 2 mM l-glutamine, 10 U of penicillin/ml, 100 mg of streptomycin/ml, and 50 μM 2-mercaptoethanol at 37°C in a humidified atmosphere with 5% CO2. Transfection assays were performed by using the Lipofectamine method according to the manufacturer's recommendations (Invitrogen).
Proteins were extracted from cells or tissues in Triton lysis buffer (TLB; 20 mM Tris [pH 7.4], 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 10% glycerol, 1 mM orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml). Extracts were clarified by centrifugation (14,000 × g for 10 min at 4°C). The concentration of soluble proteins in the supernatants was quantified by the Bradford method (Bio-Rad).
Immunoblot analysis.
Cell and tissue extracts (50 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10% or 8% polyacrylamide gel) and electrophoretically transferred to an Immobilon-P membrane (Millipore, Inc.). The membranes were incubated with 5% nonfat dry milk or 3% bovine serum albumin at 4°C overnight and then probed with polyclonal antibodies to ERK5, tubulin (Sigma), β-Gal (Promega), or MEK5. The antibodies to ERK5 and MEK5 were obtained by immunizing rabbits against the synthetic peptides SGPPPPDPGGLTPQPST and NEQDIRYRDTLGHGN, corresponding to amino acids 680 to 694 and 162 to 176 of the ERK5 and the MEK5 proteins, respectively (Eurogentec). Immune complexes were detected by enhanced chemiluminescence (Pierce) with rabbit anti-mouse immunoglobulin G coupled to horseradish peroxidase as the secondary antibody (Amersham-Pharmacia).
Protein kinase assay.
JNK, ERK1/2, and ERK5 protein kinase activity was measured in cell lysates following incubation with glutathione S-transferase (GST)-c-Jun and glutathione-Sepharose beads or with polyclonal antibodies to ERK1/2 or ERK5 and protein A-agarose beads for 2 to 3 h at 4°C, respectively. Complexes were washed three times with TLB and twice with kinase buffer (25 mM HEPES [pH 7.4], 25 mM β-glycerophosphate, 25 mM MgCl2, 2 mM dithiothreitol, 0.1% orthovanadate) prior to being incubated at 30°C for 20 min in kinase buffer containing 50 μM [γ-32P]ATP (10 Ci/mmol) and 1 μg of GST-Myc or GST-MEF2C for the ERK1/2 and ERK5 assays, respectively. SB203580 (1 μM; Promega) was added to the ERK5 assay to prevent the phosphorylation of MEF2C by p38 MAPKs. The reactions were terminated by the addition of Laemmli sample buffer. Proteins were resolved by SDS-PAGE (12% polyacrylamide gel) and identified by autoradiography. The incorporation of [32P]phosphate was quantitated by PhosphorImager analysis.
Reporter gene expression assay.
The reporter plasmid pG5E1bLuc (32) was transiently cotransfected together with a construct encoding the fusion proteins GAL4-MEF2A, GAL4-MEF2D, and GAL4-cJun (10) with or without expression vectors encoding MEKK2 or MEKK3. A pRL-Tk plasmid encoding Renilla luciferase was employed for monitoring transfection efficiency. Aliquots of cell lysates were assayed for firefly and Renilla luciferase activities according to the manufacturer's instructions (Promega). GAL4-MEF2A and GAL4-MEF2D and MEKK2 and MEKK3 constructs were kindly provided by Hung-Ying Kao (Case Western University, Cleveland, Ohio) and Christian Widmann (Université de Lausanne, Lausanne, Switzerland), respectively.
Histological and immunohistochemistry analysis.
Freshly isolated embryos and placentas were fixed at room temperature either in Bouin's fluid overnight or in 4% paraformaldehyde for 2 h, dehydrated, and embedded in paraffin. Four- to six-micrometer-thick sections were cut. For histological analysis, sections were stained with hematoxylin and eosin (17). For β-Gal staining, sections were analyzed by the ABC peroxidase method (Vector) with a primary polyclonal antibody to β-Gal (1:500 dilution; Promega) and a secondary biotinylated anti-rabbit antibody (1:200 dilution; Vector). For bromodeoxyuridine (BrdU) staining, pregnant female mice were injected with 400 mg of BrdU/kg of body weight and sacrificed 1.5 h later prior to analysis. Immunohistochemistry was performed by the boric acid buffer method (30), with the exception that the rat monoclonal antibody to BrdU was obtained from Immunologicals Direct, Oxford Biotechnology Ltd. For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining, sections were processed by using an in situ cell death detection kit (POD; Roche) according to the manufacturer's instructions.
Flow cytometry.
For cell cycle analysis, 70% confluent cells were labeled with 1 μM BrdU for 30 min, collected by trypsinization, and stained with an anti-BrdU antibody (DAKO) and propidium iodide as previously described (18). For apoptosis assay, 70% confluent MEFs were collected by trypsinization. Caspase 3 activity was measured by using the carboxyfluorescein FLICA apoptosis detection kit (Immunochemistry Technologies) according to the manufacturer's instructions. Analysis was performed on a Becton Dickinson fluorescence-activated cell sorter vantage model with the CellQuest program.
Reverse transcription (RT)-PCR.
Total RNA was isolated from pools of E9.5 hearts by using the Trizol reagent as instructed by the manufacturer. RNA concentration and quality were assessed visually by ethidium bromide-agarose gel electrophoresis (1%) under UV illumination and comparison with known amounts of mouse embryonic total RNA (Ambion). cDNA synthesis was carried out in a final volume of 20 μl of first-strand buffer (Invitrogen) containing around 3 μg of total RNA, 20 U of SuperScript II reverse transcriptase (Invitrogen), 0.025 μg of oligo(dT)/μl, and 0.5 mM concentrations of deoxynucleoside triphosphates. The primers used were as follows: cripto forward primer, 5′-GCTGTCTGAATGGAGG-3′; cripto reverse primer, 5′-AAGGCAGGCGCCAGCTAG-3′; champ forward primer, 5′-TCCTTCTGCGAAATGTG-3′; champ reverse primer, 5′-GAGAGCCTGGAGTTCAG-3′; β-actin forward primer, 5′-CCAACTTGATGTATGAAGGCTTTG-3′; β-actin reverse primer, 5′-GCCTGTACACTGACTTGAGACCAAT-3′. The other primers have been described previously (22). PCR cycles were determined under conditions of linearity by sampling the reactions from 20 to 30 cycles for each primer set. PCR products were visualized by ethidium bromide-agarose gel electrophoresis (1.2%) under UV illumination.
RESULTS
Targeted deletion of mek5 gene causes early embryonic death.
To make the MEK5 mutant mice, a targeting vector was designed to modify the mek5 gene by homologous recombination in ES cells so that exon 2 was interrupted by a neomycin resistance cassette (Fig. 1A). The linearized construct was electroporated into ES cells. The 180 colonies that survived the standard Geneticin (G418) selection were analyzed for homologous recombination by Southern blotting. An example of 1 positive clone is shown in Fig. 1B. Homologous recombination occurred with a frequency of 10%. Chimeric mice were generated from ES cells possessing the targeted allele. These mice were subsequently bred onto the C57BL/6 background to generate heterozygous mek5+/− animals. The genotyping of the mice was performed by two separate PCRs on the genomic DNA extracted from tail snips with specific primers (Fig. 1C).
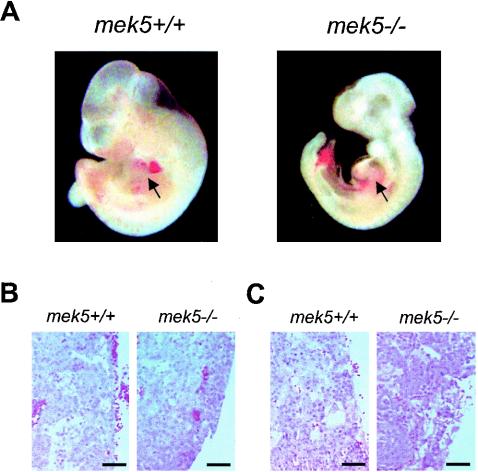
mek5+/− mice appeared to be healthy and were fertile. Genotyping analysis of multiple litters of mek5+/− intercrosses at 3 weeks after birth indicated that no mek5−/− mice were born (Table 1). To determine the age at which mek5−/− embryos die, we performed a timed mating analysis. Our experiments clearly demonstrated that up to E9.5 mek5+/+, mek5+/−, and mek5−/− fetuses were present at the expected Mendelian ratios (Table 1). However, the proportion of mek5−/− embryos started to decline around E10.5, and none could be found beyond E11.5. E10.5 mek5−/− embryos appeared abnormal (Fig. 2A). They were smaller than their wild-type or heterozygous counterparts and displayed retarded development of the head and limbs, and many exhibited dilated pericardial sacs. This abnormal phenotype was strikingly similar to the one described for erk5−/− embryos (31, 34, 42). No marked difference in the extent of vascular invasion and in the general architecture of the placenta was observed between E9.5 and E10.5 wild-type and mutant embryos (Fig. 2B and C). We concluded that a placental defect was unlikely to be the cause for the early lethality displayed by the mek5−/− embryos. This is consistent with studies that report that the phenotype of the erk5−/− embryos is manifest prior to placental abnormality (34, 42). Together, these results indicate that MEK5 is required for normal early embryonic development. The phenotypic similarity with the erk5−/− mice suggests that MEK5 is a critical component of the ERK5 signaling pathway.
TABLE 1.
Genotyping analysis of multiple litters of mek5+/− intercrosses
| Time of gestation | % (no.) of mice with genotypea:
|
Total no. of mice | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| E9.5 | 23 (77) | 51 (174) | 26 (88) | 339 |
| E10.5 | 34 (45) | 50 (67) | 16 (21) | 133 |
| E11.5 | 36 (8) | 59 (13) | 5 (1) | 22 |
| E13.5 | 42 (8) | 58 (11) | 0 (0) | 19 |
| Adult | 35 (131) | 65 (241) | 0 (0) | 372 |
Percentages were determined by PCR. As determined by observing heart beating, 4 of 21 homozygous (−/−) E10.5 embryos were viable. No mek5−/− embryos survived after 11.5 days.
FIG. 2.
Analysis of MEK5-deficient embryos. (A) A lateral view of freshly isolated wild-type (mek5+/+) and homozygous (mek5−/−) E10.5 embryos taken under a light microscope demonstrates retarded development of the head, limbs, and heart in mutant embryos. Arrows point to the heart regions. (B and C) Hematoxylin and eosin staining of sagittal sections of E9.5 (B) and E10.5 (C) mek5+/+ and mek5−/− placentas. Scale bar, 50 μm.
Tissue-specific expression of MEK5 protein in embryos and adult mice.
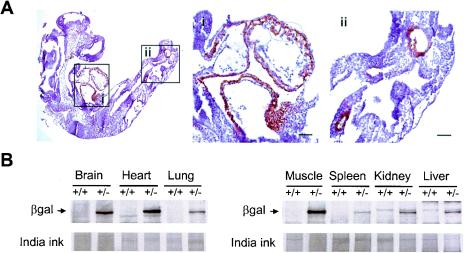
To shed some light on the embryonic lethality caused by the deletion of the mek5 gene and thus increase our knowledge of the biological function of the MEK5 signaling pathway, we examined the pattern of MEK5 protein expression in mice. The targeting vector was designed so that the lacZ gene present in the neomycin cassette inserted in exon 2 was in frame with the starting codon of the mek5 gene. This strategy allowed expression of β-Gal in place of MEK5 in cells where the homologous recombination event had occurred (Fig. 3). β-Gal staining of mutant fetuses demonstrated that MEK5 was specifically expressed in the heart and in two areas in the tip of the tail of E9.5 embryos (Fig. 3A). The areas in the tail region seem to correspond to embryonic blood vessels. A control experiment showed no β-Gal staining of wild-type embryos (data not shown). The level and distribution of MEK5 protein expression in adult mice was assessed by immunoblot analysis of tissues extracted from wild type (+/+) and heterozygous (+/−) mice. The membrane was stained with India ink to monitor protein loading. The experiment showed that MEK5 expression followed by β-Gal expression in +/− tissues was ubiquitous, with the highest levels in the heart, skeletal muscle, and brain tissue (Fig. 3B). One protein with a similar pattern of expression and an apparent molecular weight of 50 kDa was detected with the anti-MEK5 antibody in wild-type extracts (data not shown). The anti-MEK5 antibody did not detect the lowest-molecular-mass MEK5β isoform (40 kDa) that has previously been detected in rat tissues (5). The reason is not yet obvious but could be due to the different methods employed to prepare the extracts. Together with the dilated pericardial sacs exhibited by the mek5−/− embryos, the specific and high levels of expression of MEK5 in both embryonic and adult hearts led us to investigate the effect of MEK5 deletion on cardiac development.
FIG. 3.
Expression of MEK5 in embryos and adult tissues. (A) Whole-mount immunohistochemistry to β-Gal in an E9.5 mek5−/− embryo demonstrates the specific expression of MEK5 at this early stage of development. Enlarged views of the heart (i) and the tips of the tails (ii) are shown. Scale bars, 50 μm. (B) MEK5 expression is highest in brain, heart, and skeletal muscle tissue. Homozygous (+/+) and heterozygous (+/−) adult mouse tissue extracts (50 μg) were analyzed for β-Gal expression by immunoblot analysis with a specific polyclonal antibody to β-Gal. Protein loading was monitored by India ink staining.
mek5−/− embryos display abnormal development of the heart.
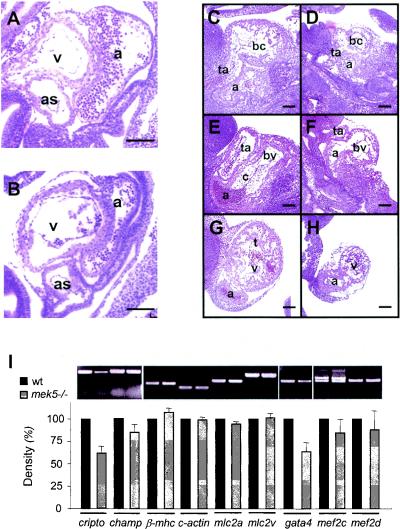
No marked morphological differences were observed between the hearts of wild-type and mek5−/− embryos up to E9.5 (Fig. 4A and B). In contrast, E10.5 mek5−/− embryos displayed striking cardiovascular defects. Histological analysis showed that, at this stage in embryonic development, the hearts of mek5−/− embryos were clearly less well developed than those in their wild-type littermates; they showed severely retarded growth, a lack of septal development, and disrupted trabecula formation (Fig. 4C to H). The spiral septum of the outflow tract, which will form the aortic and pulmonary trunks, were clearly developing in wild-type hearts (Fig. 4C) while mek5−/− embryos displayed little or no septal development in this region of the heart (Fig. 4D). There was also little evidence of the formation of the endocardial cushion tissue in the atrioventricular canal compared to the well-defined endocardial cushions in this region in wild-type littermates (Fig. 4E and F). By E10.5, trabeculation of both the bulbus cordis and the common ventricular chamber was well organized in wild-type mice (Fig. 4C and G) but highly disorganized in mek5−/− mice (Fig. 4D and H).
FIG. 4.
mek5−/− embryos display abnormal development of the heart. Hematoxylin and eosin staining of sagittal sections of E9.5 mek5+/+ (A) and mek5−/− (B) and E10.5 mek5+/+ (C, E, and G) and mek5−/− (D, F, and H) embryos. The spiral septum of the outflow tract displays little or no septal development in mek5−/− embryos (D) compared to wild-type embryos (C). The mutant heart displays little evidence of endocardial cushion tissue formation in the atrioventricular canal (compare panels E and F). The trabeculation of both the bulbus cordis and the common ventricular chamber is highly disorganized in mek5−/− embryos (D and H) compared to wild-type embryos (C and G). Abbreviations: a, common atrial chamber; as, aortic sac; bc, bulbus cordis; bv, bulbo-ventricular canal; c, cushion tissue; t, trabeculae; ta, truncus arteriosus; v, common ventricular chamber. Scale bars, 10 μm. (I) Semiquantitative RT-PCR analysis of cardiac-related transcripts expressed in wild type (+/+) and mutant (−/−) heart embryos. PCR amplifications were performed with cDNAs synthesized from RNA isolated from E9.5 embryonic hearts. PCR products were quantitated under UV illumination. The results normalized to the levels of amplification of β-actin are expressed as the percentage of expression of the gene in wild-type heart embryos. The data correspond to the means ± standard errors of two independent experiments. Abbreviations are as follows: β-mhc, β-myosin heavy chain; c-actin, cardiac α-actin; mlc2, myosin light chain 2.
To identify the molecular mechanism by which MEK5 regulates cardiac development, we compared the levels of expression of a number of cardiac structural transcripts including the MEF2 target genes cripto, champ, cardiac α-actin (c-actin), and myosin light chain 2 (mlc2) in wild-type and mutant E9.5 hearts (19, 21). The expression of the transcription factors GATA4 and MEF2C that are required for normal heart development was also investigated (19, 25). Consistent with the erk5−/− (31) phenotype, RT-PCR analysis revealed a decrease of cripto (40%) and, to a lesser extent, of champ (15%) expression in the mek5−/− hearts compared with the mek5+/+ hearts (Fig. 4I). Absence of MEK5 expression also significantly inhibited (35%) the levels of the gata4 transcript. In contrast, MEK5 was not required for normal expression of the β-myosin heavy chain (β-mhc), c-actin, mlc2a, mlc2v, mef2c, or mef2d (Fig. 4I).
These results indicate that MEK5 is required for normal cardiac development. Similar cardiovascular defects displayed by the erk5−/− mice suggest that the physiological role of MEK5 is mediated by its ability to regulate ERK5 activity. The functional consequence of the defect in cripto, champ, and gata4 expression in the mek5−/− embryos remains to be identified.
Effect of MEK5 deletion on cell growth and cell survival.
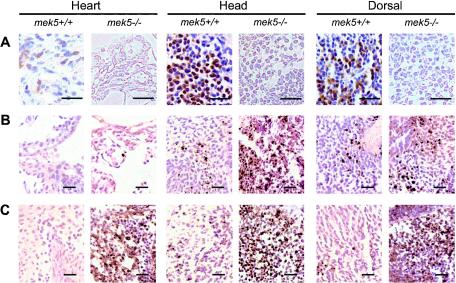
Consistent with its role in stimulating early gene expression through the regulation of MEF2 and AP-1 activity, ERK5 has been shown to contribute to cell proliferation and the survival response of neurons in the brain (16, 20, 33, 39). We examined whether MEK5 was required for mediating these biological processes in vivo. Freshly collected E9.5 and E10.5 embryos were processed for immunohistochemistry to detect cell proliferation by BrdU incorporation (Fig. 5A) or DNA fragmentation, one of the hallmarks of late stage apoptosis, by TUNEL staining (Fig. 5B and C). The heart, head, and dorsal regions of the mutant E10.5 embryos exhibited a marked decrease in proliferation and an increase in apoptosis compared to their wild-type littermates (Fig. 5A and C). Increased apoptosis in mek5−/− embryos was observed as early as E9.5, suggesting that the proliferation defect may be a consequence of abnormal cell survival in the mutant fetuses (Fig. 5B). To further test this hypothesis, we examined the effect of MEK5 deletion on the ability of MEFs to progress through the cell cycle and to undergo apoptosis (Fig. 6).
FIG. 5.
mek5−/− embryos exhibit defects in cell proliferation and cell death. Sections of the heart, head, and dorsal regions stained with BrdU (A) reveal decreased cell proliferation in homozygous (mek5−/−) compared to wild-type (mek5+/+) E10.5 embryos. TUNEL staining indicates increased apoptosis in the heart, head, and dorsal regions of the mek5−/− E9.5 (B) and E10.5 (C) embryos. Scale bars, 25 μm.
FIG. 6.
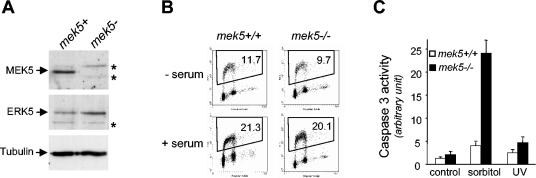
MEK5 is implicated in cell survival. (A) MEF extracts (50 μg) were analyzed for ΜΕΚ5 and ERK5 expression by immunoblot analysis with specific polyclonal anti-MEK5 and anti-ERK5 antibodies. The detection of tubulin expression was performed to monitor protein loading. Asterisks indicate nonspecific bands. (B) Seventy percent confluent MEFs were serum starved for 48 h prior to being stimulated with 10% fetal bovine serum for 24 h. Cell proliferation was assessed by fluorescence-activated cell sorter analysis to follow BrdU incorporation. The percentage of fibroblasts present in the S phase of the cell cycle is indicated. The figure shows data representative of the results from three independent experiments. − serum, without serum; + serum, with serum. (C) Wild-type and mek5−/− fibroblasts were incubated for 6 h with 500 mM sorbitol or exposed to UV radiation (60 J/m2) followed by incubation for 16 h. Caspase 3 activity was monitored by using the carboxyfluorescein FLICA apoptosis detection kit. The data, expressed as units of fluorescence, correspond to the means ± standard errors of the results from two independent experiments.
Immunoblot analysis using an anti-MEK5 antibody confirmed the absence of MEK5 expression in homozygous MEFs (Fig. 6A). The level of ERK5 expression was not significantly affected by the targeted deletion of the mek5 gene. The results showed that following 48 h of starvation, the readdition of 10% serum for 24 h stimulated the reentry of mek5−/− MEFs into the cell cycle similar to the wild-type cells (Fig. 6B). In contrast, mek5−/− MEFs exhibited increased levels of sorbitol-induced caspase 3 activity compared to wild-type cells (Fig. 6C). The protective role of MEK5 against the toxic effect of sorbitol was specific, since the absence of MEK5 expression did not significantly affect the ability of UV radiation to activate caspase 3 (Fig. 6C). UV does not activate ERK5 in MEFs (data not shown). These data confirm our hypothesis that MEK5 is not essential for normal cell cycle progression but is a critical component of the survival signaling pathways in mitotic cells.
MEK5 is required for mediating ERK5 activation and MEF2-dependent transcription.
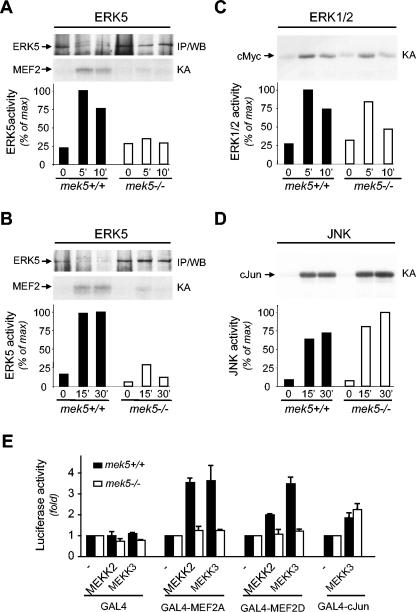
In vitro protein kinase assays and transfection studies with a constitutively activated mutant of MEK5 have demonstrated that MEK5 is a potent activator of ERK5 (45). However, the in vivo role of MEK5 in the ERK5 signaling pathway needs to be further established, as additional ERK5 activators may exist. To determine the physiological significance of MEK5 as an ERK5 activator, we examined the effect of mek5 gene elimination on ERK5 activation in MEFs in response to mitogenic and stress stimuli (Fig. 7). Epidermal growth factor (EGF) and sorbitol treatment caused a marked increase in ERK5 protein kinase activity in wild-type but not in mek5−/− MEFs (Fig. 7A and B). A control experiment showed that the residual phosphorylation of GST-MEF2C in knockout mek5 fibroblast extracts was independent of the presence of the anti-ERK5 antibody in the protein kinase assay. Consistent with the absence of ERK5 activation in the mek5−/− MEFs, immunoblot analysis of the immune complexes demonstrated that MEK5 was required for mediating ERK5 phosphorylation. Wild-type extracts displayed retarded migration of ERK5 following SDS-PAGE analysis, and this was absent from homozygous knockout mek5 extracts (Fig. 7A and B). Control experiments showed similar activation of ERK1/2 (Fig. 7C) and JNK (Fig. 7D) in mek5−/− compared to wild-type MEFs in response to EGF and sorbitol treatment, respectively.
FIG. 7.
Disruption of the mek5 gene prevents ERK5 activation and the transcriptional regulation of MEF2 factors. Wild type (+/+) and homozygous knockout (−/−) mek5 MEFs were treated with EGF (50 ng/ml) (A, C) or sorbitol (300 mM) (B, D) for the times indicated (in minutes). Endogenous ERK5 (A, B), ERK1/2 (C), and JNK (D) activity was measured by protein kinase assay (KA) in the presence of [γ-32P]ATP. The radioactivity incorporated into GST-MEF2C, GST-cMyc, or GST-cJun was quantitated after SDS-PAGE by PhosphorImager analysis. The presence of ERK5 in the immune complexes was detected by immunoblot analysis (IP/WB). Data representative of the results from three independent experiments are shown. (E) Fibroblasts were transiently transfected as described in Materials and Methods. MEF2 and cJun transcriptional activity was measured by the dual-luciferase reporter assay system. Firefly luciferase activity was normalized to that of Renilla luciferase and expressed as change relative to the control (−). The data correspond to the means ± standard errors of three independent experiments performed in duplicate.
To confirm the requirement of MEK5 in regulating ERK5 protein kinase activity, we investigated the effect of MEK5 deletion on the transcriptional regulation of MEF2, which is a well-characterized substrate of ERK5 (13, 15). Fibroblasts were cotransfected with the reporter plasmid pG5E1bLuc together with constructs encoding GAL4, GAL4-MEF2A, or GAL4-MEF2D, with or without expression vectors encoding MEKK2 or MEKK3. A control experiment was performed with GAL4-cJun, since cJun is not a direct downstream substrate of ERK5. MEKK2- and MEKK3-induced MEF2A, MEF2D, and cJun transcriptional activity was determined by the luciferase reporter assay. The results demonstrated that the absence of MEK5 protein expression prevented both MEKK2 and MEKK3 from increasing the transcriptional activity of MEF2 (Fig. 7E). In contrast, no marked differences in the activation of cJun were detected under these conditions. Consistent with in vitro evidence that MEK5 is upstream of ERK5, these data provide clear genetic evidence that MEK5 is the only ERK5 activator in cells.
DISCUSSION
The lack of available pharmacological and genetic reagents that specifically alter MEK5 activity has prevented further progress into the understanding of the role of MEK5 in vivo. We have addressed this issue by engineering a novel genetically modified mouse model deficient in MEK5 expression.
The mek5−/− embryos exhibited heart defects, including the lack of septal development and disrupted trabecula formation. Similar phenotypic abnormalities displayed by the erk5−/−, mef2c−/−, and mekk3−/− embryos identify MEK5 as an essential physiological component of the MEKK3/ERK5/MEF2 cascade (19, 31, 34, 42, 43). In vitro, the ERK5 signaling pathway has been shown to protect endothelial cells (EC) from apoptosis by phosphorylating Bad (29). Consistent with this study, the analysis of mutant mice in which the erk5 gene can be conditionally deleted suggests that the requirement of ERK5 for the survival of EC is responsible for the cardiovascular defect observed in erk5−/− and mek5−/− embryos (8). Therefore, decreased cripto, champ, and gata4 expression associated with impaired differentiation of cardiomyoblasts may be a consequence rather than a cause for the abnormal development of the mek5−/− heart.
In addition to protecting EC, ERK5 has been shown to promote neuronal survival (20, 33, 39). Blocking ERK5 activation by expressing dominant-negative MEK5 increased the death of neurons that were supported only by neurotrophin stimulation of distal axons (39). Based on these studies, we hypothesized that, similar to the heart defect, the retarded development of the brain of mek5−/− embryos (Fig. 2A) was caused by increased cellular apoptosis. MEK5, like ERK5, is expressed in the embryonic brain but at a level barely detectable by β-Gal staining (20, 33, 39) (Fig. 3A). Our results confirmed that E9.5 and E10.5 mek5−/− embryos displayed a marked increase of cell death in the brain compared to their wild-type littermates (Fig. 5B and C). Consistent with this study, we showed that MEK5 was essential for mediating the caspase-dependent apoptotic response of mitotic cells (Fig. 6C). Analysis of a genetically modified mouse model in which the mek5 gene can be conditionally deleted is necessary to further address in vivo the contribution of the MEK5 survival pathway for the normal development of the brain.
In contrast to apoptosis, our results showed no marked difference in the ability of mek5−/− compared to wild-type fibroblasts to progress through the S phase (Fig. 6B). This suggests that the cell proliferation defect observed in different areas of the mek5−/− embryos is a secondary effect of increased apoptosis in the mutant embryos (Fig. 5 and 6). However, it is important to emphasize that, although our study indicates that MEK5 is not a general regulator of the cell cycle, it does not rule out the possibility that the MEK5/ERK5 signaling pathway may be important for promoting or regulating the proliferation of certain cell types, such as cancer cells. Indeed, our work is based on primary cultures of fibroblasts, but studies with immortalized cell lines have provided evidence for a role of the ERK5 signaling pathway in mediating mitogen-induced cell cycle progression (16). This conclusion supports the idea that MEK5 may be implicated in mediating the transforming effect of oncogenes such as Ras, Src, Erb2, and Cot. Consistent with a role of MEK5 in oncogenic signaling, it is reported that foci induced by a dominant active mutant of Raf (Raf-BxB) are decreased in number by disruption of MEK5 function (7). More recently, MEK5 overexpression has been demonstrated to be associated with metastatic prostate cancer (23).
The control of gene expression is one mechanism by which the ERK5 signaling pathway regulates cell function. For example, ERK5-dependent regulation of the transcriptional activity of MEF2 has been implicated in the promotion of smooth muscle cell differentiation and neuronal survival (4, 20, 33, 39). The MEF2 family belongs to the group of MADS (MCM1, agamous, deficiens, serum response factor) box transcription factors. Their transcriptional activity is regulated by phosphorylation by various protein kinases. These include the ERK5 and p38 MAPKs that phosphorylate both MEF2A and MEF2C (13, 15, 27, 44). In contrast, MEF2D is a specific substrate of ERK5 (44). Our results confirmed that MEK5 was required for regulating the transcriptional activity of MEF2D (Fig. 7E). They also demonstrated that, although both ERK5 and p38 MAPK regulate MEF2A activity, expression of p38 MAPK was not sufficient to compensate for the inability of the MEK kinases MEKK2 and MEKK3 to increase MEF2A activity in mek5−/− fibroblasts. MEKK2 and MEKK3 have both been shown to increase p38 MAPK activity in cells (3). Together, these data indicate that the requirement of the ERK5 and p38 MAPK signaling pathways in the regulation of gene expression via MEF2 factors may be cell type specific. The functional consequence of MEF2 regulation by the MEK5/ERK5 cascade during myogenesis and brain development is yet to be determined.
Overall, our results provide the first genetic evidence that MEK5 is a critical component of the ERK5/MEF2 signaling pathway that is required for mediating normal heart development. Disruption of the mek5 gene causes increased cell death in embryos and fibroblasts, indicating that MEK5 is required for the regulation of cell survival.
Acknowledgments
We are indebted to A. Nagy for kindly providing the R1 ES cells and to H.-Y. Kao and C. Widmann for generosity in providing plasmids. We thank L. Knowles for blastocyst injections, G. Morrissey and A. Robinson for animal facility maintenance, M. Jackson for flow cytometry analysis, and A. Whitmarsh for critically reviewing the manuscript.
This work was supported in part by the MRC and the AICR and principally by the BBSRC, the Royal Society, and a Lister Institute of Preventive Medicine Research Fellowship to C.T.
REFERENCES
- 1.Abe, J., M. Kusuhara, R. J. Ulevitch, B. C. Berk, and J.-D. Lee. 1996. Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J. Biol. Chem. 271:16586-16590. [DOI] [PubMed] [Google Scholar]
- 2.Chao, T.-H., M. Hayashi, R. I. Tapping, Y. Kato, and J.-D. Lee. 1999. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J. Biol. Chem. 274:36035-36038. [DOI] [PubMed] [Google Scholar]
- 3.Deacon, K., and J. L. Blank. 1999. MEK kinase 3 directly activates MKK6 and MKK7, specific activators of the p38 and c-Jun NH2-terminal kinases. J. Biol. Chem. 274:16604-16610. [DOI] [PubMed] [Google Scholar]
- 4.Dinev, D., B. W. M. Jordan, B. Neufeld, J.-D. Lee, D. Lindemann, U. R. Rapp, and S. Ludwig. 2001. Extracellular signal regulated kinase 5 (ERK5) is required for the differentiation of muscle cells. EMBO Rep. 2:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.English, J. M., C. A. Vanderbilt, S. Xu, S. Marcus, and M. H. Cobb. 1995. Isolation of MEK5 and differential expression of alternatively spliced forms. J. Biol. Chem. 270:28897-28902. [DOI] [PubMed] [Google Scholar]
- 6.English, J. M., G. Pearson, R. Baer, and M. H. Cobb. 1998. Identification of substrates and regulators of the mitogen-activated protein kinase ERK5 using chimeric protein kinases. J. Biol. Chem. 273:3854-3860. [DOI] [PubMed] [Google Scholar]
- 7.English, J. M., G. Pearson, T. Hockenberry, L. Shivakumar, M. A. White, and M. H. Cobb. 1999. Contribution of the ERK5/MEK5 pathway to Ras/Raf signaling and growth control. J. Biol. Chem. 274:31588-31592. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi, M., S. W. Kim, K. Imanaka-Yoshida, T. Yoshida, E. D. Abel, B. Eliceiri, Y. Yang, R. J. Ulevitch, J. D. Lee. 2004. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J. Clin. Investig. 113:1138-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, G. L., and R. Lapadat. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911-1912. [DOI] [PubMed] [Google Scholar]
- 10.Kallunki, T., B. Su, I. Tsigelny, H. K. Sluss, B. Dérijard, G. Moore, R. J. Davis, and M. Karin. 1994. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 8:2996-3007. [DOI] [PubMed] [Google Scholar]
- 11.Kamakura, S., T. Moriguchi, and E. Nishida. 1999. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J. Biol. Chem. 274:26563-26571. [DOI] [PubMed] [Google Scholar]
- 12.Kasler, H. G., J. Victoria, O. Duramad, and A. Winoto. 2000. ERK5 is a novel type of mitogen-activated protein kinase containing a transcriptional activation domain. Mol. Cell. Biol. 20:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, Y., V. Kravchenko, R. I. Tapping, J. Han, R. J. Ulevitch, and J.-D. Lee. 1997. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16:7054-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, Y., R. I. Tapping, S. Huang, M. H. Watson, R. J. Ulevitch, and J.-D. Lee. 1998. Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature 395:713-716. [DOI] [PubMed] [Google Scholar]
- 15.Kato, Y., M. Zhao, A. Morikawa, T. Sugiyama, D. Chakravortty, N. Koide, T. Yoshida, R. I. Tapping, Y. Yang, T. Yokochi, and J.-D. Lee. 2000. Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem. 275:18534-18540. [DOI] [PubMed] [Google Scholar]
- 16.Kato, Y., T. H. Chao, M. Hayashi, R. I. Tapping, and J.-D. Lee. 2000. Role of BMK1 in regulation of growth factor-induced cellular responses. Immunol. Res. 21:233-237. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, M. H. 1990. Morphological stages of postimplantation embryonic development, p. 81-91. In A. J. Copp and D. L. Cockroft (ed.), Postimplantation mammalian embryos: a practical approach. IRL Press, Oxford, United Kingdom.
- 18.Khochbin, S., A. Chabanas, P. Albert, J. Albert, and J. J. Lawrence. 1988. Application of bromodeoxyuridine incorporation measurements to the determination of cell distribution within the S phase of the cell cycle. Cytometry 9:499-503. [DOI] [PubMed] [Google Scholar]
- 19.Lin, Q., J. Schwartz, C. Bucana, and E. N. Olson. 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, L., J. E. Cavanaugh, Y. Wang, H. Sakagami, Z. Mao, and Z. Xia. 2003. ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc. Natl. Acad. Sci. USA 100:8532-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, Z. P., O. Nakagawa, M. Nakagawa, H. Yanagisawa, R. Passier, J. A. Richardson, D. Srivastava, and E. N. Olson. 2001. CHAMP, a novel cardiac-specific helicase regulated by MEF2C. Dev. Biol. 234:497-509. [DOI] [PubMed] [Google Scholar]
- 22.Makino, S., K. Fukuda, S. Miyoshi, F. Konishi, H. Kodama, J. Pan, M. Sano, T. Takahashi, S. Hori, H. Abe, J.-I. Hata, A. Umezawa, and S. Ogawa. 1999. Cardiomyocytes can be generated from marrow stromal cells in vitro. J. Clin. Investig. 103:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehta, P. B., B. L. Jenkins, L. McCarthy, L. Thilak, C. N. Robson, D. E. Neal, and H. Y. Leung. 2003. MEK5 overexpression is associated with metastatic prostate cancer, and stimulates proliferation, MMP-9 expression and invasion. Oncogene 22:1381-1389. [DOI] [PubMed] [Google Scholar]
- 24.Mody, N., J. Leitch, C. Armstrong, J. Dixon, and P. Cohen. 2001. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 502:21-24. [DOI] [PubMed] [Google Scholar]
- 25.Molkentin, J. D., Q. Lin, S. A. Duncan, and E. N. Olson. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11:1061-1072. [DOI] [PubMed] [Google Scholar]
- 26.Nicol, R. L., N. Frey, G. Pearson, M. Cobb, J. Richardson, and E. N. Olson. 2001. Activated MEK5 induces serial assembly of sarcomeres and eccentric cardiac hypertrophy. EMBO J. 20:2757-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ornatsky, O. I., D. M. Cox, P. Tangirala, J. J. Andreucci, Z. A. Quinn, J. L. Wrana, R. Prywes, Y.-T. Yu, and J. C. McDermott. 1999. Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res. 27:2646-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson, G., J. M. English, M. A. White, and M. H. Cobb. 2001. ERK5 and ERK2 cooperate to regulate NF-kappaB and cell transformation. J. Biol. Chem. 276:7927-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pi, X., C. Yan, and B. C. Berk. 2004. Big mitogen-activated protein kinase (BMK1)/ERK5 protects endothelial cells from apoptosis. Circ. Res. 94:362-369. [DOI] [PubMed] [Google Scholar]
- 30.Potten, C. S., G. Owen, and D. Booth. 2002. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci. 115:2381-2388. [DOI] [PubMed] [Google Scholar]
- 31.Regan, C. P., W. Li, D. M. Boucher, S. Spatz, M. S. Su, and K. Kuida. 2002. Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc. Natl. Acad. Sci. USA 99:9248-9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seth, A., F. A. Gonzalez, S. Gupta, D. L. Raden, and R. J. Davis. 1992. Signal transduction within the nucleus by mitogen-activated protein kinase. J. Biol. Chem. 267:24796-24804. [PubMed] [Google Scholar]
- 33.Shalizi, A., M. Lehtinen, B. Gaudilliere, N. Donovan, J. Han, Y. Konishi, and A. Bonni. 2003. Characterization of a neurotrophin signaling mechanism that mediates neuron survival in a temporally specific pattern. J. Neurosci. 23:7326-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sohn, S. J., B. K. Sarvis, D. Cado, and A. Winoto. 2002. ERK5 MAPK regulates embryonic angiogenesis and acts as a hypoxia-sensitive repressor of vascular endothelial growth factor expression. J. Biol. Chem. 277:43344-43351. [DOI] [PubMed] [Google Scholar]
- 35.Sun, W., K. Kesavan, B. C. Schaefer, T. P. Garrington, M. Ware, N. Johnson, E. W. Gelfand, and G. L. Johnson. 2001. MEKK2 associates with the adapter protein Lad/RIBP and regulates the MEK5-BMK1/ERK5 pathway. J. Biol. Chem. 276:5093-5100. [DOI] [PubMed] [Google Scholar]
- 36.Talts, J. F., C. Brakebusch, and R. Fassler. 1999. Integrin gene targeting. Methods Mol. Biol. 129:153-187. [DOI] [PubMed] [Google Scholar]
- 37.Terasawa, K., K. Okazaki, and E. Nishida. 2003. Regulation of c-Fos and Fra-1 by the MEK5-ERK5 pathway. Genes Cells 8:263-273. [DOI] [PubMed] [Google Scholar]
- 38.van Drogen, F., and M. Peter. 2002. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 12:R53-R55. [DOI] [PubMed] [Google Scholar]
- 39.Watson, F. L., H. M. Heerssen, A. Bhattacharyya, L. Klesse, M. Z. Lin, and R. A. Segal. 2001. Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat. Neurosci. 4:981-988. [DOI] [PubMed] [Google Scholar]
- 40.Xia, Y., Z. Wu, B. Su, B. Murray, and M. Karin. 1998. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 12:3369-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan, C., M. Takahashi, M. Okuda, J.-D. Lee, and B. C. Berk. 1999. Fluid shear stress stimulates big mitogen-activated protein kinase 1 (BMK1) activity in endothelial cells. Dependence on tyrosine kinases and intracellular calcium. J. Biol. Chem. 274:143-150. [DOI] [PubMed] [Google Scholar]
- 42.Yan, L., J. Carr, P. R. Ashby, V. Murry-Tait, C. Thompson, and J. S. C. Arthur. 2003. Knockout of ERK5 causes multiple defects in placental and embryonic development. BMC Dev. Biol. 3:11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, J., M. Boerm, M. McCarty, C. Bucana, I. J. Fidler, Y. Zhuang, and B. Su. 2000. Mekk3 is essential for early embryonic cardiovascular development. Nat. Genet. 24:309-313. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, M., L. New, V. V. Kravchenko, Y. Kato, H. Gram, F. Di Padova, E. N. Olson, R. J. Ulevitch, and J. Han. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, G., Z. Q. Bao, and J. E. Dixon. 1995. Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 270:12665-12669. [DOI] [PubMed] [Google Scholar]