Abstract
Background
During primitive hematopoiesis in Xenopus, cebpa and spib expressing myeloid cells emerge from the anterior ventral blood island. Primitive myeloid cells migrate throughout the embryo and are critical for immunity, healing, and development. Although definitive hematopoiesis has been studied extensively, molecular mechanisms leading to the migration of primitive myelocytes remain poorly understood. We hypothesized these cells have specific extracellular matrix modifying and cell motility gene expression.
Results
In situ hybridization screens of transcripts expressed in Xenopus foregut mesendoderm at stage 23 identified seven genes with restricted expression in primitive myeloid cells: destrin; coronin actin binding protein, 1a; formin-like 1; ADAM metallopeptidase domain 28; cathepsin S; tissue inhibitor of metalloproteinase-1; and protein tyrosine phosphatase nonreceptor 6. A detailed in situ hybridization analysis revealed these genes are initially expressed in the aVBI but become dispersed throughout the embryo as the primitive myeloid cells become migratory, similar to known myeloid markers. Morpholino-mediated loss-of-function and mRNA-mediated gain-of-function studies revealed the identified genes are downstream of Spib.a and Cebpa, key transcriptional regulators of the myeloid lineage.
Conclusions
We have identified genes specifically expressed in migratory primitive myeloid progenitors, providing tools to study how different gene networks operate in these primitive myelocytes during development and immunity.
Keywords: myeloid, Xenopus, migrating, spib, CCAAT/enhancer binding protein a(cebpa), actin, destrin (dstn), coronin, actin binding protein 1a (coro1a), formin-like 1 (fmnl1), ADAM metallopeptidase domain 28 (adam28), cathepsin S (ctss), tissue inhibitor of metalloproteinase-1 (timp-1), protein tyrosine phosphatase non-receptor 6 (ptpn6)
Introduction
The innate immune system is evolutionarily ancient and is specified before the adaptive immune system (Nagata, 1977; Tochinai, 1980; Kau and Turpen, 1983; Maeno et al., 1985). Myeloid cells are capable of protecting embryos from 98% of all pathogens encountered (Jones, 2000) and develop in 2 distinct temporal waves. The first wave called primitive hematopoiesis often occurs in blood islands and results in a transient population of erythroid and myeloid cells (Costa et al., 2008; Chen et al., 2009b). The second wave, known as definitive hematopoiesis, produces hematopoietic progenitors that ultimately provide all adult blood lineages. Though much is known about the latter wave in higher vertebrates, the former remains poorly understood in all vertebrates.
Research in the lower vertebrates, zebrafish and Xenopus, has revealed that primitive hematopoiesis occurs in 2 separate embryonic compartments (Palis et al., 1999; McGrath and Palis, 2005; Lux et al., 2008). Myelopoiesis occurs in the Xenopus anterior blood island (rostral blood island derived from the anterior lateral plate mesoderm in zebrafish) while erythropoiesis occurs in the posterior ventral blood island in Xenopus (posterior lateral plate mesoderm in zebrafish) (Warga et al., 2009; Ciau-Uitz et al., 2010; Ciau-Uitz et al., 2014).
Primitive myeloid cells are the first blood cells to differentiate and become functional in the Xenopus embryo and along with neural crest are some of the earliest migratory cells. A critical function of primitive myeloid cells is their ability to move within and between tissues where they are quickly and efficiently recruited to sites such as embryonic wounds even before a functional vasculature is established (Chen et al., 2009b). Myeloid cells have been implicated in diverse contexts of organ repair and regeneration among higher vertebrates: skin (Mirza et al., 2009; Goren et al., 2009) where their depletion results in delayed re-epithelialization, reduced collagen deposition, impaired angiogenesis, and decreased cell proliferation in healing wounds; muscle where two populations of monocytes sequentially phagocytose then accumulate myofibroblasts, promote angiogenesis, and deposit collagen (Nahrendorf et al., 2007; Arnold et al., 2007); kidney where wnt7b is produced by macrophages which invade the injured tissues and reestablish a developmental program beneficial for repair and regeneration (Lin et al., 2010), liver where macrophages play critical roles in both the injury and recovery phases of inflammatory scarring (Takeishi et al., 1999; Meijer et al., 2000; Duffield et al., 2005), and colon where macrophages migrate to a wound and promote epithelial proliferation at the injury site (Pull et al., 2005; Seno et al., 2009). Genes conferring myeloid cell motility, repair, and regeneration functions remain to be identified in all vertebrates.
More recent research suggests that myeloid cells are also likely to have important functions during normal embryogenesis (Rae et al., 2007; Stefater et al., 2011). In Drosophila, haemocytic macrophages phagocytose apoptotic cells from the developing embryo (Tepass et al., 1994; Franc et al., 1999). Also, macrophages actively induce apoptosis of endothelial cells in the pupillary membrane of the developing mammalian eye (Diez-Roux et al., 1999). Of interest, homozygous null mutation of Colony stimulating factor 1 (Csf1), effectively ablating macrophages from the mouse embryo, results in a major insulin mass deficit in fetuses and adults, abnormal postnatal islet morphogenesis, and impaired pancreatic cell proliferation at weaning and late pregnancy (Banaei-Bouchareb et al., 2004). Most recently migratory primitive myeloid cells have been reported to be essential for normal Xenopus cardiac morphogenesis (Smith and Mohun, 2011). These findings suggest that myeloid cells play important roles during normal embryonic development (Savill and Fadok, 2000). Exactly what functions within the myeloid cells confer such developmental roles has proved difficult to examine in mouse and higher vertebrates because few molecular markers are available either to identify embryonic primitive myeloid cells or to trace their ontogeny.
The earliest known markers of the primitive myeloid lineage in Xenopus include cebpa and spib.a transcripts. Cebpa is a basic helix-loop-helix transcription factor critical for the differentiation of murine myeloid progenitors into granulocytemonocyte progenitors (Zhang et al., 2004). cebpa mutations are often found in human patients with myeloid leukemias (Nerlov, 2004; Mueller and Pabst, 2006). Gain- and loss-of-function studies reveal that cebp a is necessary and sufficient for myeloid differentiation in Xenopus embryos (Chen et al., 2009b). spib.a encodes an ETS domain transcription factors that marks the primitive myeloid cell lineage in Xenopus and is required for its development where it acts upstream of spi1 (also known as pu.1) in the molecular hierarchy of primitive myeloid development (Costa et al., 2008).
Although some of the target genes regulated by these key myeloid transcription factors have been identified (Smith and Mohun, 2011), in general the downstream genetic program activated in primitive myeloid cells that confer their ability to migrate and mediate immunity are still poorly understood (Smith and Mohun, 2011). This study identified a group of genes expressed in the early primitive myeloid lineage of Xenopus embryos. The temporal and spatial expression patterns suggest they emerge after myeloid specification and with the onset of migratory activity. We show that the expression of these genes is regulated by Spib.a and Cebpa. These genes encode proteins that are implicated in mediating different aspects of myeloid cell migration and should facilitate elucidating the cell biology underlying the essential developmental and immunologic functions of the migrating primitive myeloid lineage in the Xenopus embryo.
Results and Discussion
Developmental Expression of Primitive Myeloid Genes From Microarrays
In a previously published microarray experiment we identified several hundred genes expressed in early foregut (Stage 23) of Xenopus embryos (Kenny et al., 2012), GSE38654. In the course of validation by in situ hybridization we identified seven genes with punctate expression within the mesodermal layer of the anterior ventral blood island, reminiscent of primitive myeloid cells (Fig. 1). These genes included destrin (dstn); coronin, actin binding protein, 1a (coro1a); formin-like 1 (fmnl1); ADAM metallopeptidase domain 28 (adam28); cathepsin S (ctss); tissue inhibitor of metalloproteinase-1 (timp-1); and protein tyrosine phosphatase nonreceptor 6 (ptpn6). Of interest, research in mammals has suggested a role for many of these genes in adult myeloid cells (Yan et al., 2005; Shimoda et al., 2007; Mersich et al., 2010; Seto et al., 2012; Arora et al., 2012; Rochelle et al., 2013; Zajac et al., 2013; McComb et al., 2014; Masmas et al., 2014). However whether or not these genes are expressed in embryonic primitive hematopoiesis-derived myeloid cells has not previously been documented in any vertebrate species.
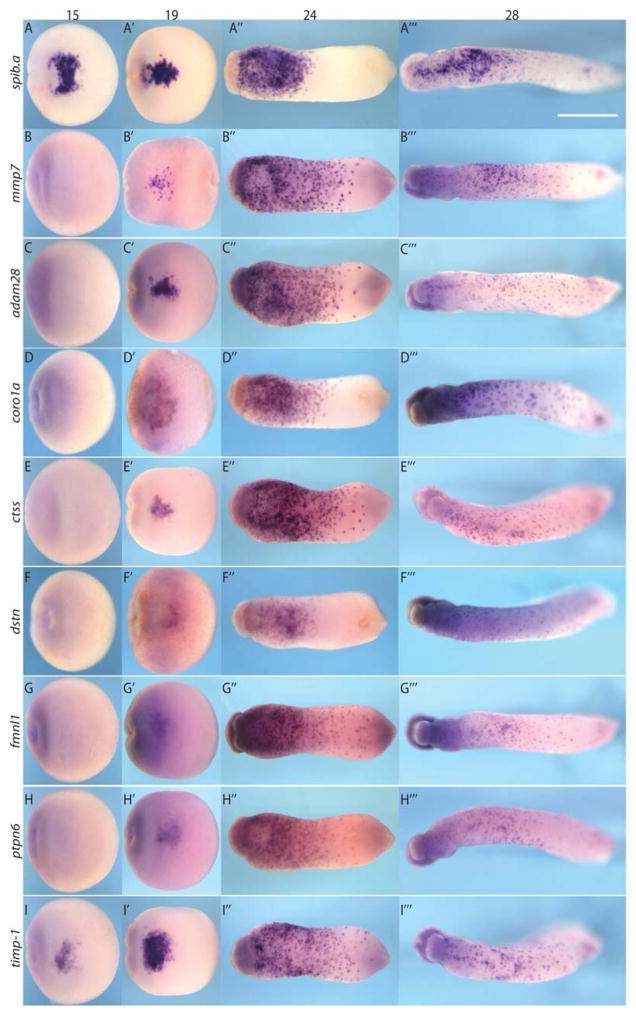
Fig. 1.
Stage 15, 19, 24, and 28 expression of identified genes progress from anterior ventral blood island to dispersed distribution. They are temporally downstream of transcriptional regulators spib.a (A–A‴ ) but coincident with other, later expressing myeloid functional genes like mmp7 (B–B‴ ). C–C‴ , adam28. D–D‴ , coro1a. E–E‴ , ctss. F–F‴, dstn. G–G‴ , fmnl1. H–H‴ , ptpn6. I–I‴ , timp-1. White scale bar = 1 mm.
To test whether these genes are expressed in the primitive myeloid lineage, we compared their developmental expression with spib.a, and its downstream target mmp7, which are known to be specifically expressed in the primitive myeloid cells during stages 15 through 28 (Costa et al., 2008). Spib.a preceded the expression of these genes in the anterior ventral blood island (aVBI) at stage 15 (presomitic stage) with the exception of the emerging expression of timp-1 (Nieuwkoop and Faber, 1994) (Fig. 1A–I). All seven genes exhibited an expression profile very similar to spib.a, although they were less abundant compared with spib.a before stage 24. At stage 32, all genes tested were dispersed throughout the embryo excluding the area of the branchial arches (data not shown), a phenomenon observed in previous primitive myeloid gene expression (Ciau-Uitz et al., 2010). The in situ analysis suggested that the seven genes were expressed in subsets of the spib.a+ cells and that expression between the genes appears nonoverlapping, possibly representing different cell subpopulations within the spib.a-expressing myeloid lineage (Fig. 1 and data not shown). Thus, overall the seven genes that we identified were expressed in the anterior ventral blood island in a pattern very similar to other known primitive myeloid marker, but in general they were expressed later than spib.a and cebpa (data not shown) which is consistent with them being myeloid-specific genes temporally downstream of spib.a.
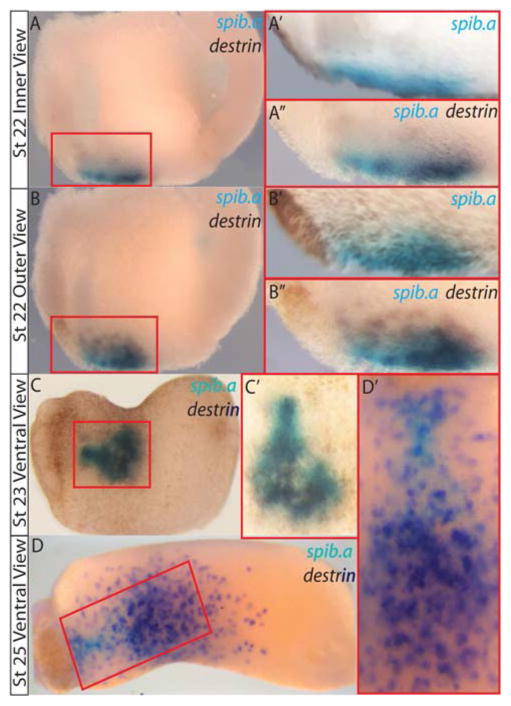
We further examined the myeloid-specific co-expression of spib.a and one of the identified genes, destrin (Fig. 2). Double whole-mount in situ hybridization (WMISH) at stage 22 revealed significant overlap of expression for these two genes (Fig. 2A–C). Later stage 25 WMISH revealed spib.a(−) destrin(+) cells migrating posteriorly and laterally out of the blood island (Fig. 2D). This is consistent with gene expression coinciding with increasing myeloid cell motility and migration away from the anterior ventral blood island. An alternative explanation of this data is the possibility of independent sequential transient waves of expression.
Fig. 2.
Sequential double in situ hybridization reveals target gene expression corresponds with increased myeloid cell motility. A: Stage 22 inner view. A′ , spib.a only (BCIP blue) initial in situ. A″ : follow-up image after second probe for destrin reveals significant overlap of two genes. B–B″ : outer view of same embryo. C and D, increased motility in destrin-expressing cells compared with spib.a-expressing cells (C′ and D′ , ventral views).
Gene Ontology Analysis Shows Role in Primitive Myeloid Activities of Cell Motility and Extracellular Matrix
Using gene ontology analysis we assessed the predicted molecular and cellular functions of the genes we isolated based on information from their human and mouse orthologs (Table 1) (on Web site ToppGene.cchmc.org; Table 1) and by literature review. Molecular function analysis revealed DSTN, CORO1A, FMNL1 are involved in actin binding and that ADAM28 and CTSS have endopeptidase activity. The biological process analysis revealed that actin filament severing activity was mediated through DSTN and FMNL1; also extracellular matrix disassembly activity was mediated through CTSS and TIMP-1. Furthermore, ToppGene coexpression analysis has revealed these genes to be expressed in blood development and cancer (Table 1).
TABLE 1.
Gene ontology analysis reveals that identified genes have potential myeloid cell activity
| dstn | corola | adam28 | ctss | fmnll | timp-1 | ptpn6 | ||
|---|---|---|---|---|---|---|---|---|
| Molecular Function | Actin Binding | X | X | X | ||||
| Endopeptidase Activity | X | X | ||||||
| Biological Process | Actin Filament Severing | X | X | |||||
| Extracellular Matrix Disassembly | X | X | ||||||
| Coexpression | Human Leukemia | X | X | X | X | X | X | |
| B lymphocyte Development | X | X | ||||||
| Human Immune Genes | X | X | X | X | ||||
| Differentiating Bipotential Myeloid Cell | X | X |
DSTN is an actin-depolymerizing factor present in actin micro-filaments and can break down actin filaments (Hatanaka et al., 1996). It functions in actin skeleton dynamics affecting cell shape and motility downstream of LIM-kinase (Suetsugu and Takenawa, 2003) and in mammalian myeloid cells (Ichikawa et al., 1982). CORO1A is an actin binding protein that interacts with microtubules and in some cell types associated with microtubules. CORONIN1 has been found to exert an inhibitory effect on cellular steady-state F-actin formation by means of an ARP2/3-dependent mechanism. It is required in humans for chemokine-mediated migration. It plays important roles in cell signaling, cell migration, phagocytosis, and vesicle trafficking (Castro-Castro et al., 2011) in human myeloid and other cells (Federzoni et al., 2013). FMNL1 homologues are implicated in BMP-mediated sprouting in endothelial cells (Wakayama et al., 2015) and ame-boid invasive cell motility and actin filament binding downstream of RHOC in human tumor cells (Kitzing et al., 2010). FMNL1 has been found in the actin-rich cores of primary human macrophage podosomes (Mersich et al., 2010).
ADAM homolog overexpression in nonsmall cell lung cancer in humans correlates with cell migration and invasion through NOTCH1 signaling in humans (Guo et al., 2012) and has been implicated in lymphocyte development (Gibb et al., 2011). ADAM10 has been found to cleave various cell surface molecules, including adhesion molecules, chemokines, and growth factor receptors. ADAM10-deficient macrophages showed reduced migration and extracellular matrix degradation as well as altered IL-10, IL-12, NO, and TNF signaling in mice (van der Vorst et al., 2015). CTSS is a lysosomal cysteine protease that has been implicated as essential in macrophage migration and development by cleavage of Rip1 kinase in mice (Verollet et al., 2011; McComb et al., 2014). TIMP-1 homologues inhibit metalloproteases which regulate immune cell development and migration by signaling downstream of TNF receptor, IL-6 receptor, EGF, and NOTCH (Khokha et al., 2013) and are found in the monocyte/macrophage population in mice (Guedez et al., 2012). SH2 domain-containing PTP homologues have intracellular protein tyrosine phosphatase activity implicated in cell migration during normal cardiac development (Lauriol et al., 2014) and when knocked out in murine myeloid cells result in attenuated renal fibrosis after unilateral ureter obstruction (Teng et al., 2015).
All these observations and findings derived from work in higher vertebrates are consistent with the identified genes having putative roles in either actin cytoskeleton or extracellular matrix functional pathways in differentiating Xenopus primitive myeloid cells (Bonnans et al., 2014). These functions could mediate the essential developmental and immunologic roles of this myeloid population. Future work will investigate which cellular functions fulfill such roles.
spib.a is Necessary and cebpa is Sufficient for Expression of Novel Primitive Myeloid Genes
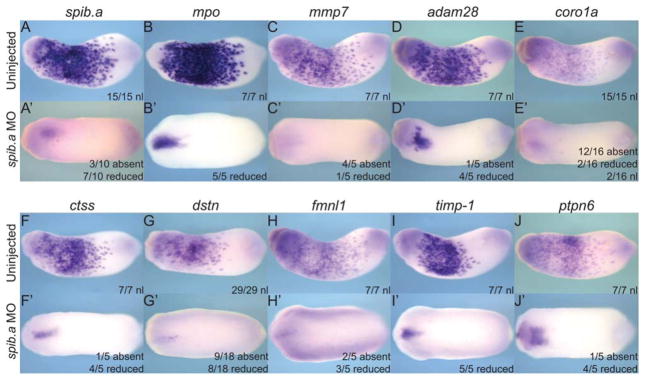
To confirm that the genes we have identified are expressed in the myeloid lineage downstream of the key transcription factor Spib.a, we examined their expression in Spib.a-depleted embryos that are known to lack primitive myeloid cells. (Fig. 3) (Costa et al., 2008). Using a well-established morpholino WMISH analysis of the known Spib.a-target genes, mpo and mmp7 confirmed that injection of a well-characterized Spib.a antisense morpholino oligo resulted in a loss of primitive myeloid cells (Smith et al., 2002; Harrison et al., 2004; Costa et al., 2008) (Fig. 3A–C′). As expected, Spib.a depletion resulted in a substantial reduction of all of the candidate genes (Fig. 3D–J′), confirming that they are expressed in the myeloid lineage downstream of Spib.a.
Fig. 3.
Knockdown of primitive myeloid lineage spib.a transcription factor results in loss of myeloid gene candidate expression in the anterior ventral blood island (aVBI) of stage 23 embryos. A–J: Normal expression of genes in the aVBI of uninjected control embryos. A′ –J′ : Reduced aVBI expression of genes in the embryos injected dorsally at 4-cell stage with 40 ng e1i1 spib.a morpholino. A+A′ , spib.a. B+B′ , mpo. C+C′ , mmp7. D+D′ , adam28. E+E′ , coro1a. F+F′, ctss. G+G′ , dstn. H+H′, fmnl1. I+I′ , timp-1. J+J′ , ptpn6.
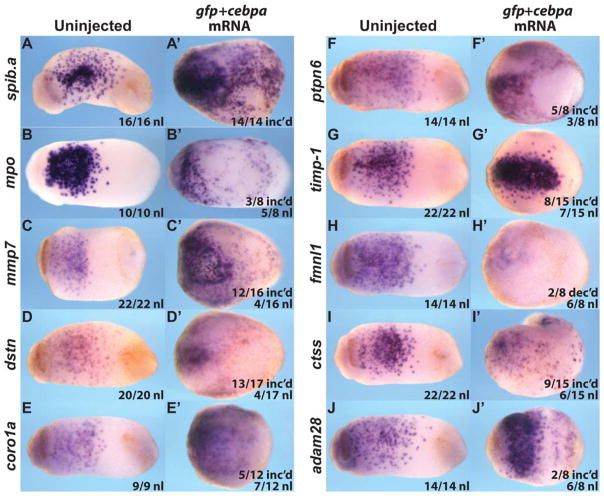
Ectopic expression of cebpa mRNA in the ectoderm (“animal cap”) of the early Xenopus embryo is sufficient to induce ectopic myeloid gene expression and generate migrating primitive myeloid cells that can be in transplanted into host embryos (Chen et al., 2009b). We therefore examined whether injection of Xenopus tropicalis cebpa into the animal blastomeres was sufficient to induce the ectopic expression of our seven candidate myeloid genes. We found increased or ectopic expression in all of the tested genes except fmnl1 (which showed normal or decreased expression; Fig. 4 and data not shown). Thus, we conclude that six of our seven genes are downstream of the essential primitive myeloid transcriptional regulator, Cebpa.
Fig. 4.
Animal cap-injected X.t. cebpa mRNA induces exogenous myeloid gene expression in Stage 21 X.l. embryos. A–J: Uninjected embryos ventral view. A′ –J′: 200 pg cebpa and 300 pg gfp mRNA coinjected embryos ventral view. A, A′ : spib.a. B, B′ : mpo. C, C′ : mmp7. D, D′ : dstn. E, E′ : coro1a. F, F′ : ptpn6. G, G′ : timp–1. H, H′ : fmnl1. I, I′ : ctss. J, J′ : adam28.
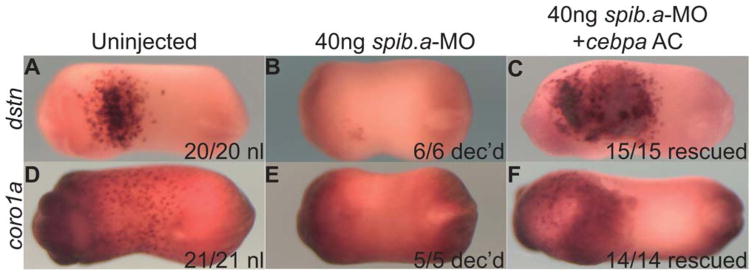
Finally we combined the spib.a-MO-mediated loss-of-function and cebpa mRNA-mediated gain-of-function approaches. We transplanted Cebp-expressing animal cap cells into the blastocoel of Spib.a-depleted embryos (a procedure known as an Einsteck experiment) (Sive et al., 2000), which rescued dstn and coro1a expression in the Spib.a morphant host embryos anterior ventral blood island (Fig. 5A–C′). We conclude that these genes are specific to the primitive myeloid lineage.
Fig. 5.
cebpa animal cap Einsteck rescue of dstn (A–C) and coro1a (D–F) expression spib.a-MO myeloid-depleted host embryos (Stage 21 ventral views). A,D: uninjected control embryos. B,E: spib.a-MO myeloid depleted host embryos. C,F: cebpa animal cap transplanted by means of Einsteck into stage 10 blastocoeles of myeloid-depleted hosts.
Conclusions
In this “Patterns and Phenotypes” work, we report seven genes that are expressed specifically in the mesoderm-derived primitive myeloid cells in Xenopus embryos. This is significant because molecular markers specific to primitive myeloid cells in higher vertebrates remain unknown, and the expression of genes in myeloid cells produced from definitive hematopoiesis are not expressed in the primitive myeloid lineage (Palis et al., 1999; James Palis, personal communication). CD34 has been identified in murine primitive myeloid cells, although its expression is not specific to the myeloid lineage (Gottgens et al., 1997; Sidney et al., 2014). We have used a combination of loss-of-function and gain-of-function experiments supporting that these primitive myeloid genes are epistatically downstream of key primitive myeloid transcriptional regulators, spib.a and cebpa.
Given the highly conserved roles of the primitive myeloid lineage, characterization of the genetic signature of this cell type is critical to better understand their function in developmental mechanisms including repair, regeneration, patterning (blood vessels), morphogenesis (bone and early heart), survival and/or apoptosis (neuronal, adipose), and cell fate decisions (pancreatic β cell) (Banaei-Bouchareb et al., 2004, 2006; Stefater et al., 2011; Smith and Mohun, 2011). Future work will determine if these genes function in a coordinated pathway for myeloid cell migration and function. For instance, loss of lurp1 gene function in Xenopus perturbed myeloid gene migration but not specification, perturbing cardiac morphogenesis (Smith and Mohun, 2011). Future work will determine the role each of these primitive myeloid progenitor genes plays in primitive myeloid cell migration, intracellular signaling, and extracellular matrix remodeling functions and determine exactly which of these functions impart the known important developmental mechanisms of this cell type (Stefater et al., 2011).
Experimental Procedures
Embryo Manipulations
Xenopus embryo manipulation and culture were performed as described previously (McLin et al., 2007).
Gene Ontology Analysis
Gene ontology analysis software (available through Web site ToppGene.cchmc.org) was used to assess the predicted molecular and cellular functions of the isolated genes based on information available from their human and mouse orthologs (Chen et al., 2009a).
Morpholino and mRNA Microinjection
Embryos with clear dorsal–ventral pigmentation differences were selected for four- to eight-cell stage injections targeting Xenopus laevis spib.a morpholino to the dorsal blastomeres that contribute to the embryo’s dorsoanterior. Eight-cell animal blastomeres were targeted for misexpression of Xenopus tropicalis cebpa mRNA.
The MO used in this study has been previously published to generate specific loss-of-function phenotypes: Xenopus laevis spib.a-e1i1-MO (40 ng) (Smith and Mohun, 2011). For animal cap Einsteck rescue experiments, we injected Xenopus tropicalis cebpa mRNA synthesized from pFTX11.HA.xtCEBPa expression plasmid (150–300 pg/embryo; kind gift from Dr. Enrique Amaya) (Chen et al., 2009b).
Einsteck Rescues
Donor fragments were generated as follows. Stage 10–10.5 gfp-and cebpa-mRNA coinjected embryos had their vitelline membranes removed. Fluorescing, mesoderm-free donor animal caps were cut using hair knives and hair loops in 0.5 × MBS with 1:500 gentamicin. Stage 10 recipient spib.a morpholino-injected embryos had a slit generated by a hair knife and the animal cap donor tissue was inserted into the blastocoele. The Einsteck embryos were cultured to stage 22 and harvested for in situ analysis (Sive et al., 2000).
In Situ Hybridization
In situ hybridization was performed as previously described (Kenny et al., 2012). A sequential procedure for double whole-mount in situ hybridization was adopted, where embryos were photographed successively after the chromogenic reactions for each probe were developed. Digoxigenin- or fluorescein-labeled Xenopus laevis probes were synthesized from: pExpress-1 ceb-paaaaaaa-SPORT6-spib.a, pCMV-SPORT6-mmp7, pCMV-SPORT6-mpo2, pCMV-SPORT6-adam28, pCMV-SPORT6-coro1a, pCMV-SPORT6-ctss, pCMV-SPORT6-dstn, pCMV-SPORT6-fmnl1, pCMV-SPORT6-ptpn6, and pExpress-1-timp-1.
Acknowledgments
We thank Dr. Enrique Amaya for the pFTX11.ha.xtcebpa expression plasmid. This project was supported in part by a CCHMC Procter Scholarship and K08 HL105661 to A.P.K., a T32 HD07463 award to E.T.S., and PHS Grants P30 DK078392 and R01 DK070858 to A.M.Z.
Grant sponsor: CCHMC Procter Scholarship; Grant numbers: K08 HL105661 and T32 HD07463; Grant sponsor: PHS; Grant numbers: P30 DK078392 and R01 HD73179.
References
- Arnold L, Henry A, Poron F, Baba-Amer Y, Van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora D, Kothe S, Van Den Eijnden M, Hooft Van Huijsduijnen R, Heidel F, Fischer T, Scholl S, Tolle B, Bohmer SA, Lennartsson J, Isken F, Muller-Tidow C, Bohmer FD. Expression of protein-tyrosine phosphatases in Acute Myeloid Leukemia cells: FLT3 ITD sustains high levels of DUSP6 expression. Cell Commun Signal. 2012;10:19. doi: 10.1186/1478-811X-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaei-Bouchareb L, Gouon-Evans V, Samara-Boustani D, Castellotti MC, Czernichow P, Pollard JW, Polak M. Insulin cell mass is altered in Csf1op/Csf1op macrophage-deficient mice. J Leukoc Biol. 2004;76:359–367. doi: 10.1189/jlb.1103591. [DOI] [PubMed] [Google Scholar]
- Banaei-Bouchareb L, Peuchmaur M, Czernichow P, Polak M. A transient microenvironment loaded mainly with macrophages in the early developing human pancreas. J Endocrinol. 2006;188:467–480. doi: 10.1677/joe.1.06225. [DOI] [PubMed] [Google Scholar]
- Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Castro A, Ojeda V, Barreira M, Sauzeau V, Navarro-Lerida I, Muriel O, Couceiro JR, Pimentel-Muinos FX, Del Pozo MA, Bustelo XR. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. EMBO J. 2011;30:3913–3927. doi: 10.1038/emboj.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aranow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Research. 2009a;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Costa RM, Love NR, Soto X, Roth M, Paredes R, Amaya E. C/EBPalpha initiates primitive myelopoiesis in pluripo-tent embryonic cells. Blood. 2009b;114:40–48. doi: 10.1182/blood-2008-11-189159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciau-Uitz A, Liu F, Patient R. Genetic control of hematopoietic development in Xenopus and zebrafish. Int J Dev Biol. 2010;54:1139–1149. doi: 10.1387/ijdb.093055ac. [DOI] [PubMed] [Google Scholar]
- Ciau-Uitz A, Monteiro R, Kirmizitas A, Patient R. Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp Hematol. 2014;42:669–683. doi: 10.1016/j.exphem.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Costa RM, Soto X, Chen Y, Zorn AM, Amaya E. spib is required for primitive myeloid development in Xenopus. Blood. 2008;112:2287–2296. doi: 10.1182/blood-2008-04-150268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Argilla M, Makarenkova H, Ko K, Lang RA. Macrophages kill capillary cells in G1 phase of the cell cycle during programmed vascular regression. Development. 1999;126:2141–2147. doi: 10.1242/dev.126.10.2141. [DOI] [PubMed] [Google Scholar]
- Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, Wu S, Lang R, Iredale JP. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115:56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federzoni EA, Humbert M, Valk PJ, Behre G, Leibundgut EO, Torbett BE, Fey MF, Tschan MP. The actin-binding protein CORO1A is a novel PU.1 (SPI1)- and CEBPA-regulated gene with significantly lower expression in APL and CEBPA-mutated AML patients. Br J Haematol. 2013;160:855–859. doi: 10.1111/bjh.12170. [DOI] [PubMed] [Google Scholar]
- Franc NC, Heitzler P, Ezekowitz RA, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- Gibb DR, Saleem SJ, Chaimowitz NS, Mathews J, Conrad DH. The emergence of ADAM10 as a regulator of lymphocyte development and autoimmunity. Mol Immunol. 2011;48:1319–1327. doi: 10.1016/j.molimm.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren I, Allmann N, Yogev N, Schurmann C, Linke A, Holdener M, Waisman A, Pfeilschifter J, Frank S. A transgenic mouse model of inducible macrophage depletion: effects of diphtheria toxin-driven lysozyme M-specific cell lineage ablation on wound inflammatory, angiogenic, and contractive processes. Am J Pathol. 2009;175:132–147. doi: 10.2353/ajpath.2009.081002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottgens B, Mclaughlin F, Bockamp EO, Fordham JL, Begley CG, Kosmopoulos K, Elefanty AG, Green AR. Transcription of the SCL gene in erythroid and CD34 positive primitive myeloid cells is controlled by a complex network of lineage-restricted chromatin-dependent and chromatin-independent regulatory elements. Oncogene. 1997;15:2419–2428. doi: 10.1038/sj.onc.1201426. [DOI] [PubMed] [Google Scholar]
- Guedez L, Jensen-Taubman S, Bourboulia D, Kwityn CJ, Wei B, Caterina J, Stetler-Stevenson WG. TIMP-2 targets tumor-associated myeloid suppressor cells with effects in cancer immune dysfunction and angiogenesis. J Immunother. 2012;35:502–512. doi: 10.1097/CJI.0b013e3182619c8e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, He L, Yuan P, Wang P, Lu Y, Tong F, Wang Y, Yin Y, Tian J, Sun J. ADAM10 overexpression in human non-small cell lung cancer correlates with cell migration and invasion through the activation of the Notch1 signaling pathway. Oncol Rep. 2012;28:1709–1718. doi: 10.3892/or.2012.2003. [DOI] [PubMed] [Google Scholar]
- Harrison M, Abu-Elmagd M, Grocott T, Yates C, Gavrilovic J, Wheeler GN. Matrix metalloproteinase genes in Xenopus development. Dev Dyn. 2004;231:214–220. doi: 10.1002/dvdy.20113. [DOI] [PubMed] [Google Scholar]
- Hatanaka H, Ogura K, Moriyama K, Ichikawa S, Yahara I, Inagaki F. Tertiary structure of destrin and structural similarity between two actin-regulating protein families. Cell. 1996;85:1047–1055. doi: 10.1016/s0092-8674(00)81305-7. [DOI] [PubMed] [Google Scholar]
- Ichikawa Y, Nagata K, Sagara J. Differentiation of myeloid leukemia cells and changes in their contractile proteins. Acta Pathol Jpn. 1982;32(suppl 1):187–196. [PubMed] [Google Scholar]
- Jones GE. Cellular signaling in macrophage migration and chemotaxis. J Leukoc Biol. 2000;68:593–602. [PubMed] [Google Scholar]
- Kau CL, Turpen JB. Dual contribution of embryonic ventral blood island and dorsal lateral plate mesoderm during ontogeny of hemopoietic cells in Xenopus laevis. J Immunol. 1983;131:2262–2266. [PubMed] [Google Scholar]
- Kenny AP, Rankin SA, Allbee AW, Prewitt AR, Zhang Z, Tabangin ME, Shifley ET, Louza MP, Zorn AM. Sizzled-tolloid interactions maintain foregut progenitors by regulating fibronectin-dependent BMP signaling. Dev Cell. 2012;23:292–304. doi: 10.1016/j.devcel.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha R, Murthy A, Weiss A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat Rev Immunol. 2013;13:649–665. doi: 10.1038/nri3499. [DOI] [PubMed] [Google Scholar]
- Kitzing TM, Wang Y, Pertz O, Copeland JW, Grosse R. For-min-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene. 2010;29:2441–2448. doi: 10.1038/onc.2009.515. [DOI] [PubMed] [Google Scholar]
- Lauriol J, Jaffre F, Kontaridis MI. The role of the protein tyro-sine phosphatase SHP2 in cardiac development and disease. Semin Cell Dev Biol. 2014;37:73–81. doi: 10.1016/j.semcdb.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Li B, Rao S, Yeo EJ, Hudson TE, Nowlin BT, Pei H, Chen L, Zheng JJ, Carroll TJ, Pollard JW, McMahon AP, Lang RA, Duffield JS. Macrophage Wnt7b is critical for kidney repair and regeneration. Proc Natl Acad Sci U S A. 2010;107:4194–4199. doi: 10.1073/pnas.0912228107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux CT, Yoshimoto M, Mcgrath K, Conway SJ, Palis J, Yoder C. All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood. 2008;111:3435–3438. doi: 10.1182/blood-2007-08-107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeno M, Tochinai S, Katagiri C. Differential participation of ventral and dorsolateral mesoderms in the hemopoiesis of Xenopus, as revealed in diploid-triploid or interspecific chimeras. Dev Biol. 1985;110:503–508. doi: 10.1016/0012-1606(85)90108-3. [DOI] [PubMed] [Google Scholar]
- Masmas TN, Ifversen M, Ek J, Schejbel L, Marquart HV, Müller K, Heilmann C, Kjaergaard S, Kirchhoff M. Application of aCGH analysis in patients with primary immunodeficiency of unknown genetic origin – identification of atypical sap deficiency and coronin-1a deficiency. J Clin Cell Immunol. 2014;5:1–7. [Google Scholar]
- McComb S, Shutinoski B, Thurston S, Cessford E, Kumar K, Sad S. Cathepsins limit macrophage necroptosis through cleavage of Rip1 kinase. J Immunol. 2014;192:5671–5678. doi: 10.4049/jimmunol.1303380. [DOI] [PubMed] [Google Scholar]
- McGrath KE, Palis J. Hematopoiesis in the yolk sac: more than meets the eye. Exp Hematol. 2005;33:1021–1028. doi: 10.1016/j.exphem.2005.06.012. [DOI] [PubMed] [Google Scholar]
- McLin VA, Rankin SA, Zorn AM. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development. 2007;134:2207–2217. doi: 10.1242/dev.001230. [DOI] [PubMed] [Google Scholar]
- Meijer C, Wiezer MJ, Diehl AM, Schouten HJ, Schouten HJ, Meijer S, Van Rooijen N, Van Lambalgen AA, Dijkstra CD, Van Leeuwen PA. Kupffer cell depletion by CI2MDP-liposomes alters hepatic cytokine expression and delays liver regeneration after partial hepatectomy. Liver. 2000;20:66–77. doi: 10.1034/j.1600-0676.2000.020001066.x. [DOI] [PubMed] [Google Scholar]
- Mersich AT, Miller MR, Chkourko H, Blystone SD. The formin FRL1 (FMNL1) is an essential component of macrophage podo-somes. Cytoskeleton (Hoboken) 2010;67:573–585. doi: 10.1002/cm.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza R, Dipietro LA, Koh TJ. Selective and specific macrophage ablation is detrimental to wound healing in mice. Am J Pathol. 2009;175:2454–2462. doi: 10.2353/ajpath.2009.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller BU, Pabst T. C/EBPalpha and the pathophysiology of acute myeloid leukemia. Curr Opin Hematol. 2006;13:7–14. doi: 10.1097/01.moh.0000190110.08156.96. [DOI] [PubMed] [Google Scholar]
- Nagata S. Electron microscopic study on the early histogenesis of thymus in the toad, Xenopus laevis. Cell Tissue Res. 1977;179:87–96. doi: 10.1007/BF00278464. [DOI] [PubMed] [Google Scholar]
- Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin): a systemical and chronological survey of the development from the fertilized egg till the end of metamorphosis. New York: Garland Publishing, Inc; 1994. [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae F, Woods K, Sasmono T, Campanale N, Taylor D, Ovchinnikov DA, Grimmond SM, Hume DA, Ricardo SD, Little MH. Characterisation and trophic functions of murine embryonic macrophages based upon the use of a Csf1r-EGFP transgene reporter. Dev Biol. 2007;308:232–246. doi: 10.1016/j.ydbio.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Rochelle T, Daubon T, Van Troys M, Harnois T, Waterschoot D, Ampe C, Roy L, Bourmeyster N, Constantin B. p210bcr-abl induces amoeboid motility by recruiting ADF/destrin through RhoA/ROCK1. FASEB J. 2013;27:123–134. doi: 10.1096/fj.12-205112. [DOI] [PubMed] [Google Scholar]
- Savill J, Fadok V. Corpse clearance defines the meaning of cell death. Nature. 2000;407:784–788. doi: 10.1038/35037722. [DOI] [PubMed] [Google Scholar]
- Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, Stappenbeck TS. Efficient colonic mucosal wound repair requires Trem2 signaling. Proc Natl Acad Sci U S A. 2009;106:256–261. doi: 10.1073/pnas.0803343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Tsujimura K, Koide Y. Coronin-1a inhibits autophago-some formation around Mycobacterium tuberculosis-containing phagosomes and assists mycobacterial survival in macrophages. Cell Microbiol. 2012;14:710–727. doi: 10.1111/j.1462-5822.2012.01754.x. [DOI] [PubMed] [Google Scholar]
- Shimoda M, Hashimoto G, Mochizuki S, Ikeda E, Nagai N, Ishida S, Okada Y. Binding of ADAM28 to P-selectin glycoprotein ligand-1 enhances P-selectin-mediated leukocyte adhesion to endothelial cells. J Biol Chem. 2007;282:25864–25874. doi: 10.1074/jbc.M702414200. [DOI] [PubMed] [Google Scholar]
- Sidney LE, Branch MJ, Dunphy SE, Dua HS, Hopkinson A. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells. 2014;32:1380–1389. doi: 10.1002/stem.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor. New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Smith SJ, Kotecha S, Towers N, Latinkic BV, Mohun TJ. XPOX2-peroxidase expression and the XLURP-1 promoter reveal the site of embryonic myeloid cell development in Xenopus. Mech Dev. 2002;117:173–186. doi: 10.1016/s0925-4773(02)00200-9. [DOI] [PubMed] [Google Scholar]
- Smith SJ, Mohun TJ. Early cardiac morphogenesis defects caused by loss of embryonic macrophage function in Xenopus. Mech Dev. 2011;128:303–315. doi: 10.1016/j.mod.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefater JA, III, Ren S, Lang RA, Duffield JS. Metchnikoff’s policemen: macrophages in development, homeostasis and regeneration. Trends Mol Med. 2011;17:743–752. doi: 10.1016/j.molmed.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Takenawa T. Regulation of cortical actin networks in cell migration. Int Rev Cytol. 2003;229:245–286. doi: 10.1016/s0074-7696(03)29006-9. [DOI] [PubMed] [Google Scholar]
- Takeishi T, Hirano K, Kobayashi T, Hasegawa G, Hatakeyama K, Naito M. The role of Kupffer cells in liver regeneration. Arch Histol Cytol. 1999;62:413–422. doi: 10.1679/aohc.62.413. [DOI] [PubMed] [Google Scholar]
- Teng JF, Wang K, Li Y, Qu FJ, Yuan Q, Cui XG, Wang QX, Xu DF. Conditional knockout of Src homology 2 domain-containing protein tyrosine phosphatase-2 in myeloid cells attenuates renal fibrosis after unilateral ureter obstruction. Chin Med J (Engl) 2015;128:1196–1201. doi: 10.4103/0366-6999.156121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. 1994;120:1829–1837. doi: 10.1242/dev.120.7.1829. [DOI] [PubMed] [Google Scholar]
- Tochinai S. Direct observation of cell migration into Xenopus thymus rudiments through mesenchyme. Dev Comp Immunol. 1980;4:273–282. doi: 10.1016/s0145-305x(80)80031-0. [DOI] [PubMed] [Google Scholar]
- Van Der Vorst EP, Jeurissen M, Wolfs IM, Keijbeck A, Theodorou K, Wijnands E, Schurgers L, Weber S, Gijbels MJ, Hamers AA, Dreymueller D, Rose-John S, De Winther MP, Ludwig A, Saftig P, Biessen EA, Donners MM. Myeloid A disintegrin and metalloproteinase domain 10 deficiency modulates atherosclerotic plaque composition by shifting the balance from inflammation toward fibrosis. Am J Pathol. 2015;185:1145–1155. doi: 10.1016/j.ajpath.2014.11.028. [DOI] [PubMed] [Google Scholar]
- Verollet C, Charriere GM, Labrousse A, Cougoule C, Le Cabec V, Maridonneau-Parini I. Extracellular proteolysis in macrophage migration: losing grip for a breakthrough. Eur J Immunol. 2011;41:2805–2813. doi: 10.1002/eji.201141538. [DOI] [PubMed] [Google Scholar]
- Wakayama Y, Fukuhara S, Ando K, Matsuda M, Mochizuki N. Cdc42 mediates Bmp-induced sprouting angiogenesis through Fmnl3-driven assembly of endothelial filopodia in zebra-fish. Dev Cell. 2015;32:109–122. doi: 10.1016/j.devcel.2014.11.024. [DOI] [PubMed] [Google Scholar]
- Warga RM, Kane DA, Ho RK. Fate mapping embryonic blood in zebrafish: multi- and unipotential lineages are segregated at gastrulation. Dev Cell. 2009;16:744–755. doi: 10.1016/j.devcel.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Collins RF, Grinstein S, Trimble WS. Coronin-1 function is required for phagosome formation. Mol Biol Cell. 2005;16:3077–3087. doi: 10.1091/mbc.E04-11-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac E, Schweighofer B, Kupriyanova TA, Juncker-Jensen A, Minder P, Quigley JP, Deryugina EI. Angiogenic capacity of M1- and M2-polarized macrophages is determined by the levels of TIMP-1 complexed with their secreted proMMP-9. Blood. 2013;122:4054–4067. doi: 10.1182/blood-2013-05-501494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, Akashi K, Tenen DG. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]