Abstract
Early recognition of pre-mRNA during spliceosome assembly in mammals proceeds through the association of U1 small nuclear ribonucleoprotein particle (snRNP) with the 5′ splice site as well as the interactions of the branch binding protein SF1 with the branch region and the U2 snRNP auxiliary factor U2AF with the polypyrimidine tract and 3′ splice site. These factors, along with members of the SR protein family, direct the ATP-independent formation of the early (E) complex that commits the pre-mRNA to splicing. We report here the observation in U2AF-depleted HeLa nuclear extract of a distinct, ATP-independent complex designated E′ which can be chased into E complex and itself commits a pre-mRNA to the splicing pathway. The E′ complex is characterized by a U1 snRNA-5′ splice site base pairing, which follows the actual commitment step, an interaction of SF1 with the branch region, and a close association of the 5′ splice site with the branch region. These results demonstrate that both commitment to splicing and the early proximity of conserved sequences within pre-mRNA substrates can occur in a minimal complex lacking U2AF, which may function as a precursor to E complex in spliceosome assembly.
The removal of intervening sequences from pre-mRNAs is catalyzed by the spliceosome, a 60S biochemical machine that consists of both protein and RNA components (19). The spliceosome directs the removal of introns and ligation of exons in a concerted fashion through two sequential transesterification reactions (6, 36). The spliceosome consists of the U1, U2, and U4/U5/U6 small nuclear ribonucleoprotein particles (snRNPs), each containing a unique snRNA and associated proteins bound to the pre-mRNA substrate (6, 19, 36). Assembly of the spliceosome active site on the pre-mRNA proceeds through several stages and is directed by conserved sequences at the splice sites and within the intron as well as other sequences in the pre-mRNA.
Early recognition of pre-mRNA substrates by the splicing machinery proceeds through the formation of the ATP-independent commitment complex (CC) in yeast or early (E) complex in mammals (15, 20, 34). In both yeast and mammals, the 5′ splice site is initially recognized by U1 snRNP; the branch region and 3′ splice site are defined by the association of the branch binding protein (BBP) and Mud2 (yeast) or SF1 and the heterodimer U2 snRNP auxiliary factor (U2AF; mammals). In the presence of ATP, U2 snRNP becomes stably associated with the pre-mRNA through a base pairing interaction between U2 snRNA and the branch region; subsequent recruitment of the U4/U6/U5 tri-snRNP and several snRNA rearrangements result in the formation of the mature spliceosome (36).
The mammalian E complex can be visualized either by gel filtration or by native gel electrophoresis (11, 26). This complex contains U1 snRNP tightly associated with the 5′ splice site and SF1 and U2AF bound to the branch region, polypyrimidine tract, and 3′ splice site. As well, the E complex contains members of the SR protein family of splicing factors (13, 14), including SC35, which has been shown to bridge U1 snRNP and U2AF35, the small subunit of U2AF (41). In studies of mammalian prespliceosome assembly, it has been shown that SR proteins facilitate the earliest recognition of the 5′ and 3′ splice sites and promote formation of ATP-independent prespliceosomal complexes (14, 35).
In yeast, two CCs, CC1 and CC2, have been identified in U2 snRNP-depleted extracts by gel electrophoresis (20, 34, 44). Formation of the faster-mobility complex CC1 is dependent on the presence of a 5′ splice site and U1 snRNP. The lower-mobility complex CC2 is dependent on the presence of a 5′ splice site as well as a functional branch point sequence (BPS). Several components of CC2 have been identified. These include the splicing factor Mud2p, which has been shown to interact with the highly conserved yeast BPS and U1 snRNP (1, 44). Mud2p has been identified as the yeast homolog of U2AF65, the large subunit of U2AF that interacts with the polypyrimidine tract 3′ to the highly degenerate mammalian BPS (43). The yeast BBP has been found to interact directly with Mud2p, which parallels the association of the mammalian splicing factor SF1 with U2AF65 (4, 33). Taken together, these results suggest similar structural organizations of the yeast and mammalian commitment complexes.
It has been demonstrated that the initial recognition of the pre-mRNA and formation of the CC in both the yeast and mammalian systems are dependent on U1 snRNP. Depletion experiments have demonstrated that the stable association of U2 snRNP with the pre-mRNA requires the presence of U1 snRNP (3). There is good evidence that U1 snRNP is involved in recruiting U2AF to the polypyrimidine tract, which in turn promotes the association of U2 snRNP with the branch region (8, 21). The stable association of U1 snRNP with the 5′ splice site involves base pairing interactions between the pre-mRNA and the 5′ end of U1 snRNA. Recently, it has been suggested that in yeast this RNA-RNA interaction is preceded by the sequence-specific recognition of the 5′ splice site by the U1 snRNP protein U1-C (12, 39).
Here, we report the identification of a novel prespliceosomal complex designated E′, which forms in U2AF-depleted HeLa nuclear extracts. The E′ complex formation is dependent on SF1 and on U1 snRNA-5′ splice site base pairing and commits the pre-mRNA to the splicing pathway. RNA-protein cross-linking combined with immunoprecipitation demonstrates that the branch binding protein SF1 is associated with the pre-mRNA within the E′ complex. The structure of complexes formed in wild-type and U2AF-depleted extracts has been probed with RNAs site-specifically derivatized with the hydroxyl radical source (S)-1-[p-(bromoacetamido)benzyl]-EDTA-Fe (Fe-BABE) (16). These experiments indicate that the proximity of the 5′ splice site and branch region that is observed in the E complex is also found in the E′ complex; this proximity is dependent upon the presence of SF1 but not a functional branch point sequence and supports a model whereby U1 snRNP alone or in conjunction with SF1 directly recruits U2AF to the pre-mRNA substrate.
MATERIALS AND METHODS
Synthesis of pre-mRNA substrates.
Wild-type and mutant pre-mRNAs were synthesized by in vitro T7 transcription using double-stranded PCR products derived from the PIP85.B plasmid (28) as templates. Transcriptions were carried out under standard conditions in the presence of 100 μCi of [α-32P]UTP; products were purified directly by 8% polyacrylamide gel electrophoresis (PAGE) (19:1).
In order to prepare derivatized pre-mRNAs, the 10-nucleotide oligomers 5′-GG(C*)UGCUGAC-3′ and 5′-UUCCCUG(C*)AG-3′, corresponding to the pre-mRNA branch site and the 3′ splice site, respectively, containing a site-specific N4-functionalized cytosine (C*), were synthesized, ligated into full-length pre-mRNAs, and derivatized with benzophenone or Fe-BABE as described previously (16, 24). Branch sequence mutants were created in an analogous fashion with a 10-nucleotide oligomer, 5′-GGGUCGUCAG-3′, corresponding to a scrambled branch region.
E and E′ complex formation.
Pre-mRNAs (50 × 103 to 100 × 103 cpm) were incubated in 10-μl reactions containing 25% HeLa nuclear extract or U2AF-depleted HeLa nuclear extract (23, 43), 60 mM KCl, and 4 U of RNase inhibitor. Following incubation at 30°C for 25 to 60 min, reactions were immediately loaded onto a 1.2% agarose-0.5× Tris-borate-EDTA gel. Dried gels were exposed to a Molecular Dynamics phosphor screen and scanned with a Molecular Dynamics Storm 840 Phosphorimager. Chase experiments were performed by preforming E′ complex in U2AF-depleted extract as described above followed by addition of an aliquot of the reaction to wild-type nuclear extract and further incubation at 30°C for various times. Competition experiments were performed as described above, except that an aliquot of the preformed complex was added to a reaction containing wild-type nuclear extract and 0.8, 1.4, or 5 pmol of competitor RNA, and the whole was further incubated at 30°C. Reactions blocking U1 or U2 snRNA were performed in extracts preincubated in the presence of 5 to 20 μM 2′-O-methyl antisense oligonucleotide (Dharmacon) as described previously (5). Add-back experiments were performed in U2AF-depleted extract with 60 μM recombinant U2AF65 or U2AF65/35 dimer. SF1 add-back experiments contained 60 μM recombinant SF1-C4 mutant (29). His6-tagged U2AF65, U2AF35, and SF1-C4 were overexpressed in Escherichia coli cells and purified by Ni-nitrilotriacetic acid chromatography as described elsewhere (17).
Gel filtration column purification of E′ complex.
Pre-mRNAs (50 × 103 to 100 × 103 cpm) were incubated under the E′ complex-forming conditions described above in 100-μl reactions. Reactions were loaded directly onto a Sephacryl S500 gel filtration column (40-ml column volume; Amersham Pharmacia Biotech) equilibrated with 20 mM Tris, pH 7.8, 0.1% Triton X-100, 60 mM KCl, and 2.5 mM EDTA (as described in reference 31). Columns were run at 50 μl min−1, and 40 to 50 1-ml fractions were collected. An aliquot of each fraction was used for scintillation counting to generate an elution profile. Fractions were then concentrated to 20 μl by using Ultrafree-MC centrifugal filters (molecular weight cutoff, 10,000; Millipore) and subjected to native agarose gels as described above.
Splicing assays.
Pre-mRNAs (50 × 103 to 100 × 103 cpm) were incubated in 20-μl reactions containing 40% HeLa nuclear extract, 2 mM MgCl2, 60 mM KCl, 1 mM ATP, 5 mM creatine phosphate (CP), and 4 U of RNase inhibitor. Competition assays were performed in the presence of competitor RNA as described above. Commitment assays were performed by incubating pre-mRNA (50 × 103 to 100 × 103 cpm) under E or E′ complex-forming conditions as described above. Aliquots were then added to a splicing mix containing ATP/CP and MgCl2 with or without competitor RNA. Following incubation at 30°C, splicing reactions were digested with 40 μg of proteinase K in the presence of 20 μg of tRNA at 55°C for 20 min. Reactions were extracted with phenol-chloroform-isoamyl alcohol, ethanol precipitated, and then subjected to denaturing PAGE (15% acrylamide; 19:1). Dried gels were exposed to a Molecular Dynamics phosphor screen and scanned with a Molecular Dynamics Storm 840 Phosphorimager.
UV cross-linking and immunoprecipitation.
Cross-linking was performed in 20-μl reactions containing 25% HeLa nuclear extract or U2AF-depleted nuclear extract, which were incubated under E or E′ complex-forming conditions and then irradiated at 302 nm on ice for 20 min. RNase A (2.5 U) was added, and reactions were digested at 37°C for 30 min. For immunoprecipitations, photolyzed samples were then added to 10 μl of protein G-Sepharose beads bound to anti-SF1-1D5 antibodies in 400 μl of buffer containing 10 mM Tris-HCl, pH 8, 150 mM KCl, 20 mM MgCl2, and 0.5 mM dithiothreitol. Samples were rotated at 4°C for 2 h, washed with buffer three times, and subjected to sodium dodecyl sulfate (SDS)-12% PAGE. For analysis of cross-links in E and E′, complexes were purified by native agarose gel electrophoresis as described above. Cross-linked proteins were isolated from the gel by electroelution (200 V for 3 h), RNA was digested with RNase A, and the samples were subjected to SDS-12% PAGE. Dried gels were exposed to a Molecular Dynamics phosphor screen and scanned with a Molecular Dynamics Storm 840 Phosphorimager.
Hydroxyl radical probing of E and E′ structure.
Hydroxyl radical cleavage experiments were performed in 20-μl reactions containing 25% HeLa nuclear extract or U2AF-depleted extract that had been dialyzed into glycerol-free buffer D (20 mM HEPES, pH 7.9, 0.1 M KCl, 0.5 mM dithiothreitol). Wild-type and U2AF-depleted extracts were cleared of SF1 by passing the extract over protein G-Sepharose bound to anti-SF1-1D5 antibodies in buffer D. Clearing of the extract was performed twice with fresh protein G-Sepharose. Following SF1 depletion, extracts were dialyzed into glycerol-free buffer D; depletion was confirmed by Western analysis. Reactions probing E′ and E complexes (60 min at 30°C) contained 60 mM KCl and 4 U of RNase inhibitor but lacked ATP/CP and MgCl2. Cleavage reactions were initiated by the addition of H2O2 and ascorbic acid as described elsewhere (40). Reactions were quenched with thiourea-glycerol (final concentrations, 30 mM and 1%, respectively), treated with proteinase K, extracted with phenol-chloroform-isoamyl alcohol, and ethanol precipitated. Primer extension reactions were performed as described elsewhere (37) by using SuperScript reverse transcriptase (RT) (Gibco BRL). For analysis of the 5′ splice site, the primer (5′-TCGAGACGAGCTGACATC-3′) that was complementary to the region immediately 5′ of the branch site was used. Primer extension reactions contained approximately 1 to 5 pmol of RNA substrate and 1 μM DNA primer. Following extension, reactions were extracted with phenol-chloroform-isoamyl alcohol, ethanol precipitated, and then subjected to denaturing PAGE (8% acrylamide; 19:1). Dried gels were exposed to a Molecular Dynamics phosphor screen and scanned with a Molecular Dynamics Storm 840 Phosphorimager.
RESULTS
Identification of a novel prespliceosomal complex.
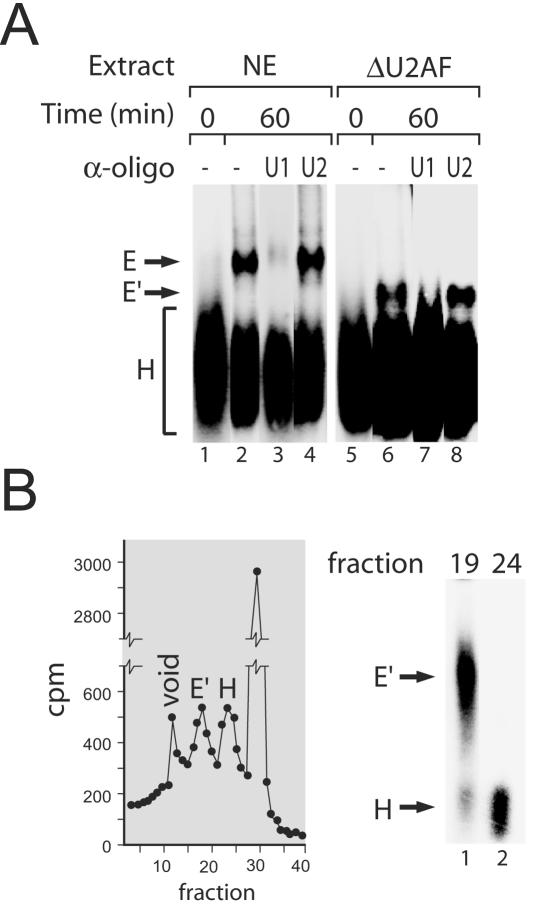
Work by Reed and coworkers has demonstrated that a complex formed on pre-mRNAs corresponding to the previously characterized E complex can be resolved from the nonspecific H complex by native agarose gel electrophoresis (11, 26). As part of a series of investigations of the earliest steps of spliceosome assembly, we specifically depleted HeLa nuclear extracts of U2AF (23, 43) and analyzed complex formation on the PIP85.B pre-mRNA by native agarose gel. Following incubation of pre-mRNA in depleted extract in the absence of ATP for 60 min at 30°C, we were able to observe the formation of a complex of slower mobility than H, distinct from E, which we designated E′ (Fig. 1A; compare lanes 2 and 6). Formation of both the E and the E′ complexes involves U1 snRNA-5′ splice site base pairing, since preincubation of extract with an antisense 2′-O-methyl oligonucleotide complementary to the 5′ end of U1 snRNA abolished complex formation (Fig. 1A, lanes 3 and 7). Similarly, pretreatment of extract with RNase H and a DNA oligonucleotide directed against the 5′ end of U1 snRNA abolished the formation of both E and E′ complexes (data not shown). Blocking the interaction of U2 snRNP with the pre-mRNA using an antisense oligonucleotide had no effect on either E or E′ formation (Fig. 1A, lanes 4 and 8). To obtain further evidence of E′ complex identity and stability, complexes formed on pre-mRNA substrates incubated in U2AF-depleted extract were isolated by gel filtration (31), and fractions that were obtained were analyzed by native agarose gel (Fig. 1B). We observed two distinct peaks that represent the isolation of the E′ complex from the previously identified H complex.
FIG. 1.
Identification of a novel prespliceosome complex (E′). (A) Native agarose gel shift analysis of complexes formed in nuclear extract (NE) and U2AF-depleted nuclear extract (ΔU2AF). Complex formation was analyzed in the presence and absence of anti-U1 and anti-U2 oligonucleotides. (B) Gel filtration column purification of the E′ complex from U2AF-depleted extract. (Left panel) Pre-mRNA elution profile of column-purified prespliceosome reaction. The void volume and E′ and H complexes are indicated. (Right panel) Native agarose gel analysis of an aliquot of fraction 19 (E′, lane 1) and fraction 24 (H, lane 2).
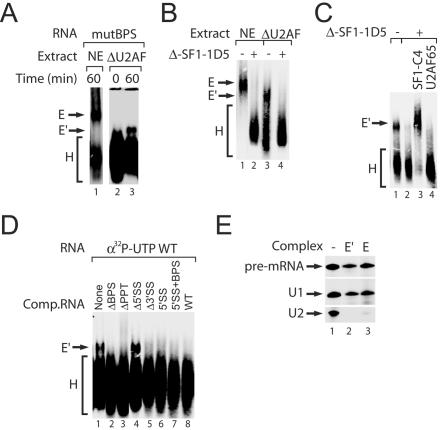
In order to examine the effect of the pre-mRNA branch region on E′ complex formation, mutant RNAs were prepared in which the BPS was scrambled. E′ complex was detected on a pre-mRNA substrate that contained a mutant BPS (Fig. 2A). This result is consistent with the observation that the formation of the E complex is not dependent on a functional BPS (Fig. 2A) (7). We next tested whether SF1 was required for E′ complex formation by analyzing complex formation in SF1-depleted extracts. Depletion of SF1 from wild-type or U2AF-depleted nuclear extracts prevented formation of both the E and E′ complexes, as assayed by native gel (Fig. 2B, lanes 2 and 4). We next asked whether recombinant SF1 could reconstitute E′ formation in U2AF- and SF1-depleted extract (Fig. 2C). Pre-mRNAs incubated under E′ complex-forming conditions in U2AF- and SF1-depleted nuclear extract were unable to form E′ complex (Fig. 2C, lane 2), but recombinant SF1-C4 (a functional SF1 mutant) (29) was able to restore E′ formation (Fig. 2C, lane 3). Addition of recombinant U2AF65 had no effect on the doubly depleted extract (Fig. 2C, lane 4).
FIG. 2.
Formation of the E′ complex requires SF1, and the complex contains U1 snRNP. (A) E′ complex formation in the absence of a functional BPS within the pre-mRNA. Complexes were formed on pre-mRNAs containing a scrambled BPS (mutBPS) in wild-type nuclear extract (NE) or U2AF-depleted nuclear extract (ΔU2AF). (B) E′ complex formation requires the branch binding protein SF1. Native agarose gel shift of complexes formed in wild-type (NE) or U2AF-depleted (ΔU2AF) nuclear extract which had been SF1 immunodepleted (+). (C) SF1 reconstitutes formation of E′ in depleted extracts. Native agarose gel shift of complexes formed in U2AF-depleted nuclear extract which had been SF1 immunodepleted (+) and reconstituted with recombinant SF1-C4 mutant (lane 3) or U2AF65 (lane 4). (D) E′ complex formation is dependent on the presence of a 5′ splice site within the pre-mRNA. Complexes were formed on a body-labeled ([α-32P]UTP) wild-type (WT) pre-mRNA in extract saturated with unlabeled mutant competitor RNA. (E) E′ complex contains U1 snRNP. Shown are results from RT primer extension of RNAs purified from isolated complexes (lanes 2 and 3) and RNA isolated from an aliquot of precipitated nuclear extract (lane 1). The pre-mRNA, U1 snRNA, and U2 snRNA are indicated.
In order to examine the effect of the pre-mRNA sequence elements on E′ complex formation, mutant RNAs were prepared in which one or more sequence elements were eliminated. We challenged E′ complex formation on wild-type body-labeled pre-mRNA in U2AF-depleted nuclear extract that was preincubated with a 500-fold excess of cold mutant RNAs (Fig. 2D). Competitor RNAs containing a 5′ splice site inhibited E′ formation, but no inhibition was observed with a substrate that lacked this sequence element (Δ5′SS), indicating that a 5′ splice site is both necessary and sufficient for formation of the E′ complex.
Since the E′ complex requires a 5′ splice site to form, we wanted to confirm that the E′ complex contains U1 snRNP. Therefore, both E and E′ complexes were purified from agarose gels, and the isolated RNA was subjected to reverse transcription with the appropriate primers. U1 snRNA was found to be present in both the E and E′ complexes (Fig. 2E), indicating that the E′ complex contains U1 snRNP; we were unable to detect U2 snRNA in either complex (Fig. 2E).
Relationship of E′ to the prespliceosome.
The formation of the E complex commits the bound pre-mRNA to the splicing pathway. Thus, prior formation of E complex at 30°C on a 32P-labeled substrate results in efficient splicing of the labeled RNA even after the addition of a 500-fold excess of competitor RNA (20). We tested the commitment ability of both wild-type and U2AF-depleted extracts by preincubation of pre-mRNA in extract followed by a chase with extract containing an excess of competitor RNA. Direct incubation of a radiolabeled pre-mRNA substrate in extract in the presence of increasing concentrations of unlabeled competitor RNA resulted in the decrease of spliced products (Fig. 3A, lanes 1 to 4, and B). When E complex was formed by preincubation of substrate RNA in wild-type extract, followed by addition of an extract mix containing competitor, no inhibition of splicing was observed (Fig. 3A, lanes 5 to 8). Commitment correlated with E complex formation, since preincubation of a pre-mRNA in wild-type or U2AF-depleted extract on ice followed by a chase with wild-type extract mix containing competitor RNA resulted in significantly decreased splicing (Fig. 3A, lanes 9 to 12 and 13 to 16). We tested whether the formation of E′ commits pre-mRNA to the splicing pathway by preincubation of labeled substrate RNA in U2AF-depleted extract under E′ complex-forming conditions followed by a chase with wild-type extract containing competitor (Fig. 3A, lanes 17 to 20). Under these conditions, normal splicing was observed, indicating that the E′ complex is a functional commitment complex (Fig. 3B).
FIG. 3.
Commitment of pre-mRNA to splicing in the absence of U2AF. (A) The E′ complex contains commitment ability. RNAs were preincubated in nuclear extract (NE) or U2AF-depleted nuclear extract (ΔU2AF) and chased with wild-type nuclear extract saturated with increasing competitor RNA (0.8, 1.4, or 5 pmol). Splicing substrates, products, and intermediates are indicated. (B) Quantification of splicing products (from panel A) generated after the formation of commitment complexes at 30°C under E complex-forming conditions in wild-type extract [NE(30)] or E′ complex-forming conditions in U2AF-depleted extract [ΔU2AF(30)] alongside a control reaction in wild-type extract incubated on ice [NE(0)]. (C) The E′ complex is a precursor to the E complex. Preformed E′ complex chased with nuclear extract (NE; lanes 1 and 2), U2AF65 (lanes 3 and 4) or U2AF65/35 heterodimer (lanes 5 and 6) shifts E′ complex to E complex.
We investigated the relationship between the E′ and E complexes by preforming E′ on a substrate pre-mRNA, chasing with either wild-type extract or recombinant U2AF, and then analyzing the complexes by native agarose gel electrophoresis (Fig. 3C). Following the formation of the E′ complex, addition of wild-type nuclear extract efficiently chased E′ into the E complex (Fig. 3C, lane 2); however, addition of an aliquot of U2AF-depleted extract failed to shift E′ (data not shown). Furthermore, addition of U2AF65 alone or recombinant U2AF efficiently chased E′ into the E complex (Fig. 3C, lanes 4 and 6), suggesting that E′ is a functional precursor to E in the commitment of this pre-mRNA to the splicing pathway.
Detection of BBP in the E′ complex.
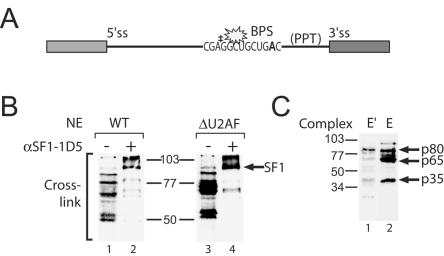
Since pre-mRNAs containing a mutant BPS were incorporated into both the E and E′ complexes (Fig. 2A), we wanted to determine whether SF1 associated with the pre-mRNA branch region in the absence of U2AF. We prepared a pre-mRNA modified at the BPS with the photo-cross-linker benzophenone (24) and containing a unique 32P label to facilitate the analysis of cross-links (Fig. 4A). E′ and E complexes were formed on derivatized pre-mRNAs with the same efficiency as that observed with unmodified pre-mRNAs (data not shown), and the resulting complexes were photolyzed to cross-link proteins bound proximal to the branch region (Fig. 4B). Immunoprecipitation with an antibody specific to SF1 enriched an 80-kDa cross-link, confirming the association of the BBP with the pre-mRNA in U2AF-depleted as well as wild-type HeLa extract (Fig. 4B, lanes 2 and 4). We determined the cross-links specific to E and E′ by performing SDS-PAGE with cross-linked proteins eluted from purified complexes (Fig. 4C). In the E complex, three proteins with molecular weights of 80, 65, and 35 kDa were efficiently cross-linked to the branch region: these are SF1 and most likely the two subunits of U2AF, U2AF65 and U2AF35 (Fig. 4C, lane 2). The cross-link to the 3′ splice site binding protein U2AF35 is consistent with a U2AF-mediated bending of the pre-mRNA (17). In the E′ complex, only the cross-link to the BBP SF1 was detected (Fig. 4C, lane 1). This result suggests that SF1 can associate with the pre-mRNA in the absence of U2AF.
FIG. 4.
E′ complex contains SF1 at the branch point. (A) Pre-mRNA containing a unique 32P label (‡) was site-specifically modified with benzophenone (star) at the BPS. (B) Cross-linking SF1 to the branch region under E′ complex conditions. Modified pre-mRNAs were incubated in wild-type (WT) or U2AF-depleted (ΔU2AF) nuclear extracts under E (lanes 1 and 2) and E′ (lanes 3 and 4) complex-forming conditions, irradiated with UV light, treated with RNase A, and analyzed by SDS-PAGE before (lanes 1 and 3) and after (lanes 2 and 4) immunoprecipitation with anti-SF1 antibody (anti-SF1-1D5). The position of SF1 is indicated. (C) E′ complex contains SF1. Proteins cross-linked to the BPS were purified from agarose gel-isolated E′ complex (lane1) and E complex (lane 2). Indicated are molecular weight markers and cross-links to p80, p65, and p35 proteins.
Directed hydroxyl radical probing of pre-mRNA structure in the E′ complex.
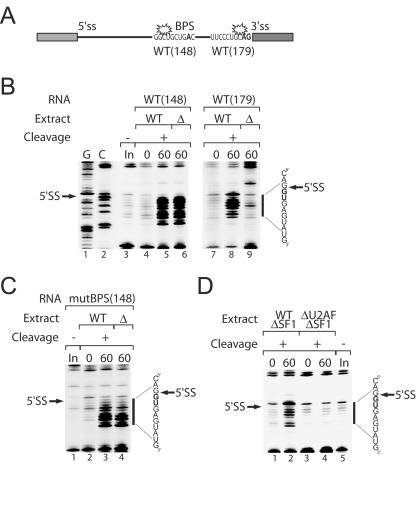
We probed the structure of RNA bound in the E′ complex using pre-mRNA that was site-specifically derivatized with a directed hydroxyl radical probe, Fe-BABE, tethered to the pre-mRNA (16). Diffusible radicals produced from a tethered iron-EDTA moiety are excellent probes of local structure, since they are capable of cleaving only the phosphodiester backbone within ∼10 to 20 Å from their site of generation. Using a wild-type RNA construct that had been derivatized with Fe-BABE at either the branch region (WT148) or the 3′ splice site (WT179), we probed complexes formed in wild-type and U2AF-depleted extracts (Fig. 5A and B). Reactions were incubated under E or E′ complex formation conditions (lacking ATP and MgCl2), followed by Fe-BABE-mediated cleavage and analysis of cleavage products by RT primer extension. As we have previously reported, under E complex-forming conditions, probes tethered to either the branch region or 3′ splice site generated strong cleavages at the 5′ splice site (Fig. 5B, lanes 5 and 8), indicating the proximity of these sequence elements in the E complex. No cleavages at the 5′ splice site were observed from either derivatized pre-mRNA when incubated in nuclear extract on ice (Fig. 5B, lanes 4 and 7). Interestingly, strong 5′ splice site cleavages were observed under E′ complex-forming conditions in U2AF-depleted extract using pre-mRNAs derivatized at the branch region (Fig. 5B, lane 6). In contrast, no 5′ splice site cleavage was observed under the same conditions using the probe derivatized at the 3′ splice site (Fig. 5B, lane 9). The same panel of cleavages was also generated independently of a functional branch region (Fig. 5C). Pre-mRNA derivatized analogously to WT148 but with a scrambled branch point sequence was incubated under E or E′ complex-forming conditions and probed by Fe-BABE-mediated cleavage. As reported previously, 5′ splice site cleavages are still observed in substrates lacking a functional branch sequence (Fig. 5C, lane 3) (16). Similarly, the same pattern of cleavages was observed from the mutant branch region pre-mRNA that was incubated under E′ complex-forming conditions (Fig. 5C, lane 4), suggesting that a functional branch region is not required for the proximity of the 5′ splice site-branch region and consistent with the weak dependence of E complex formation on the branch sequence (7).
FIG. 5.
Directed hydroxyl radical cleavage of pre-mRNA within the ATP-independent prespliceosome complexes. (A) Pre-mRNA substrate derivatized with Fe-BABE (star) at the branch region [WT(148)] or 3′ splice site [WT(179)]. (B) Analysis of pre-mRNA cleavage at the 5′ splice site in E and E′ complexes. Reactions were performed in nuclear extract (WT) and U2AF-depleted (Δ) nuclear extract probed with WT148 (lanes 4 to 6) or WT179 (lanes 7 to 9) pre-mRNAs. Fe-BABE cleavage reactions were initiated after incubation at 30°C for 0 or 60 min. Reverse transcription of cleavage reactions were compared to input RNA (In; lane 3) for location of reverse transcription stops. G and C sequencing is shown (lanes 1 and 2). (C) Analysis of pre-mRNA cleavage at the 5′ splice site with mutant BPS pre-mRNAs. Pre-mRNAs derivatized at position 148 containing a scrambled branch region were probed in wild-type (WT) and U2AF-depleted (Δ) nuclear extracts. Fe-BABE cleavage reactions were initiated after incubation at 30°C for 0 or 60 min. Cleavage reactions (lanes 2 to 4) were compared to input RNA (In; lane 1) for location of reverse transcription stops. (D) Analysis of pre-mRNA cleavage at the 5′ splice site in SF1-depleted extracts. Reactions were performed in SF1-depleted nuclear extract (WT ΔSF1) or ΔU2AF SF1-depleted extract (ΔU2AF ΔSF1) and probed with WT148 pre-mRNA. Fe-BABE cleavage reactions were initiated after incubation at 30°C for 0 or 60 min. Reverse transcription lanes of cleavage reactions were compared to input RNA (In; lane 5) for location of reverse transcription stops. The location of and sequence around the 5′ splice site are indicated. Regions of significant cleavage are represented by the vertical bar.
We wanted to determine whether the observed cleavages at the 5′ splice site were dependent on the presence of the BBP SF1. Previously, it was found that the depletion of U1 snRNP from nuclear extract abolished the cleavages at the 5′ splice site that were observed with pre-mRNAs derivatized at either the branch region or the 3′ splice site (16). This result correlates well with the observation that U1 snRNP is required for E complex formation. Consistent with this result, experiments with branch region-derivatized RNAs in U1 snRNP- and U2AF-depleted extracts did not show cleavages at the 5′ splice site, and assembly of the E′ complex was also abolished in this extract (data not shown). We then specifically depleted SF1 from wild-type and U2AF-depleted nuclear extracts and probed Fe-BABE-modified pre-mRNAs for complex formation and 5′ splice site cleavage. Although the E complex could not be detected on native gels in SF1-depleted extract, strong cleavages were still observed at the 5′ splice site (Fig. 5D, lane 2). However, depletion of both SF1 and U2AF abolished E′ complex formation and severely diminished the observed cleavages at the 5′ splice site (Fig. 5D, lane 4). This result may be interpreted as a redundant requirement for SF1/U2AF in tethering the branch region and 5′ splice site; either of these factors bound to the branch region can interact with U1 snRNP at the 5′ splice site, resulting in the proximity of these two pre-mRNA sequence elements.
DISCUSSION
We have detected the formation of a novel CC, designated E′, on pre-mRNA substrates that were incubated in U2AF-depleted extracts. The E′ complex forms in the absence of ATP and contains U1 snRNP and SF1 (Fig. 6). E′ complex formation requires a 5′ splice site and functional U1 snRNP as well as SF1. Our results suggest that E′ is analogous to the yeast commitment complex CC1 and may be similar to the yeast CC1 in several ways: formation of both the E′ complex and CC1 requires a functional 5′ splice site and the presence of U1 snRNP. The formation of the E′ complex does not require a functional branch sequence mirroring the requirement for formation of CC1. Furthermore, since the overall pathways of spliceosome assembly in yeast and mammals are highly conserved (1), it is likely that formation of the E complex and the yeast CC follow similar assembly pathways. The observation of E′ in HeLa extracts suggests a further parallel with the yeast splicing system: the assembly of the mammalian spliceosome may proceed through the formation of two ATP-independent CCs.
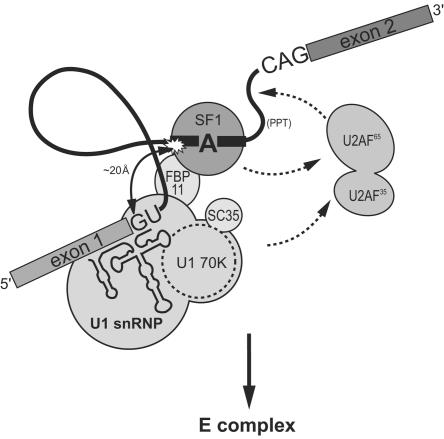
FIG. 6.
Assembly of the E complex proceeds through early recognition of the 5′ splice site-branch region followed by recruitment of U2AF to the polypyrimidine tract. Pre-mRNAs are committed to prespliceosome assembly by the association of U1 snRNP with the 5′ splice site. SF1 interacts with the branch point sequence and may contact FBP11, providing a bridging interaction between SF1 and U1 snRNP. This interaction results in the proximity of the BPS and the 5′ splice site (curved arrow). U1 snRNP and/or SF1 recruit U2AF to the polypyrimidine tract (dotted arrows) resulting in structural reorganization of the 3′ splice site and E complex formation.
Experiments with the yeast system have demonstrated that the commitment of pre-mRNAs to splicing is dependent on U1 snRNP and that the complexes formed in U2 snRNP-depleted extracts are intermediates in spliceosome assembly (20, 34). Similarly, our results suggest that commitment of mammalian pre-mRNAs to splicing is dependent upon the interaction of U1 snRNP with the 5′ splice site and the interaction of SF1 at the branch region followed by the recruitment of U2AF to the polypyrimidine tract and 3′ splice site to give the E complex (Fig. 6).
Although no sequences 3′ to the BPS in yeast have been shown to be essential for CC formation, the formation of the mammalian E complex is dependent on the presence of a functional polypyrimidine tract. However, the roles of SF1/U2AF and yeast BBP/Mud2p in CC formation may be more similar in the mammalian and yeast systems than previously reported. It has been reported that yeast commitment complex formation (CC2) can be detected in ΔMud2p extracts but not in extracts lacking BBP (1, 2). This result and other data (1) suggest that BBP is more important to commitment complex formation than Mud2p. Our results demonstrate the formation of the E′ complex in U2AF-depleted extracts but not in SF1-depleted extracts. These observations of HeLa nuclear extract parallel the previously reported observations of yeast extract and suggest that not only are bridging interactions conserved between the yeast and mammalian systems (1), but the temporal recognition of the pre-mRNA and the formation of the CCs may also be conserved. These observations are somewhat surprising, given that yeast splicing is dependent on a functional branch sequence and mammalian splicing is dependent on a functional polypyrimidine tract.
The precise determinants of commitment in the prespliceosome are not fully understood. The studies outlined here indicate that commitment occurs in the absence of U2AF but confirm that the commitment ability of E′ requires U1 snRNP association with the 5′ splice site. Although the U1 snRNP-pre-mRNA association in E′ requires base pairing between the U1 snRNA and the 5′ splice site, actual commitment probably precedes this association. For example, a pre-mRNA that was incubated in extract where the 5′ end of U1 snRNA has been removed by RNase H digestion does not splice when added to a reaction containing wild-type U1 snRNP, suggesting “commitment” in the digested extract (data not shown).
U2 snRNP is recruited to the CC through its association with U2AF independent of U1 snRNP at the 5′ splice site. U2 snRNP can be recruited to a minimal RNA substrate that contains a branch region and polypyrimidine tract forming a complex in nuclear extract, Amin, that resembles the A complex buts lacks U1 snRNP (27). However, this Amin complex rapidly dissociates in the presence of ATP, and therefore stable U2 snRNP recruitment to the assembling spliceosome must involve other factors that are not present in the Amin complex. A recent report has demonstrated that although commitment to splicing involves ATP-independent CC formation, commitment to splice site choice takes place in the prespliceosome A complex (22). Therefore, commitment to spliceosome formation is a separate event from commitment to splice site choice, which may take place concomitant with U2 snRNP stabilization at the branch region.
The role of U1 snRNP in CC formation remains unclear. In the canonical pathway, prespliceosome formation is dependent on U1 snRNP to form the E complex (and the E′ complex described here). However, a class of pre-mRNAs containing cis-acting elements that promote spliceosome formation in the absence of U1 snRNP has been identified (10). In these pre-mRNAs, the recruitment of U2 snRNP to the branch region involves additional factors: specifically, members of the SR protein family which can reconstitute splicing in U1 snRNP-depleted extracts (9).
The SR proteins are essential splicing factors with characteristic RNA binding domains and a serine/arginine-rich motif which associates with the pre-mRNA at early stages and throughout spliceosome assembly (14, 32, 35). The SR proteins have been shown to influence splice site selection (13, 41) as well as commit pre-mRNAs to the splicing pathway (14). Indeed, it has been shown that different SR proteins are capable of committing pre-mRNAs to splicing with pronounced substrate specificity (14). Furthermore, the role of SR proteins may be to provide the initial recognition of the 5′ and 3′ splice sites (35). For example, SF2/ASF has been shown to increase U1 snRNP binding to the 5′ splice site (13, 18) and SC35, which bridges U1 snRNP and U2AF35 cross-links to the 3′ splice site in an E3′ complex (35, 41). As well, SC35 has also been shown to activate 5′ splice site usage independent of functional U1 snRNP (38). SR proteins may contribute to the formation and commitment ability of the E′ complex, such as initial U1 snRNP recruitment and later U2AF recruitment to the polypyrimidine tract to form the E complex.
The branch binding protein SF1 interacts specifically with the pre-mRNA branch region; however, its role in mammalian prespliceosome assembly has been difficult to define. Work from several groups suggests that SF1 contributes to branch point definition and acts cooperatively with U2AF to define not only the branch region but also the 3′ splice site (4, 33). Our results indicate that SF1 is involved in the formation of the E′ complex and may help promote the stable association and proximity of the 5′ splice site and branch region.
A proximity of both the branch region and 3′ splice site with the 5′ splice site in the E complex has been previously observed by use of pre-mRNA probes that were modified with Fe-BABE (16). Similar experiments, under E′ complex-forming conditions in the absence of U2AF, demonstrated that U2AF is required for 5′ and 3′ splice site association in the E complex. However, our data indicate that the 5′ splice site and branch region are close to one another in E′, and this proximity probably reflects bridging interactions between U1 snRNP and the BBP SF1 (Fig. 6) (30). Interactions between SF1, the mammalian Prp40-like protein FBP11, and U1 snRNP have been proposed to bridge the 5′ splice site and the branch region in the E complex (1, 30); a similar structural organization is likely present in the E′ complex.
The fact that no 5′ splice site cleavages are observed in E′ with a pre-mRNA probe that was derivatized at the 3′ splice site is consistent with the role of U2AF as a 3′ splice site recognition factor which brings that portion of the RNA into direct proximity with the branch region and hence the 5′ splice site (17). It has been demonstrated previously that U2AF35 interacts with the U1 snRNP specific protein U1-70K via an interaction with the SR protein SC35 that serves to bridge the two ends of the intron (41). It is possible that this interaction serves to recruit U2AF to the polypyrimidine tract or that the proximity of the 5′ and 3′ splice sites is a consequence of the 5′ splice site-branch interaction that was first established in the E′ complex.
U1 snRNP association with the pre-mRNA defines the 5′ splice site and the interaction of U2AF65 with the polypyrimidine tract, and specifically, the direct recognition of the AG dinucleotide by U2AF35 defines the 3′ splice site (25, 42, 44, 45). The proximity of the 5′ splice site and branch region in the E′ complex are consistent with a U1 snRNP- and SF1-facilitated recruitment of U2AF to the pre-mRNA (8, 21) and argue for a very early definition of the intron that precedes this association.
Acknowledgments
We thank M. Green and D. Rio for providing U2AF65 and U2AF35 expression plasmids, respectively, and Angela Krämer for providing the SF1-C4 expression plasmid and anti-SF1 antibodies. We thank Steven Chaulk, Mark Glover, Kevin Wilson, and members of the MacMillan lab for helpful discussions and comments.
O.A.K. was supported by a graduate fellowship from the Alberta Heritage Foundation for Medical Research (AHFMR). This work was supported by the Natural Sciences and Engineering Research Counsel of Canada and an establishment grant from AHFMR to A.M.M.
REFERENCES
- 1.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403-412. [DOI] [PubMed] [Google Scholar]
- 2.Abovich, N., X. C. Liao, and M. Rosbash. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8:843-854. [DOI] [PubMed] [Google Scholar]
- 3.Barabino, S. M. L., B. J. Blencowe, U. Ryder, B. S. Sproat, and A. I. Lamond. 1990. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell 63:293-302. [DOI] [PubMed] [Google Scholar]
- 4.Berglund, J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blencowe, B. J., and A. I. Lamond. 1999. Purification and depletion of RNP particles by antisense affinity chromatography. Methods Mol. Biol. 118:275-287. [DOI] [PubMed] [Google Scholar]
- 6.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Champion-Arnaud, P., O. Gozani, L. Palandjian, and R. Reed. 1995. Accumulation of a novel spliceosomal complex on pre-mRNAs containing branch site mutations. Mol. Cell. Biol. 10:5750-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cote, J., R. T. Beaudoin, and B. Chabot. 1995. The U1 small nuclear ribonucleoprotein/5′ splice site interaction affects U2AF65 binding to the downstream 3′ splice site. J. Biol. Chem. 270:4031-4036. [DOI] [PubMed] [Google Scholar]
- 9.Crispino, J. D., B. J. Blencowe, and P. A. Sharp. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265:1866-1869. [DOI] [PubMed] [Google Scholar]
- 10.Crispino, J. D., J. E. Mermoud, A. I. Lamond, and P. A. Sharp. 1996. Cis-acting elements distinct from the 5′ splice site promote U1-independent pre-mRNA splicing. RNA 2:664-673. [PMC free article] [PubMed] [Google Scholar]
- 11.Das, R., and R. Reed. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, H., and M. Rosbash. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419:86-90. [DOI] [PubMed] [Google Scholar]
- 13.Eperon, I. C., D. C. Ireland, R. A. Smith, A. Mayeda, and A. R. Krainer. 1993. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 12:3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, X. D. 1993. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature 365:82-85. [DOI] [PubMed] [Google Scholar]
- 15.Jamison, S. F., A. Crow, and M. A. Garcia-Blanco. 1992. The spliceosome assembly pathway in mammalian extracts. Mol. Cell. Biol. 12:4279-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent, O. A., and A. M. MacMillan. 2002. Early organization of pre-mRNA during spliceosome assembly. Nat. Struct. Biol. 9:576-581. [DOI] [PubMed] [Google Scholar]
- 17.Kent, O. A., A. Reayi, L. Foong, K. A. Chilibeck, and A. M. MacMillan. 2003. Structuring of the 3′ splice site by U2AF65. J. Biol. Chem. 278:50572-50577. [DOI] [PubMed] [Google Scholar]
- 18.Kohtz, J. D., S. F. Jamison, C. L. Will, P. Zuo, R. Luhrmann, M. A. Garcia-Blanco, and J. L. Manley. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119-124. [DOI] [PubMed] [Google Scholar]
- 19.Krämer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 20.Legrain, P., B. Seraphin, and M. Rosbash. 1988. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol. Cell. Biol. 8:3755-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y., and B. J. Blencowe. 1999. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J. Biol. Chem. 274:35074-35079. [DOI] [PubMed] [Google Scholar]
- 22.Lim, S. R., and K. J. Hertel. 2004. Commitment to splice site pairing coincides with A complex formation. Mol. Cell 15:477-483. [DOI] [PubMed] [Google Scholar]
- 23.MacMillan, A. M., P. S. McCaw, J. D. Crispino, and P. A. Sharp. 1997. SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc. Natl. Acad. Sci. USA 94:133-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacMillan, A. M., C. C. Query, C. R. Allerson, S. Chen, G. L. Verdine, and P. A. Sharp. 1994. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 8:3008-3020. [DOI] [PubMed] [Google Scholar]
- 25.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcarcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 26.Michaud, S., and R. Reed. 1993. A functional association between the 5′ and 3′ splice sites is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 7:1008-1020. [DOI] [PubMed] [Google Scholar]
- 27.Query, C. C., P. S. McCaw, and P. A. Sharp. 1997. A minimal spliceosomal complex A recognizes the branch site and polypyrimidine tract. Mol. Cell. Biol. 17:2944-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Query, C. C., M. J. Moore, and P. A. Sharp. 1994. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 8:587-597. [DOI] [PubMed] [Google Scholar]
- 29.Rain, J. C., Z. Rafi, Z. Rhani, P. Legrain, and A. Kramer. 1998. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA 4:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 31.Reed, R., and M. D. Chiara. 1999. Identification of RNA-protein contacts within functional ribonucleoprotein complexes by RNA site-specific labeling and UV crosslinking. Methods 18:3-12. [DOI] [PubMed] [Google Scholar]
- 32.Roscigno, R. F., and M. A. Garcia-Blanco. 1995. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA 1:692-706. [PMC free article] [PubMed] [Google Scholar]
- 33.Rutz, B., and B. Seraphin. 1999. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA 5:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59:349-358. [DOI] [PubMed] [Google Scholar]
- 35.Staknis, D., and R. Reed. 1994. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell. Biol. 14:7670-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 37.Stern, S., D. Moazed, and H. Noller. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481-489. [DOI] [PubMed] [Google Scholar]
- 38.Tarn, W. Y., and J. A. Steitz. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 8:2704-2717. [DOI] [PubMed] [Google Scholar]
- 39.Will, C. L., S. Rumpler, J. Klein Gunnewiek, W. J. van Venrooij, and R. Luhrmann. 1996. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 24:4614-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, K. S., and H. F. Noller. 1998. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell 92:131-139. [DOI] [PubMed] [Google Scholar]
- 41.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 42.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 43.Zamore, P. D., J. G. Patton, and M. R. Green. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355:609-614. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zorio, D. A., and T. Blumenthal. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402:835-838. [DOI] [PubMed] [Google Scholar]