Abstract
A central question in histone code theory is how various codes are recognized and utilized in vivo. Here we show that TBL1 and TBLR1, two WD-40 repeat proteins in the corepressor SMRT/N-CoR complexes, are functionally redundant and essential for transcriptional repression by unliganded thyroid hormone receptors (TR) but not essential for transcriptional activation by liganded TR. TBL1 and TBLR1 bind preferentially to hypoacetylated histones H2B and H4 in vitro and have a critical role in targeting the corepressor complexes to chromatin in vivo. We show that targeting SMRT/N-CoR complexes to the deiodinase 1 gene (D1) requires at least two interactions, one between unliganded TR and SMRT/N-CoR and the other between TBL1/TBLR1 and hypoacetylated histones. Neither interaction alone is sufficient for the stable association of the corepressor complexes with the D1 promoter. Our data support a feed-forward working model in which deacetylation exerted by initial unstable recruitment of SMRT/N-CoR complexes via their interaction with unliganded TR generates a histone code that serves to stabilize their own recruitment. Similarly, we find that targeting of the Sin3 complex to pericentric heterochromatin may also follow this model. Our studies provide an in vivo example that a histone code is not read independently but is recognized in the context of other interactions.
In eukaryotic cells, genetic information is organized in a highly conserved structural polymer, termed chromatin, which is composed of repeating subunits called nucleosomes. Emerging evidence suggests that covalent modifications of histones have pronounced roles in chromatin structure and function (3, 34, 48). For instance, acetylation and deacetylation of the lysine residues at the histone N-terminal tails correlate in general with transcriptional activation and repression, respectively (30). The recent identification of enzyme systems carrying out histone modifications, together with the discovery of binding proteins that “read” covalent marks on histones, has led to the proposal that the pattern of modifications acts as an information code that influences gene transcription (12, 20, 31, 33, 35).
While it is evident that histone modifications can have profound effects on transcription, it is much less clear as to how different histone codes are recognized and utilized. Current studies appear to suggest that once a code is generated, it can serve as an independent signal that allows the recruitment of a downstream regulatory protein(s). For instance, the bromodomain of TAFII250 and the chromodomain of HP1 are capable of binding acetylated histone tails and the K9 methylated H3 tails in vitro, respectively (2, 19, 21). Furthermore, K9 methylation is required for heterochromatin association of HP1 in cells (28). As a histone code involved in transcriptional activation, a recent study showed that acetylation of H4 on lysines 8 and 12 is sufficient for recruitment of TFIID at least in vitro, presumably through the double bromodomains in TAFII250 (1). However, exactly how these various codes are recognized and utilized in vivo is not clear.
Identified initially as corepressors for nuclear receptors such as thyroid hormone receptors (TR) and retinoic acid receptors (6, 17), N-CoR and SMRT are related proteins and have also been implicated in repression by many other transcription factors, including Mad/Mxi, BCL6/LAZ3, ETO, and CBF {for a review, see reference 14). Recent biochemical studies reveal that both SMRT and N-CoR exist in large protein complexes with an estimated size of 1.5 to 2 MDa and containing a set of core subunits, including histone deacetylase 3 (HDAC3), GPS2, TBL1 (transducin beta-like protein 1), and TBLR1 (TBL1-related protein) (16, 23, 36, 37, 41, 44). Human TBL1 and TBLR1 are highly related WD-40 repeat proteins, sharing 89% sequence identity. A redundant function of TBL1/TBLR1 in repression was revealed by a recent study using small interfering RNA (siRNA) to TBL1 and TBLR1 (41). Both TBL1 and TBLR1 can bind histones H2B and H4 in vitro (41), raising question as to whether these proteins are involved in potential histone code recognition during repression by SMRT/N-CoR complexes. The presence of TBL1 and TBLR1 in the HDAC3-containing SMRT/N-CoR complexes is reminiscent of the RbAp46 and RbAp48 (also highly related WD-40 repeat proteins) in the HDAC1/2-containing Sin3 and NURD complexes (40, 45, 46). In vitro reconstitution experiments indicated that in the NURD complex RbAp46/48 interacts directly with HDAC1/2 to form a core complex required for HDAC1/2 enzymatic activity (47). However, TBL1 and TBLR1 neither interact with HDAC3 directly nor are required for in vitro deacetylation of histones by HDAC3/SMRT or N-CoR complexes (15, 41, 44). Thus, although the presence of two related WD-40 repeat histone-binding proteins appears to be a conserved feature of the class I HDAC complexes, the functional significance of such conservation is not clear.
TBL1 and TBLR1 seem to be multiple functional proteins. Their association with SMRT/N-CoR complexes and involvement in transcriptional repression is underscored by the finding of the yeast SET3 complex as the homologous complex of SMRT/N-CoR (29). The yeast SET3 complex was shown to be involved in repression of the sporulation gene program and to contain Snt2, Sif2, and Hos2, which are the yeast homologs of the mammalian SMRT/N-CoR, TBL1/TBLR1, and HDAC3, respectively (29). On the other hand, the Drosophila TBL1 homolog, ebi, was genetically identified as a putative F-box protein involved in ubiquitin-dependent degradation of Tramtrack88 (8, 24). In addition, mammalian TBL1 (Ebi) was shown to collaborate with Siah1 in a novel pathway for β-catenin degradation (25). More recently, both TBL1 and TBLR1 were reported to be required for transcriptional activation by nuclear receptors and other regulated transcription factors (27). A role in cofactor exchange through ubiquitin-dependent protein degradation was proposed to explain such unexpected functions for TBL1 and TBLR1 in transcriptional activation (27).
Here we present evidence for a novel role of TBL1/TBLR1 in histone code reading and targeting of the HDAC3-containing SMRT/N-CoR to chromatin. In addition, we show that RbAp46/48 in the Sin3A complex have a similar function. We also present evidence that TBL1 and TBLR1 are not absolutely required for T3-dependent activation by TR.
MATERIALS AND METHODS
Cloning and plasmid constructions.
The glutathione S-transferase (GST)-TBL1 and GST-TBLR1 constructs were as described previously (41). The constructs for expression of the tail-less Xenopus H2B (amino acids 32 to 125) and H4 (amino acids 26 to 102) were generated first by PCR followed by cloning as NdeI fragments and into the pRSETA vector. The construct for GST-RbAp46 was a gift from Yi Zhang (University of North Carolina).
Cell culture and siRNA experiments.
Cell culture and siRNA treatment were essentially as described previously (41). For transfection of siRNAs, HeLa α2 cells were first cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% charcoal-stripped serum for 3 days and then transfected at a cell confluency of ∼40 to 50% with the indicated amounts of siRNA by using the TransIT-TKO transfection reagent (Mirus) according to the manufacturer's instructions. Two days after transfection, cells were collected and processed for Western analysis, immunostaining, and reverse transcription-PCR (RT-PCR) or chromatin immunoprecipitation (ChIP) as indicated. For experiments with T3 and TSA, α2 cells first were seeded at a density 4 × 105 cells/100-mm-diameter tissue culture dish. After 24 h of incubation, the culture medium was replaced by DMEM with 10% charcoal-stripped fetal calf serum (CS-FCS) (Gemini Bio-Products) for 3 days, followed by replacement of the medium with fresh CS-FCS supplemented with 10 nM thyroid hormone (T3) or 300 nM trichostatin A (TSA) for up to 60 min. For the experiments involving both siRNAs and T3 or TSA, T3 or TSA was added 2 days after siRNA transfection and incubated for 1 h or as indicated for ChIP assays and for 6 h for RT-PCR analysis. For the treatment with lysine-coenzyme A (Lys-CoA), cells were permeabilized with transport buffer (20 mM HEPES-KOH [pH 7.3], 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM dithiothreitol) containing digitonin (35 μg/ml) on ice for 5 min. After 1 h of incubation with 50 μM of Lys-CoA, the transport buffer was changed to CS-FCS containing 10 nM T3 for 1 h. All siRNAs were synthesized by Dharmacon Research (Lafayette, Colo.). The siTBL1 used contains two siRNAs, 5′-AAGAGAATGGAGCACATGAAA-3′ and 5′-AAGATGAGCATAACCAGTGAC-3′. The siTBLR1 sequence is 5′-AAGGCCCTATATTTGCATTAA-3′. The siHDAC3 used was a SMART pool purchased from Dharmacon.
RNA extraction, RT-PCR, and ChIP.
RNA extraction, RT-PCR, and ChIP were performed as described previously (42). The antibodies against various modified histones were purchased from Upstate Biotechnology (22). The antibodies against SMRT/N-CoR complexes were as described previously (42). Primers used for ChIP analysis were as follows: P1 pair primers, 5′-GGAGGCCAAGGCGGGTAGGTCATCT-3′ and 5′-CCGGGTCAGGGGAAGGAGTCAGGTCA-3′; P2 pair primers, 5′-AGGCCACAGCACCCAATCAAGA-3′ and 5′-AAAGACCGTGTGCAGGGAATGTG-3′.
Western blotting and immunostaining.
Western blotting analyses were performed primarily as described previously (42), using antibodies as indicated. For immunostaining, α2 cells were grown on coverslips in six-well plates and then transfected with 5 nM siTBL1, siTBLR1, or both by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's suggestions. Following 24 h of incubation, α2 cells on coverslips were transferred to a new plate with fresh DMEM containing 10% fetal bovine serum (Gemini Bio-Products). After an additional 2 days of incubation, the transfected cells were washed in phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde in PBS for 15 min, washed, permeabilized in 0.25% Triton X-100 for 5 min, and blocked with 10% normal goat serum. Next, cells were incubated with anti-TBL1 or -TBLR1 for 2 h at 37°C. After being washed with PBS, the cells were incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Molecular Probes) as the secondary antibody (1:200) for 2 h at room temperature. The slides were mounted on a microscope stage and visualized with an LSM 510 fluorescence microscope (Zeiss). Images were analyzed by using LSM imaging software (Zeiss).
In vitro pull-down assay.
GST pull-down experiments for analysis of protein-protein interaction were as described previously (41), using GST fusion proteins and in vitro-translated [35S]methionine-labeled proteins as indicated. For the competition assay using purified histone, 200 ng, 600 ng, or 1.8 μg of hypo- or hyperacetylated histone was preincubated with 35S-H2B or -H4 at 4°C for 1 h before incubation with GST-TBL1. For histone tail peptide binding and competition assays, all peptides were synthesized and purified by Genemed Synthesis Inc. (South San Francisco, Calif.). The sequence of each peptide was as indicated in Fig. 5A. For biotinylated histone peptide binding assay, 1 μg of biotinylated H4 or acetylated H4 tail peptides was immobilized on streptavidin-agarose beads (Invitrogen), and then pull-down assays were performed with in vitro-translated proteins as indicated. For peptide competition assay, 1, 5, or 10 μg of each lysine residue-modified peptide was preincubated with 1 μg of either biotinylated H4 peptide or 35S-H4, followed by incubation with either 35S-TBL1 or GST-TBL1. The binding assays were performed at 4°C for 2 h in binding buffer (20 mM Tris-HCl [pH 7.1], 120 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, 0.1% NP-40, 10% glycerol) containing 1 mM phenylmethylsulfonyl fluoride. Each experiment was repeated at least three times to ensure reproducibility, and a representative result was scanned and quantified by using a UMAX ASTRA 2400S scanner and NIH IMAGE 6.2 software.
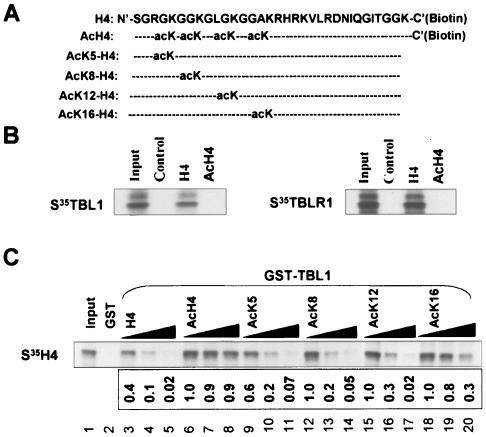
FIG. 5.
The recognition of histone H4 tail by TBL1 is not dependent on a specific lysine. (A) H4 N-terminal tail peptides. The acetylated lysine residue (AcK) in each peptide is indicated. (B) Binding of TBL1 and TBLR1 to a biotinylated H4 tail peptide with or without acetylation on lysines 5, 8, 12, and 16. (C) The binding of [35S]methionine-labeled H4 to GST-TBL1 was competed with various H4 tails as indicated. The increasing amounts of peptides represent 1, 5, and 10 μg, respectively. Note that while AcH4 peptide competed poorly, the rest of the peptides competed nearly as efficiently as unacetylated H4 peptide, except for AcK16. A representative result from three different experiments was scanned and analyzed by using NIH IMAGE 6.2 software. The binding in the presence of the smallest amount of acH4 peptide (lane 6) was arbitrarily set as 1, and the rest of the data are shown as the relative binding in comparison to this binding.
RESULTS
Transcriptional repression by unliganded TR is correlated with the recruitment of the HDAC3-containing SMRT/N-CoR complexes.
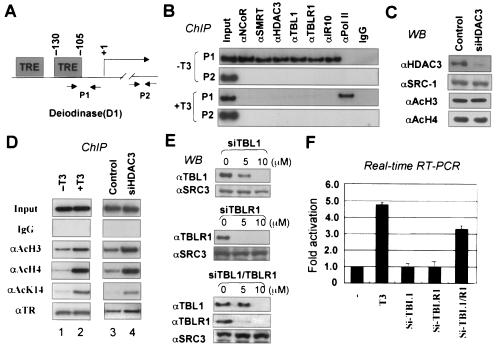
To unravel the roles of TBL1 and TBLR1, we first sought to determine whether the HDAC3-containing SMRT/N-CoR complexes were recruited to an endogenous TR target gene, deiodinase 1 (D1), in a HeLa α2 cell line which constitutively expresses a FLAG-tagged human TRα (32). The D1 gene promoter contains a well-characterized thyroid hormone response element (TRE) located at positions −105 to −130 relative to the transcriptional start site (Fig. 1A). In HeLa α2 cells, this gene is actively repressed by TRα in the absence of T3 and activated in the presence of T3 (32, 41). We used ChIP assays to determine the association of the SMRT/N-CoR complexes with the D1 promoter. We found that SMRT, N-CoR, and their associated proteins HDAC3, TBL1, TBLR1, and IR10 (associated only with N-CoR) were associated with the D1 promoter in untreated HeLa α2 cells but not in those treated with 50 nM T3 for 1 h (Fig. 1B, P1). As controls, the SMRT and N-CoR complexes were not found to associate with the PS2 promoter (data not shown) or with the coding region of the D1 genes in the same experiments (Fig. 1B, P2). Furthermore, in agreement with the presence of SMRT/N-CoR complexes, ChIP assays using antibodies specifically against various modified histones indicated that the D1 promoter was associated with hypoacetylated histones H3 and H4 in the absence of T3 (Fig. 1D, compare lane 1 with lane 2). Consistent with the idea that TR binds constitutively to TRE in chromatin, ChIP assay using a TRα-specific antibody detected the presence of TR under both conditions. These results therefore established that repression of the D1 gene by unliganded TR is correlated with the recruitment of SMRT/N-CoR complexes.
FIG. 1.
TBL1 and TBLR1 have a redundant but essential function in repression of the D1 gene by unliganded TR. (A) Diagram of the D1 promoter showing the positions of TREs and primers used for PCR amplification in ChIP assays. The P2 pair primers amplify a DNA fragment ∼2.5 kb downstream of the transcriptional start site. (B) ChIP assays showing the association of SMRT/N-CoR complexes with the D1 promoter in the absence but not in the presence of T3 (1 h). Note that SMRT/N-CoR complexes were not associated with the P2 region. IgG, immunoglobulin G. (C) Western blotting (WB) showing that siHDAC3 treatment knocked down HDAC3 but did not affect the global levels of H3 and H4 acetylation. (D) ChIP assays showing the changes of histone modifications in response to T3 treatment for 1 h or after siHDAC3 treatment for 72 h. (E) Specific knockdown of TBL1, TBLR1, or both by siRNA as shown by Western blotting. (F) Quantitative RT-PCR analysis of the effect of different siRNA treatments on D1 gene expression. The effect of T3 treatment (6 h) was included as a reference. Error bars indicate standard deviations.
Since T3 treatment not only relieved corepressor complexes but also was expected to recruit coactivators (see Fig. 8B), the change in histone modifications described above was likely the combined result of releasing the HDAC3-containing SMRT/N-CoR complexes and recruiting coactivators such as CBP/p300. To evaluate the effect of corepressor complexes more specifically, we made use of an HDAC3-specific small interfering RNA (siHDAC3) to knock down the expression of HDAC3. HeLa α2 cells were first transfected with siHDAC3 or a control siRNA for 3 days and then processed for ChIP assays to determine the histone modifications over the D1 promoter. The ChIP results showed that treatment of α2 cells with siHDAC3 did not affect TR binding but led to a significant increase of the levels of acetylated H3 and H4 (Fig. 1D, compare lane 3 with lane 4). The control Western analysis showed that siRNA treatment led to a more than 90% reduction of HDAC3 protein and that knocking down HDAC3 did not affect the global acetylation levels of histones H3 and H4 (Fig. 1C). These results reveal a crucial role for HDAC3 in mediating deacetylation by SMRT/N-CoR complexes, in full agreement with the fact that HDAC3 is the major HDAC associated with SMRT/N-CoR complexes (16, 23). Taking these results together, we conclude that SMRT/N-CoR complexes are targeted to the D1 gene promoter by unliganded TR and contribute to histone hypoacetylation primarily through its associated HDAC3 activity.
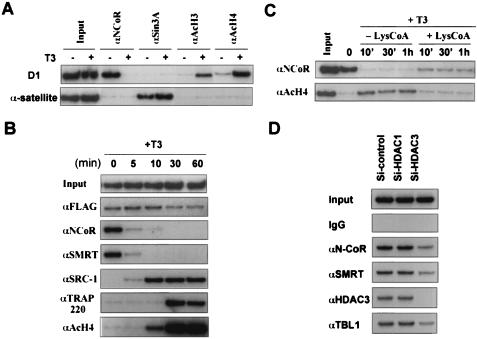
FIG. 8.
Histone hypoacetylation alone is sufficient neither for recruiting SMRT/N-CoR nor for maintaining chromatin association in the absence of unliganded TR-SMRT/N-CoR interaction. (A) ChIP assays showing that although both loci contained hypoacetylated H3 and H4, N-CoR was targeted only to the D1 promoter, whereas Sin3A was found only in the α-satellite sequence of chromosome 4. (B) ChIP assay was used to assess the association of corepressors and coactivators with the D1 promoter after incubation with T3 for different amounts of time. The time course experiment revealed that T3 treatment led to a rapid dissociation of the SMRT/N-CoR complexes from the D1 promoter, recruitment of the coactivators SRC-1 and TRAP220, and changes in H4 acetylation. (C) Inhibition of histone acetylation is not sufficient to prevent dissociation of N-CoR from the D1 promoter in the presence of T3. Cells were first permeabilized with digitonin and incubated with or without Lys-CoA (50 μM) for 1 h before addition of T3. The residual association (∼20%) of N-CoR was likely due to the effect of Lys-CoA on the ability of TR to bind T3 and/or coactivators. (D) ChIP assay showing that siRNA against HDAC3 but not HDAC1 impaired the targeting of SMRT/N-CoR complexes to D1 promoter. IgG, immunoglobulin G.
Removal of TBL1/TBLR1 does not affect the expression of SMRT/N-CoR or disrupt the entire SMRT/N-CoR complexes.
To investigate the roles of TBL1 and TBLR1 in SMRT/N-CoR complexes, we made use of siRNAs specific for TBL1 or TBLR. We first established the conditions in which treatment of HeLa α2 cells with siTBL1, siTBLR1, or both led to specific knockdown of TBL1, TBLR1, or both (Fig. 1E). We then examined the effect of these siRNAs on the repression function of SMRT/N-CoR complexes by analyzing D1 gene expression. Quantitative real-time RT-PCR (qPCR) analysis (Fig. 1F) showed that treatment with either siTBL1 or siTBLR1 alone had little, if any, effect on D1 gene expression. However, the combination of siTBL1 and siTBLR1 led to a substantial derepression of the D1 gene (∼3.3-fold increase). A similar level of de-repression was observed when siHDAC3 was used (data not shown; see Fig. 6E). Under the same conditions, a 6-h T3 treatment resulted in ∼5-fold activation. These results establish that TBL1 and TBLR1 are functionally redundant but together are essential for D1 gene repression by unliganded TR. As expected, no effect on D1 gene expression was observed when the regular HeLa cells lacking FLAG-TRα were used in this experiment (data not shown).
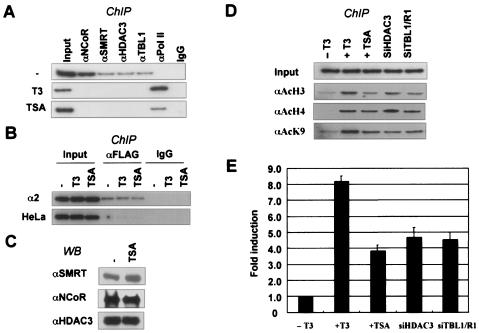
FIG. 6.
Histone hypoacetylation is essential for targeting SMRT/N-CoR to the D1 promoter. (A) ChIP assays showing that like T3 treatment, TSA treatment dissociated SMRT/N-CoR complexes from the D1 promoter and led to the recruitment of Pol II. IgG, immunoglobulin G. (B) ChIP assays showing that TSA treatment did not affect binding of TR (FLAG-TRα) to the D1 promoter. Note that no signal was detected when the parental HeLa cells lacking FLAG-TRα were used for ChIP. (C) Western blotting (WB) analysis showing that TSA treatment did not affect the levels of N-CoR and HDAC3 in HeLa α2 cells. (D) ChIP assays showing that like T3 treatment, TSA treatment led to increased histone acetylation. Similarly, siHDAC3 and double siTBL1/TBLR1 treatments (72 h) also resulted in an increase in histone acetylation. (E) qPCR analysis comparing the effects on D1 gene expression of treatment with T3 (6 h), TSA (6 h), and siRNAs against HDAC3 or against TBL1 and TBLR1 (72 h). Error bars indicate standard deviations.
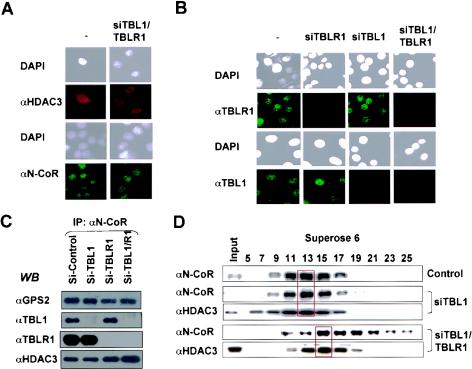
To understand why TBL1 and TBLR1 are redundant but essential for repression of the D1 gene by unliganded TR, we first tested whether knocking down TBL1/TBLR1 affected the stability, subcellular location, and/or assembly of the remaining complexes. Since they are putative F-box proteins (25, 27), knocking down TBL1 and/or TBLR1 may lead to increased rather than decreased levels of SMRT and N-CoR. Interestingly, in multiple attempts we had not observed any significant effect of the combined siTBL1 and siTBLR1 treatment on the levels of TRα, SMRT, N-CoR, and HDAC3 (data not shown) (41). By immunofluorescence staining we also observed no effect on the nuclear location of N-CoR and HDAC3 upon knockdown of both TBL1 and TBLR1 (Fig. 2A), whereas the specificity and knockdown of TBL1 and TBLR1 were confirmed (Fig. 2B). Furthermore, coimmunoprecipitation experiments showed that siTBL1 and siTBLR1 treatment did not affect the association of HDAC3 and GPS2 with N-CoR (Fig. 2C) and SMRT (data not shown). In further support, gel filtration analysis of the cellular extracts derived from double-siRNA-treated cells showed cofractionation of HDAC3 with N-CoR, although the complex became smaller (Fig. 2D), a result expected with the removal of TBL1 and TBLR1. Thus, removal of TBL1 and TBLR1 does not appear to affect the expression and subcellular localization of N-CoR and HDAC3 or the association of the core subunits HDAC3 and GPS2 with SMRT and N-CoR, although we cannot exclude the possibility that the association of other components in the SMRT and N-CoR complexes may be affected.
FIG. 2.
TBL1 and TBLR1 are not required for stability, nuclear localization, and association of HDAC3 with SMRT/N-CoR. (A) Immunostaining showing that knocking down both TBL1 and TBLR1 did not affect the levels of expression as well as nuclear localization of N-CoR and HDAC3. DAPI, 4′,6′-diamidino-2-phenylindole. (B) Immunostaining showing the specificity of siRNAs against TBL1 and TBLR1. (C) Immunoprecipitation (IP) and Western blotting (WB) showing that N-CoR remains associated with HDAC3 and GPS2 in the absence of TBL1 and TBLR1. (D) Gel filtration experiment demonstrating that the N-CoR still cofractionated with HDAC3 and became smaller after removal of TBL1/TBLR1 by siRNAs. Note that knocking down TBL1 alone had little effect on the gel filtration pattern of N-CoR.
TBL1 and TBLR1 are functionally redundant but are required for targeting SMRT/N-CoR complexes to chromatin by unliganded TR.
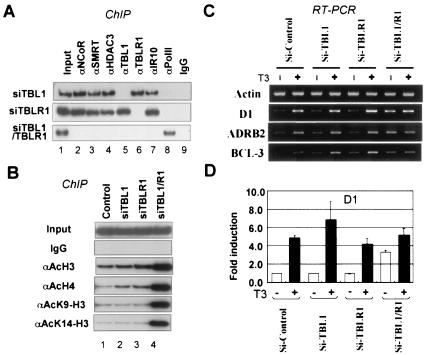
We next tested whether TBL1 and TBLR1 could be required for targeting of the SMRT/N-CoR complexes to the D1 gene promoter by unliganded TR. The α2 cells were first treated with siRNAs against TBL1 and TBLR1 alone or in combination, and the association of the corepressor complexes with the D1 promoter was determined by ChIP assays. Treatment of cells with siRNA against TBL1 or TBLR1 led to a specific inhibition of its own association with the D1 promoter but had little effect on the association of other components of the complexes (Fig. 3A). Significantly, simultaneous reduction of TBL1 and TBLR1 led to inhibition of the association of the entire complexes with the D1 promoter (Fig. 3A). Thus, these results uncover an essential role for TBL1/TBLR1 in targeting SMRT/N-CoR complexes to the D1 promoter by unliganded TR.
FIG. 3.
Knocking down of TBL1/TBLR1 together impaired the chromatin targeting of the SMRT/N-CoR complexes by unliganded TR. (A) ChIP results showing that while knocking down TBL1 or TBLR1 individually had a limited effect, knocking down both TBL1 and TBLR1 abolished the association of SMRT/N-CoR complexes with the D1 promoter. IgG, immunoglobulin G. (B) ChIP assays showing that knocking down both TBL1 and TBLR1 led to a significant increase in the levels of histone acetylation. (C) Semiquantitative RT-PCR results showing the effect of knockdown of TBL1, TBLR1, or both on expression of three different TR target genes, D1, ADRB2, and BCL3. (D) qPCR analysis of the D1 data in panel C. Error bars indicate standard deviations.
In support of the results that removal of TBL1/TBLR1 led to the dissociation of the SMRT and N-CoR complexes from the promoter, ChIP assay using a Pol II-specific antibody revealed the presence of Pol II after siTBL1 and siTBLR1 treatment (Fig. 3A). Furthermore, ChIP assay (Fig. 3B) showed that double siRNA treatment led to substantial increases in acetylation of histones H3 and H4, whereas single siRNA treatment had only a slight effect. Control Western analysis showed that knocking down both TBL1 and TBLR1 did not affect the global histone acetylation (data not shown). Finally, control experiments confirmed that neither the expression of FLAG-TRα nor its binding to the D1 promoter was affected by any of these siRNA treatments (data not shown). Collectively these results demonstrate that TBL1 and TBLR1 are functionally redundant but essential for targeting of SMRT and N-CoR complexes to the D1 promoter by unliganded TR.
TBL1 and TBLR1 are not absolutely required for T3-dependent activation.
Because TBL1 and TBLR1 were recently shown to be required for transcriptional activation by various nuclear receptors (27), we next examined the effect of siTBL1 and siTBLR1 on T3-dependent activation. For this purpose, HeLa α2 cells were first treated with the indicated siRNA for 72 h and then induced with 50 nM T3 for 6 h. Total RNA was prepared from each sample, and the responses of three TR target genes (11), namely, D1, ADRB2 (β-2 adrenergic receptor), and BCL3 (B-cell lymphoma 3-encoded protein) to T3 were assessed by semiquantitative RT-PCR. The results in Fig. 3C show that treatment with siTBL1 or siTBLR1 alone did not appreciably affect activation induced by T3 for all three genes. However, double siRNA treatment led to a substantial derepression of all three TR target genes. Under this condition, no significant T3-dependent activation was observed, most likely as a result of loss of repression rather than loss of activation. Indeed, qPCR analysis confirmed that siTBL1 and siTBLR1 alone did not affect the T3 induction of the D1 gene and that T3 treatment led to a further increase of D1 transcription even in the case of double siRNA treatment (from ∼3.6- to ∼5.1-fold) (Fig. 3D). Thus, in our experimental setting, TBL1 or TBLR1 does not appear to be required for T3-dependent activation of all three TR target genes we have tested.
TBL1 and TBLR1 recognize and bind preferentially to hypoacetylated H2B and H4.
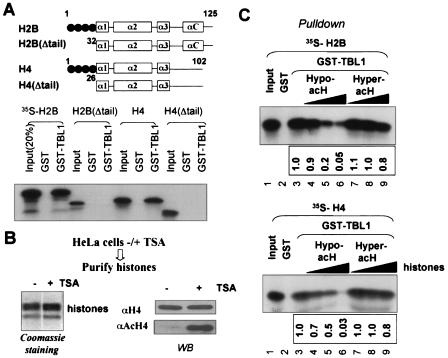
We recently showed that among four core histones, TBL1 and TBLR1 bind preferentially to histones H2B and H4 (41). Thus, we hypothesized that TBL1 and TBLR1 could play a role in targeting SMRT/N-CoR complexes to chromatin via their interaction with histones. We first sought to determine whether binding of TBL1 and TBLR1 to histones requires the histone N-terminal tails. For this purpose, we performed in vitro pull-down assays using GST-TBL1 and in vitro-translated full-length or tail-less H2B and H4. The results in Fig. 4A indicate that the N-terminal tail is required for TBL1 to bind H2B and H4. Similar results were obtained when GST-TBLR1 was used (data not shown).
FIG. 4.
Binding of TBL1 and TBLR1 to histones H2B and H4 requires histone N-terminal tails and is affected by acetylation. (A) In vitro GST pull-down assays showing that the N-terminal tails of histones H2B and H4 are required for their interaction with TBL1 and TBLR1. (B) Core histones purified from TSA-treated and untreated HeLa cells. Left panel, Coomassie blue staining of purified core histones; right panel, Western blot (WB) analysis using antibodies against H4 or acetylated H4. (C) Competition experiments showing that TBL1 and TBLR1 bound preferentially to hypoacetylated histones. The binding of [35S]methionine-labeled H2B or H4 to GST-TBL1 was challenged with increasing amounts of purified core histones (200 ng, 600 ng, and 1.8 μg). The experiments were repeated at least three times, and data were highly reproducible. The representative result in this figure was scanned and analyzed by using NIH IMAGE 6.2 software. The binding in the absence of competitive histones (lane 3) was arbitrarily set as 1, and the rest of the data are shown as the relative binding in comparison to the binding in lane 3.
We next sought to determine whether acetylation affects the binding of TBL1 and TBLR1 to histones H2B and H4. We purified hypoacetylated and hyperacetylated core histones from HeLa cells treated with or without TSA. The resulting core histones were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis and visualized by Coomassie blue staining (Fig. 4B, left panel). The Western analysis (Fig. 4B) confirmed that core histones derived from TSA-treated cells were hyperacetylated. To evaluate the effect of histone acetylation on binding of TBL1, we used a competition assay in which the binding of in vitro-translated, [35S]methionine-labeled H2B or H4 to GST-TBL1 was challenged with increasing amounts of hypo- or hyperacetylated core histones derived from TSA-treated or untreated cells. A representative result (Fig. 4C, upper panel) shows that core histones from TSA-treated cells competed approximately 16 times less efficiently for binding of H2B (compare lanes 4 and 9). An even more pronounced difference was observed for the binding of H4 (∼26.7 times less efficient) (Fig. 4C, lower panel). Thus, TBL1 and TBLR1 bind preferentially to hypoacetylated histones, presumably H2B and H4.
We next wished to substantiate the above binding results by an independent assay. Because acetylation on H4 has a more pronounced effect on binding of TBL1, we focused our effort on H4. We first compared the binding of TBL1 and TBLR1 to the chemically synthesized histone H4 tail peptide (amino acids 1 to 30) without acetylation (H4) or with acetylation at lysines 5, 8, 12, and 16 (acH4). To facilitate the pull-down assay, a biotin residue together with a short linker (GGK) was added at the C termini of the peptides. These peptides were immobilized to streptavidin-agarose beads and used to pull down in vitro-translated, [35S]methionine-labeled TBL1 and TBLR1. As shown in Fig. 5B, TBL1 and TBLR1 bound readily to the H4 tail but not to the acetylated H4 tail. These results confirmed a marked effect of acetylation on binding of TBL1 and TBLR1 to histone H4 tails.
To test whether the binding of TBL1/TBLR1 is determined by a specific deacetylated lysine reside in the H4 tail, we set up another competition assay. In this experiment we challenged the binding of in vitro-translated [35S]methionine-labeled H4 to GST-TBL1 with synthetic H4 peptides containing no acetylation, acetylation at all four lysine residues, or acetylation at each individual lysine (Fig. 5C). The results show that the AcK5, AcK8, and AcK12 peptides competed almost as efficiently as the unacetylated H4 tail and that the AcK16 peptide competed less efficiently (Fig. 5C). As expected, the AcH4 tail competed poorly. Thus, among four lysines tested, acetylation on lysine 16 has a clear effect on binding of TBL1. However, multiple lysines appear to contribute to the binding, as the acK16 peptide was still much more efficient in competing binding of H4 than the acH4 peptide.
Histone hypoacetylation is required for stable association of SMRT/N-CoR complexes to chromatin by unliganded TR.
Given the above results that TBL1 and TBLR1 are required for targeting SMRT/N-CoR complexes to chromatin and that TBL1 and TBLR1 bind preferentially to hypoacetylated histones, we wished to determine the role of histone acetylation, if any, in targeting SMRT/N-CoR complexes to the D1 promoter by unliganded TR. Toward this end, we tested the ability of unliganded TR to recruit SMRT/N-CoR complexes to the D1 promoter under the condition where histone deacetylation was blocked by TSA. HeLa α2 cells were treated with or without T3 or TSA for 1 h, and the association of SMRT/N-CoR complexes with the D1 promoter was analyzed by ChIP assays. The results in Fig. 6A showed that, like T3 treatment, TSA treatment resulted in dissociation of the SMRT/N-CoR complexes from the D1 promoter. To test whether TSA treatment also affected binding of TR, we performed ChIP assay with an anti-FLAG antibody. The results in Fig. 6B showed that the binding of TR to the D1 promoter was not affected by TSA treatment. The signal detected with FLAG antibody reflected the binding of TR, because ChIP assay with the parental HeLa cells (without FLAG-TRα) yielded only background signal (Fig. 6B). Furthermore, TSA treatment did not appear to induce degradation of the N-CoR complex, as Western blotting showed that TSA did not affect the levels of N-CoR and HDAC3 in the HeLa α2 cells (Fig. 6C).
To better understand the effect of TSA on release of SMRT/N-CoR complexes from the D1 promoter, we compare the histone modification status under various conditions. As shown in Fig. 6D, TSA treatment led to increased levels of acetylation on both H3 and H4. The increase in histone acetylation is comparable to that observed after siHDAC3 or siTBL1/siTBLR1 treatment. Together, these results suggest that induction of histone acetylation is likely sufficient to dissociate the SMRT/N-CoR complexes from the chromatin, although the potential involvement of other histone modifications could not be formally excluded. Finally, assuming that TSA did not affect the direct interaction between unliganded TR and SMRT/N-CoR, these results imply that the interaction between unliganded TR and SMRT/N-CoR alone is insufficient for the stable recruitment of SMRT/N-CoR complexes. In support of this, we observed no effect of TSA on the interaction between unliganded TR and SMRT/N-CoR in an in vitro GST-TR pull-down assay, as described previously (data not shown) (23). It is noteworthy that a more pronounced effect on histone acetylation was repeatedly observed for T3 treatment in comparison to TSA (Fig. 6B, compare lane 2 to the other lanes), presumably as a combined result of corepressor complex dissociation and subsequent active recruitment of coactivators by liganded TR (see Fig. 7D).
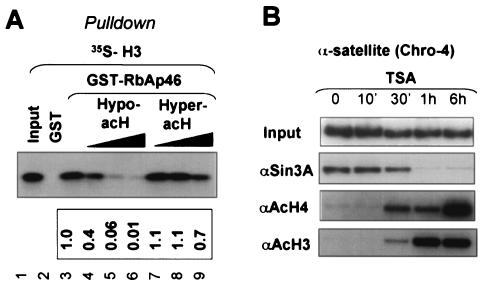
FIG. 7.
RbAP46 also binds preferentially to hypoacetylated histones, and hypoacetylated histones are required for association of the Sin3A complex with pericentromeric heterochromatin. (A) The binding of [35S]methionine-labeled histone H3 to RbAP46 was competed with hypo- or hyperacetylated core histones. The amount of histones used was the same as for Fig. 4C. (B) To assess the effect of histone acetylation on binding of the Sin3A complex to the α-satellite sequence in the chromosome 4 pericentric heterochromatin, HeLa α2 cells were treated with TSA in a time course as indicated. ChIP assay was then carried out to determine the binding of the Sin3A complex and the status of histone acetylation.
Consistent with the result that TSA treatment resulted in dissociation of the SMRT/N-CoR complexes from the D1 promoter, qPCR analysis showed increased transcription of the D1 gene after TSA treatment (Fig. 6E). The increased level of transcription is consistent with the ChIP data in Fig. 6A, showing the recruitment of RNA Pol II after T3 and TSA treatment.
RbAp46 also binds preferentially to hypoacetylated histones.
Since RbAp46 and RbAp48 in Sin3 and Mi-2/NuRD complexes are analogous to TBL1 and TBLR1 in SMRT/N-CoR complexes, we next tested whether RbAp46 also binds preferentially to the hypoacetylated histones. We have shown previously that RbAp46 binds to histone H3 rather than H4 (41). Using a competition experiment set up as for Fig. 4C, we found that core histones derived from TSA-untreated cells competed much more efficiently (∼40-fold) than those from TSA-treated cells (Fig. 7A). Thus, RbAp46, much like TBL1 and TBLR1, also binds preferentially hypoacetylated histones (presumably H3).
Targeting of the Sin3A complex to pericentromeric chromatin is dependent on histone hypoacetylation.
To test whether histone acetylation also influences targeting of the Sin3 complex to chromatin, we made use of our recent finding that the Sin3A complex is present and actively involved in the maintenance of the hypoacetylated status of the pericentric heterochromatin (39). As shown in Fig. 7B, the association of Sin3A with the α-satellite sequence in the chromosome 4 centromere was diminished after TSA treatment, which led to increases in acetylation of H3 and H4. Similar results were observed when the association of Sin3A with the pericentromeric sequences of chromosomes 10 and 11 was analyzed (data not shown). These results suggest that interaction between HDAC complexes and hypoacetylated histones may have a general role in stabilizing the association of HDAC complexes with chromatin. Given their in vitro interaction with hypoacetylated histones (Fig. 7A), we suggest that RbAp46/48 in the Sin3A complex are likely to be the proteins that recognize and bind to the hypoacetylated histone tails in pericentric heterochromatin.
Histone hypoacetylation alone is not sufficient for stable association of SMRT/N-CoR complexes with chromatin.
Recent studies suggest that H3-K9 methylation serves as an epigenetic marker to recruit heterochromatin protein I (HP1) for long-term repression (2, 21, 26, 28). We next tested whether the interaction of SMRT/N-CoR and Sin3A complexes with hypoacetylated histones alone is sufficient to recruit and/or maintain the association of the corepressor complexes with chromatin. We find that the histone hypoacetylation alone is not sufficient to recruit or maintain the binding of the corepressor complexes, based on the following evidence. First, as shown in Fig. 8A, ChIP assays revealed the absence of Sin3A in the D1 promoter and, conversely, the absence of SMRT/N-CoR in the α-satellite of chromosome 4, although both loci were associated with hypoacetylated H3 and H4. This result implies that histone hypoacetylation itself is not sufficient for recruiting either corepressor complex. If histone hypoacetylation is sufficient for binding of SMRT/N-CoR corepressor complexes, we would expect to see the presence of SMRT/N-CoR in the α-satellite of chromosome 4. Second, in a time course experiment where we followed the dissociation of SMRT and N-CoR and changes in histone acetylation upon T3 treatment (Fig. 8B), we found that the dissociation of SMRT and N-CoR from the D1 promoter was a rapid event, occurring within 5 min upon addition of T3. However, the increase in histone acetylation was not detected until 10 min after T3 treatment, indicating that the dissociation of SMRT/N-CoR complexes occurred prior to an increase in histone acetylation. This result implies that in the absence of unliganded TR and SMRT/N-CoR interaction, the interaction between SMRT/N-CoR and hypoacetylated histones is not sufficient to maintain their chromatin association. It is noteworthy that the recruitment of coactivator SRC-1 could be detected as early as 5 min, which may recruit CBP/p300 and account for the subsequent increase in histone acetylation. Consistent with a previous publication (32), the recruitment of TRAP220, a subunit of the TRAP/DRIP/SMCC complex was not detected until 30 min after T3 treatment.
Next we tested whether addition of T3 would lead to dissociation of SMRT/N-CoR under conditions where T3-induced histone acetylation is inhibited. Previous studies suggest that histone acetylation upon T3 treatment is most likely a result of T3-dependent recruitment of CBP/p300 (18). To inhibit T3-induced histone acetylation, we first permeated HeLa α2 cells with digitonin and preincubated the cells with Lys-CoA, a potent CBP/p300-selective hypoxanthine-aminopterin-thymidine inhibitor (50 μM), for 1 h before the addition of T3. The cells were then taken at various time points after addition of T3, and the association of N-CoR with the D1 promoter was determined by ChIP assay. As shown in Fig. 8C, addition of Lys-CoA indeed blocked the T3-dependent increase of H4 acetylation as revealed by ChIP assay. However, much of the N-CoR was dissociated from the chromatin under these conditions, suggesting that blocking histone acetylation in the absence of unliganded TR and N-CoR interaction is by itself insufficient to fully maintain the association of the N-CoR complex with chromatin. Together these data indicate that the interaction with hypoacetylated histones alone is neither sufficient to recruit nor able to maintain the chromatin association of the SMRT/N-CoR complexes.
Role of histone deacetylation by HDAC3 in targeting SMRT/N-CoR complexes to chromatin.
Thus far our results indicated that targeting of SMRT/N-CoR complexes to the D1 promoter by unliganded TR also requires TBL1/TBLR1 (Fig. 3) and hypoacetylated histones (Fig. 6). As HDAC3 is critically important for the observed histone deacetylation over the D1 promoter (Fig. 1D), we next tested the role of HDAC3 in targeting SMRT/N-CoR complexes to the D1 promoter. As shown in Fig. 8D by ChIP assays, treatment with siHDAC3 significantly impaired the association of SMRT, N-CoR, and TBL1 with the D1 promoter, whereas treatment with siHDAC1 had no effect. Thus, deacetylation by HDAC3 has a critical role in targeting SMRT/N-CoR complexes to the D1 promoter. Control coimmunoprecipitation experiments showed that siHDAC3 treatment did not affect expression or formation of the remaining SMRT/N-CoR complexes (data not shown) (41).
DISCUSSION
Role of TBL1/TBLR1 in targeting corepressor SMRT/N-CoR complexes for repression in chromatin.
In contrast to the general notion that the interaction between unliganded TR and SMRT/N-CoR is sufficient to target the corepressor complexes for transcriptional repression, we provide evidence that while such interaction is necessary, alone it is not sufficient for targeting SMRT/N-CoR complexes to an endogenous TR target gene (D1). We show that TBL1 and TBLR1 have an essential role in targeting SMRT/N-CoR complexes to the D1 promoter. In the absence of TBL1/TBLR1, SMRT/N-CoR complexes were no longer associated with the D1 promoter (Fig. 3A), whereas the binding of unliganded TR to the D1 promoter was not affected. This effect on SMRT/N-CoR chromatin association is unlikely to be indirect, because knockdown of TBL1/TBLR1 by siRNAs has no effect on the stability and subcellular localization of SMRT, N-CoR, and HDAC3 (Fig. 2). In addition, knockdown of TBL1/TBLR1 did not affect the association of HDAC3 with SMRT/N-CoR (Fig. 2C), suggesting that the core complex containing HDAC3 and SMRT/N-CoR can form in the absence of TBL1/TBLR1. This result is consistent with previous studies showing that SMRT/N-CoR directly interacts with HDAC3 to promote HDAC3 enzymatic activity and that there is no direct interaction between TBL1/TBLR1 and HDAC3 (15, 16, 44).
Several lines of evidence indicate that the role of TBL1/TBLR1 in targeting SMRT/N-CoR complexes to the D1 promoter likely lies in their interaction with hypoacetylated histones. First, TBL1 and TBLR1 are histone-binding proteins that bind preferentially to hypoacetylated histones (Fig. 4 and 5). Second, histone hyperacetylation induced by TSA treatment resulted in dissociation of the SMRT/N-CoR complexes from the D1 promoter in the absence of T3 (Fig. 6A). Third, knockdown of HDAC3 impaired the targeting of SMRT/N-CoR to the D1 promoter (Fig. 8D). Together these results suggest that the interaction between TBL1/TBLR1 and hypoacetylated histones is important for targeting SMRT/N-CoR complexes to chromatin.
Two-interaction, feed-forward working model for targeting of SMRT/N-CoR complexes to chromatin by unliganded TR.
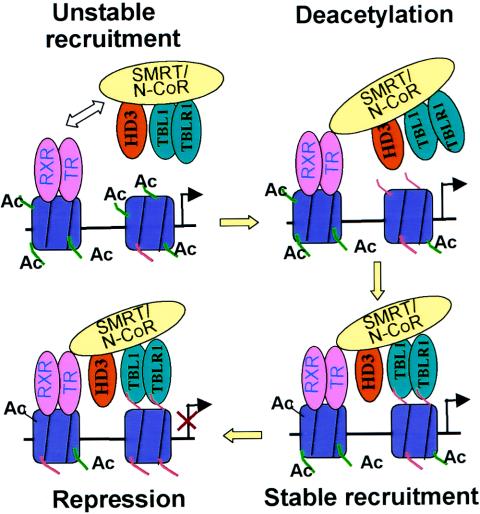
While both TBL1/TBLR1 and hypoacetylated histones are essential, they are insufficient for targeting SMRT/N-CoR to chromatin in the absence of the interaction between unliganded TR and SMRT/N-CoR (Fig. 8A, B, and C). Similarly, unliganded TR alone cannot recruit SMRT/N-CoR complexes to the D1 promoter in the absence of TBL1/TBLR1 (Fig. 3A) or hypoacetylated histones (Fig. 6A). Taken together, our data support a two-interaction, feed-forward working model (Fig. 9) for targeting SMRT/N-CoR complexes to chromatin. First, unliganded TR interacts directly with SMRT/N-CoR. This interaction, although not sufficient for the stable recruitment of the complexes, initiates limited histone deacetylation. TBL1/TBLR1 then recognizes and binds the resultant deacetylated histone tails. This binding stabilizes the association of the SMRT/N-CoR complexes with chromatin and allows deacetylation of additional histones. The subsequent stable (but able to be dynamic) association of SMRT/N-CoR and extensive deacetylation finally lead to repression and maintenance of repression. The result that knockdown HDAC3 impaired the targeting of SMRT/N-CoR complexes to the D1 promoter (Fig. 7D) provides further support for this working model.
FIG. 9.
A two-interaction, feed-forward model for targeting SMRT/N-CoR complexes to chromatin by unliganded TR. First, as TR/RXR heterodimers, unliganded TR binds constitutively to its target genes in chromatin and unstably recruits SMRT/N-CoR. This unstable recruitment initiates limited histone deacetylation through associated HDAC3. Histone deacetylation generates limited hypoacetylated histone tails (H2B and H4) and allows TBL1/TBLR1 to bind. The binding of TBL1/TBLR1 to hypoacetylated histones H2B and H4 stabilizes the recruitment of SMRT/N-CoR complexes by unliganded TR, and the stable recruitment of SMRT/N-CoR complexes in turn leads to further deacetylation and finally transcriptional repression. It should be pointed out that SMRT and N-CoR can also bind to hypoacetylated histone H3 (43), and this interaction, although not sufficient (see Discussion), could also contribute to the stable binding of SMRT/N-CoR complexes to the hypoacetylated histones in a feed-forward mode (43).
It is noteworthy that SMRT and N-CoR were recently shown to interact with hypoacetylated histones, and this interaction was proposed to act through a feed-forward mechanism to promote and maintain histone deacetylation (43). The interaction with hypoacetylated H3 involves a SANT domain in SMRT and N-CoR (43). Thus, like TBL1/TBLR1, SMRT/N-CoR could potentially bind directly to hypoacetylated histones and contribute to recruitment of the SMRT/N-CoR complexes through the feed-forward model as illustrated in Fig. 9. However, given our data that knockdown of TBL1/TBLR1 impaired the association of the SMRT/N-CoR complexes with the D1 promoter, the interaction between SMRT/N-CoR and hypoacetylated histones alone seems insufficient to support chromatin targeting of the SMRT/N-CoR complexes by unliganded TR. The functional significance of this direct binding of hypoacetylated histones by SMRT/N-CoR in targeting SMRT/N-CoR complexes to chromatin remains to be demonstrated.
Role of TBL1/TBLR1 in protein degradation and transcriptional activation.
In addition to their association with SMRT/N-CoR and role in repression, TBL1/TBLR1 and their Drosophila homolog, ebi, have also been implicated as putative F-box proteins involved in degradation of Tramtrack88 and β-catenin (8, 24, 25). More recently, TBL1 and TBLR1 were shown to be required for transcriptional activation by nuclear receptors and other transcription factors (27). In this case, TBL1 and TBLR1 were proposed to be required for corepressor-coactivator exchange that is essential for transcription. In our hands, TBL1 and TBLR1 do not appear to have any significant effect on the stability of SMRT and N-CoR proteins (data not shown). One likely explanation is that TBL1 and TBLR1 are context-dependent multifunctional proteins. If they are associated with proteins such as Siah-1 and SIP or an adaptor protein like Phyllopod (24, 25), TBL1 and TBLR1 can function as F-box proteins involved in degradation. Within the SMRT/N-CoR complexes, TBL1/TBLR1 may not be able to interact with these adaptor proteins, which are required for ubiquitin-dependent degradation, and thus are not involved in protein degradation. Indeed, during our multiple efforts at purification of SMRT and N-CoR complexes, we have not identified the presence of Siah-1 or SIP. However, we showed previously by gel filtration analysis that while SMRT and N-CoR proteins in HeLa nuclear extracts all existed in large protein complexes (1.5 to 2 MDa), there were fractions containing TBL1 but without SMRT/N-CoR (23). We are currently testing whether this smaller TBL1 complex(es) may contain Siah-1 and/or SIP.
In contrast to the results in a recent report (27), we found that knockdown of TBL1 or TBLR1 individually by siRNA had no significant effect on T3-dependent activation of three TR target genes that we tested (Fig. 3C and D). These results could not be explained by inefficiency of the siRNAs we used, because knockdown was confirmed by both Western analysis (Fig. 1E) and ChIP assay (Fig. 3A), and the same results were observed when multiple different siRNAs against TBL1 or TBLR1 were tested. In support of this, we did not observe any significant effect of knockdown of TBL1 or TBLR1 on androgen-dependent activation of several AR target genes in LNCaP cells (H.-G. Yoon and J. Wong, unpublished data). In addition, we did not observe any significant association of TBL1 or TBLR1 with the D1 promoter upon T3 treatment (Fig. 1B). Together, our data argue against an essential role for TBL1 or TBLR1 in transcriptional activation by TR. Whether the requirement for TBL1 or TBLR1 in transcriptional activation is a cell type- or context-dependent phenomenon remains to be solved in the future.
A common role for WD-40 repeat proteins in various corepressor complexes?
A common feature of all three major mammalian class I HDAC-containing complexes (Sin3A, NURD, and SMRT/N-CoR) is the presence of two highly related WD-40 repeat proteins (RbAp46/48 in Sin3A and NURD and TBL1/TBLR1 in SMRT/N-CoR). The association of WD-40 repeat proteins with HDACs extends to Drosophila and mammalian Groucho (5) and yeast TUP1 (10, 38). For these two proteins, their repression function correlates with their histone binding activity (5, 9). The role of this family of proteins in targeting corepressor complexes to chromatin for repression in vivo was first revealed by studies on yeast TUP1 by Davie et al. They showed that the proper chromatin targeting of TUP1/SSN6 in yeast is sensitive to both histone tail mutations and histone deacetylase mutations (7). In this study, we show that targeting of SMRT/N-CoR complexes to the D1 promoter by unliganded TR requires TBL1/TBLR1 (Fig. 3A) and is also sensitive to TSA treatment (Fig. 6A). Similarly, we show that the association of the Sin3A complex with pericentromeric heterochromatin is also sensitive to TSA (Fig. 7B) and that RbAp46 binds preferentially to hypoacetylated histones, presumably H3, in vitro (Fig. 7A). Thus, interaction with hypoacetylated histones seems to be a conserved function among WD-40 repeat proteins that are present in various corepressor complexes. It is therefore tempting to suggest that, like TBL1/TBLR1 in SMRT/N-CoR complexes, all of these WD-40 repeat proteins may function in a feed-forward mode to stabilize the chromatin association of their corresponding corepressor complexes. It will be interesting in the future to determine whether Sif2, the yeast homolog of TBL1/TBLR1 (29), also binds histones and has a similar role in targeting the yeast SET3 complex for repression.
Reading and function of the histone code.
A key issue in the histone code hypothesis is exactly how each code or modification is recognized and utilized. One possibility is that once a histone code is generated, it serves as an independent signal for the binding of a downstream regulatory protein(s), which in turn specifies the function of the code. The findings that the bromodomain of TAFII250 binds specifically acetylated H4 tails and that the chromodomain of HP1 and polycomb differentially recognizes K9- versus K27-methylated H3 tail provide evidence for this idea (2, 4, 13, 19, 21). In this study, we show that while TBL1/TBLR1 can bind hypoacetylated H4 tail in vitro, in vivo such an interaction occurs only in the context of unliganded TR-SMRT/N-CoR interaction and histone hypoacetylation alone is not sufficient to recruit the SMRT/N-CoR complexes to chromatin. Thus, in this case the histone code involved does not seem to serve as an independent signal in a signaling cascade to specify the interaction with a downstream regulatory protein(s). Rather, it functions in a feed-forward mode (Fig. 9) to provide additional interactions to stabilize the recruitment of the corepressor complexes. This feed-forward, two-interaction mode provides at least the following two advantages for regulation: cooperation and specificity. As indicated previously (31), sophisticated functional pathways are often assembled through multiple weak protein-protein interactions that together provide sufficient stability and duration for a biological response. Such a scheme also lends itself well to rapid transcriptional activation in response to hormone, since breaking the interaction between TR and SMRT/N-CoR is sufficient to dislodge the corepressor complexes from the TR target gene (Fig. and 7C 8B). Furthermore, such a mode of interaction would provide specificity by excluding the binding of the SMRT/N-CoR complexes to other hypoacetylated loci. If TBL1/TBLR1 or RbAp46/48 can read hypoacetylated histones independently, one would expect SMRT/N-CoR to associate with pericentromeric heterochromatin and the Sin3A complex to associate with the D1 promoter. Our results show that while the Sin3A complex was found to associate with pericentromeric heterochromatin, the association of SMRT/N-CoR was not detected (Fig. 8A). Conversely, Sin3A was not detected in the hypoacetylated D1 promoter (Fig. 8A). These results suggest that the Sin3A complex may follow the same feed-forward mechanism for targeting to a specific locus for repression. In this regard, the reading of the histone code in the context of other interactions is unlikely to be unique to the SMRT/N-CoR complexes but rather is likely to be a common mechanism for how various histone codes are read and utilized.
Acknowledgments
We thank Jun Qin and Sharon Dent for critical discussion and reading of the manuscript. We also thank Joseph Fondell for providing the HeLa α2 cell line and Andrew Dennis for technical assistance in qPCR analysis.
This work is supported by NIH grants DK58679 to J.W. and GM62437 to P.A.C.
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 4.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 5.Chen, G., and A. J. Courey. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1-16. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454-457. [DOI] [PubMed] [Google Scholar]
- 7.Davie, J. K., R. J. Trumbly, and S. Y. Dent. 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22:693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, X., L. Tsuda, K. H. Zavitz, M. Lin, S. Li, R. W. Carthew, and S. L. Zipursky. 1999. ebi regulates epidermal growth factor receptor signaling pathways in Drosophila. Genes Dev. 13:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 10.Edmondson, D. G., W. Zhang, A. Watson, W. Xu, J. R. Bone, Y. Yu, D. Stillman, and S. Y. Roth. 1998. In vivo functions of histone acetylation/deacetylation in Tup1p repression and Gcn5p activation. Cold Spring Harbor Symp. Quant. Biol. 63:459-468. [DOI] [PubMed] [Google Scholar]
- 11.Feng, X., Y. Jiang, P. Meltzer, and P. M. Yen. 2000. Thyroid hormone regulation of hepatic genes in vivo detected by complementary DNA microarray. Mol. Endocrinol. 14:947-955. [DOI] [PubMed] [Google Scholar]
- 12.Fischle, W., Y. Wang, and C. D. Allis. 2003. Binary switches and modification cassettes in histone biology and beyond. Nature 425:475-479. [DOI] [PubMed] [Google Scholar]
- 13.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 15.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The smrt and n-cor corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 17.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Z. Q., J. Li, L. M. Sachs, P. A. Cole, and J. Wong. 2003. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription. EMBO J. 22:2146-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 20.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 21.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 22.Li, J., Q. Lin, H. G. Yoon, Z. Q. Huang, B. D. Strahl, C. D. Allis, and J. Wong. 2002. Involvement of histone methylation and phosphorylation in regulation of transcription by thyroid hormone receptor. Mol. Cell. Biol. 22:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, S., C. Xu, and R. W. Carthew. 2002. Phyllopod acts as an adaptor protein to link the sina ubiquitin ligase to the substrate protein tramtrack. Mol. Cell. Biol. 22:6854-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuzawa, S. I., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7:915-926. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511-526. [DOI] [PubMed] [Google Scholar]
- 28.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 29.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber, S. L., and B. E. Bernstein. 2002. Signaling network model of chromatin. Cell 111:771-778. [DOI] [PubMed] [Google Scholar]
- 32.Sharma, D., and J. D. Fondell. 2002. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl. Acad. Sci. USA 99:7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 34.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 35.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 36.Vermeulen, M., M. J. Carrozza, E. Lasonder, J. L. Workman, C. Logie, and H. G. Stunnenberg. 2004. In vitro targeting reveals intrinsic histone tail specificity of the Sin3/histone deacetylase and N-CoR/SMRT corepressor complexes. Mol. Cell. Biol. 24:2364-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen, Y. D., V. Perissi, L. M. Staszewski, W. M. Yang, A. Krones, C. K. Glass, M. G. Rosenfeld, and E. Seto. 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]
- 39.Xin, H., H. G. Yoon, P. B. Singh, J. Wong, and J. Qin. 2004. Components of a pathway maintaining histone modification and HP1 binding at the pericentric heterochromatin in mammalian cells. J. Biol. Chem. 279:9539-9546. [DOI] [PubMed] [Google Scholar]
- 40.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 41.Yoon, H. G., D. W. Chan, Z. Q. Huang, J. Li, J. D. Fondell, J. Qin, and J. Wong. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon, H. G., D. W. Chan, A. B. Reynolds, J. Qin, and J. Wong. 2003. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12:723-734. [DOI] [PubMed] [Google Scholar]
- 43.Yu, J., Y. Li, T. Ishizuka, M. G. Guenther, and M. A. Lazar. 2003. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 22:3403-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]