Abstract
Molecular cytogenetic techniques, such as in situ hybridization methods, are admirable tools to analyze the genomic structure and function, chromosome constituents, recombination patterns, alien gene introgression, genome evolution, aneuploidy, and polyploidy and also genome constitution visualization and chromosome discrimination from different genomes in allopolyploids of various horticultural crops. Using GISH advancement as multicolor detection is a significant approach to analyze the small and numerous chromosomes in fruit species, for example, Diospyros hybrids. This analytical technique has proved to be the most exact and effective way for hybrid status confirmation and helps remarkably to distinguish donor parental genomes in hybrids such as Clivia, Rhododendron, and Lycoris ornamental hybrids. The genome characterization facilitates in hybrid selection having potential desirable characteristics during the early hybridization breeding, as this technique expedites to detect introgressed sequence chromosomes. This review study epitomizes applications and advancements of genomic in situ hybridization (GISH) techniques in horticultural plants.
1. Introduction
In plant genomic sciences, since the mid-1950s, various molecular cytogenetic approaches have been developed for chromosome research. Since Mendelian genetics that guides about genetic movement of chromosome from generation to generation, scientific techniques for chromosome observation have meliorated significantly. During mitosis, staining of chromosome is the basic technique for chromosome observation [1].
Cytogenetic research and chromosome analysis are the main aspects in genomics and genetic sciences. Molecular cytogenetic techniques, such as in situ hybridization methods, are admirable tools to analyze the genomic structure and function, chromosome constituents, recombination patterns, alien gene introgression, genome evolution, aneuploidy, and polyploidy [2, 3].
After in situ hybridization technique development by John et al. [4] and Gall and Pardue [5], various approaches were achieved such as radioactively labeled probes improved into nonradioactive probes labeled with biotin [6] and detection by indirect (antibody-fluorochrome conjugate) and direct (fluorochrome detection) staining. Genomic in situ hybridization (GISH) was first used to discriminate the genomes of the intergeneric hybrid between parental genomes, that is, Hordeum chilense and Secale africanum [7].
Genomic in situ hybridization (GISH) is an efficacious technique, that is, used for genome differentiation of one parent from the other by utilizing special chromosome-labeling techniques. GISH has a gratuity role in cytogenetics for investigation of evolutionary relationship of crops and identification of inserted region in the parent from the alien species. GISH technique follows the same protocol as in the fluorescent in situ hybridization (FISH) technique. However, genomic and blocking DNA utilization in GISH differentiate it from FISH analysis [8].
In plant species, genome organization and homology study are carried out by the use of genomic in situ hybridization (GISH). In addition, karyotype analysis of many plant species has been performed by the GISH technique [9]. Another GISH approach is precise dissection of chromosome-pairing behaviors in interspecific hybrids during meiosis. Closely related genomes in interspecific hybrids can be cytological discriminated by well-developed genomic in situ hybridization analysis [10].
Structural rearrangements, evolution of chromosome, and phylogenetic as well as genomic relationships can be studied extensively by modern cytogenetic techniques, that is, in situ hybridization by fluorescent and genomic probes (FISH and GISH) [11, 12]. GISH has an ability to distinguish genomic relationship of polyploids. In polyploids, GISH effectively confirmed the presumed parental genomes and in addition this technique also provides their origin information, that is, whether polyploids are alloploids or autoploids (Table 1) [13].
Table 1.
Genomic in situ hybridization (GISH) application in various horticultural plants.
| Plant | Chromosome number | Genomic DNA | Blocking DNA | Working | Reference |
|---|---|---|---|---|---|
| Fruit | |||||
| Cherry | 2n = 16 | P. avium/P. fruticosa | P. fruticosa/P. avium | Genomic evaluation | [30] |
| Citrus | 2n = 18 | C. reticulata (dig labeled) | C. maxima (biotin labeled) | Karyotype analysis of chromosomes | [17] |
| Diospyros | 2n = 30 | D. kaki + D. glandulosa | No blocking DNA | Multicolor GISH study of somatic hybrid chromosomes | [18] |
| Mango | 2n = 40 | M. caloneura/M. cochinchinensis/M. flava/M. gracilipes/M. griffithii/M. sylvatica/M. indica/M. foetida/M. macrocarpa | No blocking DNA | Phylogenetic division using karyotype tool | [30] |
| Vegetable | |||||
| Onion | 2n = 40 | A. galanthum | A. fistulosum | Gene identification on chromosomal regions | [19] |
| 2n = 40 | A. roylei | A. cepa | Karyotype study and hybridity status in F1 hybrids | [41] | |
| 2n = 40 | A. fistulosum | A. cepa | Genomic analysis of advanced interspecfic generations with relative resistance to downy mildew | [62] | |
| Onion & garlic | 2n = 40 | A. sativum (garlic) | A. cepa (onion) | Chromosome evaluation in onion and garlic somatic hybridization | [35] |
| Potato | 2n = 24 | S. verrucosum, S. hougasii, S. andreanum | S. jamesii | Auto and allopolyploid genomic origin in potato species | [29] |
| Tomato | 2n = 24 | L. esculentum/S. lycopersicoides/S. sitiens | L. esculentum/S. lycopersicoides/S. sitiens | Genomic discrimination in interspecific and intergeneric hybrids | [40] |
| Tomato & potato | 2n = 24 | L. pennellii (tomato) | S. tuberosum (potato) | Genomic analysis of trigenomic hybrids | [38] |
| Ornamental | |||||
| Begonia | 2n = 28 | Tuberous Begonia | B. socotrana | Genomic constituents of begonia hybrids | [39] |
| Clivia | 2n = 22 | C. nobilis/C. caulescens/C. gardenii/C. miniata | C. nobilis/C. caulescens/C. gardenii/C. miniata | Hybrid confirmation | [48] |
| Festulolium (Festuca × Lolium) | 2n = 4x = 28 | Lolium multiflorum/Festuca pratensis | No blocking DNA | Genomic constitution determination in hybrids | [63] |
| Lycoris | 2n = 14–22 | L. aurea/L. radiata | L. aurea/L. radiata | Chromosome complements variation in interspecific hybrids | [51] |
| Orchids | 2n = 38 | P. aphrodite/P. sanderiana/P. mannii/P. vialacea/P. ambainensis/P. stuartiana | P. aphrodite/P. sanderiana/P. mannii/P. vialacea/P. ambainensis/P. stuartiana | Genomic composition and species relationship by genomic analysis | [64] |
| 2n = 26 | P. delenatii/P. rothschildianum | P. delenatii/P. rothschildianum | Phylogenetic classification on the basis of chromosome pairing resemblance | [55] | |
| Grass (Poa jemtlandica, Poa flexuosa, Poa alpina) | 2n = 38 2n = 42 2n = 32 |
Poa jemtlandica/Poa alpina/Poa flexuosa | Poa alpina/Poa flexuosa | Genome composition and hybrid origin confirmation | [65] |
| Primula egaliksensis | 2n = 40 | Primula mistassinica, Primula nutans | Salmon sperm DNA | Genomic composition and evolution of allopolyploid | [66] |
| Rhododendron | 2n = 26 | R. aureum, R. brachycarpum, R. catawbiense “Catharine van Tol,” R. yakushimanum | R. aureum, R. catawbiense “Nova Zembla,” R. yakushimanum | Paternity description of interspecific hybrids | [50] |
| Tulipa | 2n = 24 | T. fosteriana, T. gesneriana | T. tarda | Genomic recombination in three generations (F1, BC1, and BC2) | [59] |
| T. gesneriana and T. fosteriana | Darwin hybrid “Yellow Dover” counter stained with DAPI | Origin investigation by karyotype genomic method | [36] | ||
| T. gesneriana and T. fosteriana | T. sacstatila | Genomic information of interspecific hybrids | [43] | ||
| Lilium | 2n = 24 | Sorbonne and L. regale | Herring sperm | Introgression determination in interpoloid hybrids | [57] |
| Oriental/Asiatic/Martagon | Oriental/Asiatic | Backcross progeny analysis | [60] | ||
| L. longiflorum “Snow queen” | Herring sperm | L. rubellum Baker introgression into L. longiflorum Thunb. | [67] | ||
| L. longiflorum | Herring sperm | Genomic evaluation of backcross progenies | [68] | ||
In situ hybridization by genomic DNA (GISH) is an approach to identify alien chromosomes as well as chromatin and chromosomal rearrangements, that is, formed due to mosaic chromosomes [14]. Genome constitution visualization and chromosome discrimination from different genomes in allopolyploids are carried out by GISH practices [15].
The purpose of this study is to report the applications and advancements of GISH technique on the genome of horticultural crops.
2. Chromosomal Evaluation
In situ hybridization method, using genomic DNA of two species as a probe, is an effective way to approach the individual chromosome identification of nonsomatic hybrids [16]. Similarly, [17] analyzed satsuma mandarin chromosome for evolution and identification of individual chromosome by double-target GISH method of in situ technique. After staining, 18 chromosomes of satsuma mandarin were classified into eight groups on the basis of position and relative size of CMA (chromomycin A3) region as well as relative length of chromosome. Citrus reticulata Blanco (Dig-rhodamine labeling) and Citrus maxima Burm (biotin-fluorescein isothiocyanate) were utilized as probe DNAs. GISH clearly identified 6 individual heterozygous chromosomes and 6 pairs of speculated homozygous chromosomes. In ten chromosomes, containing clear GISH signals on the CMA (+) sites, Dig-rhodamine-labeled regions were detected on 9 chromosomes which showed close evolutionary relationship of satsuma mandarin with C. reticulata Blanco. Further, GISH results also proved the involvement of C. maxima Burm in the satsuma mandarin origin. Therefore, the GISH karyotype technique has potential for homologous and individual chromosomal identification of citrus species.
Identification and differentiation of parental chromosomes in somatic hybrids of fruiting plants can be analyzed in detail by the GISH technique such as in Diospyros (persimmon) hybrids. Multicolor GISH analysis of Diospyros kaki (containing 90 total chromosomes) × Diospyros glandulosa (containing 30 total chromosomes) somatic hybrids proved this statement. Under fluorescence microscope, GISH findings showed 90 D. kaki chromosomes and 30 D. glandulosa chromosomes in somatic hybrids. Using GISH advancement as multicolor detection is a significant approach to analyze the small and numerous chromosomes such as in Diospyros species. Another important point is that after five years of subculturing, somatic hybrid plants of Diospyros can hold chromosomes of their parents [18].
Chromosome region detection is a remarkable application of GISH in cytogenetics. In introgression breeding, discrimination of different genomes and chromosome locus identification is carried out by this method. Yamashita et al. [19] applied the GISH technique to find out chromosome site of Rf (a restoring gene of pollen fertility) locus by comparing GISH results of male sterile and male fertile plants of introgressed progenies. To determine Rf locus location on chromosome, Allium galanthum genomic DNA was used as probe DNA, while for blocking DNA, Allium fistulosum genomic DNA was utilized for in situ hybridization of F1 hybrids of both species and backcross generations. In F1 hybrids, 8 chromosomes from A. galanthum were markedly differentiated from A. fistulosum. Furthermore, chromosome introgression by continuous backcrossing was possible. Important breakthrough in these findings is that A. galanthum chromosomal region (especially from male fertile plants) in four backcross generations, that is, BC4 to BC7 were detected simultaneously through GISH in one chromosome. GISH findings confirmed that the location of Rf locus is signalized on the 5F chromosome of male fertile plant, that is, A. fistulosum.
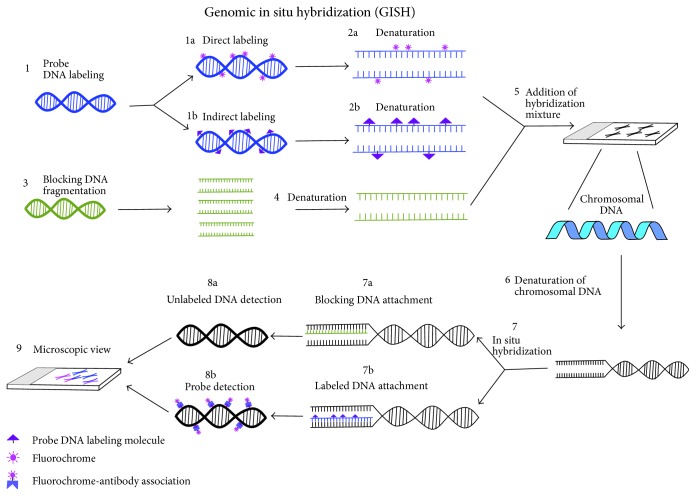
Advancement in GISH is double GISH technique, in which two genomic DNA probes differentially labeled are used, which provides an improvement in the simple GISH method (Figure 1). Therefore, parental genomic DNA of both candidates (having approximately equal concentration) participated in hybridization with homologous sequences [20].
Figure 1.
Genomic in situ hybridization (GISH) mechanism diagram.
In mandarins and lemon-lime group, heteromorphic chromosome-pair evaluations on the basis of rDNA sites and CMA banding patterns were performed to assess the evolutionary relationships [21, 22]. However, these methods obtained some improvement regarding evolutionary relationships of citrus, but more accurate methods were required for comprehensive karyological study. The genomic in situ hybridization (GISH) technique is a useful mean for chromosome and genome characterization in somatic hybrids and polyploids [23]. In many somatic hybrids of fruit trees such as in Citrus aurantium L. + Poncirus trifoliata (L.) Raf. in citrus and D. kaki L. × D. glandulosa Lace, parent chromosome identification was completed by the GISH method [24, 25].
3. Cytogenetical Classification
Using genomic in situ hybridization (GISH), [26] demonstrated noteworthy genomic differentiation among the diploid North and Central American species of potato. GISH results indicated the first evidence that the North and Central American tetraploid species (belongs to longipedicellata series, Solanaceae) are allotetraploids. GISH findings clearly discriminated parental genome in potato species. This GISH genomic classification potential, on the basis of discrimination, was also in accordance with [27, 28] DNA-sequencing description.
North and Central American potato species, containing hexaploid level, are genomically originated from Solanum demissum and Solanum hougasii. GISH investigation was carried out by using their presumed diploid (AA, BB, PP) or tetraploid (AABB) parental genomic DNA, to determine ploidy recombination in Solanum demissum and Solanum hougasii. GISH information reveled that S. hougasii has an allotropic behavior, that is, one genome from AA and the other belonged to BB. In addition, S. hougasii third genome is more intimately related to P genome or species related to P genome. S. demissum, in comparison, containing three chromosome sets are closely related to the basic genome A. GISH-based classification of potato species was in agreement with taxonomic division of Mexican hexaploid species. In which, S. hougasii belonged to the allopolyploid iopetala group while S. demissum belonged to the autopolyploid acaulia group. Therefore, GISH, a DNA technique, plays a key role to classify the species by using modern cytogenetical approaches [29].
Genomic in situ hybridization (GISH), a molecular technique, can be a helpful tool for the species classification. In GISH analysis, chromosome labeling is uniform and intense with same species probes. On the other hand, such labeling is inadequate and irregular if probe DNA is from different species. Reference [30] applied the GISH method on mango (Mangifera indica). He selected 8 wild Mangifera species for using as probes with M. indica as a chromosome sample. His main aim was to investigate a relationship between 8 wild species with M. indica. A basic focusing criterion was on signal strength of genomic in situ hybridization on M. indica metaphase chromosomes. On the basis of number and intensity of hybridization signals, eight wild species were effectively phylogenetically classified into four groups. GISH results clarified that Mangifera sylvatica Roxb probes showed highest signal intensity on M. indica chromosomes. Hence, GISH findings showed a closed relationship between M. indica and M. sylvatica [31], using AFLP markers, already confirmed the phylogenetic relationship among other species. However, GISH phylogenetic classification further improved this relationship. Consequently, GISH technique is a precise way to classify Mangifera species.
4. Genomic Constitution
Horticultural crop genomic compositions and parental chromosomes can be investigated by utilizing GISH methods such as in strawberry [32] and D. kaki [24].
GISH analysis can significantly distinguish the different genomes, foremost the evaluation on the basis of chromosome size in each genome. Discrimination of the different genomes proved the incidence of rearrangements after interspecific hybridization [33]. Karyotype and genomic study of sour cherry proved that GISH can differentiate chromosomes between parental species chromosomes. On the basis of size and centromere position, karyotype results of Prunus avium and Prunus cerasus clearly distinguish chromosomes from one another. Utilization of genomic DNA as a probe helps in hybridization with species-specific repetitive sequences which are dispersed over the genome. Hybridization distribution in 32 chromosomes of sour cherry showed 16 chromosomes came from P. avium, while the rest of the 16 chromosomes from P. fruticosa. These finding ensured that P. cerasus genome constituents are composed of P. fruticosa and P. avium [34].
In somatic hybrids, chromosome and cytoplasm study has a key role in molecular analysis. Chromosome constitution helps in identification of parental chromosomes. Important chromosomal composition was figured out by [35], using genomic in situ hybridization analysis, in somatic hybrids derived from onion (Allium cepa L.) crossing with garlic (Allium sativum L.). Results clarify that one line containing 40 chromosomes is composed of 17 garlic chromosomes and 20 onion chromosomes, while the rest of the three were chimeric chromosomes. On the other hand, one line containing 41 chromosomes having similar composition pattern is composed of 21 onion chromosomes. Important finding was that chimeric chromosome composition origin was due to onion and garlic chromosome fusion. Therefore, chromosome deletion caused fragmentation of chromosomes. Structural amendments in chromosomes may be a reason of chromosome number variations between the hybrid lines. PCR-RFLP analysis of both hybrid lines was agreeable with that of GISH results that onion has more parental contribution in somatic hybrids.
In molecular cytogenetics, clear and unambiguous genomic distinction can be obtained by GISH. A phenomenon of genomic composition and genomic distinction is widely used in plants containing large chromosomes such as in Tulipa [36] and Lilium [37]. However, advances of GISH are that it has differentiated parental genomes in small chromosome-containing plants such as tomato [38].
GISH analysis was carried out in begonia for Begonia socotrana and tuberous hybrid chromosome identification in various ‘Elatior'-begonia hybrids. This study helped out to obtain information related to chromosome number and parental origin of these cultivars. Tuberous Begonia genomic DNA was used as probe DNA (labeled with digoxigenin-11-dUTP) while B. socotrana genomic DNA as a blocking DNA. Clear distinctions of tuberous Begonia genome were visualized when probe concentration was 150 ng per slide, and blocking DNA concentration was with 30 times more than that of B. socotrana. According to GISH distinction protocol, ‘Elatior'-begonia hybrids divided into 2 groups, that is, B. socotrana containing short chromosomes (0.6 μm to 1.03 μm in length) and tuberous Begonia containing long chromosomes (1.87 μm to 3.88 μm in length), respectively. In ‘Elatior'-begonia hybrids, chromosome numbers that came from tuberous Begonia ranged from 14 to 56 while those from B. socotrana ranged from 7 to 28. Consequently, such GISH findings recommended variations in ploidy levels among ‘Elatior'-begonia hybrids. Intergenomic recombination has not been observed in these hybrids. In begonia breeding, genome composition and ploidy estimation are key points for further approaches and this can be accomplished by GISH molecular cytogenetic tools [39].
Genomic in situ hybridization (GISH) provides comparatively efficient, however less accurate, mean to observe genome constitution at whole chromosome or recombinant segment stage. In sexual and somatic hybrids, GISH technique is a significant way to make a difference between parental genomes. This method was applied in many plant species such as tomato. Yuanfu et al. [40] analyzed intergenic and interspecific hybrids of Solanum lycopersicoides, Solanum sitiens, and Lycopersicon esculentum by using the genomic in situ hybridization method for chromosome differentiation and relationship assessment among three genomes. L. esculentum is a cultivated tomato while Solanum lycopersicoides and Solanum sitiens are wild nightshade species. Cultivated tomato genome, in hybrids, was clearly distinguishable from two nightshade wild species by using standard protocol conditions. More specific and precise conditions applied in the GISH method can also distinguish nightshade species genome. This indicated a phylogenetically distant relationship between L. esculentum and two wild species. Sequence homology sharing was high from S. lycopersicoides and S. sitiens. During meiosis, chromosomal associations of intergeneric and interspecific hybrids were consistent. During diakinesis, F1 hybrids of L. esculentumum × S. lycopersicoides and L. esculentum × S. sitiens express univalent formation behavior. However, S. lycopersicoides × S. sitiens F1 hybrids formed bivalents during diakinesis. F1 hybrids of L. esculentum × S. sitiens express lower pairing frequency between homologous chromosomes as compared to L. esculentum × S. sitiens hybrid plants. This was due to the development of an allotetraploid and a monosomic addition. Homologous chromosome constituents were observed in trigenomic hybrid containing 12 additional chromosomes from S. sitiens and 2 from Solanum lycopersicoides.
Genomic in situ hybridization (GISH) potential to confirm constitution phenomenon in somatic hybrids was elaborated by [41]. Findings revealed that all Allium cepa × Allium roylei F1 hybrids had a chromosome number 16, that is, diploid (2n) while chromosome morphology was not differed among hybrids. A. roylei genomic DNA was used as a probe while A. cepa served as blocking DNA. According to GISH information, 8 chromosomes in each hybrid expressed green fluorescence which indicated A. roylei genomic DNA whereas 8 chromosomes exhibited blue fluorescence, a sign of blocking DNA. In addition, metaphase chromosome spreads did not show any genomic recombination in somatic hybrids. Therefore, GISH analysis verified the maternal origin in onion hybrids.
5. Polyploidy Confirmation
Genomic in situ hybridization (GISH) is a powerful tool in confirming the origin of polyploid taxon origination and can also offer initial insights into the genomic rearrangement rate. Camellia reticulata, an ornamental horticultural plant, has a polyploidy behavior. Variation rate, with basic chromosome number x = 15, is from diploid (30) to hexaploid (90). To determine the evolutionary history and genome composition of C. reticulata, GISH analysis was applied. C. pitardii and C. saluenensis genomic DNA was applied as a probe DNA for labeling and hybridization of C. reticulata metaphase chromosomes. C. pitardii-dyed section was observed in tetraploid (4n) and hexaploid (6n) camellia plants, respectively. However, C. saluenensis-painted part was only expressed in hexaploid (6n) genome. Clear evidence is provided by GISH about allopolyploid evolutionary origin of Camellia reticulata. GISH proved that polyploid progenitors are diploid C. saluenensis, C. reticultata, and C. pitardii. Development of allotetraploids were due to C. reticultata hybridization with C. pitardii while allohexaploid development was because of these allotetraploid hybridization with C. saluenensis [42].
Genome composition in polyploids, that is, diploid (2n), triploid (3n), and tetraploid (4n), can be illustrated by using GISH mechanism. GISH helps in understanding the polyploidy formation due to interspecific crosses between Tulipa gesneriana and Tulipa fosteriana. Diploid progeny (2n = 24) showed equal distribution of parental chromosomes, that is, one set of chromosomes from one parent and one from the other. However, in triploid plants, 24 chromosomes from one parent (T. gesneriana) while 12 from other parent (T. fosteriana) contributed. In addition, tetraploid genome comprised of 36 T. gesneriana chromosomes and 12 T. fosteriana chromosomes. In tulip hybrids, localization of GISH signals on chromosomal regions of T. gesneriana were observed predominately in intercalary and telomeric or subtelomeric positions. Moreover, in triploid and tetraploid, putative rRNA gene positions were found in telomeric and intercalary regions [43].
6. Hybrid Verification
For parental genome discrimination, visualization in somatic hybrids may require hybridization mixture stringency, post hybridization washes, and most important ratio of genomic and blocking DNA [44]. Chromosome-banding technique can not differentiate the chromosomes related to parental species. In this regard, the GISH method of detection has a priority for hybrid analysis and verification [45]. GISH verified hybridity status in various horticultural crops such as in citrus [25], cucumber [46], and Buddleja [47].
For hybrid verification in Clivia (Amaryllidaceae), GISH and giemsa C-banding analyses were applied. Initial information clarifies that “Belgian hybrids” and “German hybrids” were similar to Clivia miniata on the basis of karyotypic and genomic investigation. C. cyrtanthiflora confirmed its hybridity status. GISH needed more stringency level and high blocking DNA ratio as compared to probe DNA. Parental genome location in mitotic metaphase chromosomes of five artificially developed hybrids was identified by GISH and C-banding methods. On metaphase plate, there was a considerable ability for different parental genome centromeres to inhabit 2 different concentric domains. Genome association did not correlate with centromeric heterochromatin presence or absence. Therefore, Clivia hybrids can be identified easily through chromosomal study, even during vegetative phase [48].
Hybrids, developed from interspecific crosses, should be verified either contained hybrid characters or not. GISH analytical technique has proved to be the most exact and effective way for hybrid status confirmation [49].
In plants, acquiring large chromosomes, GISH is an informative tool to remarkably distinguish donor parental genomes in hybrids [38].
Interspecific hybrids were obtained from interspecific crosses between 5 Rhododendron species. Positive GISH results can only be obtained by using mitotic chromosome spreads through anthers. Significant distinction of paternal and maternal chromosomes in hybrid chromosomes was observed when 50 ng probe DNA concentration was used with blocking DNA, that is, collectively 3 mg per milliliter hybridization genomic mixture. In alien genome and chromosome constitution detection in Rhododendron hybrids, GISH has a practical importance. Therefore, this method is a reliable improvement in breeding analysis [50].
Interspecific hybrids of Lycoris taxa are one-third in natural habitat. Most of them are sterile and morphologically highly diverse. Partial fertility helps in observing meiotic process in L. aurea × L. radiata hybrids. In backcross progenies, recovery of functional gametes can be successfully obtained. GISH describes modifications in chromosome number and constitution of such functional gametes. High genome homology was observed between Lycoris MT- and A-genomes. This indicated homoeologous recombination and partial fertility of interspecific hybrids during meiosis. Variation in recombination pattern and chromosome complements in functional gametes recommended that interspecific hybridization is a major reason of Lycoris species diversification [51].
7. Introgression Breeding
Introgression breeding is used to transfer horticultural traits successfully in the progeny. In tulip, this approach was applied in Darwin hybrids and Tulipa gesneriana. Extent of intergenomic recombination was demonstrated by GISH technique. Total genomic DNA used for this purpose was T. fosteriana cv. Princeps and T. gesneriana cv. Ile de France. Complete distinction of parental genomes and intergenomic recombination identification was approached by GISH. T. fosteriana chromosome contribution and recombinations in genome differed in all progenies. Maximum recombination by T. fosteriana was five. In recombination, one fragment of T. gesneriana and one fragment of T. fosteriana were present. A number of recombinant segments in each chromosome was two, and their allocation was ranged from distal to highly interstitial regions. Darwin hybrids served an important role as intermediate parent for T. fosteriana germplasm introgression into the T. gesneriana distinction [52].
In molecular cytogenetics, GISH efficiency is applied to visualize homoeologous chromosome pairing during meiosis, parental genome assortment, and genome recombination [12]. The GISH method was used in genomic constitution investigation to determine downy mildew resistance potential through introgression breeding in (A. cepa × A. roylei) hybrids by backcrossing with A. cepa. The GISH study revealed that location of A. roylei fragments, in backcross generations, containing downy mildew resistance gene is at chromosome number 3's long arm (distal end) [53, 54].
Parental genome detection and differentiation during meiotic chromosome pairing in Paphiopedilum F1 hybrids was carried out by GISH. Genome homology between the parental species elucidates the close relationship between the parental species. This demonstrated regular chromosome association during P. delenatii interspecific crossing with P. bellatulum and P. rothschildianum. However, hybrids developed from distant parents carrying karyotype verification such as P. delenatii and P. rothschildianum interspecific crossing with P. callosum, P. glaucophyllum, P. micranthum, and P. moquetteanum showed irregular chromosome pairing. Multivalents and autosyndesis presence showed that discrimination of Paphiopedilum species chromosomes was due to some structural modifications and microrearrangements during interspecific hybridization. In Paphiopedilum hybrids, genetic interaction and genome homology demonstrate the chromosome pairing phenomena [55].
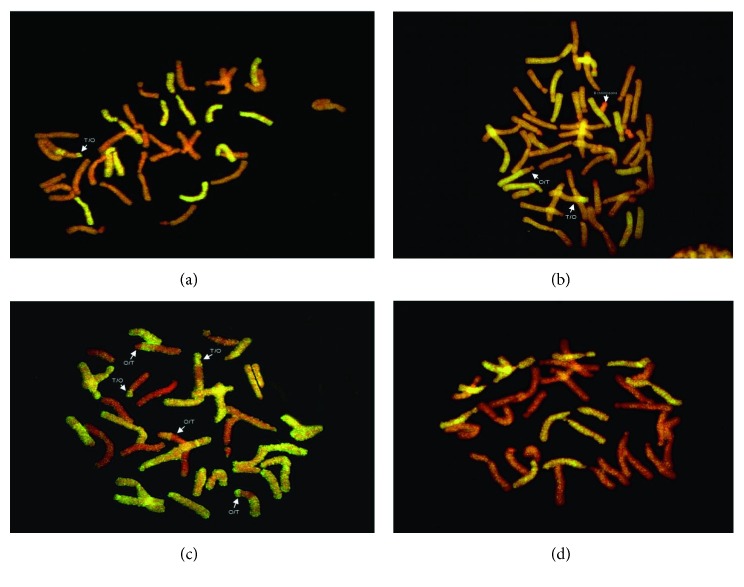
In Lilium, hybridization of allotriploid lily with diploid lily is a successful method of introgression breeding [56]. The GISH study explained that OTO lilies (allotriploid) introgressing with OO genome lilies (diploid) showed aneuploidy behavior. In addition, allotriploid lilies played an important role in T-genome chromosome variation in the progenies. GISH indicated that male sterile plants as maternal parents can be used for developing aneuploids [57]. In Lilium (Oriental × Trumpet) hybrids, GISH analysis identified the genomic constituents and parental recombination. Results revealed that most of OT (Oriental × Trumpet) hybrids are developed by backcrossing the F1 hybrid with oriental parent. Figures 2(a), 2(b), and 2(d) showed more oriental chromosomes as compared to trumpet chromosomes. OT hybrid “Motown” showed 24 Oriental and 12 Trumpet chromosomes with one T/O genomic recombination. Similarly, a tetraploid (4x) OT Lilium hybrid “Stentor” showed 36 Oriental chromosomes with one O/T recombinant chromosome and only 12 Trumpet chromosomes were signalized containing one T/O parental recombination. In one triploid hybrid “Morini,” chromosomal composition were remarkably deviate from the normal genomic composition, that is, Trumpet chromosomes were more as compared to Oriental chromosomes which proved that this hybrid was developed by backcrossing with Trumpet parent. Furthermore, GISH results confirmed no recombinant chromosomes in “Trudy” (OT) Lilium hybrid. In addition, 24 oriental and 12 Trumpet chromosomes were observed in this hybrid. Consequently, for genomic investigation and parental genomic contribution, GISH is a consistent method.
Figure 2.
Genomic in situ hybridization (GISH) images of OT (Oriental × Trumpet) hybrids. (a) Motown (3x), (b) Stentor (4x), (c) Morini (3x), and (d) Trudy (3x), with Oriental (red) and Trumpet (yellow) chromosomes, respectively.
Tetraploid interspecific hybrids (OA) produced by somatic chromosome doubling can be used for breeding purposes. Such tetraploids crossing with diploid Asiatic and tetraploid Asiatic or tetraploid OA hybrid developed triploid and tetraploid progenies, respectively. GISH authentic analysis of F1 OA hybrids explained that each parent (Oriental and Asiatic) contributed 12 chromosomes. However, 24 Asiatic chromosomes and 12 Oriental chromosomes developed triploid plants in backcross progeny. Important point illustrated by GISH was that tetraploid progeny developed by tetraploid OA crossing with OA hybrids consisted of equal number of maternal chromosomes, that is, oriental and Asiatic contributing 24 chromosomes, respectively. In O × OA (2x–4x) progenies, some progenies showed complete or double contribution of Oriental parental chromosomes [58].
Detail genomic constitution and parental genomic recombination can be identified in hybrids and polyploids through modern genomic techniques such as GISH method of in situ cytogenetic analysis [36].
In tulip, F1, BC1, and BC2 progenies developed from Darwin hybrids backcrossing with Tulipa gesneriana were analyzed cytogenetically through GISH. GISH can measure the nature and extent of intergenomic recombination. One tetraploid (2n = 4x = 48) and one aneuploid BC2 (2n = 2x = 25 + 1) were obtained in BC1 and BC2, respectively. F1b hybrid morphometric analysis discriminates T. fosteriana and T. gesneriana parental chromosomes on the basis of total length of chromosomes. In addition, some heterochromatin segments in the intercalary and telomeric sites expressed higher fluorescence intensity. The FISH study, confirming the GISH results, showed these sites rich with rDNA. Noticeable information about genome was that there was a significant amount of intergenomic rearrangement between parental genomes of two species. This genomic recombination ranged from 3 to 8 in numbers in BC1 progeny while 1 to 7 in BC2 plants. Recombinant chromosomes carried mainly a single recombinant segment developed from single or in few situation double crossover phases. This makes the information clear that, unlike the condition, most F1 hybrids of other plant species and certain genotypes of Darwin hybrid tulips showed normal diploid behavior, that is, haploid gamete and diploid sporophyte development [59].
Lilium hybrids have extensive intergenomic recombination in their backcross progenies. Trumpet or Martagon chromosome fragments participated a lot during backcrossing breeding program. Therefore, it enhances the fertility of new developed progeny. In this regard, backcross progenies of OT and MA hybrids were analyzed through the GISH technique. In OT hybrids, BC1 progenies developed 15 euploids (2x and 3x) and 6 aneuploids while BC2 developed only aneuploids. First division restitution (FDR) produces 2n eggs which was the basic reason of triploid progeny production whereas aneuploid progeny production was due to viable aneuploid gametes. In GISH analysis of MA hybrid, two BC1 progenies showed aneuploid behavior containing chromosome number 35 and 32, respectively. One BC1 progeny carrying triploid nature was the result of indeterminate meiotic restitution (IMR) [60].
Parental chromosome discrimination in intergenomic or interspecific hybrids required a molecular tool such as GISH. Mode of 2n gametes origin and intergenomic recombination in lily hybrids can be illustrated easily using such technique [57]. Potential value of intergenomic hybrids (L. auratum × L. henryi) can be confirmed by this method. GISH confirmed equal participation of both parents in L. auratum × L. henryi hybrid progeny. Viable 2n gamete producing ability of progeny gives an opportunity to produce Oriental Auratum Henryi hybrids after crossing with oriental hybrids. OAuH progeny showed triploid behavior comprising of 12 Oriental chromosomes and 24 AuH hybrid chromosomes. Cytogenetic study explained that FDR (first division restitution) mode in F1 hybrids and through sexual polyploidization recombinant chromosome fragments can be transferred to further generations for obtaining horticultural characters [61].
8. Conclusion
As a cytogenetic tool, GISH is a primitive and prominent technique in plant genome analysis. GISH is a key advancement to identify and characterize the genomes of hybrids and progenies developed by classical breeding methods. Chromosome structure, genetic organization, genomic constituents, and genomic recombinations are easily approachable by the use of this technique. Although GISH provides a large range of genomic study, genome sequencing provides more clear genetic information. Therefore, further improvement in genetic information can be performed due to advancements in GISH such as multicolor GISH analysis.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Project no. NRF-2016R1D1A1B04932913).
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper. Also, the funding agency has no conflict of interests regarding the publication of this manuscript.
References
- 1.Halfhill M. D., Warwick S. I. Mendelian genetics and plant reproduction. In: Neal Stewart C. Jr, editor. Plant Biotechnology and Genetics: Principles, Techniques and Applications. New York, USA: Wiley and Sons; 2016. p. p. 34. [Google Scholar]
- 2.Andres R. J., Kuraparthy V. Development of an improved method of mitotic metaphase chromosome preparation compatible for fluorescence in situ hybridization in cotton. Journal of Cotton Science. 2013;17(2):149–156. [Google Scholar]
- 3.Younis A., Ramzan F., Hwang Y. J., Lim K. B. FISH and GISH: molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Reports. 2015;34(9):1477–1488. doi: 10.1007/s00299-015-1828-3. [DOI] [PubMed] [Google Scholar]
- 4.John H. A., Birnstile M. L., John K. W. RNA-DNA hybrids at the cytological level. Nature. 1969;223(5206):582–587. doi: 10.1038/223582a0. [DOI] [PubMed] [Google Scholar]
- 5.Gall J. G., Pardue M. L. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proceedings of the National Academy of Sciences of the United States of America. 1969;63(2):378–383. doi: 10.1073/pnas.63.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayburn A. L., Gill B. S. Use of of biotin-labeled probes to map specific DNA sequences on wheat chromosomes. The Journal of Heredity. 1985;76(2):78–81. doi: 10.1093/oxfordjournals.jhered.a110049. [DOI] [Google Scholar]
- 7.Schwarzacher T., Leitch A. R., Bennett M. D., Heslop-Harrison J. S. In situ localization of parental genomes in a wide hybrid. Annals of Botany. 1989;64(3):315–324. doi: 10.1093/oxfordjournals.aob.a087847. [DOI] [Google Scholar]
- 8.Singh R. J. Plant Cytogenetics. 2. London, UK: Boca Raton, CRC Press; 2003. [Google Scholar]
- 9.Ksiazczyk T., Taciak M., Zwierzykowski Z. Variability of ribosomal DNA sites in Festuca pratensis, Lolium perenne, and their intergeneric hybrids, revealed by FISH and GISH. Journal of Applied Genetics. 2010;51(4):449–460. doi: 10.1007/BF03208874. [DOI] [PubMed] [Google Scholar]
- 10.Benavente E., Cifuentes M., Dusautoir J. C., David J. The use of cytogenetic tools for studies in the crop-to-wild gene transfer scenario. Cytogenetic and Genome Research. 2008;120(3-4):384–395. doi: 10.1159/000121087. [DOI] [PubMed] [Google Scholar]
- 11.Levin D. A. The role of chromosome change in plant evolution. Systematic Botany. 2004;29:460–461. [Google Scholar]
- 12.Robledo G., Lavia G. I., Seijo G. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theoretical and Applied Genetics. 2009;118(7):1295–1307. doi: 10.1007/s00122-009-0981-x. [DOI] [PubMed] [Google Scholar]
- 13.Wong C., Murray B. G. In situ hybridization with genomic and rDNA probes reveals complex origins for polyploid New Zealand species of Plantago (Plantaginaceae) New Zealand Journal of Botany. 2014;52(3):315–327. doi: 10.1080/0028825X.2014.898664. [DOI] [Google Scholar]
- 14.Sun Y., Xu C. H., Wang M. Q., Zhi D. Y., Xia G. M. Genomic changes at the early stage of somatic hybridization. Genetics and Molecular Research. 2014;13(1):1938–1948. doi: 10.4238/2014.March.17.21. [DOI] [PubMed] [Google Scholar]
- 15.Nakazawa D., Kishimoto T., Sato T., et al. Genomic in situ hybridization (GISH) analysis of intergeneric hybrids in Colchicaceae. Euphytica. 2011;181(2):197–202. doi: 10.1007/s10681-011-0393-2. [DOI] [Google Scholar]
- 16.Maluszynska J., Hasterok R. Identification of individual chromosomes and parental genomic in Brassica juncea using GISH and FISH. Cytogenetic and Genome Research. 2005;109(1–3):310–314. doi: 10.1159/000082414. [DOI] [PubMed] [Google Scholar]
- 17.Kitajima A., Yamasaki A., Habu T. Chromosome identification and karyotyping of satsuma mandarin by genomic in situ hybridization. Journal of the American society for Horticultural Science. 2007;132(6):836–841. [Google Scholar]
- 18.Choi Y., Tao R., Yonemori K., Sugiura A. Multi-color genomic in situ hybridization identifies parental chromosomes in somatic hybrids of Diospyros kaki and D. glandulosa. Hortscience. 2002;37(1):184–186. [Google Scholar]
- 19.Yamashita K., Takatori Y., Tashiro Y. Chromosomal location of a pollen fertility-restoring gene, Rf, for CMS in Japanese bunching onion (Allium fistulosum L.) possessing the cytoplasm of A. galanthum Kar. et Kir. revealed by genomic in situ hybridization. Theoretical and Applied Genetics. 2005;111(1):15–22. doi: 10.1007/s00122-005-1941-8. [DOI] [PubMed] [Google Scholar]
- 20.Seijo G., Lavia G. I., Fernandez A., et al. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. American Journal of Botany. 2007;94(12):1963–1971. doi: 10.3732/ajb.94.12.1963. [DOI] [PubMed] [Google Scholar]
- 21.Carvalho R., Soares Filho W. S., Guerra M. Chromosomal relationships among cultivars of Citrus reticulate Blanco, its hybrids and related species. Plant Systematics and Evolution. 2003;240(1):149–161. [Google Scholar]
- 22.Carvalho R., Soares W. S., Brasileoro-Vidal A. C., Guerra M. The relationships among lemons, limes and citron: a chromosomal comparison. Cytogenetic and Genome Research. 2005;109(1–3):276–282. doi: 10.1159/000082410. [DOI] [PubMed] [Google Scholar]
- 23.Raina S. N., Mukai Y., Yamamoto M. In situ hybridization identifies the diploid progenitor species of Coffea arabica (Rubiacea) Theoretical and Applied Genetics. 1998;97(8):1204–1209. doi: 10.1007/s001220051011. [DOI] [Google Scholar]
- 24.Choi Y., Tao R., Yonemori K., Sugiura A. Multi-color genomic in situ hybridization identifies parental chromosomes in somatic hybrids of Diospyros kaki and D. glandulosa. Hortscience. 2007;37(1):184–186. [Google Scholar]
- 25.Fu C. H., Chen C. L., Guo W. W., Deng X. X. GISH, AFLP and PCRRFLP analysis of an intergeneric somatic hybrid combining Goutou sour orange and Poncirus trifoliate. Plant Cell Reports. 2004;23(6):391–396. doi: 10.1007/s00299-004-0828-5. [DOI] [PubMed] [Google Scholar]
- 26.Pendinen G., Gavrilenko T., Jiang J., Spooner D. M. Allopolyploid speciation of the Mexican tetraploid potato species Solanum stoloniferum and S. hjertingii revealed by genomic in situ hybridization. Genome. 2008;51(9):714–720. doi: 10.1139/G08-052. [DOI] [PubMed] [Google Scholar]
- 27.Spooner D. M., Rodríguez F., Polgár Z., Ballard H. E., Jr, Jansk S. H. Genomic origins of potato polyploids: GBSSI gene sequencing data. Crop Science. 2008;48(Supplement 1):27–36. [Google Scholar]
- 28.Rodríguez F., Spooner D. M. Nitrate reductase phylogeny of potato (Solanum sect. Petota) genomes with emphasis on the origins of the polyploid species. Systematic Botany. 2009;34(1):207–219. doi: 10.1600/036364409787602195. [DOI] [Google Scholar]
- 29.Pendinen G., Spooner D. M., Jiang J., Gavrilenko T. Genomic in situ hybridization reveals both auto and allopolyploid origins of different North and Central American hexaploid potato (Solanum sect. Petota) species. Genome. 2012;55(6):407–415. doi: 10.1139/g2012-027. [DOI] [PubMed] [Google Scholar]
- 30.Nishiyama K., Choi Y. A., Honsho C., Eiadthong W., Yonemori K. Application of genomic in situ hybridization for phylogenetic study between Mangifera indica L. and eight wild species of Mangifera. Scientia Horticulturae. 2006;110(1):114–117. doi: 10.1016/j.scienta.2006.06.005. [DOI] [Google Scholar]
- 31.Eiadthong W., Yonemori K., Kanzaki S., Sugiura A., Utsunomiya N., Subhadrabandhu S. Amplified fragment length polymorphism analysis for studying genetic relationships among Mangifera species in Thailand. Journal of the American Society for Horticultural Science. 2000;125(2):160–164. [Google Scholar]
- 32.Nathewet P., Yanagi T. Chromosome observation methods in the genus Fragaria. Acta Horticulturae. 2014;1049:201–206. [Google Scholar]
- 33.Reis G. B., Mesquita A. T., Torres G. A., Andrade-Vieira L. F., Pereira A. V., Davide L. C. Genomic homeology between Pennisetum purpureum and Pennisetum glaucum (Poaceae) Comparative Cytogenetics. 2014;8(3):199–209. doi: 10.3897/CompCytogen.v8i3.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuster M., Schreiber H. Genome investigation in sour cherry, P. cerasus L. Acta Horticulturae (International Society for Horticultural Science) 2000;538:375–379. [Google Scholar]
- 35.Yamashita K., Hisatsune Y., Sakamoto T., Ishizuka K., Tashiro Y. Chromosome and cytoplasm analysis of somatic hybrids between onion (Allium cepa L.) and garlic (A. sativum L.) Euphytica. 2002;125(2):163–167. doi: 10.1023/A:1015826702550. [DOI] [Google Scholar]
- 36.Marasek A., Mizuochi H., Okazaki K. The origin of Darwin hybrid tulips analyzed by flow cytometry, karyotype analyses and genomic in situ hybridization. Euphytica. 2006;151(3):279–290. doi: 10.1007/s10681-006-9147-y. [DOI] [Google Scholar]
- 37.Barba-Gonzalez R., Van Silfhout A. A., Ramanna M. S., Visser R. G. F., Van Tuyl J. M. Progenies of allotriploids of Oriental x Asiatic lilies (Lilium) examined by GISH analysis. Euphytica. 2006;151(2):243–250. doi: 10.1007/s10681-006-9148-x. [DOI] [Google Scholar]
- 38.Haider A. S. N., Ramanna M. S., Jacobsen E., Visser R. G. F. Genome differentiation between Lycopersicon esculentum and L. pennellii as revealed by genomic in situ hybridization. Euphytica. 2002;127(2):227–234. doi: 10.1023/A:1020282311843. [DOI] [Google Scholar]
- 39.Marasek-Ciolakowska A., Ramanna M. S., ter Laak W. A., van Tuyl J. M. Genome composition of ‘Elatior’-begonias hybrids analyzed by genomic in situ hybridization. Euphytica. 2010;171(2):273–282. doi: 10.1007/s10681-009-0057-7. [DOI] [Google Scholar]
- 40.Yuanfu J., Pertuze R., Chetelat R. T. Genome differentiation by GISH in interspecic and intergeneric hybrids of tomato and related nightshades. Chromosome Research. 2004;12(2):107–116. doi: 10.1023/b:chro.0000013162.33200.61. [DOI] [PubMed] [Google Scholar]
- 41.Chuda A., Adamus A. Hybridization and molecular characterization of F1Allium cepa × Allium roylei plants. Acta Biologica Cracoviensia Series Botanica. 2012;54(2):25–31. [Google Scholar]
- 42.Liu L. Q., Gu Z. J. Genomic in situ hybridization identifies genome donors of Camellia reticulata (Theaceae) Plant Science. 2011;180(3):554–559. doi: 10.1016/j.plantsci.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Marasek A., Okazaki K. GISH analysis of hybrids produced by interspecific hybridization between Tulipa gesneriana and T. fosteriana. Acta Horticulturae. 2007;743:133–137. [Google Scholar]
- 44.Schwarzacher T., Heslop-Harrison J. S. Practical In Situ Hybridization. London, UK: Oxford: BIOS Scientific Publishers; 2000. [Google Scholar]
- 45.Bennett M. D. The development and use of genomic in situ hybridization (GISH) as a new tool in plant biosystematics. In: Brandham P. E., Bennett M. D., editors. Kew Chromosome Conference IV. Kew: Royal Botanic Gardens; 1995. pp. 167–183. [Google Scholar]
- 46.Chen J. F., Staub J. E., Qian C. T., Jiang J. M., Luo X. D., Zhuang F. Y. Reproduction and cytogenetic characterization of interspecific hybrids derived from Cucumis hystrix Chakr. × C. sativus L. Theoretical and Applied Genetics. 2003;106(4):688–695. doi: 10.1007/s00122-002-1118-7. [DOI] [PubMed] [Google Scholar]
- 47.Van Laere K., Van Huylenbroeck J., Van Bockstaele E. Introgression of yellow flower colour in Buddleja davidii by means of polyploidisation and hybridisation. HortScience. 2011;38:96–103. [Google Scholar]
- 48.Ran Y., Hammett K. R. W., Murray B. G. Hybrid identification in Clivia (Amaryllidaceae) using chromosome banding and genomic in situ hybridization. Annals of Botany. 2001;87(4):457–462. doi: 10.1006/anbo.2000.1365. [DOI] [Google Scholar]
- 49.Barba-Gonzalez R., Ramanna M. S., Visser R. G. F., Van Tuyl J. M. Intergenomic recombination in F1 lily hybrids (Lilium) and its significance for genetic variation in the BC1 progenies as revealed by GISH and FISH. Genome. 2005;48(5):884–894. doi: 10.1139/g05-057. [DOI] [PubMed] [Google Scholar]
- 50.Czernicka M., Mścichowska A., Klein M., Muras P., Grzebelus E. Paternity determination of interspecific rhododendron hybrids by genomic in situ hybridization (GISH) Genome. 2010;53(4):277–284. doi: 10.1139/g10-007. [DOI] [PubMed] [Google Scholar]
- 51.Chung M. Y., Chung J. D., Ramanna M., van Tuyl J. M., Lim K. B. Production of polyploids and unreduced gametes in Lilium auratum × L. henryi hybrid. International Journal of Biological Sciences. 2013;9(7):693–701. doi: 10.7150/ijbs.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marasek-Ciolakowska A., Ramanna M. S., van Tuyl J. M. Introgression of chromosome segments of Tulipa fosteriana into T. gesneriana detected through GISH and its implications for breeding virus resistant Tulips. Acta Horticulturae. 2011;886:175–182. [Google Scholar]
- 53.Van Heusden A. W., SHIGYO M., Tashiro Y., Vrielink-Van G. R., Kik C. AFLP linkage group assignment to the chromosomes of Allium cepa L. via monosomic addition lines. Theoretical and Applied Genetics. 2000;100(3-4):480–486. doi: 10.1007/s001220050062. [DOI] [Google Scholar]
- 54.Scholten O., Van Heusden A. W., Khrustaleva L. I., et al. The long and winding road leading to the successful introgression of downy mildew resistance into onion. Euphytica. 2007;156(3):345–353. doi: 10.1007/s10681-007-9383-9. [DOI] [Google Scholar]
- 55.Lee Y. I., Chang F. C., Chung M. C. Chromosome pairing affinities in interspecific hybrids reflect phylogenetic distances among lady’s slipper orchids (Paphiopedilum) Annals of Botany. 2011;108(1):113–121. doi: 10.1093/aob/mcr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee H. I., Younis A., Hwang Y. J., Kang Y. I., Lim K. B. Molecular cytogenetic analysis and phylogenetic relationship of 5S and 45S ribosomal DNA in Sinomartagon Lilium species by fluorescence in situ hybridization (FISH) Horticulture, Environment and Biotechnology. 2014;55(6):514–523. doi: 10.1007/s13580-014-0066-x. [DOI] [Google Scholar]
- 57.Zhou S., Yuan G., Xu P., Gong H. Study on lily introgression breeding using allotriploids as maternal parents in interploid hybridizations. Breeding Science. 2014;64(1):97–102. doi: 10.1270/jsbbs.64.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chung M. Y., Chung J. D., Van Tuyl J. M., Lim K. B. Progeny analysis of hybrid lilies crossed between several genotypes and tetraploid oriental-asiatic hybrids. Korean Journal of Breeding Science. 2010;42(1):100–108. [Google Scholar]
- 59.Marasek-Ciolakowska M., He H., Bijman P., Ramanna M. S., Arens P., van Tuyl J. M. Assessment of intergenomic recombination through GISH analysis of F1, BC1 and BC2 progenies of Tulipa gesneriana and T. fosteriana. Plant Systematics and Evolution. 2012;298(5):887–899. doi: 10.1007/s00606-012-0598-4. [DOI] [Google Scholar]
- 60.Luo J. R., Ramanna M. S., Arens P., Niu L. X., Van Tuyl J. M. GISH analyses of backcross progenies of two Lilium species hybrids and their relevance to breeding. The Journal of Horticultural Science and Biotechnology. 2012;87(6):654–660. [Google Scholar]
- 61.Younis A., Hwang Y. J., Lim K. B. Exploitation of induced 2n-gametes for plant breeding. Plant Cell Reports. 2014;33(2):215–223. doi: 10.1007/s00299-013-1534-y. [DOI] [PubMed] [Google Scholar]
- 62.Budylin M. V., Yu Kan L., Romanov V. S., Khrustaleva L. I. GISH study of advanced generation of the interspecific hybrids between Allium cepa L. and Allium fistulosum L. with relative resistance to downy mildew. Russian Journal of Genetics. 2014;50(4):387–394. doi: 10.1134/S1022795414040036. [DOI] [PubMed] [Google Scholar]
- 63.Akiyama Y., Kubota A., Yamada-Akiyama H., Ueyama Y. Development of a genomic in situ hybridization (GISH) and image analysis method to determine the genomic constitution of festulolium (Festuca × Lolium) hybrids. Breeding Science. 2010;60(4):347–352. doi: 10.1270/jsbbs.60.347. [DOI] [Google Scholar]
- 64.Lin C., Chen Y., Chen W., Chen C., Kao Y. Genome organization and relationships of Phalaenopsis orchids inferred from genomic in situ hybridization. Botanical Bulletin of Academia Sinica. 2005;46:339–345. [Google Scholar]
- 65.Brysting A. K., Holst-Jensena A., Le Itch I. Genomic origin and organization of the hybrid Poa jemtlandica (Poaceae) verified by genomic in situ hybridization and chloroplast DNA sequences. Annals of Botany. 2000;85(4):439–445. doi: 10.1006/anbo.1999.1088. [DOI] [Google Scholar]
- 66.Guggisberg A., Baroux C., Grossniklaus U., Conti E. Genomic origin and organization of the allopolyploid Primula egaliksensis investigated by in situ hybridization. Annals of Botany. 2008;101(7):919–927. doi: 10.1093/aob/mcn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lim K. B., Chung J. D., Van Kronenburg B. C., Ramanna M. S., De Jong J. H., Van Tuyl J. M. Introgression of Lilium rubellum Baker chromosomes into L. longiflorum Thunb.: a genome painting study of the F1 hybrid, BC1 and BC2 progenies. Chromosome Research. 2000;8(2):119–125. doi: 10.1023/A:1009290418889. [DOI] [PubMed] [Google Scholar]
- 68.Lim K. B., Ramanna M. S., Jacobsen E., Van Tuyl J. M. Evaluation of BC2 progenies derived from 3x-2x and 3x-4x crosses of Lilium hybrids: a GISH analysis. Theoretical and Applied Genetics. 2003;106(3):568–574. doi: 10.1007/s00122-002-1070-6. [DOI] [PubMed] [Google Scholar]