Abstract
U-box-containing Prp19p is an integral component of the Prp19p-associated complex (the nineteen complex, or NTC) that is essential for activation of the spliceosome. Prp19p makes numerous protein-protein contacts with other NTC components and is required for NTC stability. Here we show that Prp19p forms a tetramer in vitro and in vivo and we map the domain required for its oligomerization to a central tetrameric coiled-coil. Biochemical and in vivo analyses are consistent with Prp19p tetramerization providing an interaction surface for a single copy of its binding partner, Cef1p. Electron microscopy showed that the isolated Prp19p tetramer is an elongated particle consisting of four globular WD40 domains held together by a central stalk consisting of four N-terminal U-boxes and four coiled-coils. These structural and functional data provide a basis for understanding the role of Prp19p as a key architectural component of the NTC.
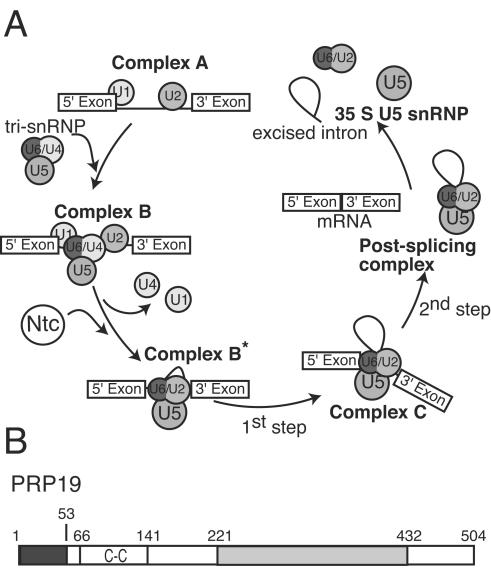
The spliceosome is a megadalton-sized complex composed of four ribonucleoprotein particles (snRNPs; U1, U2, U5, and U4/U6) and additional pre-mRNA splicing factors. This dynamic macromolecular machine catalyzes the excision of introns from pre-mRNA to generate protein-coding transcripts, a process requiring a complex series of steps that involve assembly and disassembly of large multiprotein complexes (reviewed in references 19, 25, and 32). Spliceosome assembly begins when the U1 and U2 snRNPs recognize the 5′ and 3′ splice sites, respectively (Fig. 1A, complex A). After binding of the U5.U4/U6 tri-snRNP (Fig. 1A, complex B), the spliceosomal complex rearranges, releasing both the U1 and U4 snRNPs concomitant with the arrival of the Prp19p-associated complex (the nineteen complex, or NTC) (33, 34). Both the release of the two snRNPs and the arrival of the NTC mark the transition from the inactive to the active spliceosome, which is composed of the U5 and U2/U6 snRNPs (Fig. 1A, complex C). 5′ splice site cleavage and lariat formation, followed by 3′ splice site cleavage and exon ligation, occur within the activated spliceosome.
FIG. 1.
(A) Schematic model of pre-mRNA splicing. (B) Organization of characterized domains within Prp19p. The dark shaded portion represents the U-box domain, while the light shaded portion represents the WD40 repeats. The predicted coiled-coil domain is labeled C-C.
Regulation of these coordinated structural rearrangements among both RNA and protein components of the spliceosome is not fully understood; however, the transition from an inactive to active complex is directly linked to the addition of the NTC. Recent work has shown that the NTC is required for stabilizing both U5 and U6 snRNA interactions with pre-mRNA and for destabilizing the interactions of the Sm-like (Lsm) proteins with the U6 snRNA during the formation of complex C (3). NTC components may also facilitate the rearrangement of the U5 snRNP from a 30S complex into a 45S complex during the transition from complex B to complex C (22, 23). Although these biochemical data provide insight into NTC function, it is unknown how the NTC mediates and/or stabilizes structural changes during this dynamic stage in spliceosome assembly.
In Saccharomyces cerevisiae, the NTC can be isolated as a distinct unit that is composed of at least 11 proteins, many of which have been identified (6, 27, 33). NTC proteins include Prp19p, Syf1p (Ntc90), Cef1p (Ntc85), Clf1p (Ntc77), Prp46p (Ntc50), Cwc2p (Ntc40), Syf2p (Ntc31), Isy1p (Ntc30), Snt309p (Ntc25), and Ntc20p. NTC components, highly conserved across species, are also present in Schizosaccharomyces pombe and human spliceosomal complexes (reviewed in reference 17).
Accumulating evidence suggests that Prp19p is required for the structural integrity of the NTC and its subsequent association with the spliceosome (3, 27, 33). Prp19p interacts directly with a number of NTC components, including Cef1p, Snt309p, and Cwc2p, and is critical for their assembly into a multiprotein complex (5, 6, 27, 35). Prp19p contains three recognized protein motifs: an N-terminal U-box, a predicted coiled-coil (21), and a WD40 repeat domain at its C terminus (Fig. 1B). The WD40 repeat domain interacts with Cwc2p, whereas the N-terminal part of the protein interacts with Cef1p and Snt309p (27). The U-box, despite lacking zinc-chelating residues, is structurally similar to RING finger domains (28). Like many proteins that contain RING domains, both human and budding yeast Prp19ps exhibit E3 ubiquitin ligase activity in vitro (15, 28). Mutations in the U-box that do not disrupt the structural integrity of Prp19p are still unable to rescue function in vivo, suggesting that this enzymatic activity of Prp19p is essential for some aspect of NTC structure or function (28).
Although the above-mentioned studies have suggested models of NTC organization, a fundamental understanding of the composition and assembly of the complex is lacking. We address details of NTC organization here by examining the stoichiometries of Prp19p and Cef1p within the complex. We demonstrate that while Cef1p is monomeric, Prp19p is tetrameric, and we map the domain required for oligomerization and test the functional consequences of disrupting Prp19p tetramerization in vivo. Our data suggest how Prp19p might provide flexibility for structural rearrangements within the spliceosome while still serving as a binding scaffold for numerous proteins.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae strains used in this study (Table 1) were grown either in synthetic minimal medium with the appropriate nutritional supplements or yeast extract-peptone-dextrose (14). Transformations were performed by the lithium acetate method (11). The ability of prp19 mutations to rescue growth of the prp19-1 strain was scored by serial dilutions grown under the permissive condition of 25 to 36°C for 3 days. S. pombe strains (Table 1) were grown in yeast extract medium or minimal medium with appropriate supplements (24). Crosses were performed on glutamate medium (minimal medium lacking ammonium chloride and containing 0.01 M glutamate; pH 5.6).
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| S. pombe strains | ||
| KGY246 | ade6-M210 ura4-D18 leu1-32 h− | Lab stock |
| KGY1318 | cwf8::cwf8-HA::Kanrade6-M216 leu1-32 ura4-D18 h+ | Lab stock |
| KGY1607 | cdc5::cdc5-myc13::Kanrade6-M210 leu1-32 ura4-D18 h− | Lab stock |
| KGY1608 | cwf8::cwf8-myc13::Kanrade6-M210 leu1-32 ura4-D18 h− | Lab stock |
| KGY2105 | cdc5::cdc5-HA::Kanrade6-M216 leu1-32 ura4-D18 h+ | Lab stock |
| KGY2108 | cdc5::cdc5-myc13::Kanr/cdc5::cdc5-HA::Kanrade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 h−/h+ | This study |
| KGY4476 | cwf8::cwf8-myc13::Kanr/cwf8::cwf8-HA::Kanrade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 h−/h+ | This study |
| KGY4985 | cdc5::cdc5-myc13::Kanrcwf8::cwf8-myc13::Kanrcwf7::cwf7-HA3::Kanrade6-M216 leu1-32 ura4-D18 h+ | This study |
| KGY4986 | cdc5::cdc5-TAP::Kanr/cdc5::cdc5-TAP::Kanrcwf8::cwf8-myc13::Kanr/cwf8::cwf8-HA3::Kanrade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 h−/h+ | This study |
| S. cerevisiae strains | ||
| KGY1811 | MATaprp19-1 ura3-52 lys2-801 ade2-101 leu2-Δ1 trp1-Δ1 | J. Abelson |
| KGY1294 | MATα CEF1::CEF1-HA3::Kanr | Lab stock |
| KGY1296 | PJ69-4A, MATa; trp1-190 leu2-3,112 ura3-52 his3-200 gal4 ΔLYS2 replaced by GAL1-HIS3 GAL2-ADE2, met2 replaced by GAL-LacZ | 16 |
| KGY1356 | MATα PRP19::PRP19-HA3::Kanr | Lab stock |
Molecular biology techniques.
PCR amplifications were performed using Pfu Turbo (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. Mutations in PRP19 were made by the two-step PCR overlap extension method. Truncations of PRP19 were generated by PCR. To facilitate cloning into pRS415 GAL1, a 5′ BamHI site and a 3′ PstI site were included within the oligonucleotides used for PCR. For cloning PCR products into pET15b (Novagen) for expression in Escherichia coli, a 5′ NdeI site and a 3′ BamHI site were included within the oligonucleotides. DNA sequencing confirmed the presence of only the desired mutations.
Immunoprecipitations and immunoblotting.
Native protein lysates were prepared from approximately 2.4 ×108 cells as detailed previously (13). To the resultant 1.0 ml of lysate, 2 μg of antihemagglutinin (anti-HA; 12CA5) or anti-myc (9E10) antibody was added to precipitate tagged proteins. Immunoprecipitates were resolved on Novex NuPAGE 4-to-12% bis-Tris gels using NuPAGE MOPS SDS running buffer (Invitrogen, Carlsbad, Calif.) and transferred by electroblotting to polyvinylidene difluoride membrane (Immobilon P; Millipore Corp., Bedford, Mass.). Anti-HA (12CA5) antibodies and anti-myc (9E10) antibodies were used at 2 μg/ml, and antibodies were detected using horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (0.8 mg/ml; Jackson Immunoresearch Laboratories, West Grove, Pa.) at a dilution of 1:50,000. Immunoblots were visualized using enhanced chemiluminescence reagents (Amersham Pharmacia Biotech). For immunoblots that were quantitated, detection was performed with an Odyssey instrument (Licor Corp., Lincoln, Nebr.) using the protocol and reagents supplied by the manufacturer. Quantitation was performed with Odyssey version 1.2.
Yeast two-hybrid assays.
The yeast two-hybrid system used in this study was described previously (16). Various portions of the PRP19 cDNAs were cloned into the bait plasmid pGBT9 and/or the prey plasmid pGAD424 (Clontech, Palo Alto, Calif.) and sequenced to ensure the absence of PCR-induced mutations and that the correct reading frame had been retained.
Two-hybrid screens were performed as described previously (16). To test for protein interactions, both bait and prey plasmids were cotransformed into S. cerevisiae strain PJ69-4A. β-Galactosidase reporter enzyme activity in the two-hybrid strains was measured using the Galacto-Star chemiluminescent reporter assay system according to the manufacturer's instructions (Tropix Inc., Bedford, Mass.), with the exception that cells were lysed by glass bead disruption. Each sample was measured in triplicate. Reporter assays were recorded either on the BMG luminometer (Bartlett-Williams Scientific, Chapel Hill, N.C.) or the Mediator Phl luminometer (Aureon Biosystems, Vienna, Austria).
Expression of recombinant fusion proteins.
Three constructs spanning the different domains of Prp19p were produced: PRP19 1-58, PRP19 66-141, and PRP19 165-503, along with the full-length protein and the W88A and W88P mutants in the context of the full-length protein. Proteins were expressed in Rosetta(DE3) cells (Novagen). Cells were grown to an optical density at 600 nm of 0.7 and induced with 1 mM isopropyl-β-d-thiogalactopyranoside. Cells were lysed in 20 mM Tris HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, and 5 mM BME, and proteins were purified using Ni2+-nitrilotriacetic acid columns (QIAGEN) following the manufacturer's instructions. Proteins were eluted from the column by using an imidazole gradient from 20 to 900 mM. In the case of Prp19p 66-141, the His6 tag was cleaved by incubation with thrombin (25 U/∼10 mg of protein) for 14 h at 4°C. Proteins were further purified using a Mono-Q 10/10 column (20 mM Tris [pH 8.0], elution 10 to 600 mM NaCl, and 1 mM dithiothreitol [DTT]) (Amersham Pharmacia Biotech).
Gel filtration.
Gel filtration was performed using a 10/30 S200 column (Amersham Pharmacia Biotech). Running buffer was 20 mM Tris (pH 8.0), 100 mM NaCl, and 1 mM DTT. The protein running concentration was ∼5 mg/ml.
CD and secondary structure estimation.
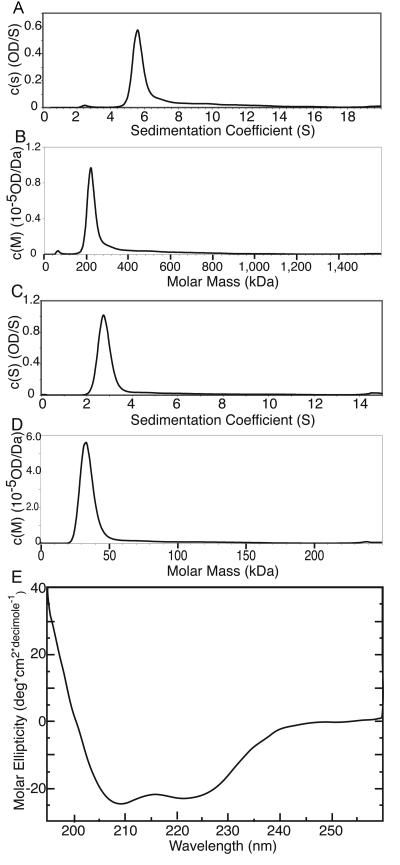
Circular dichroism (CD) measurements were recorded using a Jasco J-810 (Jasco Inc.) spectropolarimeter. Protein was dialyzed into 5 mM sodium phosphate (pH 8.0), 10 mM NaCl, and 1 mM DTT. Spectra were obtained using the average of three scans over the range of 190 to 260 nm with a step size of 0.2 nm and a bandwidth of 1 nm. Secondary structure estimates were obtained using the program K2d (1).
Analytical ultracentrifugation.
Sedimentation velocity experiments were conducted with an Optima XLA apparatus (Beckman-Coulter, Fullerton, Calif.), with a four-hole An60Ti rotor and double sector cells with charcoal-filled Epon centerpieces (path length, 1.2 cm) and quartz windows. The value of the radial distance of the bottom of the cell was between 7.13 and 7.15 cm. For sedimentation velocity experiments, samples (397 μl) and reference solutions (400 μl) were loaded into cells. The experiments were conducted at 25°C at a rotor speed of 129,024 × g. Concentration profiles were measured at 280 nm. Partial specific volumes and molecular masses for Prp19p and Prp19p fragments were calculated based on their amino acid compositions using the program Sednterp (20). The velocity scans were analyzed with the program Sedfit (version 8.7) (31). For each analysis, 300 scans collected approximately 2 min apart were analyzed. All size distributions were solved on a radial grid of 1,000 radius values between the meniscus and bottom, a confidence level of P = 0.95, a resolution of n = 300, and sedimentation coefficients between 0.1 and 20 s.
Electron microscopy and image processing.
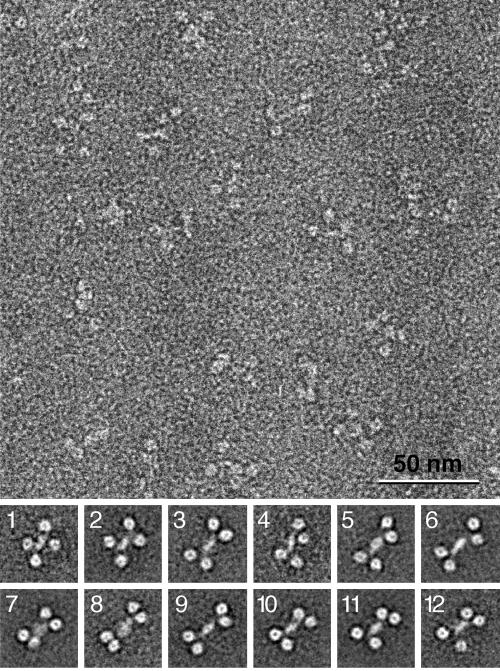
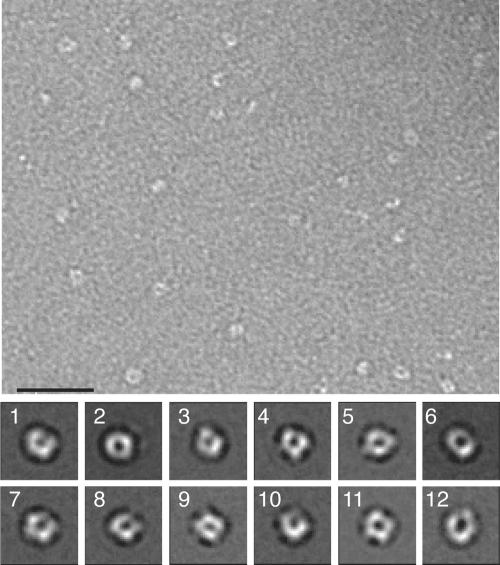
Uranyl formate-stained samples were prepared for electron microscopy as described previously (26). Images were taken with a Philips Tecnai T12 electron microscope at an acceleration voltage of 120 kV with a magnification of ×52,000 and a defocus of −1.5 μm using low-dose procedures. After inspection with a JEOL JFO-3000 laser diffractometer, drift-free images were digitized with a Zeiss SCAI scanner using a step size of 7 μm. Three- by three-pixel images were averaged to yield a pixel size of 0.4 nm at the specimen level. A total of 9,725 His6-Prp19p particles were selected from 28 micrographs, and 4,507 His6-Prp19p 165-503 particles were selected from 9 micrographs using WEB, the display program associated with the SPIDER program suite (9), which was used for the subsequent image-processing steps. The His6-Prp19p particles were windowed into 128- by 128-pixel images and subjected to 10 rounds of multireference alignment and K-means classification specifying 200 output classes. Due to the extensive flexibility of the Prp19p tetramers, many class averages only showed three WD40 repeats, whereas the fourth WD40 repeat was averaged out. Therefore, only a subset of class averages is shown below in Fig. 5, in which all four WD40 repeats are clearly resolved. The His6-Prp19p 165-503 particles were windowed into 40- by 40-pixel images and were subjected to 10 rounds of multireference alignment and K-means classification specifying 20 output classes.
FIG. 5.
Electron micrograph and representative projection averages of negatively stained His6-Prp19p. (Upper panel) Typical micrograph area of negatively stained His6-Prp19p. Bar, 50 nm. (Lower panel) Twelve representative averages obtained by multireference alignment and classification of 9,725 His6-Prp19p particle images. The averages reveal the variability of the position of the globular domains in relation to the central stalk. Side length of the average images is 40 nm. Number of particles in each projection average: panel 1, 50; panel 2, 51; panel 3, 35; panel 4, 30; panel 5, 35; panel 6, 39; panel 7, 41; panel 8, 22; panel 9, 64; panel 10, 7; panel 11, 46; panel 12, 22.
RESULTS
Prp19p, but not Cef1p, self-associates in vivo.
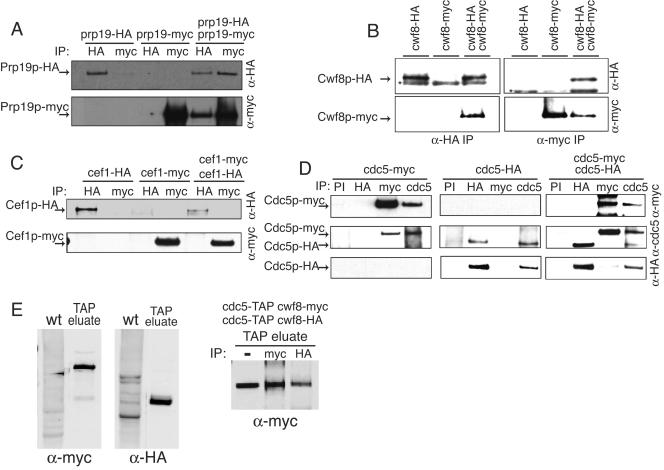
The stoichiometry of Prp19p (or any other NTC component) within the NTC in vivo has not been analyzed. To determine if there are multiple Prp19p molecules within a single NTC unit, coimmunoprecipitation analyses were performed on a diploid strain in which one of the PRP19 loci was modified to encode a protein with 13 copies of the myc epitope (myc13) at its C terminus, while the other PRP19 locus was tagged with sequences encoding three copies of the HA epitope (HA3). These modifications did not affect Prp19p function, as all epitope-tagged strains exhibited wild-type growth rates and morphologies (data not shown). Anti-HA antibody pulled down both Prp19p-HA3 and Prp19p-myc13 (Fig. 2A), while Prp19p-myc13 was not immunoprecipitated from the single-tagged strain with anti-HA antibody, indicating specificity of the anti-HA antibody (Fig. 2A). In the reciprocal experiment, anti-myc immunoprecipitates from the diploid but not the single HA-tagged strain contained Prp19-HA3 in addition to Prp19p-myc13 (Fig. 2A). These results establish that Prp19p can associate with itself in vivo, suggesting that Prp19p exists in cells as an oligomer.
FIG. 2.
Prp19p, but not Cef1p, self-associates in vivo. (A) S. cerevisiae Prp19p-HA3 coimmunoprecipitates Prp19p-myc13 in vivo. Anti-HA (upper panel) and anti-myc (lower panel) immunoblots of immunoprecipitates (IP) from prp19-HA3, prp19-myc13, and prp19-HA3 prp19-myc13 strains are shown. Immunoprecipitations were performed with anti-HA antibodies or anti-myc antibodies. (B) S. pombe Cwf8p-HA3 and Cwf8p-myc13 associate in vivo. Anti-HA (upper panel) and anti-myc (lower panel) immunoblots of IP from cwf8-HA3, cwf8-myc13, and cwf8-HA3 cwf8-myc13 strains are shown. Immunoprecipitations were performed with anti-HA antibodies or anti-myc antibodies. An asterisk indicates the position of the immunoglobulin G heavy band. (C) S. cerevisiae Cef1p-HA3 does not associate with Cef1p-myc13 in vivo. Anti-HA (upper panel) and anti-myc (lower panel) immunoblots of IP from cef1-HA3, cef1-myc13, and cef1-HA3 cef1-myc13 strains are shown. Immunoprecipitations were performed with anti-HA antibodies or anti-myc antibodies. (D) S. pombe Cdc5p-HA3 does not coimmunoprecipitate Cdc5p-myc13 in vivo. An anti-myc (upper panel), an anti-Cdc5 (middle panel), and an anti-HA (lower panel) immunoblot of IP from cdc5-HA3, cdc5-myc13, and cdc5-HA3 cdc5-myc13 strains are shown. Immunoprecipitations were performed with preimmune sera (PI), anti-HA antibodies, anti-myc antibodies, or anti-Cdc5p immune sera (cdc5). (E) The Cdc5p-TAP complex contains more than one copy of Cwf8p. Anti-myc and anti-HA immunoblots of a protein lysate prepared from S. pombe wild-type strain (wt) (KGY246) or of an eluate from a tandem affinity purification from the diploid cdc5-TAP/cdc5-TAP cwf8-HA3/cwf8-myc13 strain (KGY4986) (left panels) are shown. Immunoprecipitations (IP) were performed on the TAP eluate from the diploid strain KGY4986 with anti-HA antibodies and anti-myc antibodies and then immunoblotted with anti-myc antibodies (right panel).
To determine if the NTC of S. pombe contains multiple copies of the Prp19p ortholog, Cwf8p, we constructed an S. pombe diploid strain where each locus of cwf8+ was engineered to have either a myc13 or an HA3 tag, and immunoprecipitations were performed as described above. Through this analysis we found that S. pombe Cwf8p-HA3 was able to coprecipitate Cwf8p-myc13 in the diploid strain but not in the single-tagged haploid strains, and vice versa (Fig. 2B). These results establish that the oligomeric state of Prp19p is conserved.
Because Cef1p binds directly to Prp19p, we asked whether it, too, was present in more than a single copy in protein complexes in vivo. We constructed a diploid strain containing both Cef1p-HA3 and Cef1p-myc13 and did immunoprecipitations using either anti-HA or anti-myc antibodies that were then blotted with either anti-HA or anti-myc antibodies. In these experiments, Cef1p-HA3 was not able to coimmunoprecipitate Cef1p-myc13 and Cef1p-myc13 could not precipitate Cef1p-HA3 from the diploid strain (Fig. 2C). Therefore Cef1p, unlike Prp19p, does not appear to be present in multiple copies within the NTC in vivo. This situation is conserved in S. pombe, since different epitope-tagged versions of the S. pombe homolog of S. cerevisiae Cef1p, Cdc5p, also failed to interact with one another in the appropriate diploid strain (Fig. 2D).
The analyses performed above left open the possibility that oligomers of Prp19p/Cwf8p exist only outside of the Cef1p/Cdc5p-containing spliceosomal complex. To examine this possibility, an S. pombe diploid strain was constructed with the relevant genotype cdc5-TAP/cdc5-TAP cwf8-HA3/cwf8-myc13. After tandem affinity purification was performed on this strain, the eluate was found to contain both tagged proteins as expected (Fig. 2E, left panels). To determine whether each Cdc5p molecule bound to one molecule of Cwf8p or whether a single molecule of Cdc5p could bind multiple molecules of Cwf8p, the eluate was diluted and subjected to immunoprecipitation with anti-myc or anti-HA antibodies, and the immunoprecipitates were blotted for the presence of Cwf8p-myc13. As would be predicted if each Cdc5p-TAP complex contained multiple copies of Cwf8p, both anti-myc and anti-HA immunoprecipitates contained Cwf8p-myc13 (Fig. 2E, right panel). We could not examine the reciprocal immunoprecipitate by immunoblotting because Cwf8p-HA3 comigrates with the immunoglobulin G heavy chain. However, because Cdc5p is present in single copy in the NTC (Fig. 2D), this result could only be explained if at least two molecules of Cwf8p interacted within the same Cdc5p-containing complex.
Prp19p binds to itself.
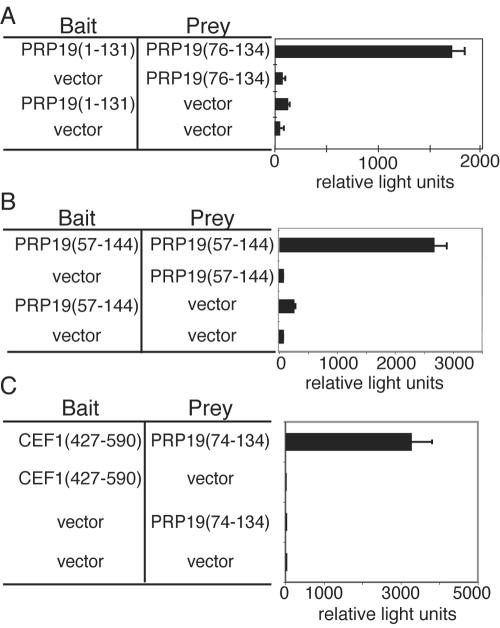
To further delineate protein-protein interactions involving Prp19p, we performed a stringent two-hybrid screen using an N-terminal fragment of Prp19p containing both its U-box and predicted coiled-coil region (Prp19p 1-131) as bait. The ability of Prp19p 1-131 to fold correctly was confirmed by CD (data not shown). In this screen PRP19 76-134, which encompasses a large portion of the predicted coiled-coil domain, was the only clone isolated multiple times (Fig. 3A), suggesting that this region of Prp19p is capable of interacting directly with itself and Prp19p self-association might be mediated by this domain. To test this we used the entire predicted coiled-coil region of Prp19p 57-144 as both bait and prey and asked if they could interact in the two-hybrid system. As we predicted, Prp19p 57-144 interacts with itself (Fig. 3B). These results are consistent with previous gel filtration chromatography experiments that showed recombinant Prp19p exists in an oligomeric state (data not shown) (33).
FIG. 3.
Mapping Prp19p interactions with itself and Cef1p. (A) PJ69-A was transformed with pGBT9PRP19 1-131 and screened with the two-hybrid library YHL-1. The bar graph shows β-galactosidase activity (represented by relative light units) of the strains containing the bait and prey constructs indicated. (B) The PJ69-4A strain was transformed with pGBT9PRP19 57-144 and pGAD424 carrying PRP19 57-144. The bar graph shows β-galactosidase activity (represented by relative light units) of the strains containing the bait and prey constructs indicated. (C) The PJ69-4A strain was transformed with pGBT9CEF1 427-590 and pGAD424PRP19 74-134, which are shown schematically above the bar graph. The bar graph shows β-galactosidase activity (represented by relative light units) of the strains containing the bait and prey constructs indicated.
Prp19p binds directly to the essential pre-mRNA splicing factor Cef1p. We previously showed that Cef1p interacts with the N terminus of Prp19p (27). In the same screen using Prp19p 1-131, we obtained a clone of Cef1p. To determine whether the region of Prp19p that was isolated in the above screen was sufficient for interacting with Cef1p, we tested whether Prp19p 76-134 could interact with Cef1p (Fig. 3C). Indeed, the predicted coiled-coil in Prp19p was sufficient to support Cef1p interaction (Fig. 3C).
Prp19p forms a stable tetramer.
To define what type of oligomer Prp19p formed, recombinant His6-Prp19p (mass, 58.6 kDa) was purified and analyzed using sedimentation velocity analytical ultracentrifugation. Analysis of the velocity data with the program Sedfit (31) showed that 78% of His6-Prp19p exhibited a sedimentation value (s) of 5.8 (Fig. 4A; root mean square deviation of the fit, 0.0073). This value is consistent with a complex of 230 kDa (Fig. 4B), the same molecular mass as a Prp19p tetramer. The rest of the protein sedimented either as a monomer (s = 2.3; 2.5%) or as a nonspecific aggregate (∼20%). His6-Prp19p has a frictional ratio value of 1.7, indicating that the tetramer is elongated in shape.
FIG. 4.
Continuous size distribution analysis of sedimentation velocity data of His6-Prp19p and Prp19p 66-141. (A) The calculated s [c(s)] is plotted versus the sedimentation coefficient (s) for His6-Prp19p. The sedimentation velocity profiles were fitted to a continuous sedimentation distribution. (B) Calculated mass [c(M)] is plotted versus molar mass (in kilodaltons) for His6-Prp19p. (C) c(s) plotted versus sedimentation coefficients (s) for Prp19p 66-141. The sedimentation velocity profiles were fit to a continuous sedimentation distribution. (D) c(M) plotted versus molar mass (in kilodaltons) for Prp19p 66-141. (E) CD spectrum of Prp19p 66-141.
The coiled-coil region of Prp19p is sufficient for tetramerization.
To determine which domain is required for Prp19p tetramerization, we used sedimentation velocity analytical ultracentrifugation to analyze the ability of the Prp19p U-box, the coiled-coil region, and the WD40 repeats to oligomerize. As expected from the two-hybrid results, neither the U-box (His6-Prp19p 1-58) nor the C terminus containing the WD40 repeats (His6-Prp19p 165-503) was able to tetramerize (data not shown). However, Prp19p 66-141 (mass, 8.5 kDa), spanning the coiled-coil region, did form a tetramer. This Prp19p fragment sedimented with an s value of 2.8 (Fig. 4C; root mean square deviation of the fit, 0.0126), consistent with a tetramer of 35-kDa molecular mass (Fig. 4D). Significantly, Prp19p 66-141 has a frictional ratio value of 2.7, indicating that this domain is very elongated. Indeed, analysis by CD showed this region of Prp19p to be almost entirely helical in nature (Fig. 4E). The CD spectrum of Prp19p 66-141 fits a model where the protein is 70% helical and 30% coil (1), consistent with a domain with a coiled-coil fold (21). Taken together, we conclude that Prp19p 66-141 contains a coiled-coil sufficient for directing tetramerization of Prp19p.
Structural analysis of the Prp19p tetramer by electron microscopy.
In order to understand the architecture of Prp19p tetramers, full-length protein was purified and examined using electron microscopy. Micrographs of negatively stained His6-Prp19p revealed a particle of variable shape, but with each containing an elongated central stalk that was flanked by two globular domains on each side (Fig. 5). A total of 9,725 particles were selected from digitized electron micrographs and subjected to multireference alignment and classification specifying 200 output classes. A representative subset of these two-dimensional projection averages (Fig. 5, panels 1 through 12) emphasizes the variability in the shape of Prp19p. The two globular domains that flank the central stalk on either side show central stain accumulations, giving them a donut-like appearance. These domains appear to be tethered to the central stalk by flexible linkers, as they can either be close together or spread wide apart (Fig. 5, compare panels 8 and 2).
As noted above, Prp19p contains three structural domains, an N-terminal U-box, a central coiled-coil, and C-terminal WD40 repeats that likely adopt a typical-bladed β-propeller structure (10) (Fig. 1B). The donut-shaped domains, with dimensions of ∼6.5 by 6.5 nm, are consistent in size and shape with the expected β-propeller structure, while the central stalk, with its rope-like appearance, is consistent with the expected projection structure of a coiled-coil domain. The small size of the U-box (7.8 kDa) would make it difficult to identify by negative stain electron microscopy.
To confirm our prediction regarding the identity of the donut-shaped domains, we analyzed the C terminus of Prp19p by negative stain electron microscopy. Micrographs of His6-Prp19p 165-503 revealed well-dispersed donut-shaped particles (Fig. 6, upper panel). These monomeric particles looked identical to the domains seen tethered to the central stalk of the full-length Prp19p tetramers (Fig. 5). We selected 4,572 particles from digitized micrographs and used multireference alignment and classification to group them into 20 classes. The resulting projection averages (Fig. 6, lower panel) confirmed that the four donut-like domains seen in the full-length Prp19p tetramers correspond to its WD40 repeats.
FIG. 6.
Electron micrograph and representative projection averages of negatively stained His6-Prp19p 165-503. (Upper panel) Typical micrograph area of negatively stained His6-Prp19p. Bar, 25 nm. (Lower panel) Twelve representative averages obtained by multireference alignment and classification of 4,507 His6-Prp19p 165-503 particle images. Side length of the average images is 16.8 nm. Number of particles in each projection average: panel 1, 168; panel 2, 554; panel 3, 185; panel 4, 224; panel 5, 244; panel 6, 409; panel 7, 205; panel 8, 187; panel 9, 193; panel 10, 216; panel 11, 179; panel 12, 197.
Prp19p oligomerization is required for function in vivo.
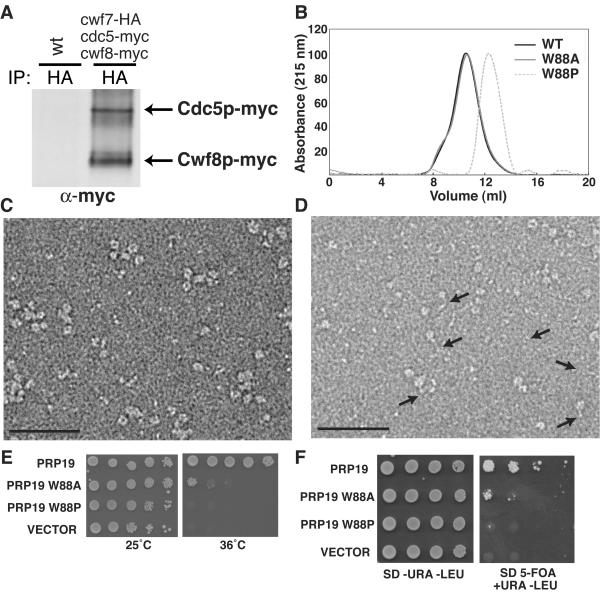
Thus far, we have shown that Prp19p/Cwf8p is oligomeric within the NTC in vivo (Fig. 2), and our in vitro results suggest that it is tetrameric. In contrast, Cef1p is not oligomeric within the NTC (Fig. 2). To determine the stoichiometry of Prp19p/Cwf8p within the NTC relative to Cef1p, a cwf7-HA3 cdc5-myc13 cwf8-myc13 strain was generated. Cwf7p is a component of the NTC that, like Cdc5p, binds directly to Cwf8p. This allowed us to isolate the NTC through anti-HA immunoprecipitation and determine the ratio of Cwf8p to Cdc5p within it by anti-myc immunoblotting (Fig. 7A). By quantitating the results of three such coimmunoprecipitations, we determined that Cwf8p-myc13 was, on average, 3.94 times more abundant than Cdc5p-myc13 (Fig. 7A). These data provide compelling evidence that Prp19p is present within the NTC as a tetramer.
FIG. 7.
Mutational analysis of the Prp19p coiled-coil region. (A) Cwf8p/Prp19p is present within the NTC in a 4:1 ratio with Cdc5p/Cef1p. An anti-myc immunoblot of an anti-HA immunoprecipitate (IP) from either a wild-type S. pombe strain (KGY246) or from cwf7-HA3, cdc5-myc13, or cwf8-myc13 (KGY4985). (B) Gel filtration elution profiles of His6-Prp19p (black line), His6-Prp19p-W88A (grey line), and His6-Prp19p-W88P (dashed line). (C) Typical micrograph area of negatively stained His6-Prp19p-W88A. Bar, 50 nm. (D) Typical micrograph area of negatively stained His6-Prp19p-W88P. Arrows point to the flexible coiled-coil region of Prp19p that can no longer oligomerize. Bar, 50 nm. (E) Haploid strain containing a temperature-sensitive copy of PRP19 (prp19-1) (KGY1811) was transformed with pRS415 plasmids containing wild-type PRP19 and prp19 coil-coil mutations under the control of the GAL1 promoter. The ability of prp19 mutations to rescue growth of the prp19-1 strain was scored by serial dilutions grown on medium lacking glucose at the permissive temperature (25°C) or at the restrictive temperature (36°C) for 3 days. (F) A prp19::HIS3 ura3-52 leu2-Δ1 haploid strain carrying a URA3-selectable vector expressing wild-type PRP19 was transformed by a standard lithium acetate method with PRP19 cDNA and prp19 mutations under control of the GAL1 promoter in a LEU2-based vector. Ura+ Leu+ transformants were serially diluted on plates containing 5-fluoroorotic acid and uracil to score the ability of prp19 mutations to rescue growth of the prp19Δ strain.
To determine whether the ability of Prp19p to tetramerize is required for its function in vivo, mutations predicted to disrupt tetramerization of the coiled-coil domain were introduced into the full-length PRP19 gene. In particular, Trp88, which is found in the critical A position of the coiled-coil heptad and is conserved in Prp19p orthologs, was altered to Ala88 and Pro88. Prp19p W88A and Prp19p W88P were produced in vitro and tested for their ability to tetramerize. The W88A mutant retained the ability to tetramerize in vitro while the W88P mutant did not, as determined by gel filtration analysis and electron microscopy (Fig. 7B, C, and D). These prp19 mutants, along with wild-type PRP19, were then assayed for their ability to complement the prp19 temperature-sensitive allele, prp19-1, at the restrictive temperature and the null allele of prp19 (PRP19 is an essential gene) by a conventional plasmid shuffle approach. The W88A mutation that did not lose its ability to tetramerize in vitro was able to complement both prp19 mutations, indicating that W88 is not strictly required for Prp19p function (Fig. 7E). In contrast W88P was unable to rescue either prp19-1 at the restrictive temperature or prp19Δ (Fig. 7F). These results indicate that the ability of Prp19p to tetramerize through its central coiled-coil stalk is required for its in vivo function.
DISCUSSION
The NTC is an essential subcomplex within the spliceosome. Numerous protein-protein interactions have been mapped between NTC components using two-hybrid analysis, far Western analysis, and direct binding assays (2, 4, 5, 6, 8, 27, 30, 35). However, the molecular details of NTC organization and how the NTC associates with other splicing complexes are not known. Here, we have found that Prp19p forms a tetramer and that the ability of Prp19p to oligomerize is essential for its in vivo function. The characterization of the Prp19p tetramer provides the first structure-based model for NTC organization.
The precise shape and relative orientation of the domains in Prp19p tetramers vary with the four globular WD40 domains flexibly tethered to a central coiled-coil tetramer. Class averages of Prp19p tetramers illustrated the myriad positions and angles that the WD40 repeats can adopt in relation to the stalk-like coiled-coil. It is likely that the flexibility of the WD40 repeats will decrease when other NTC binding partners bind. However, the variability of the Prp19p tetramer structure may be important for numerous transitions that occur during the formation of the active spliceosome. Such flexibility could allow Prp19p to accommodate substantial structural rearrangements during dynamic reorientation of spliceosome components without compromising strong binding interactions with other components of the NTC.
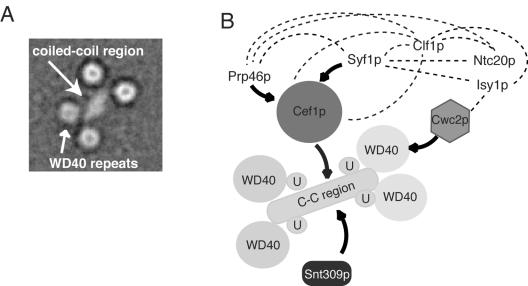
Indeed, structural characterization of the Prp19p tetramer by electron microscopy has provided significant clues as to how Prp19p might act as a central organizing unit within the NTC. Taking into account the plethora of described NTC protein-protein interactions, a model of NTC organization can be constructed highlighting the many protein-protein interdependencies within the NTC (2, 7, 8, 27, 30) (Fig. 8). In addition to creating a binding platform for Snt309p and Cef1p, the distinct spatial environment sampled by each of the four WD40 domains creates four independent protein-protein interaction modules surrounding the coiled-coil domains. As yet, only Cwc2p, an essential RNA recognition motif containing splicing factor, has been shown to interact directly with the C-terminal region of Prp19p (Fig. 8) (27). It is likely that other NTC components also interact with the WD40 repeats, although these interactions could depend on the presence of the full complement of complex members. The ability of Prp19p to interact with numerous NTC components combined with its essential role in NTC stability suggest that the Prp19p tetramer acts as an essential scaffold for the organization of the entire NTC and perhaps other associated factors. It may be for this reason that human PRP19 was identified as the nuclear matrix protein hNMP200 (12). Extension of our structural analysis to include other NTC components should be revealing in this regard.
FIG. 8.
Model for NTC organization. (A) Representative projection average of the Prp19p tetramer. Side length of the average image is 40 nm. (B) Model of protein-protein interactions mapped between Prp19p, Cef1p, Cwc2p, Prp46p, Syf1p, Isy1p, Snt309p, and Ntc20. U, U-box domain in Prp19p; C-C, coiled-coil region in Prp19p. Solid line with arrow, protein-protein interactions detected by in vitro binding assays and far Western analysis (5, 7, 27, 35); dotted lines, protein-protein interactions found by two-hybrid analysis (27). Proteins and domains are not drawn to scale.
Interestingly, Cef1p binds directly to the Prp19p coiled-coil (Fig. 3C) (27). Our in vivo investigations suggest that while four copies of Prp19p exist within the NTC, only a single molecule of S. cerevisiae Cef1p or S. pombe Cdc5p is present. The association of Cef1p with the NTC requires Prp19p oligomerization (33), and Cef1p stability requires Prp19p function (27). Therefore, an appealing model for NTC organization is that Prp19p tetramerization occurs before Cef1p, and possibly other NTC components can bind. The same Prp19p coiled-coil region that binds Cef1p also interacts with Snt309p, although there is no evidence that Snt309p and Cef1p directly interact or compete for an interaction site on Prp19p (6, 27). However, Snt309p is required to maintain Prp19p-Cef1p interaction at elevated temperatures (7), suggesting that Snt309p may help to stabilize the Prp19p tetramer and subsequently the Cef1p binding site. The proximity of the Cef1p and Snt309p binding sites in Prp19p makes it possible that Snt309p and Cef1p might interact with each other when bound to the Prp19p coiled-coil domain. Alternatively, they could bind different faces of the central stalk. Future studies on the structure of this group of proteins would distinguish between these possibilities.
Interestingly, one common feature of the otherwise-diverse family of RING/U-box-containing proteins is that they are often integral components of macromolecular assemblies (18, 29). It has been postulated that the spatial organization of the RING structure within these E3 ligase complexes may be essential for positioning the substrate in the correct orientation to optimize the transfer of ubiquitin from the E2 (36, 37). Although highly speculative, proteins that interact with either the Prp19p WD40 repeats or the coiled-coil region might also contact the U-box and be modified to promote changes in NTC organization and/or function during the splicing reaction. If, in fact, Prp19p exhibits E3 ligase activity within the context of the spliceosome, the flexible Prp19p tetramer might be well suited for facilitating the correct orientation of substrates for U-box-directed modification.
Acknowledgments
We thank Hilal Lashuel for valuable advice and assistance regarding analytical ultracentrifugation and Ryoma Ohi for critical reading of the manuscript.
This work was supported by the Giovanni Armenise-Harvard Foundation (T.W.), the Howard Hughes Medical Institute (K.L.G.), and National Institutes of Health grant RO1 GM62112 (W.J.C). J.A.R. received training grant support from T32CA09385. C.W.V.K. received training grant support from T32GM08320. M.D.O. is an Agouron Institute Fellow of the Jane Coffin-Childs Foundation, and K.L.G. is an Investigator of the Howard Hughes Medical Institute. The molecular EM facility at Harvard Medical School was established by a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained by funds from the National Institutes of Health, GM62580 (to T.W.).
REFERENCES
- 1.Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Yehuda, S., I. Dix, C. S. Russell, M. McGarvey, J. D. Beggs, and M. Kupiec. 2000. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, S. P., D. I. Kao, W. Y. Tsai, and S. C. Cheng. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279-282. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. H., W. Y. Tsai, H. R. Chen, C. H. Wang, and S. C. Cheng. 2001. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J. Biol. Chem. 276:488-494. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. H., W. C. Yu, T. Y. Tsao, L. Y. Wang, H. R. Chen, J. Y. Lin, W. Y. Tsai, and S. C. Cheng. 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 30:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H. R., S. P. Jan, T. Y. Tsao, Y. J. Sheu, J. Banroques, and S. C. Cheng. 1998. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol. 18:2196-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H. R., T. Y. Tsao, C. H. Chen, W. Y. Tsai, L. S. Her, M. M. Hsu, and S. C. Cheng. 1999. Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 96:5406-5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dix, I., C. S. Russell, R. T. O'Keefe, A. J. Newman, and J. D. Beggs. 1998. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA 4:1675-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Higuera, I., J. Fenoglio, Y. Li, C. Lewis, M. P. Panchenko, O. Reiner, T. F. Smith, and E. J. Neer. 1996. Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein beta subunit. Biochemistry 35:13985-13994. [DOI] [PubMed] [Google Scholar]
- 11.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 12.Gotzmann, J., C. Gerner, M. Meissner, K. Holzmann, R. Grimm, W. Mikulits, and G. Sauermann. 2000. hNMP 200: a novel human common nuclear matrix protein combining structural and regulatory functions. Exp. Cell Res. 261:166-179. [DOI] [PubMed] [Google Scholar]
- 13.Gould, K. L., S. Moreno, D. J. Owen, S. Sazer, and P. Nurse. 1991. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10:3297-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif.
- 15.Hatakeyama, S., M. Yada, M. Matsumoto, N. Ishida, and K. I. Nakayama. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276:33111-33120. [DOI] [PubMed] [Google Scholar]
- 16.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 18.Kentsis, A., R. E. Gordon, and K. L. Borden. 2002. Self-assembly properties of a model RING domain. Proc. Natl. Acad. Sci. USA 99:667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 20.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 21.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 22.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Luhrmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205-2208. [DOI] [PubMed] [Google Scholar]
- 23.Makarova, O. V., E. M. Makarov, H. Urlaub, C. L. Will, M. Gentzel, M. Wilm, and R. Luhrmann. 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23:2381-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 25.Murray, H. L., and K. A. Jarrell. 1999. Flipping the switch to an active spliceosome. Cell 96:599-602. [DOI] [PubMed] [Google Scholar]
- 26.Ohi, M., Y. Li, Y. Cheng, and T. Walz. 2004. Negative staining and image classification: powerful tools in modern electron microscopy. Biol. Proced. Online 6:23-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohi, M. D., and K. L. Gould. 2002. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8:798-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohi, M. D., C. W. Vander Kooi, J. A. Rosenberg, W. J. Chazin, and K. L. Gould. 2003. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 30.Russell, C. S., S. Ben-Yehuda, I. Dix, M. Kupiec, and J. D. Beggs. 2000. Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA 6:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 33.Tarn, W. Y., C. H. Hsu, K. T. Huang, H. R. Chen, H. Y. Kao, K. R. Lee, and S. C. Cheng. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarn, W. Y., K. R. Lee, and S. C. Cheng. 1993. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl. Acad. Sci. USA 90:10821-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai, W. Y., Y. T. Chow, H. R. Chen, K. T. Huang, R. I. Hong, S. P. Jan, N. Y. Kuo, T. Y. Tsao, C. H. Chen, and S. C. Cheng. 1999. Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem. 274:9455-9462. [DOI] [PubMed] [Google Scholar]
- 36.Wu, G., G. Xu, B. A. Schulman, P. D. Jeffrey, J. W. Harper, and N. P. Pavletich. 2003. Structure of a β-TrCP1-Skp1-β-catenin complex: destruction motif binding and lysine specificity of the SCFβ-TrCP1 ubiquitin ligase. Mol. Cell 11:1445-1456. [DOI] [PubMed] [Google Scholar]
- 37.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703-709. [DOI] [PubMed] [Google Scholar]