Abstract
Until recently, the only biological function attributed to the 3′→5′ exonuclease activity of DNA polymerases was proofreading of replication errors. Based on genetic and biochemical analysis of the 3′→5′ exonuclease of yeast DNA polymerase δ (Pol δ) we have discerned additional biological roles for this exonuclease in Okazaki fragment maturation and mismatch repair. We asked whether Pol δ exonuclease performs all these biological functions in association with the replicative complex or as an exonuclease separate from the replicating holoenzyme. We have identified yeast Pol δ mutants at Leu523 that are defective in processive DNA synthesis when the rate of misincorporation is high because of a deoxynucleoside triphosphate (dNTP) imbalance. Yet the mutants retain robust 3′→5′ exonuclease activity. Based on biochemical studies, the mutant enzymes appear to be impaired in switching of the nascent 3′ end between the polymerase and the exonuclease sites, resulting in severely impaired biological functions. Mutation rates and spectra and synergistic interactions of the pol3-L523X mutations with msh2, exo1, and rad27/fen1 defects were indistinguishable from those observed with previously studied exonuclease-defective mutants of the Pol δ. We conclude that the three biological functions of the 3′→5′ exonuclease addressed in this study are performed intramolecularly within the replicating holoenzyme.
DNA replication errors are an important source of genetic change. Several biochemical activities have evolved in order to prevent errors from becoming mutations. One is the 3′→5′ exonuclease (Exo) activity present in many DNA polymerases (Pol). A well-established function for this exonuclease activity is the proofreading of errors made by the DNA polymerase (6, 27). Mismatch repair (MMR) is a second fidelity system that can correct replication errors which escape proofreading. Characteristically, mutations that inactivate the exonuclease activity of the replicative DNA polymerase combined with those which inactivate MMR confer a very strong mutator phenotype, far exceeding the sum of individual mutator effects. Such synergistic hypermutability caused by a combination of proofreading and MMR defects can also lead to accumulation of lethal mutations sufficient to block propagation of a double-mutant strain (31, 35).
Another synergistic interaction between exonuclease deficiency in Pol δ and mutations in RAD27, which encodes 5′-flap endonuclease FEN1, highlights the role for the Pol δ-Exo in creating or maintaining a ligatable nick during Okazaki fragment maturation. These two genetic defects are often synthetic lethal (15, 25). In vivo evidence that Pol δ-Exo supplements FEN1 in Okazaki maturation was obtained with viable double mutants that that involved a rad27-p allele with a partial defect. These mutants exhibited hyperrecombination and an unusual pattern of hypermutability. The most frequent class of mutations were extended duplications (up to 100 bp) flanked by short direct repeats (4 to 10 bp) (24). Such mutations are usually observed when the removal of 5′-flaps generated by DNA polymerase displacement between Okazaki fragments is impaired (43). In agreement with the proposed defect in Okazaki maturation, biochemical experiments have demonstrated that strand displacement by the Exo-deficient Pol δ is increased to such an extent that full activity of FEN1 was required in order to create a ligatable nick (22).
An additional mutation avoidance function of Pol δ-Exo that is distinct from proofreading has been suggested based upon the strong mutator synergy with mutations in EXO1 (45). Exonuclease I (Exo1) was discovered in yeast as a 5′→3′ double-stranded DNA (dsDNA)-exonuclease (12, 41, 42). However, its human homolog is capable of supporting both 5′ and 3′ excision during MMR in extracts and in a reconstituted system (11, 16, 17). Exo1 has been implicated in double-strand break repair and recombination, telomere maintenance, Okazaki fragment maturation, and in MMR (reviewed in reference 48). Exo1 physically interacts with eukaryotic MutS and MutL complexes and, based on genetic data, was also proposed to play structural role in MMR (1, 2, 47). In the yeast Saccharomyces cerevisiae, mutator defects caused by deletion of EXO1 or by mutations inactivating its exonuclease are much milder than those due to elimination of mismatch recognition. Weak mutator effects of Exo1 deficiency in yeast depend in part on Pol ζ lesion bypass polymerase as determined from epistatic interactions with rev3 (47, 49). It was suggested that Exo1 participates in an MMR-independent mutation avoidance pathway as well as in mismatch removal during MMR (48). By analogy with Escherichia coli, it was suggested that mismatch removal in eukaryotic MMR could be accomplished by several redundant nucleases, including Exo1 (42). While a weak mutator on its own, exo1 is a very strong mutator in double-mutant yeast strains that also lack the exonuclease activity of either Pol δ or Pol ɛ (45). Hypermutability in an exo1Δ pol3-01 (lacking the exonuclease activity of Pol δ) double mutant is comparable to the extreme hypermutability observed in pol3-01 pms1 or pol3-01 msh2 double mutants. This hypermutability could only be measured in homozygous diploid strains because the haploid double mutants were inviable, presumably due to error catastrophe. These results led us to hypothesize that Pol δ-Exo could be one of the redundant exonucleases that function in MMR. The observed hypermutability of pol3-01 exo1 double mutants is explained by their deficiency in both proofreading and MMR, because Pol δ-Exo functions in proofreading, whereas both Exo1 and Pol δ-Exo participate in MMR. For the purpose of this paper, we will assume that the function of Pol δ-Exo in the Exo1-dependent mutation avoidance pathway is through its participation in MMR. However, the less likely possibility of an unknown mutation avoidance system with a similar strong mutator effect as MMR cannot be excluded.
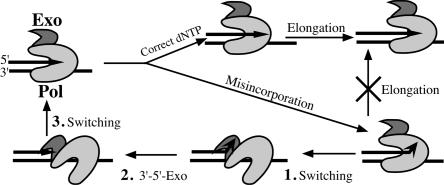
Pol δ is central to DNA replication in all eukaryotes. It carries out its polymerization functions in a complex with PCNA (for recent reviews, see references 21 and 29). However, it is not clear whether the exonuclease can also function as a separate activity in vivo or whether all biological functions are linked to Pol δ in the replicative complex. Structural studies of smaller proofreading DNA polymerases have shown that the polymerase and exonuclease active sites are localized in separate domains. The closest structural model for Pol δ is that of the bacteriophage RB69 DNA polymerase, which is very similar to the phage T4 polymerase. In this structure, the two active sites are separated by about 40 Å (13). Translocation of the nascent 3′ end from the polymerase to the exonuclease active site within the same enzyme molecule requires strand separation of 2 to 3 bp. Upon misinsertion of a base, translocation of the mismatched primer terminus from the polymerase to the exonuclease domain is as important for efficient proofreading as the catalytic exonuclease activity (9, 30) (Fig. 1). Based on these results, it appears that proofreading would be most efficient if performed intramolecularly (i.e., by the same DNA polymerase molecule that introduced the error) rather than intermolecularly (i.e., by the exonuclease active site of a separate DNA polymerase molecule).
FIG. 1.
Proofreading of incorporation errors requires both domain switching and exonuclease activity. Upon misincorporation, DNA Pol elongation is inhibited. Switching of the nascent 3′ end from Pol to the Exo domain (step 1) followed by exonuclease cleavage (step 2) and by switching back to Pol domain (step 3) leads to reestablishment of proper elongation substrate.
However, any conclusion about the mode of action in vivo cannot be determined just from structural predictions and in vitro efficiency. Is has been suggested that cellular mutation avoidance, especially in eukaryotes, can be accomplished through intermolecular activities (reviewed in reference 38). For example, during lagging strand synthesis, Pol δ must dissociate from DNA after ligation of adjacent Okazaki fragments; therefore, some Pol δ molecules in the vicinity of replication fork may not be associated with the nascent 3′ end. As a result, the exonuclease activity of Pol δ molecules not included in the replication complex could participate in proofreading and other biological functions of the exonuclease. Furthermore, there are several other 3′→5′ exonucleases in the cell which may be able to remove 3′ nucleotides and substitute for Pol δ exonuclease in its mutation avoidance functions. Genetic studies with yeast double mutants have suggested that errors generated by Pol δ could be proofread by the exonuclease activity of Pol ɛ (32, 33); however, these data could also be explained by the exonucleases of Pol δ and Pol ɛ having mutation avoidance functions distinct from proofreading (45).
Evidence for intramolecular proofreading occurring in vivo has only been obtained for T4 DNA polymerase. A mutant form of this polymerase compromised in strand separation was also affected in translocation from the polymerase to the exonuclease active site (34). Such mutants are considered to have a defect in “switching” (Fig. 1). Based on their mutator phenotype in vivo, it was concluded that switching is important for proofreading in T4, and therefore it is performed by the polymerase molecule that synthesizes the DNA strand. Thus, switching mutants appear to be a good tool for resolving the question about the in vivo mode of action for the exonuclease in a DNA polymerase. Until now it has not been clear how the 3′→5′ exonuclease of Pol δ functions in relation to its replication functions within cells. Attempts to isolate switching mutants in yeast Pol δ by sequence alignment with T4 Pol did not identify any mutants with a significant mutator effect (19). In this paper, we report the identification and characterization of yeast Pol δ mutants that are defective in switching of the nascent primer terminus between the polymerase and exonuclease sites. These mutants have enabled us to establish that Pol δ proofreading requires the wild-type capacity for switching. Switching is also essential for the Pol δ exonuclease function in MMR and Okazaki fragment maturation. We conclude that all three functions of Pol δ exonuclease are performed intramolecularly, i.e., within the replicating holoenzyme.
MATERIALS AND METHODS
Yeast strains and plasmids.
All strains were isogenic to E134 (MATα lys2-A14 ade5-1 his7-2 leu2-3,112 trp1-289 ura3-52) (23, 46). pol3 mutations were made with site-directed mutagenesis and introduced into the yeast genome via two-step gene replacement as described previously (24). All mutations were designed to change a restriction site which allowed genotyping by combination of PCR and restriction digestion. Sequences of mutant plasmids and oligonucleotides are available upon request. pBL304 contained the POL3 gene (as a 3.75-kb MluI-HindIII fragment) inserted into the SalI-HindIII sites of YCp50 (pBR322 URA3 CEN4 ARS1).
Yeast genetic methods.
Methods for modifying yeast strains, measuring mutation rates, and sequencing can1 mutant alleles were performed as described in references 15 and 23. All conclusions about differences in mutation rates were made based on nonoverlapping 95% confidence intervals calculated for each mutation rate (not shown in the experimental tables; data available upon request). In order to assess the viability of pol3 mutations in combination with null alleles of DNA metabolic genes, double mutants were obtained from single pol3 mutants in the presence of plasmid pBL304 carrying the wild-type POL3 gene and a URA3 marker. Deletions were generated by KanMX4 replacement cassettes as described previously (23). pBL304 could be readily lost from pol3 mutants, as determined by growth of plasmid-less cells on medium with 5-fluoro-orotic acid (5-FOA). Double mutants that were unable to grow on 5-FOA were considered inviable in the absence of wild-type POL3. Inability to lose pBL304 was confirmed by assessing plasmid loss after growing strains under nonselective conditions (yeast extract, 10 g/liter, peptone, 20 g/liter; glucose, 20 g/liter, adenine, 10 mg/liter [YPDA]). Following two passages of replica plating under nonselective conditions, strains were plated at low density onto YPDA to allow growth of colonies that lost the plasmid as well as colonies containing the plasmid. After 4 days of growth, about 1,000 individual colonies were replica plated on complete medium and on the medium without uracil. In the strains that were unable to lose the plasmid based on the 5-FOA assay, only one or two colonies were able to grow without uracil. Single mutants and double mutants that were determined as viable in the 5-FOA selective assay produced 10 to 50% colonies without pBL304 (POL3) plasmid in the nonselective medium assay.
Plasmid-based screen for mutator mutations in Exo III motif of the yeast POL3.
The collection of putative mutator mutations was initially obtained by oligonucleotide-directed mutagenesis of the bases corresponding to amino acids 513 to 528 within the Exo III motif in the yeast POL3 gene contained in the pBL304 plasmid. Mutagenic oligonucleotide mix (Bioserve Biotechnologies) was designed to contain 94% of a wild-type base and 2% of each of 3 possible altered bases in every position corresponding to the first and second nucleotides of mutagenized codons. The mutagenized plasmid mix was transformed into a pol3-t mutant strain containing a temperature-sensitive allele of POL3. Transformants were grown at 37°C, where the genomic pol3-t allele is not functional, and replica plated to selective media that enabled the detection of mutations within the mutation reporters, lys2-A14, his7-2, and can1. (The screening capacity of reporters is discussed in the Results section.) In control experiments with the pBL304 plasmid containing the previously characterized Exo-deficient allele pol3-01, we observed clear mutator phenotypes after growth at 37°C. The use of a yeast strain with the temperature-sensitive allele pol3-t allowed better detection of mutator mutations having exonuclease-related defects, since they are usually temperature resistant (24). The mutagenized region of the POL3 gene was sequenced in the isolated mutator strains. Forty-one mutator strains isolated from 2,982 screened transformants contained mutations in residues V515, Y516, C517, K519, D520, Y522, L523, P524, L525, and M528. The total number of different mutations was 52, since 11 strains contained double changes.
Enzymes.
Pol δ-L523H was purified from overproduction strain YH1178 (pol3-L523H pep4::KanMX strain isogenic to E134) carrying pBL336-LH (2μm ori TRP1 GAL1-pol3-L523H), and pBL341 (2μm ori URA3 GAL1-POL31 GAL10-POL32) (7). The L523H mutation was introduced into plasmid pBL336 (containing wild-type POL3) by gap repair in strain YH1178, analogous to the method described previously for other mutants (22). The resulting plasmid allele was verified by PCR amplification and sequencing of the relevant region of the POL3 gene. Pol δ-L523S and Pol δ-L523D were prepared similarly through gap repair and overproduction in strains YH1170 (as YH1178, but pol3-L523S) and YH1166 (as YH1178, but pol3-L523D), respectively. The purification of Pol δ-01 and Pol δ-5DV has been described previously (22). Replication factor C (RFC), replication protein A (RPA), and PCNA were purified as described previously (4, 18, 20).
Replication assays.
Standard 30-μl assay mixtures contained 20 mM Tris-HCl, pH 7.8, 1 mM dithiothreitol (DTT), 100-μg/ml bovine serum albumin, 8 mM MgAc2, 1 mM ATP, 80 μM each of the nonradioactive deoxynucleoside triphosphates (dNTPs) and 20 μM [α-32P]dNTP, 100 mM NaCl, 50 fmol of singly primed single-stranded M13mp18 DNA, 10 pmol of RPA, 100 fmol of RFC, 250 fmol of PCNA, and the indicated levels of wild-type or mutant Pol δ. In general, the DNA was preincubated with RPA, PCNA, and RFC for 1 min at 30°C and the reaction was started by adding Pol δ. Incubations were at 30°C. Reaction products were analyzed by electrophoresis on a 1% agarose gel or by acid precipitation followed by filtration over a glass fiber filter and scintillation counting. The gels were dried and analyzed on a phosphorimager. Quantitation was carried out with ImageQuant software. The images in the figures were contrast enhanced for visualization.
Nuclease assays.
The 50-μl pUC19 nuclease assay mixture contained 20 mM Tris-HCl, pH 7.8, 1 mM DTT, 100-μg/ml bovine serum albumin, 8 mM MgAc2, 1 mM spermidine-HCl, 50 fmol of pUC19 DNA (which had been linearized with EcoRI, followed by filling in of the overhanging ends with dATP and [3H]dTTP), and wild-type or mutant Pol δ. After 4 min at 30°C, the reactions were stopped by addition of 100 μl of 25 mM EDTA, 25 mM sodium pyrophosphate, and 50-μg/ml carrier DNA, followed by 125 μl of 10% trichloroacetic acid. After 10 min on ice, the tubes were spun in a microcentrifuge for 10 min. Two hundred microliters of the supernatant was added to a water-miscible scintillation fluid and counted in a liquid scintillation counter.
The 113-nucleotide (nt) 5′- and 3′-biotinylated template Bio-V6 (Bio-5′-AGTTGGGTTGGTTTTGTTTGGTGGAACCT25CTCCCTTCTTCTCCTCCCTCTCCCTTCCCT31-Bio) was prepared by hybridizing two half-oligonucleotides to a bridging primer followed by ligation with T4 DNA ligase and purification by preparative urea-polyacrylamide gel electrophoresis (PAGE). The template was primed with primer COKA13A (5′-32P-AGGGAAGGGAGAGGGAGGAGAAGAAGA) forming a single 3′-terminal C-A mismatch with the Bio-V6 template. All oligonucleotides were obtained from Integrated DNA Technologies and purified by polyacrylamide electrophoresis or high-performance liquid chromatography before use. After hybridization, a twofold molar excess of streptavidin was added. The nuclease assay was carried out in a 30-μl reaction mixture containing 20 mM Tris-HCl, pH 7.8, 1 mM DTT, 100-μg/ml bovine serum albumin, 8 mM MgAc2, 100 mM NaCl, 150 fmol of template primer, 500 fmol of RPA, 400 fmol of RFC, and 750 fmol of PCNA. The reaction mixture was incubated at 30°C for 1 min, and the reaction was started by addition of 600 fmol of Pol δ. Aliquots (8 μl) were withdrawn at the indicated times and analyzed by 12% denaturing PAGE. After drying of the gel, radioactivity was quantitated by phosphorimaging. Rates of hydrolysis of the 3′-terminal mismatched nucleotide were calculated from the 30-s time points with a pseudo-first-order kinetic model.
RESULTS
Genetic and biochemical defects caused by pol3-L523X mutations.
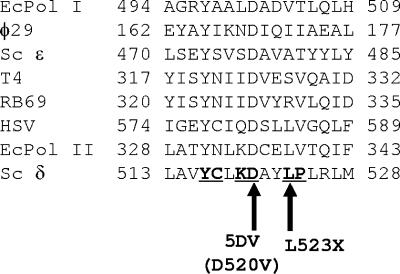
Mutations modifying L523 were recovered repeatedly in our plasmid-based screen for strong mutator mutations in the Exo III motif of yeast POL3, the catalytic subunit of Pol δ (Materials and Methods). While the L523 is invariant in Pol δ sequences (not shown), this residue is not conserved in several other DNA polymerases (Fig. 2). This contrasts with the invariant aspartate residue (D520 in S. cerevisiae Pol δ) required for exonuclease catalysis in all B-class polymerases, including yeast Pol δ (24, 39). Based on this, we reasoned that L523 is unlikely to be directly involved in catalysis but may well be important for a specific Pol δ-related function. To further understand why amino acid replacements at position 523 lead to a mutator phenotype, we created several mutations changing L523 at the chromosomal POL3 gene. In total, seven different changes were made: L523R, L523Q, and L523V, which were isolated in the initial plasmid-borne screen, as well as changes to charged residues L523D and L523H and changes to smaller residues L523A and L523S.
FIG. 2.
Alignment of Exo III motifs in different DNA polymerases. Invariant amino acids in the alignment of all 26 Pol δ from the GenBank (not shown) are underlined and boldface. Arrows point to residues mutated in the current study to create pol3-5DV and pol3-L523X alleles. Ec, E. coli; Sc, S. cerevisiae; HSV, herpes simplex virus.
The mutator activity of the pol3 alleles was assessed with three reporters: lys2-A14 reversion (−1 frameshifts in an A14 run), his7-2 reversion (+1 frameshifts in an A7 run), and forward mutations in the CAN1 gene. The lys2-A14 reversion assay is very sensitive to inactivation of MMR because premutation intermediates arising during replication escape proofreading, thus leaving the burden of mutation prevention to MMR (45, 46). The his7-2 reversion assay and the can1 forward mutation assay are sensitive to both proofreading and MMR defects (31, 37, 45).
In addition to pol3-L523X, we measured changes in mutation rates caused by two previously described alleles, pol3-01 and pol3-5DV, that completely inactivate the exonuclease. pol3-01 (D321A, E323A) is a double mutation in the Exo I motif (31), and pol3-5DV (D520V) is a mutation in the Exo III motif (24). The L523 mutations caused mutator phenotypes (Table 1) that in some cases exceeded the mutator effects of the two previously characterized exonuclease-defective pol3 mutants.
TABLE 1.
Mutation rates in pol3 mutants
| Allele | Mutation ratea
|
||
|---|---|---|---|
| lys2-A14 | his7-2 | can1 | |
| POL3+ | 1 | 1 | 1 |
| pol3-01 | 9.7 | 78 | 19 |
| pol3-5DV | 4.9 | 16 | 12 |
| pol3-L523H | 1.2 | 3.1 | 1.5 |
| pol3-L523V | 2.6 | 3.7 | 5.1 |
| pol3-L523Q | 3.7 | 13 | 8.4 |
| pol3-L523A | 6.4 | 14 | 15 |
| pol3-L523S | 7.9 | 28 | 20 |
| pol3-L523R | 32 | 37 | 90 |
| pol3-L523D | 72 | 178 | 113 |
Rates are relative to wild type: lys2-A14 reversion, 29 × 10−8; his7-2 reversion, 1.2 × 10−8; can1 forward mutation, 41 × 10−8.
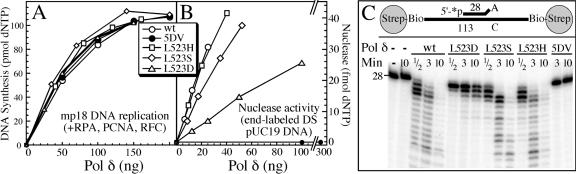
Three L523X mutant forms of Pol δ were overexpressed and purified from yeast. These mutants were chosen because they showed mutator phenotypes that were strong (L523D), medium (L523S), and marginal (L523H). Their DNA polymerase activities were comparable to that of the wild type (Fig. 3A). This is consistent with structural studies which indicate that the polymerase and exonuclease domains are autonomous. However, unlike the catalytic mutants (Pol δ-01 and Pol δ-5DV), the L523 mutants retained partial to full exonuclease activity, when measured as hydrolysis of the 3′ end of linear blunt-ended dsDNA (Fig. 3B). In order to measure exonuclease activity toward a mismatch that would be encountered by Pol δ during processive DNA replication: i.e., in a complex with PCNA, we used an oligonucleotide-based template-primer assay system. The substrate contained biotin-streptavidin blocks to prevent dissociation of PCNA by sliding off the DNA (3). PCNA was loaded by RFC onto the RPA-coated template primer prior to addition of the polymerase. Rates of hydrolysis of the 3′-terminal mismatched nucleotide were measured (Fig. 3C). Unlike the catalytic mutants, the L523 mutant enzymes retained considerable exonuclease activity: 100% (86 to 111%) with L523H, 68% (59 to 72%) with L523S, and 10% (7 to 16%) with L523D (ranges of values obtained in three independent experiments). After removal of the terminal mismatch, all enzymes continued to degrade efficiently into the dsDNA region, with the exception of Pol δ-L523D. Because of the low catalytic activity of Pol δ-L523D, both the activity on dsDNA and its processivity are expected to be seriously impaired, and this was indeed observed (Fig. 3C). In the absence of PCNA, the nuclease activity of each form of Pol δ decreased to 5 to 10% of the activity demonstrated in the presence of PCNA (data not shown).
FIG. 3.
Polymerase and exonuclease activities of the wild-type and mutant DNA polymerases. Singly primed SS M13mp18 DNA replication assays were performed (A) and exonuclease activities on terminally labeled blunt-end double-stranded (DS) pUC19 were determined (B) as described in Materials and Methods. The replication assays measured processive polymerase activity, with PCNA present, whereas the exonuclease assay on blunt-end DNA were carried out without PCNA. (C) Exonuclease activities were measured on an oligonucleotide template primer containing a C-A mismatch in the presence of RPA and PCNA, loaded by RFC. Strep-Bio, streptavidin-biotin. The streptavidin blocks prevent dissociation of PCNA by sliding off the DNA. See Materials and Methods for further details. In the absence of PCNA, the nuclease activity of each form of Pol δ was severely decreased to 5 to 10% (data not shown).
One possible explanation for a mutator phenotype with a DNA polymerase that has robust exonuclease activity is that the enzyme fails to translocate the mismatched primer terminus from the polymerase to the exonuclease domain. Exonuclease-proficient mutator mutants have been identified for E. coli DNA polymerase I and for the T4 DNA Pol (28, 30). In a biochemical analysis, these mutants showed altered partitioning of the DNA between the polymerase and exonuclease active sites, and it has been suggested that these mutants are defective in switching upon misincorporation. In this study, we developed a biochemical assay to assess defects in switching upon misincoporation by Pol δ. The premise of the assay is that DNA synthesis by either an exonuclease-defective or a switching-defective enzyme will be inhibited when misincorporation occurs. Under conditions of precursor nucleotide imbalance, misincorporation by the polymerase is increased. For the wild-type enzyme, this is not a problem because rapid translocation of the mismatch to the exonuclease domain, i.e., switching, followed by excision allows the polymerase to resume synthesis. However, because the polymerase does not readily elongate from mismatches, overall inhibition of DNA synthesis by either an exonuclease-defective or switching-defective Pol δ is expected under these conditions (Fig. 1). This inhibition assay will only work for a switching-defective polymerase if proofreading in trans is either inefficient or does not occur at all. Proofreading in trans involves dissociation of the misincorporating enzyme, followed by binding of the mismatch to another enzyme molecule in its exonuclease site. After subsequent excision of the mismatch, perhaps another cycle of dissociation and rebinding allows resumption of polymerization (Fig. 1).
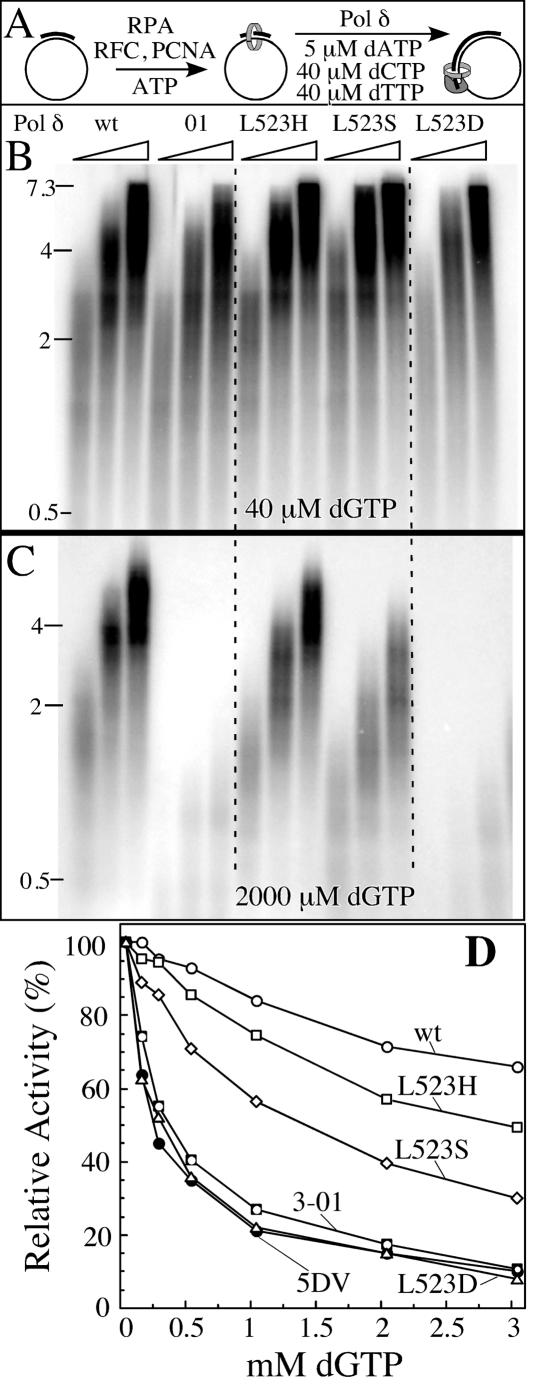
Replication of the 7.3-kb-long singly primed single stranded M13mp18 DNA by Pol δ holoenzyme was monitored by denaturing agarose gel electrophoresis (Fig. 4). All five DNA polymerases tested showed comparable activity when the concentration of the radioactive dATP was 5 μM and each of the other dNTPs was 40 μM (Fig. 4B). Selective increase of dGTP to 2 mM is expected to increase misinsertion as the dGTP/dATP ratio is now 400. Under this condition of imbalance, only minor inhibition of replication by wild-type Pol δ was observed. Some inhibition of overall DNA synthesis is expected even for the wild-type enzyme as it is forced through multiple, unproductive cycles of misincorporation followed by proofreading. However, consistent with the requirement for removal of misincorporated bases, replication by Pol δ-01 and Pol δ-5DV was almost completely inhibited (Fig. 4C and D). Replication by Pol δ-L523D was also strongly inhibited, while that by the L523S and L523H enzymes showed intermediate phenotypes. In order to obtain a quantitative estimate of the inhibition, we measured replication activity as a function of dGTP concentration and determined the concentration of dGTP required to obtain 50% inhibition. This was achieved at 0.25 to 0.35 mM dGTP for the two exonuclease-deficient forms and for Pol δ-L523D; at 1.5 mM and 3 mM for the Pol δ-L523S and -L523H mutants, respectively; and at an estimated ∼10 mM dGTP for the wild type (Fig. 4D).
FIG. 4.
Nucleotide imbalance inhibits replication in switching mutants of Pol δ. (A) Scheme of the assay. (B) Replication with 40 μM dGTP. Wt, wild type. (C) Replication with 2 mM dGTP. Results with Pol δ-5DV (not shown) were indistinguishable from those with Pol δ-01 (see also panel D). (D) Replication with various concentrations of dGTP (from 40 μM to 3 mM). Standard replication reaction mixtures as described in Materials and Methods were modified to contain 40 μM dCTP, 40 μM dTTP, 5 μM [α-32P]dATP, and the indicated concentrations of dGTP. Reactions in panels B and C were stopped after 1, 2, and 4 min of incubation and analyzed on a 1% alkaline agarose gel. Reactions in panel D were stopped after 4 min of incubation, and acid-precipitable radioactivity was determined. The activity at 40 μM dGTP was set at 100%.
Assuming that the fidelity of nucleotide insertion by Pol δ-L523H or Pol δ-L523S is not affected by the mutation in the exonuclease domain (see Discussion), the most straightforward explanation for the observed inhibition is that these two enzymes, which exhibit full or nearly full wild-type exonuclease catalytic activity, have a reduced capacity to switch the nascent 3′ end between the Pol and Exo sites. Therefore, we designated them as switching mutants. Importantly, the severity of the proposed biochemical defect in switching correlates well with the observed mutator effect caused by the pol3-L523D, -L523S, and -L523H mutations in vivo (Table 1) (data presented below). We propose that in the other pol3-L523X mutants, a defect in switching may also contribute to their biological defects.
pol3-L523X mutations exhibit synthetic lethality with msh2, exo1, and rad27 defects.
Synthetic lethality and/or synergistic hypermutability caused by combining exonuclease-defective pol3 mutations with mutations causing a defect in mismatch recognition (e.g., msh2), 5′-flap removal (rad27), and mismatch removal (exo1) highlight the biological roles of the 3′→5′ exonuclease in proofreading, Okazaki fragment maturation, and MMR, respectively (see the introduction). We asked whether the in vivo functions that depend on the exonuclease activity of Pol δ would also be impaired in the switching mutants.
Double mutants were created in the presence of a plasmid carrying wild-type POL3, and the viability of the double mutants was assessed in a plasmid-loss assay as described in Materials and Methods. Since the transformation frequency of the pol3-L523R strain with the rad27Δ::KanMX4 deletion cassette was very low and transformants grew slowly, lethality of the pol3-L523R rad27Δ double mutant was established by tetrad analysis (not shown). Similar to the catalytic pol3-01 and pol3-5DV mutants, most switching mutants were inviable in combination with msh2Δ, exo1Δ, or rad27Δ (Table 2). Viable combinations with exo1Δ and msh2Δ were formed only with the pol3-L523V and pol3-L523H alleles, which had very weak mutator activities on their own (Table 1). Interestingly, all pol3-L523X mutations, including the weakest mutator alleles, were synthetically lethal with rad27Δ. We sought to determine if at least some of them would be viable in combination with the more subtle rad27-p mutation that decreases the affinity of FEN1 for PCNA (15). Diploids were generated that were heterozygous for rad27-p and either pol3-L523H (weakest defect) or pol3-L523S (medium defect). All spores in 12 dissected tetrads from pol3-L523H/POL3 RAD27/rad27-p diploids were viable. Genotyping by PCR and restriction digestion (not shown) identified 12 pol3-L523H rad27-p viable double mutants among the meiotic progeny. However, pol3-L523S rad27-p double mutants were inviable. There were 24% dead spores in the tetrads of the pol3-L523S/POL3 RAD27/rad27-p diploid, and there were no double mutants among 36 meiotic segregants genotyped (P < 10−4).
TABLE 2.
Synthetic lethality of pol3 mutations
Switching mutation pol3-L523S and catalytic mutation pol3-5DV show similar mutator effects when combined with MMR defects.
Homozygous diploid double mutants that combine the catalytic exonuclease defect in pol3-01 with msh2 (or pms1) or with exo1 are viable, unlike the corresponding haploid double-mutant strains. This rescue of viability has been attributed to the recessive nature of the majority of lethal genetic changes that lead to error catastrophe in haploid double mutants. The hypermutability of diploid double mutants is due to the elimination of most if not all of the mutation avoidance activity that could compensate for the exonuclease defect in pol3-01. In other words, mutator defects in pol3 mutants can be masked to various degrees by the MMR machinery (31, 45). Therefore, measuring mutation rates in diploids of double mutants of msh2 or exo1 with the pol3 alleles (including pol3-L523X) allows a better evaluation of the relative severity of the pol3 defects in vivo than can be obtained by measurements in the single pol3 mutants. Specifically, the mutator effect of a pol3 allele in a double mutant with msh2 allows us to evaluate defect of proofreading caused by the pol3 mutation, whereas the mutator effect in pol3 exo1 addresses the defect caused by the pol3 mutation in a nuclease-related putative MMR function.
Diploid double mutants were created by expressing the HO-endonuclease as described previously (44) in pol3 msh2 or pol3 exo1 double-mutant haploids, in which the pol3 defect was complemented by wild-type POL3 in plasmid pBL304. Diploids were propagated on YPDA in order to allow the loss of the POL3 and HO plasmids. Based on the high frequency of loss of the POL3-containing plasmid (not shown), the diploid double mutants are viable.
Mutator synergy (excess of mutation rate in a double mutant over the sum of rates in single mutants) was observed with both catalytic and switching mutants for both reporters tested (Table 3). (Note that can1 mutation rates could not be measured in the diploids because canavanine resistance is recessive.) Both pol3-L523S and pol3-5DV double mutants with either msh2Δ or exo1Δ showed a synergistic increase in his7-2 reversion, 15 to 60 times more than would be expected for additive interaction. Thus, the catalytically defective mutation pol3-5DV and the switching-defective mutation pol3-L523S impair both the proofreading and MMR functions of the exonuclease in vivo. Importantly, for each type of double mutant, with exo1Δ or with msh2Δ, the rates of his7-2 reversion were similar for both pol3 mutations, suggesting that their in vivo functions are impaired to the same extent by either the catalytic or the switching mutations. There was also strong synergy for reversion of lys2-A14 in pol3 exo1Δ double mutants (460 to 620 times more than expected for additive interaction), supporting the conclusion that both types of pol3 defects compromise MMR.
TABLE 3.
Mutation rates in single- and double-mutant diploid strains
| Genotype | Relative mutation ratea
|
|
|---|---|---|
| lys2-A14 | his7-2 | |
| POL3 | 1 | 1 |
| exo1/exo1 | 24 | 14 |
| msh2/msh2 | 6,400 | 65 |
| pol3-5DV/pol3-5DV | 39 | 95 |
| pol3-5DV/pol3-5DV exo1/exo1 | 39,300 (63) | 4,600 (109) |
| pol3-5DV/pol3-5DV msh2/msh2 | 50,400 (6,439) | 9,300 (160) |
| pol3-L523S/pol3-L523S | 19 | 240 |
| pol3-L523S/pol3-L523S exo1/exo1 | 15,300 (43) | 3,800 (254) |
| pol3-L523S/pol3-L523S msh2/msh2 | 14,600 (6,419) | 7,900 (305) |
Rates are relative to wild type: lys2-A14 reversion, 36 × 10−8; his7-2 reversion, 1.1 × 10−8. Relative rates expected for an additive effect of two mutators in the double mutant are shown in parentheses.
As shown earlier, most errors in microsatellites, such as those leading to mutations in the lys2-A14 reporter, escape correction by the proofreading exonuclease (26, 40, 44, 46). In agreement with these observations, the mutator synergy between the pol3 mutations and msh2Δ was modest: only two- to eightfold over the rate expected for additivity.
Synergistic interactions of weak pol3-L523X mutators with msh2, exo1, and rad27 defects.
Since haploid strains combining pol3-L523V or pol3-L523H with msh2Δ, or exo1Δ as well as pol3-L523H with rad27-p, were viable (Table 2), we examined mutability in these double mutants (Table 4). Both msh2Δ and exo1Δ showed significant synergy with the pol3 mutants in a his7-2 reversion or can1 forward mutation assay. There was also significant synergy in the pol3 double mutants with exo1Δ for lys2-A14 reversions. Based on comparison of 95% confidence intervals (not shown), the pol3-L523X msh2Δ double mutants showed either no synergy or only marginal synergy in the lys2-A14 reversion assay, reflecting the escape of frameshift mutations in this target from proofreading (see the previous section). We conclude that both weak pol3-L523X mutators are defective in proofreading and MMR in vivo. However, since the mutation rates were lower than in the corresponding double mutants with pol3-L523S and pol3-5DV, the proofreading and MMR defects in pol3-L523H and -L523V are partial (Table 4).
TABLE 4.
Mutation rates in haploid strains carrying pol3-L523X, exo1Δ, and msh2Δ
| Genotype | Mutation ratea
|
||
|---|---|---|---|
| lys2-A14 | his7-2 | can1 | |
| POL3+ | 1 | 1 | 1 |
| exo1Δ | 33 | 2.0 | 6.0 |
| msh2Δ | 7,200 | 46 | 16 |
| pol3-L523V | 2.6 | 3.7 | 5.1 |
| exo1Δ pol3-L523V | 690 (36) | 810 (5.7) | 130 (11) |
| msh2Δ pol3-L523V | 16,000 (7,200) | 7,400 (50) | 68 (21) |
| pol3-L523H | 1.2 | 3.1 | 1.5 |
| exo1Δ pol3-L523H | 290 (34) | 210 (5) | 39 (7) |
| msh2Δ pol3-L523H | 19,000 (7,200) | 2,000 (49) | 500 (18) |
Rates are relative to wild type (absolute rates for wild type are given in the footnotes to Table 1). Relative rates expected for an additive effect of two mutators in the double mutant are shown in parentheses.
The haploid double mutant pol3-L523H rad27-p showed a synergistic increase in mutation rate for all three reporters (Table 5). In order to assess whether there was an increase in the rate of extended duplications that are diagnostic of a defect in Okazaki fragment maturation (43), we compared the mutation spectrum of forward can1 mutations in the pol3-L523H rad27-p double mutant with mutation spectra in single-mutant and wild-type strains (Tables 5 and 6). Importantly, the synergistic increase in can1 mutation rates can be accounted for by extended duplications flanked by short direct repeats. The absolute rate of duplications in this double mutant (1.7 × 10−5) is very close to the rate of duplications in a pol3-5DV rad27-p double mutant (1.8 × 10−5) involving a Pol δ without Exo activity. We conclude that the switching defect in the pol3-L523H mutant impedes Okazaki maturation to an extent comparable with that in the catalytically null pol3-5DV mutant.
TABLE 5.
Mutation rates in haploid strains carrying pol3-L523H and rad27-pa
| Genotype (no. of isolates)b | Mutation rate
|
can1
|
||
|---|---|---|---|---|
| lys2-A14 | his7-2 | Mutation rate | Duplication ratec | |
| Wild type (4) | 1 | 1 | 1 | 0 (0/20) |
| pol3-L523H (4) | 1.4 | 2.5 | 2.5 | 0 (0/27) |
| rad27-p (4) | 2.1 | 2.3 | 2.6 | 1.3 (9/18) |
| pol3-L523H rad27-p (8) | 9.7 [3.5]d | 33 [4.8] | 33 [5.1] | 28 (21/25) [1.3] |
Rates are relative to the wild type: lys2-A14 reversion, 30 × 10−8; his7-2 reversion, 1.5 × 10−8; can1 forward mutation rate, 61 × 10−8.
Isolates from tetrad dissection of the double heterozygous diploid pol3-L523H/POL3 rad27-p/RAD27.
Rates of extended duplications in CAN1 relative to the total rate of can1 mutations in the wild type were calculated based on the fraction of duplications among all can1 mutants sequenced (numbers of duplications and the total numbers of sequenced can1 mutants are given in parentheses as the numerator and denominator, respectively). The mutation spectra for the wild type and rad27-p are taken from reference 24. Mutation spectra determined in this work are presented in Table 6.
Relative rates expected for an additive effect of two mutators in the double mutant are shown in brackets.
TABLE 6.
Spectrum of can1 mutationsa
| Mutation type | Wild typeb
|
pol3-L523H
|
pol3-L523H rad27-p
|
|||
|---|---|---|---|---|---|---|
| Mutation | No. of occurrences/ total (%) | Mutation | No. of occurrences/ total (%) | Mutation | No. of occurrences/ total (%) | |
| Base substitution | C→A | 4/20 | G→A | 3/27 | TT→CA | 1/25 |
| C→T | 2/20 | G→T | 3/27 | C→A | 1/25 | |
| C→G | 1/20 | A→T | 1/27 | 2/25 (8) | ||
| G→A | 4/20 | T→C | 5/27 | |||
| G→T | 3/20 | T→A | 1/27 | |||
| A→G | 1/20 | C→A | 1/27 | |||
| T→A | 1/20 | 14/27 (52) | ||||
| 16/20 (80) | ||||||
| Frameshift | T4→T3 | 1/20 | T6→T7 | 3/27 | G4→G3 | 1/25 |
| T3→T2 | 1/20 | T6→T5 | 1/27 | C1→C0 | 1/25 | |
| 2/20 (10) | T5→T4 | 1/27 | 2/25 (8) | |||
| T4→T3 | 1/27 | |||||
| T3→T2 | 1/27 | |||||
| C3→C2 | 2/27 | |||||
| A2→A1 | 1/27 | |||||
| A3→A1 | 1/27 | |||||
| (AT)2→(AT)1 | 1/27 | |||||
| 12/27 (44) | ||||||
| Insertion/deletion | Δ1324-1339 (16, 8) | 1/20 (5) | Δ887-893 (7, 4) | 1/27 (4) | I224-255 (32, 5) | 1/25 |
| I289-306 (18, 4) | 1/25 | |||||
| I380-403 (24, 6) | 1/25 | |||||
| I630-689 (60, 8*) | 2/25 | |||||
| I762-810 (49, 7) | 2/25 | |||||
| I776-802 (27, 8*) | 1/25 | |||||
| I906-1049 (144, 7*) | 1/25 | |||||
| I1002-1019 (18, 6) | 2/25 | |||||
| I1163-1183 (21, 5) | 1/25 | |||||
| I1265-1333 (69, 5) | 2/25 | |||||
| I1401-1424 (24, 6) | 1/25 | |||||
| I1488-1520 (33, 5*) | 1/25 | |||||
| I1494-1552 (59, 6) | 1/25 | |||||
| I1580-1617 (38, 6) | 4/25 | |||||
| 21/25 (84) | ||||||
| Complex | TTCTC→CTCTCTG | 1/20 (5) | 0/27 (0) | 0/25 (0) | ||
All designations are the same as those in reference 24. Nucleotide coordinates of the wild-type CAN1 gene are used with the A of the ATG as nt 1. Deletion mutations are designated by Δx-y (a, b). Insertion mutations are designated by 1x-y (a, b). x and y are the nucleotide coordinates of the first and last nucleotides, respectively, of the wild-type sequence that are deleted (Δx-y) or inserted (1x-y); a is the length of the deletion/insertion; and b is the length of the associated short direct repeat sequence. *,repeat with a 1-nt mismatch.
Data are from reference 24.
DISCUSSION
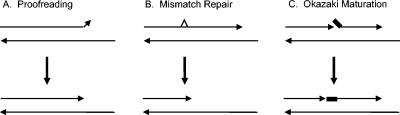
Previous genetic studies have established that the exonuclease activity of Pol δ participates in three distinct fidelity mechanisms, i.e., proofreading, maturation of Okazaki fragments, and EXO1-dependent mutation avoidance, which is proposed to involve degradation of the mismatched DNA during MMR (see Fig. 5 and the introduction). Below, we summarize the evidence suggesting that switching between the polymerase and exonuclease domains is also required in these three pathways and discuss the implications of this conclusion.
FIG. 5.
Three biological functions of the yeast Pol δ 3′→5′ exonuclease. Shown are substrates and products resulting from the action of Pol δ. (A) Proofreading. Exonuclease removes mismatched or misaligned nucleotide(s) at the 3′ terminus. (B) MMR. Exonuclease removes a long stretch of DNA including a mismatch (shown as a triangle) that has not been removed by proofreading. (C) Okazaki maturation. Exonuclease digests several nucleotides from the 3′ terminus of a nascent strand. This allows the 5′-displaced strand (thick line) to realign and create a ligatable nick.
Defects in partitioning of substrate DNA between polymerase and exonuclease sites have been documented for several DNA polymerases (28, 30). The partitioning mutations in T4 DNA polymerase are of particular interest because this enzyme is closely related to Pol δ. Kinetic studies of a G255S mutant in the exonuclease domain of the T4 enzyme have shown that the mutant is defective for strand separation at the primer terminus. This results in inhibition of translocation of the 3′ end of the nascent strand from the polymerase to the exonuclease domain (30, 34). While this defect resulted in a strong mutator phenotype in phage T4, the yeast pol3-G447S mutation that was chosen based on sequence alignment gave an antimutator phenotype for frameshifts in homopolymeric runs (19). Our data indicate that amino acid L523 adjacent to the conserved Exo III motif is important for switching in Pol δ.
Crystallographic studies of bacteriophage RB69 DNA polymerase, the closest structural homolog to Pol δ, show that the polymerase and exonuclease activities reside in separable domains with the active sites separated by about 40 Å (13). Therefore, it was not surprising that the L523X mutants of Pol δ displayed normal polymerization activity. However, in two different assay systems, Pol δ-L523H and Pol δ-L523S showed wild-type or near-wild-type exonuclease activity (Fig. 3). In order to reconcile the observed mutator phenotype of these mutants with the presence of an intact exonuclease activity, we investigated whether these mutants could be partially defective in switching the 3′ terminus between the polymerase and exonuclease domains. In a DNA replication assay under conditions of nucleotide precursor imbalance that enhance misincorporation, defects in either the exonuclease catalytic activity or in switching of the mismatch primer terminus from the Pol to Exo site would result in an overall inhibition of DNA synthesis, as indeed was observed for all three L523 mutant enzymes. The most compelling argument for the importance of the L523 residue in switching was obtained with Pol δ-L523D. This enzyme was inhibited by dNTP imbalance to the same degree as the exonuclease-deficient Pol δ-01 or Pol δ-5DV. However, its exonuclease domain retains partial activity, and therefore this inhibition cannot be attributed to the reduction in exonuclease catalysis alone, indicating that this mutant is at least partially defective in switching the mismatched 3′ terminus to the exonuclease domain (Fig. 4). The other L523 mutants were inhibited to a lesser extent under conditions of nucleotide precursor imbalance. As Pol δ-L523H showed wild-type exonuclease activity, the observed inhibition can be attributed to a partial defect in switching. A similar argument for a switching defect can be made for Pol δ-L523S, which was more strongly inhibited than the L523H mutant under dNTP imbalance, yet still exhibited near-wild-type exonuclease activity. However, the interpretation of the exonuclease measurements for Pol δ-L523S may be complicated by the likely bimodal binding of the substrate to the enzyme. Pre-steady-state kinetic studies of bacteriophage T4 DNA polymerase have shown that even a mismatched template-primer DNA binds predominantly to the polymerase site rather than the exonuclease site (5, 8). To observe catalytic activity, the DNA substrate either has to switch to the exonuclease site or dissociate and rebind to the exonuclease site. If the structurally related Pol δ shows similar binding properties, a mutation that inhibits switching would also cause a decrease in the measured rate of exonuclease activity, even though the microscopic rate of catalysis might not be affected. Therefore, the observed lower exonuclease activity for Pol δ-L523S may in part or in total be caused by a switching defect in this mutant.
While all observations are consistent with a partial defect in switching by the Pol δ-L523 mutants, the observed in vivo mutator phenotype as well as the observed inhibition of in vitro DNA replication under dNTP imbalance conditions can also be explained if the mutant polymerases showed an increased rate of misincorporation and/or frameshift misalignment compared to the wild type. We consider this unlikely because mutant L523X enzymes show wild-type levels of DNA polymerase activity and because, based on the analogies with RB69 DNA polymerase (see the introduction), the Pol and Exo activities reside in separate domains. However, we cannot currently exclude the possibility that incorporation fidelity is negatively affected through long-range perturbations in these mutants. Even if this happens, it cannot explain the defects in Okazaki maturation caused by L523X changes. Importantly, a recent study of the activity of Pol δ at a nick adds further support to our proposed switching model. When a replicating PCNA-Pol δ complex reaches downstream dsDNA, it rapidly turns over dNTP to dNMP at the nick position by repeated cycles of extension followed by exonucleolytic degradation, a process called “idling ” (14). The rate of idling is dependent on the efficiency of forth and back switching between the Pol and Exo domains. The rate of idling by Pol δ-L523H is only 32% that of the wild type, that of the L523S mutant is only 10% of that of the wild type, and that of the L523D mutant is only 1% of that of the wild type, indicating that all three mutants show defects in switching in an assay, which does not depend upon misincorporation.
The three in vivo functions of Pol δ-Exo depend on efficient switching between Pol and Exo sites.
All pol3-L523X mutations showed synthetic lethality and/or mutator synergy with msh2, exo1, and rad27 mutations, highlighting the in vivo defects in proofreading, MMR, and Okazaki fragment maturation, respectively. All mutants have wild-type DNA polymerase activity but exhibit exonuclease-related defects (Fig. 3 and 4). Based on a comparison of the in vitro switching and exonuclease activity defects of the pol3-L523X mutants with their in vivo defects, we concluded that the function of the nuclease in proofreading, MMR, and Okazaki fragment maturation requires efficient switching. This conclusion is most straightforward for the pol3-L523H mutant. In vitro Pol δ-L523H showed wild-type exonuclease activity, while exhibiting a partial switching defect. Therefore the in vivo defects in proofreading, MMR, and Okazaki fragment maturation, revealed as synergistic hypermutability in combination with msh2, exo1, and rad27-p (Tables 4 and 5), are likely to result from a defect in switching. Similar arguments can be made for the pol3-L523S mutant. The in vivo defects in pol3-L523S msh2 and pol3-L523S exo1 double mutants are comparable with those of the double mutants involving catalytically null pol3-5DV (Table 3, his7-2 reporter). However, unlike Pol δ-5DV, the Pol δ-L523S mutant enzyme retains 70% of wild-type exonuclease activity, indicating that the switching defect in this mutant contributes substantially to the observed phenotypes. In summary, the biological defects in the pol3-L523X mutants exceeded the level expected from just a partial deficiency in exonuclease activity.
We conclude that switching from the polymerase to the exonuclease domain in Pol δ is required for its multiple roles in vivo. As discussed below, this requirement leads us to propose that the exonuclease mediates its functions primarily within the holoenzyme complex that is performing DNA synthesis.
Proofreading is performed by the same Pol δ molecule that introduces an error.
In principle, the requirement for switching between the polymerase and exonuclease sites during proofreading could be bypassed if the polymerase were able to dissociate from the DNA after making an error, followed by another enzyme molecule that could bind the mismatch directly in its exonuclease domain and catalyze degradation. Such intermolecular proofreading is not in agreement with the in vivo levels of proofreading defects of the pol3 switching mutants, as compared to the catalytic null mutants. Specifically, if intermolecular proofreading involving different Pol δ molecules would be frequent, the proofreading defect in pol3-L523S should be much smaller than in the catalytically dead pol3-5DV, while pol3-L523H would not be expected to show any significant proofreading defect. We conclude that intramolecular proofreading, i.e., by the same polymerase molecule that introduced an error, is the prevailing mechanism for proofreading spontaneous replication errors made by the Pol δ in yeast. Since this conclusion is based upon the phenotypes of the L523X switching mutants, we currently cannot exclude the possibility of intermolecular proofreading by wild-type polymerase or by other pol3 mutants. For example, errors introduced by the catalytically defective pol3-01 have been proposed to be proofread in the intermolecular mode by the DNA polymerase that replicates the opposite strand (32, 33). If intermolecular Pol δ/Pol ɛ proofreading does take place, it does not fully compensate for proofreading defects in the catalytically inactive pol3-01 mutant (31, 45).
While we conclude that proofreading is primarily performed by the same Pol δ molecule that introduced an error, the mechanism by which the replication machinery extends the DNA strand containing an error remains to be established. Studies with mutant pol3-01 indicate that replication checkpoints and bypass polymerases may be involved in mutation avoidance in the absence of the Pol δ exonuclease (10). An increased capacity for extension of an error made by Pol δ-L523X mutant polymerases may explain why some of them are showing stronger mutator effects than the catalytically null pol3-01 or pol3-5DV, as well as the differences in mutator effects between these two catalytically null polymerases (Table 1). However, we note that up to 99% of the mutator effects in single pol3 mutants can be masked by replication-associated mutation avoidance functions, such as Msh2 (mismatch recognition) and Exo1 (mismatch removal). Therefore, the observed differences in mutator effects of single pol3 mutants could be due to activities distinct from normal DNA replication such as DNA repair and translesion synthesis. Studies of mutator effects and genetic interactions of exonuclease-related pol3 mutations in the absence of MMR may lead to a better understanding of factors that determine error extension during replication.
Role of the Pol δ exonuclease in MMR.
Biochemical studies have established that both prokaryotic and eukaryotic MMR systems remove up to several hundred nucleotides surrounding a mismatch. In E. coli at least four nucleases and one helicase were implicated in mismatch removal (reviewed in reference 36). So far, the only enzyme clearly implicated in mismatch removal in eukaryotes is Exo1, which in the in vitro MMR assay has been shown to catalyze both 3′ and 5′ degradation (see the introduction). The synergistic mutator phenotypes and synthetic lethality lead us to propose that in yeast, Exo1 and Pol δ-Exo provide partially redundant activities for the removal of a mismatch-containing DNA strand during MMR (45 and the introduction). Since Pol δ switching is required in Exo1-related mutation avoidance it is likely that the Pol δ exonuclease functions here also as part of the replicating holoenzyme complex, rather than as a free exonuclease. The most straightforward explanation of our results is that Exo1 and Pol δ exonuclease have overlapping roles in DNA degradation during mismatch removal. However, in order to carry out efficient mismatch removal over hundreds of nucleotides as a part of holoenzymes attached to DNA, the exonuclease activity of Pol δ should be converted into a processive exonuclease mode. This could occur upon interaction with the mismatch recognition complex, similar to the conversion of Exo1 into a processive exonuclease triggered by MutSα (17).
Role of the Pol δ exonuclease in Okazaki fragment maturation.
Our previous in vivo and in vitro studies have shown that the exonuclease activity of Pol δ is important for efficient maturation of Okazaki fragments (22, 24). This activity of Pol δ contributes to the prevention of double-strand breaks, genome rearrangements, and mutations. The exonuclease activity facilitates the formation of ligatable nicks by reducing the strand displacement capacity of Pol δ. We have hypothesized that there is a balance between strand displacement and 3′→5′ degradation within a replication complex, which allows efficient maintenance of a ligatable nick. The synthetic lethality of the pol3-L523H rad27Δ double mutant, together with the strong induction of duplication mutations observed in the viable pol3-L523H rad27-p double mutant, indicate that polymerase-to-exonuclease switching is also essential for Okazaki maturation (Tables 2 and 5). In fact, genetic data suggest that the weakest switching mutant, pol3-L523H, has only a partial defect in proofreading and MMR, while its defect in Okazaki maturation is as strong as that of catalytically null pol3-5DV. Our recent study has highlighted the importance of switching between the polymerase and exonuclease domains in order for Pol δ to maintain a ligatable nick in vitro (14). Based on the current study, we suggest that the same Pol δ molecule that synthesized an Okazaki fragment assists in maintenance of a ligatable nick in vivo.
Acknowledgments
We thank Jan Drake, Kasia Bebenek, John Majors, and Tom Darden for advice in the course of this work and for comments on the manuscript.
This work was supported in part by grant GM32431 from the National Institutes of Health (P.M.B.) and by a fellowship of the Sigma Foundation (P.G.).
REFERENCES
- 1.Amin, N. S., M.-N. Nguyen, S. Oh, and R. D. Kolodner. 2001. exo1-dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21:5142-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argueso, J. L., D. Smith, J. Yi, M. Waase, S. Sarin, and E. Alani. 2002. Analysis of conditional mutations in the Saccharomyces cerevisiae MLH1 gene in mismatch repair and in meiotic crossing over. Genetics 160:909-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayyagari, R., X. V. Gomes, D. A. Gordenin, and P. M. Burgers. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278:1618-1625. [DOI] [PubMed] [Google Scholar]
- 4.Ayyagari, R., K. J. Impellizzeri, B. L. Yoder, S. L. Gary, and P. M. J. Burgers. 1995. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell. Biol. 15:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom, L. B., X. Chen, D. K. Fygenson, J. Turner, M. O'Donnell, and M. F. Goodman. 1997. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J. Biol. Chem. 272:27919-27930. [DOI] [PubMed] [Google Scholar]
- 6.Brutlag, D., and A. Kornberg. 1972. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′→5′ exonuclease activity in deoxyribonucleic acid polymerases. J. Biol. Chem. 247:241-248. [PubMed] [Google Scholar]
- 7.Burgers, P. M., and K. J. Gerik. 1998. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 273:19756-19762. [DOI] [PubMed] [Google Scholar]
- 8.Capson, T. L., J. A. Peliska, B. F. Kaboord, M. W. Frey, C. Lively, M. Dahlberg, and S. J. Benkovic. 1992. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry 31:10984-10994. [DOI] [PubMed] [Google Scholar]
- 9.Cowart, M., K. J. Gibson, D. J. Allen, and S. J. Benkovic. 1989. DNA substrate structural requirements for the exonuclease and polymerase activities of procaryotic and phage DNA polymerases. Biochemistry 28:1975-1983. [DOI] [PubMed] [Google Scholar]
- 10.Datta, A., J. L. Schmeits, S. A. Neelam, P. J. Lau, K. Myung, and R. K. Kolodner. 2000. Checkpoint dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3-01 mutants. Mol. Cell 6:593-603. [DOI] [PubMed] [Google Scholar]
- 11.Dzantiev, L., N. Constantin, J. Genschel, R. R. Iyer, P. M. Burgers, and P. Modrich. 2004. A defined human system that supports bidirectional mismatch-provoked excision. Mol. Cell 15:31-41. [DOI] [PubMed] [Google Scholar]
- 12.Fiorentini, P., K. N. Huang, D. X. Tishkoff, R. D. Kolodner, and L. S. Symington. 1997. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 17:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin, M. C., J. Wang, and T. A. Steitz. 2001. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 105:657-667. [DOI] [PubMed] [Google Scholar]
- 14.Garg, P., C. M. Stith, N. Sabouri, E. Johansson, and P. M. Burgers. 2004. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 18:2764-2773. [DOI] [PMC free article] [PubMed]
- 15.Gary, R., M. S. Park, J. P. Nolan, H. L. Cornelius, O. G. Kozyreva, H. T. Tran, K. S. Lobachev, M. A. Resnick, and D. A. Gordenin. 1999. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 19:5373-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genschel, J., L. R. Bazemore, and P. Modrich. 2002. Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 277:13302-13311. [DOI] [PubMed] [Google Scholar]
- 17.Genschel, J., and P. Modrich. 2003. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell 12:1077-1086. [DOI] [PubMed] [Google Scholar]
- 18.Gomes, X. V., S. L. Gary, and P. M. Burgers. 2000. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J. Biol. Chem. 275:14541-14549. [DOI] [PubMed] [Google Scholar]
- 19.Hadjimarcou, M. I., R. J. Kokoska, T. D. Petes, and L. J. Reha-Krantz. 2001. Identification of a mutant DNA polymerase delta in Saccharomyces cerevisiae with an antimutator phenotype for frameshift mutations. Genetics 158:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 21.Hubscher, U., G. Maga, and S. Spadari. 2002. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71:133-163. [DOI] [PubMed] [Google Scholar]
- 22.Jin, Y. H., R. Ayyagari, M. A. Resnick, D. A. Gordenin, and P. M. Burgers. 2003. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′→5′-exonuclease activities of Pol δ in the creation of a ligatable nick. J. Biol. Chem. 278:1626-1633. [DOI] [PubMed] [Google Scholar]
- 23.Jin, Y. H., A. B. Clark, R. J. Slebos, H. Al-Refai, J. A. Taylor, T. A. Kunkel, M. A. Resnick, and D. A. Gordenin. 2003. Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 34:326-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin, Y. H., R. Obert, P. M. Burgers, T. A. Kunkel, M. A. Resnick, and D. A. Gordenin. 2001. The 3′→5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. USA 98:5122-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin, and T. D. Petes. 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t). Mol. Cell. Biol. 18:2779-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroutil, L. C., K. Register, K. Bebenek, and T. A. Kunkel. 1996. Exonucleolytic proofreading during replication of repetitive DNA. Biochemistry 35:1046-1053. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel, T. A. 1988. Exonucleolytic proofreading. Cell 53:837-840. [DOI] [PubMed] [Google Scholar]
- 28.Lam, W. C., E. J. Van der Schans, C. M. Joyce, and D. P. Millar. 1998. Effects of mutations on the partitioning of DNA substrates between the polymerase and 3′-5′ exonuclease sites of DNA polymerase I (Klenow fragment). Biochemistry 37:1513-1522. [DOI] [PubMed] [Google Scholar]
- 29.Majka, J., and P. M. Burgers. 2004. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acids Res. Mol. Biol. 78:227-260. [DOI] [PubMed] [Google Scholar]
- 30.Marquez, L. A., and L. J. Reha-Krantz. 1996. Using 2-aminopurine fluorescence and mutational analysis to demonstrate an active role of bacteriophage T4 DNA polymerase in strand separation required for 3′→5′-exonuclease activity. J. Biol. Chem. 271:28903-28911. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, A., A. L. Johnston, L. H. Johnston, and A. Sugino. 1993. Pathway correcting DNA replication errors in S. cerevisiae. EMBO J. 12:1467-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, A., and A. Sugino. 1994. The 3′-5′ exonucleases of both DNA polymerase δ and ɛ participate in correcting errors of DNA replication in S. cerevisiae. Mol. Gen. Genet. 242:289-296. [DOI] [PubMed] [Google Scholar]
- 33.Pavlov, Y. I., S. Maki, H. Maki, and T. A. Kunkel. 26. May 2004, posting date. Evidence for interplay among yeast replicative DNA polymerases alpha, delta and epsilon from studies of exonuclease and polymerase active site mutations. BMC Biol. 2:11. [Online.] http://www.biomedcentral.com. [DOI] [PMC free article] [PubMed]
- 34.Reha-Krantz, L. J., L. A. Marquez, E. Elisseeva, R. P. Baker, L. B. Bloom, H. B. Dunford, and M. F. Goodman. 1998. The proofreading pathway of bacteriophage T4 DNA polymerase. J. Biol. Chem. 273:22969-22976. [DOI] [PubMed] [Google Scholar]
- 35.Schaaper, R. M. 1993. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 268:23762-23765. [PubMed] [Google Scholar]
- 36.Schofield, M. J., and P. Hsieh. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57:579-608. [DOI] [PubMed] [Google Scholar]
- 37.Shcherbakova, P. V., and T. A. Kunkel. 1999. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol. 19:3177-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevelev, I. V., and U. Hubscher. 2002. The 3′ 5′ exonucleases. Nat. Rev. Mol. Cell. Biol. 3:364-376. [DOI] [PubMed] [Google Scholar]
- 39.Soengas, M. S., J. A. Esteban, J. M. Lazaro, A. Bernad, M. A. Blasco, M. Salas, and L. Blanco. 1992. Site-directed mutagenesis at the Exo III motif of phi 29 DNA polymerase; overlapping structural domains for the 3′-5′ exonuclease and strand-displacement activities. EMBO J. 11:4227-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauss, B. S., D. Sagher, and S. Acharya. 1997. Role of proofreading and mismatch repair in maintaining the stability of nucleotide repeats in DNA. Nucleic Acids Res. 25:806-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szankasi, P., and G. R. Smith. 1995. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267:1166-1169. [DOI] [PubMed] [Google Scholar]
- 42.Tishkoff, D. X., A. L. Boerger, P. Bertrand, N. Filosi, G. M. Gaida, M. F. Kane, and R. D. Kolodner. 1997. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. USA 94:7487-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 44.Tran, H. T., N. P. Degtyareva, D. A. Gordenin, and M. A. Resnick. 1999. Genetic factors affecting the impact of DNA polymerase delta proofreading activity on mutation avoidance in yeast. Genetics 152:47-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran, H. T., D. A. Gordenin, and M. A. Resnick. 1999. The 3′→5′ exonucleases of DNA polymerase δ and ɛ and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2000-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran, H. T., J. D. Keen, M. Kricker, M. A. Resnick, and D. A. Gordenin. 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 17:2859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran, P. T., N. Erdeniz, S. Dudley, and R. M. Liskay. 2002. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amsterdam) 1:895-912. [DOI] [PubMed] [Google Scholar]
- 48.Tran, P. T., N. Erdeniz, L. S. Symington, and R. M. Liskay. 2004. EXO1—a multi-tasking eukaryotic nuclease. DNA Repair (Amsterdam) 3:1549-1559. [DOI] [PubMed]
- 49.Tran, P. T., J. A. Simon, and R. M. Liskay. 2001. Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:9760-9765. [DOI] [PMC free article] [PubMed] [Google Scholar]