Abstract
A strand-specific imprint (break) controls mating-type switching in fission yeast. By introducing a thiamine repressible promoter upstream of the mat1 locus, we can force transcription through the imprinted region, erasing the imprint and inhibiting further mating-type switching, in a reversible manner. Starting from a synchronized, virgin M-cell population, we show that the site- and strand-specific break is formed when DNA replication intermediates appear at mat1 during the first S phase. The formation of the break is concomitant with a replication fork pause and binding of the Swi1 protein at mat1 until early G2 and then rapidly disappears. Upon its formation, the break remains stable throughout the cell cycle and triggers mating-type switching during the second S phase. Finally, we have recreated the mating-type switching pedigree at the molecular and single-cell levels, allowing for the first time separation between the establishment of imprinting and its developmental fate.
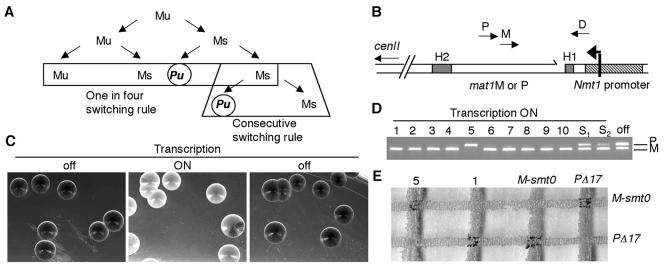
The fission yeast Schizosaccharomyces pombe possesses a remarkable system for changing its mating type (MT) (reviewed in references 4 and 13). Homothallic MT switching in fission yeast, similar to that in budding yeast, is a genetically programmed event in haploid cells initiated by a site-specific lesion at the mat1 locus. This lesion is then repaired by homologous donor sequences, mat2-P and mat3-M, located on the same chromosome. These loci are maintained in a silent chromatin state, preventing their transcription or recombination, but they can serve as donors of genetic information at mat1 by recombination (7, 15, 25). The MT switching pattern, as determined several years ago by pedigree analyses at the single-cell level, follows two strict rules, as depicted in Fig. 1A (17, 28, 34). The first, termed the one-in-four rule, indicates that two consecutive divisions are necessary to produce one switched cell among four granddaughter cells. The first division produces two cells with identical MTs, but with different switching potentials, termed unswitchable (u) and switchable (s), such that only the s cell produces one switched cell after the second division. The second, termed the consecutive rule, indicates that the sister of this switched cell is able to switch during subsequent cell divisions. The newly switched cell is unswitchable, and will follow the same fate as the unswitchable (Mu or Pu) cells (where M stands for minus and P stands for plus). Therefore, an exponential culture- or well-grown colony contains not two but four cell types, with P or M MT information, and with two switching potentials, u and s, found in equal proportions (1 Pu:1 Mu:1 Ps:1 Ms). Cells from opposite MTs are able to conjugate in G1 and sporulate when grown under sporulating conditions. The integrity of this pedigree and switching is conveniently verified by staining cells within a colony with iodine vapors, a marker of spore-containing colonies, such that wild-type homothallic cells stain darkly, whereas mutant cells exhibit a streaky or white phenotype (31).
FIG. 1.
The inducible MT switching system. (A) Pedigree of MT switching in S. pombe. M and P represent the two MTs, and the suffixes “u” and “s” designate switchable and unswitchable cells. The one-in-four and consecutive switching rules are indicated. (B) Schematic of the mat1 locus located on chromosome II. The mat1 locus is bordered by the H1 and H2 homology boxes, shown as grey boxes. The nmt1 promoter was inserted just distal to the mat1 locus, with transcription of the top strand extending towards mat1. The nmt1 transcriptional start site begins 265 bp upstream from the imprint or break (shown as a broken line distal to mat1). The location of primers D (mat1 distal), M (matM), and P (matP) used for detecting MT alleles are indicated. (C) Transcription at mat1 represses MT switching in a reversible manner. Colonies of homothallic cells containing the unexpressed (off panels) or expressed (on; middle panel) nmt1 transcript at mat1 were grown in MM and then stained by iodine vapors to detect spores and proficient MT switching. (D) Identification of pure mat1-M- or mat1-P-containing colonies. Individual white colonies were selected for multiplex colony PCR analysis and shown to contain either P or M information at mat1, using primer D (mat1-distal specific), M (matM), or P (matP). S1 and S2 indicate streaky colonies, and the off label represents a colony chosen from the transcriptionally repressed state. (E) Mating test. Two of the white colonies tested by PCR (5P and 1M) were further tested for mating ability on MM lacking thiamine. The black intersection at the mating junctions show that these virgin strains can conjugate and subsequently sporulate uniquely with tester strains of the opposite MT.
The change of MT can be measured molecularly, as well as genetically. Early attempts at identifying the intermediates of MT switching, using classic DNA extraction techniques, concluded that MT switching was initiated by a double-strand break (DSB) (7, 36), analogous to that of budding yeast (42). Using a gentle, nonshearing DNA isolation approach, it was recently shown that S. pombe possesses a site-specific but sequence-nonspecific single-stranded break (SSB) (1) or modified nick (23) and not an unprocessed Okazaki primer RNA (11) or RNase-sensitive modification (47). The strand-specific nick remains stable until the next round of replication, when it is transformed into a double-strand blunt-ended intermediate, used to initiate gene conversion (24).
Important insights into the regulation of imprint formation came through the discovery that the orientation of the mat1 locus, with respect to a centromere-distal origin of replication, is essential for proper establishment of the imprint (11). The replication polarity is established by two cis-acting elements flanking the mat1 locus, the mat1-distal replication pause site 1 (MPS1) and the mat1-proximal replication termination site 1 (RTS1) (12). The subsequent identification and disruption of the RTS1 and MPS1 elements further supported the idea that only replication coming from the mat1-distal side could lead to normal imprint formation. Polar replication pausing and termination at mat1 depends on swi1 and swi3 gene products, whereas a mutant of swi7, encoding for polymerase α, has no effect on pausing activities but reduces break formation in an unknown manner (12, 40). The authors of previous studies proposed a model in which only the sister chromatid containing the newly synthesized lagging strand is switchable and will form the break. swi1 and swi3 show similar genetic interactions and thus may work together as a complex, but so far there is no identified homolog of swi3. However, Swi1 possesses 25% sequence homology with Tof1 in budding yeast and with Timeless in flies (12). Recent studies of fission and budding yeasts have shown that Swi1 and Tof1 appear to monitor DNA damage in S phase through association with the replisome and by forming complexes with checkpoint factors at stalled replication forks (18, 22, 37).
Novel cis- and trans-acting elements, necessary for imprinting or SSB formation, have been identified, both distal to mat1 and within the H1 homology box itself. The mat1-distal elements, switch-activating sites 1 and 2 (SAS1 and SAS2), as well as the essential SAS1 binding protein Sap1, necessary for chromosome stability, are essential for normal SSB formation (3, 14). Finally, recent scanning mutagenesis studies within the H1 homology box have revealed two important elements reducing SSB formation. One site defined a minimal cis-acting region necessary for MPS1 activity and Swi1 binding to this site, in vivo. Two other sites mapped a region necessary for maintaining SSB levels throughout the cell cycle and for the first time reveal cis-acting elements, which uncouple imprint formation and maintenance, at mat1 (24).
Thus, the MT-switching system in S. pombe provides an excellent model system for studying the relationships among imprint formation, replication, pausing, repair, and recombination at one defined locus. However, as the genetic pedigree shows, a homothallic culture always possesses four different cell types, with P or M information, which are switchable (with a break) or unswitchable (no break). Therefore, we designed an inducible system in S. pombe in which a homogeneous virgin cell population experienced a SSB at mat1 simultaneously. To control imprint or SSB formation, we introduced a promoter (nmt1) upstream from the mat1 locus, forcing RNA polymerase II-dependent transcription of the broken DNA strand. When the promoter is turned on, the SSB disappears. By maintaining transcription, we have isolated and stabilized a homogeneous population of unbroken Mu (or Pu) cells. Subsequently, when the promoter is turned off, we are able to monitor the kinetics of each step required for MT switching in a sequential manner. The ability to control this gene conversion process offers a way to study the first asymmetric division separately from the second or, in other words, the establishment of a site-specific imprint from its subsequent activities.
MATERIALS AND METHODS
Strains.
All strains were constructed starting from SP62 (h90 leu1-32 ura4-Δ18). SP62 was transformed with a cassette containing the strong nmt1 promoter and the kanamycin gene for selection (6), along with homology to the mat1-distal region for one-step homologous insertion (39), producing strain AHR3. A C-terminal hemagglutinin (HA) tag of swi1 within the chromosome was performed with the same PCR-based gene targeting system (6). A purified swi1-HA PCR product was transformed into strain AHR3, producing AHR5. AHR3 and AHR5 were crossed with a cdc25 strain, creating AHR6 and AHR7. The following stable heterothallic strains were used for MT testing: SP807 (mat1-M smt-0 leu1-32 ade6-210) and a P-stable derivative of SP62 (mat1-PΔ17::LEU2 leu1-32 ura4-Δ18). An ade6-210 derivative of AHR3 (AHR8) was made and crossed with a stable ade6-216 derivative of SP807, creating the diploid strain AHR9, used for diploid pedigree analysis (mat1-nmt1::KAN/mat1-M smt-0 ade6-210/ade6-216 leu1-32/leu1-32).
General techniques.
Mating assays were performed as previously described (35). Synchronization of cells in late G2 was performed by shifting cultures containing the cdc25 temperature-sensitive allele to the nonpermissive temperature for 3 h. DNA was isolated from cells grown in Edinburgh minimal medium (MM) with or without thiamine by a classical method (35), digested with PvuII and XhoI, and analyzed by Southern blotting. A probe revealing the cut DNA at mat1 was made using a mat1-distal-specific fragment. To analyze SSB formation, DNA was prepared and digested with restriction enzymes (SspI and HindIII or NdeI and HindIII) in agarose plugs, as described previously (1). DNA was purified from agarose by gel extraction and resolved on an 8% denaturing polyacrylamide gel electrophoresis (PAGE) gel. DNA was electroblotted and revealed with a single-stranded probe specific for the mat1-distal upper strand, prepared by primer extension, and purified under denaturing PAGE conditions. Total mRNA was extracted by the glass bead phenol method and analyzed by Northern analysis with formaldehyde agarose gels. Samples were normalized to total RNA and estimated with ethidium bromide gels. A double-stranded radiolabeled probe was used to simultaneously reveal the mat1-Mc constitutive transcript and the inducible nmt1 ectopic transcript at mat1.
Two-dimensional gels.
Two-dimensional (2D) gel analysis of replication intermediates was carried out as described previously (9, 24). Strains were grown in MM with or without thiamine. DNA was prepared and digested with SspI and HindIII in agarose plugs (1). Enriched fractions for replication intermediates were obtained using benzoylated naphthoylated DEAE (BND) cellulose columns. Gels were hybridized with a mat1-distal-specific fragment.
ChrIP analysis.
Exponential cultures were grown in MM at 33°C, and extracts were prepared as previously described (24). Whole-cell extraction, chromatin immunoprecipitation (ChrIP), and DNA extraction of the immunoprecipitated material were performed as previously described (33). The Anti-HA Affinity Matrix from Roche was used for the immunoprecipitation reactions. Samples were assayed by agarose gel electrophoresis. ChrIP time course experiments were repeated twice. Primer pair sequences in the mat1 region used for ChrIP are available upon request.
Pedigree analysis.
Diploid pedigree analysis was performed as previously described (3, 28). Diploid cells from strain AHR9 were selected for the mat1::nmt1 “on” condition by being plated on MM without thiamine and were verified to contain the mat1-M::nmt1/mat1-M smt-0 alleles by PCR. Diploid cells in the “off” state were grown and maintained on MM in the presence of thiamine to turn off transcription of the nmt1 promoter and contained either M or P information at the mat1::nmt1 allele. For the on-to-off condition, virgin M cells were isolated (mat1-M::nmt1/mat1-M smt-0) on MM plates without thiamine (nmt1 on), and transferred to MM plates containing thiamine (nmt1 off) at the start of the cell pedigree manipulation.
RESULTS
Establishment of an inducible system for MT switching.
We introduced a thiamine-repressible promoter in neutral and close proximity to mat1 (200 bp away) (3, 27), such that the upper strand containing the imprint or SSB was transcribed (Fig. 1B). We reasoned that upon induction of the nmt1 promoter, the RNA polymerase would induce a repair event, perhaps in a manner analogous to transcription-coupled repair, a well-conserved excision repair process for repairing UV and oxidative damages (30; for reviews, see references 20 and 43). It was previously shown that the nmt1 promoter is repressed quickly, in less than 30 min, but induction requires three to four cell divisions to sufficiently dilute internal cellular thiamine levels (32). To determine whether transcription affected MT switching, an exponential culture of cells containing the ectopic promoter at mat1 were grown in MM containing thiamine and subsequently plated on MM plates with thiamine (Fig. 1C, left panel, “off”). Cells from the same culture were washed, resuspended, and diluted in MM lacking thiamine. After 16 h, corresponding to at least three cell divisions, the cells were plated on MM lacking thiamine (Fig. 1C, center panel, “on”). Thiamine was then added to the same culture; after 1 h, cells were plated on MM plates containing thiamine (Fig. 1C, right panel, “off”). At least 500 colonies were scored in each condition for MT efficiency by iodine staining. When transcription was turned off, all cells were darkly staining, indicating proficient MT switching. However, when transcription was turned on (in the absence of thiamine), the cells appeared either uniformly white (65%) or slightly streaky (35%), indicating a defect in MT switching. Importantly, this effect was reversible, since turning off transcription rescued the wild-type MT switching phenotype in all plated cells.
We next determined the MT status of the plated colonies by PCR analysis (Fig. 1D). Ten uniformly white colonies, as well as two streaky colonies (S1 and S2) from the transcriptionally proficient condition (on), were analyzed along with colonies from the transcriptionally repressed (off) condition. The colonies from the off state possessed both P and M information at mat1. However, the white colonies contained either M or P sequences. Interestingly, isolation of M-containing colonies was much more frequent than of P-containing colonies, occuring 96% of the time (data not shown). The streaky colonies contained a mixture of M and P in various amounts, depending on the level of streakiness. Because sterile cells form colonies that also stain white, we further analyzed the stable M and P cells for mating ability with stable mating tester strains (Fig. 1E). A P virgin colony (5) could only mate with a stable M strain, whereas an M virgin colony (1) was able to mate uniquely with a P stable strain (Fig. 1E). Altogether, these data show that transcription at mat1 produces transcriptionally competent stable P or M cells. This effect is reversible and cells are not sterile. Therefore, nmt1 transcription does not appear to disturb the mat1 transcripts governing mating and meiosis or to cause gross chromosomal rearrangements or mutations at mat1.
Loss and appearance of the break upon transcription.
We next decided to analyze the consequences of transcription through the imprinted site at mat1. Asynchronous cells were harvested at various intervals in the absence of thiamine, and genomic DNA and RNA were isolated to monitor nmt1 transcript and break levels as a function of time. DNA prepared by this classical method sheared the fragile site at mat1, creating DSBs at mat1 (1, 7). Northern analysis revealed the appearance of the nmt1 transcript (Fig. 2B, left panel, 11.5 h) just 30 min before the disappearance of the mat1-distal cut fragment (Fig. 2C, left panel, 12 h). Therefore, relative to the induction of transcription, the disappearance of the break was rapid, occurring within a fraction of the cell cycle. The probe used for Northern analysis also revealed the constitutive mat1-Mc endogenous transcript, which was unaffected by introduction and expression of the ectopic nmt1 transcript (Fig. 2A and B; Materials and Methods). The length of the mat1::nmt1 RNA was about 400 nucleotides, indicating that the RNA polymerase runs through the imprinted region, passes the repaired break site, and terminates roughly at the mat1-Mc termination site (Fig. 2A and B).
FIG. 2.
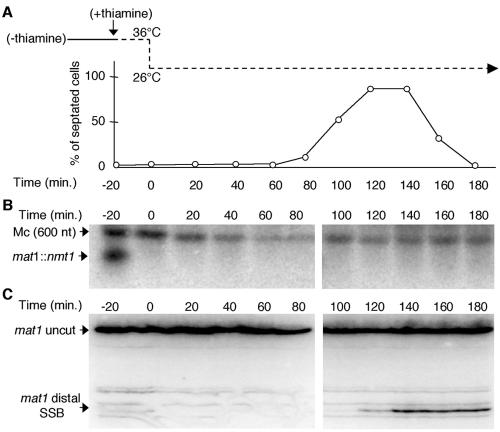
Kinetics of break repair and/or formation and nmt1 expression. (A) Schematic of the mat1 locus indicating probes used for Southern and Northern analyses. The polarity and size of the mat1-Mc and mat1::nmt1 transcripts are shown, and the location of the XhoI and PvuII restriction sites are indicated. (B) Northern analysis of the nmt1 transcript from cells grown in the absence of thiamine over time (off/ON), or in the presence of thiamine (ON/off). (C) Southern analysis of mat1 cut levels from the same time intervals. Induction of the nmt1 transcript at mat1 occurs at about 11.5 h, with concomitant loss of the break. Thus, transcription induces rapid loss of the nick at mat1. In contrast, the break reappears slowly relative to nmt1 repression, requiring almost 2 h to reach steady-state levels. (D) DNA and mRNA levels were quantified using a Molecular Dynamics phosphorimager, and intensities were plotted.
To determine the kinetics of reappearance of the break, when the nmt1 transcript is turned off, we added thiamine to the same culture after the 16-h time point and monitored the kinetics of break formation and transcription (Fig. 2B and C, right panels). In contrast to break loss, reappearance of the break reached steady-state levels after 2 h, which was relatively slow compared to the repression of nmt1, occurring within 30 min (Fig. 2D).
The SSB is formed in S phase.
Numerous studies have suggested that the de novo formation of the break occurs during S phase. To establish timing of SSB formation, we constructed a strain containing the nmt1 promoter at mat1 and the temperature-sensitive cdc25 allele, which at the nonpermissive temperature causes cell cycle arrest in late G2. Beginning with a homogeneous culture of Mu virgin cells synchronized in G2, thiamine was added to the culture to stop transcription 20 min before synchronous release into the cell cycle. Cells were harvested at 20-min intervals, and cell morphology and septum formation were monitored (Fig. 3A). Total RNA was prepared and analyzed by Northern blotting (Fig. 3B). Genomic DNA, prepared and digested from the same cell fractions in agarose plugs, was analyzed on denaturing PAGE gels and probed with a single-stranded DNA probe, revealing only the upper strand with or without the imprint (Fig. 3C). Under these conditions, the strand-specific imprint migrated as a single-strand nicked molecule (23). We observed abrupt appearance of the SSB concomitant with the peak of septation (a marker of S phase), which remains stable after S phase (Fig. 3C). Figure 3B confirms that repression of nmt1 transcription occurred rapidly, within 20 min. Turning off transcription after mitosis (40 min) or just before S phase (80 min) did not affect the timing of SSB formation (data not shown). In addition, treatment of Mu virgin cells with hydroxyurea, which prevents progression into S phase, prevented formation of the break (data not shown). Therefore, unbroken Mu cells experienced an SSB upon the first round of DNA replication, producing switchable Ms cells (Fig. 1A), and this break was subsequently maintained.
FIG. 3.
The imprint or SSB is formed during the first S phase. (A) Synchronization of cells in late G2 and septum formation. Thiamine was added to G2-synchronized, virgin Mu cells, which were then released synchronously into the cell cycle (36 to 26°C). RNA and DNA were prepared from cells harvested at regular intervals. The peak of septation (120 to 140 min) indicates S phase. (B) Northern analysis (as described in the legend to Fig. 2) to confirm thiamine addition and rapid repression of the nmt1 transcript at mat1. (C) Denaturing PAGE gel of SSB formation. DNA was prepared in agarose plugs to preserve the integrity of the lesion. The single-stranded nick appears only upon the peak of septation (120 to 140 min).
The MPS1 replication pause and SSB were formed simultaneously.
We next wanted to determine when the imprint or SSB was made in relation to the replication fork pause site at mat1, called MPS1 (11). Because pausing was still observed in a deletion mutant strain that contains no SSB (mat1-distal sequences necessary for imprint formation are deleted), an imprinting pathway was proposed in which the polar replication pause occurs before imprint or SSB formation and perhaps sets up functions necessary to establish the imprint (12).
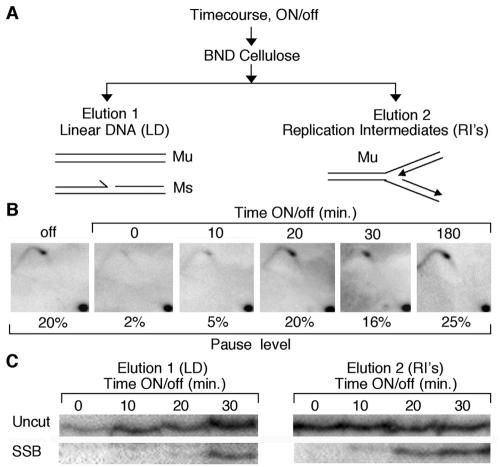
To establish the timing of SSB formation and pausing from an exponential culture of stable Mu cells, we repressed transcription and harvested cells at regular intervals. DNA was prepared, and a fraction from each point was loaded onto a BND cellulose column (24) to separate double-stranded DNA from partially single-stranded DNA molecules, most likely containing replication intermediates (Fig. 4A). Figure 4B shows the MPSI pause levels by 2D gel analysis. Just before transcription is turned off at mat1 (the 0-min point), we observed the absence of the pause and SSB. Thus, transcription at mat1 was able to destabilize the MPS1 pause site. However, within 10 min after repression of transcription, the pause (and the nick, discussed below) was observed, reaching steady-state levels within 20 min, concomitant with the timing of repression of the nmt1 promoter (Fig. 3B).
FIG. 4.
The SSB and MPS1 occur simultaneously. (A) Schematic showing how linear DNA and replication intermediates are enriched from purified time course DNA. (B) A thiamine time course was performed from isolated virgin Mu cells, and DNA harvested from cells was analyzed by 2D gel and PAGE (C, right panel), to monitor the kinetics of the MPS1 pause and imprint or SSB formation. Upon addition of thiamine (nmt1 repression), the pause structure and break are formed at the same time, within 10 min. The percentage of pausing is indicated (C). Denaturing PAGE gel analysis of the SSB shows that it is part of a replicating structure. Linear DNA (LD) and replication intermediates (RI’s) from the same time course DNA used in the results shown in panel B were collected and analyzed by denaturing PAGE. The SSB is first seen in the replication intermediate-containing fraction (elution 2).
In an attempt to better characterize the DNA containing the SSB, we asked whether the DNA molecules with a break are also found with the replicating fork or only after the replication fork has passed. Figure 4C shows the comparison between the two eluted fractions of DNA from the BND column, run on a denaturing gel (results from elution 1 are shown in Fig. 4C, left panel; results from elution 2 are shown on the right panel). The break was found in elution 2, enriched for replication intermediates 10 min before the break from elution 1, enriched for linear DNA, and also containing 10 times more DNA than elution 2. Therefore, the imprint or SSB appears during replication of the mat1 locus and is maintained after the fork passes.
Swi1 binding at MPS1 accumulates in late S phase.
In a previous study, we observed a 10-fold enrichment of Swi1 at the MPS1 site in asynchronized cells by using ChrIP (24). We decided to measure the binding of the Swi1 protein using our inducible system with a synchronized cell population. After isolation and synchronization of Mu cells in G2, we added thiamine to repress transcription and collected cell fractions for Swi1 binding by ChrIP, SSB formation by PAGE, and MT switching by PCR analyses (Fig. 5). A fraction of cells from each time interval were also analyzed for the percent of septation, an indicator of S phase (Fig. 5C), as well as for cell morphology and fluorescence-activated cell sorter analysis, to follow the status of the cells within the cell cycle (data not shown). The schematic in Fig. 5A indicates the location of the primer pairs used for PCR analysis of the DNA from ChrIP. No significant binding of Swi1 was observed at the pause site between G2/M (0 min) but was significantly enriched in S phase, reaching a maximum accumulation in late S/G2 (160 min) to 30 fold. During the subsequent G2, binding levels go down to 2 fold but rise again in late S/G2 to 20 fold. Synchronized cultures in which thiamine was never added did not show enrichment of Swi1 binding upon release, S/G2, or G2; thus, transcription at mat1 prevented accumulation of Swi1 binding in S phase (data not shown).
FIG. 5.
Swi1 binds the MPS1 pause site during S phase. Mu cells containing an HA-tagged Swi1 construct were synchronized in late G2, and transcription was repressed upon release into the cell cycle. (A) Schematic of the mat1 locus and ChrIP analysis. Primers (a, b, and c) used for PCR analysis of ChrIP DNA fractions are shown, as well as the location of the MPS1 and RTS1 pause and termination sites. (B) ChrIP analysis. Enrichment of Swi1 at MPS1 peaks in late S1 (160-min) and S2 (280-min) phases. (C) Septation index. The percent of septation is plotted over time. The two peaks represent the two consecutive S phases. (D) Denaturing PAGE gel analysis of break formation. The break coincides with the first peak of septation (S phase) and is maintained. (E) PCR analysis of MT switching intermediates from the total (input) DNA. Recombination occurs only in S2, whereas Swi1 binding is observed in both S phases.
Uncoupling of SSB formation and MT switching.
The kinetics of SSB formation and MT switching were determined from the same DNA fractions used for ChrIP (Fig. 5B) by PAGE and PCR analyses, respectively. Specific primer pairs amplifying mat1-M or mat1-P were used as indicated in Fig. 1B. Although the SSB was formed during the first replication period (Fig. 5D), only M information was still found at mat1, meaning that the cells had not yet switched their MT (Fig. 5E). It is only during the second S phase that we could see appearance of the P allele at mat1, which represents about one-fourth of the population, as consistent with the pedigree (one-in-four switching rule) (Fig. 1A). Thus, we observed Swi1 binding in cells that have not undergone MT switching, supporting the idea that Swi1 activity is necessary for pause and SSB formation, independent of recombination functions at mat1. Therefore, this inducible system was able to effectively separate imprint formation steps from recombination at mat1.
Cellular pedigree analysis of MT switching using the inducible system.
A critical prediction from our molecular observations and a requirement for virginity is that MT switching should only occur after two consecutive divisions. The pattern of MT switching can be determined by pedigree analysis at the single-cell level. We performed this analysis with a diploid strain, in which one homolog contained the nonswitchable mat1-M smt-0 deletion, while the other contained the ectopic nmt1 inducible promoter at mat1 (Materials and Methods) (3, 28). Under low-nitrogen growth conditions, diploid mat1-M::nmt1/mat1-M smt-0 cells were unable to form asci and continued growing. Once the mat1-M allele switches its MT to mat1-P, then cells will stop growing and initiate sporulation.
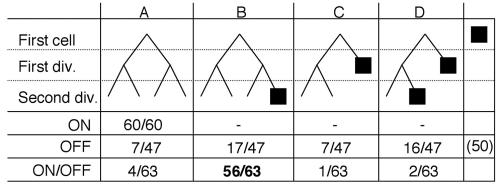
We followed the growth patterns (A, B, C, and D) of this diploid strain during at least three cell divisions, under the following three conditions: in the absence of switching (transcription on), during MT switching (transcription off), and during MT switching induction (transcription on to off) (Fig. 6). We first tested switching efficiencies of a stable virgin Mu/M smt-0 cell, when the promoter was turned on and kept on. The pedigrees of the purified virgin diploids analyzed in this situation, showed that the cells continued to divide and never formed asci (60 of 60 cells; A pattern), indicating the stable virgin state of the cell.
FIG. 6.
Cellular MT pedigree analysis. Diploid cells from the strain containing mat1::nmt1/mat1-M smt-0 were separated by micromanipulation during at least three divisions (two divisions are shown, for simplicity) and observed microscopically for growth and spore formation on solid sporulation medium in the following three different growing conditions: transcription on, off, and on/off. The four possible pedigrees are shown (A, B, C, and D), in which B and D represent the one-in-four and consecutive switching rules, respectively. The filled-in squares represent cells that formed an ascus, indicating that mat1::nmt1 contained the P allele. Virgin M-containing diploid cells (mat1-M::nmt1/mat1-M smt-0) maintained in the on condition were unswitchable. The experimental switching efficiency, in which virgin M diploid cells were then placed in the off condition for micromanipulation, is in boldface type (56 of 63 cells) and indicates that 89% of the virgin mat1-M-containing chromatids are unswitchable and that no more than two divisions are required to switch to mat1P (Miyata's one-in-four switching rule).
Without transcription (off control state), about 50% of the cells never divided and produced asci (50 of 97 cells), indicating that one-half of the total number of diploid cells analyzed contained the Pu or Ps allele. The remaining 47 cells were followed; after the first division, the cells produced one ascus 50% of the time (23 of 47 cells; C and D patterns) in a pair of dividing sister cells. This indicates that one-half of these dividing cells contained the Ms allele and efficiently switched MT after one division. Among the other half that did not sporulate after the first division (24 of 47 cells; A and B patterns), 17 of these (71%) produced one ascus among four related cousins after the second division (B pattern). Altogether, this indicates that when the nmt1 promoter was repressed, cells switched their MT efficiently, following the switching rules.
We then followed the mating efficiency, starting with Mu/M smt-0 virgin diploids, but repressed transcription at the start of pedigree analysis (on to off condition). No cells produced asci before the first division. Only 3 of 63 cells (C and D patterns) produced one ascus among sister cells after the first division. After the second division, we observed that 89% of cells contained one ascus among four related cousins (B pattern). This indicated that transcription removes the imprint at the mat1 locus, establishing a virgin cell population, but when transcription is repressed, efficient MT switching is allowed to occur only at the second division, confirming the one-in-four rule. Thus, separation of the imprinting and recombination steps can also be studied at the individual cell level.
DISCUSSION
The ability to begin with a synchronous population of cells that have suffered a DNA damage, such as a DSB, by introduction of the inducible HO or I-SceI endonucleases with their recognition sites has greatly advanced our understanding of DSB repair pathways, de novo telomere formation, adaptation and recovery responses to DSBs, and acquisition of new chromosome ends in yeast and mammalian systems (reference 44 and references therein). By controlling the initiation of the DSB at MAT in Saccharomyces cerevisiae, the kinetics of DSB repair and the replication and recombination factors important for MAT switching were analyzed (reviewed in reference 19). For the first time with S. pombe, it is possible to regulate the break at mat1 and produce a homogeneous population of cells, allowing monitoring of natural break formation, pausing, DSB formation, and MT switching in this organism.
Virgin Mu cells.
Previous studies inferred when and how the imprint is formed in a heterogeneous cell population. We describe the spontaneous resetting of the imprint in all cells. The relatively fast (within 20 min) and simple (addition of thiamine) repression of the nmt1 promoter allowed us to follow the kinetics of each molecular event, from imprinting formation to MT switching, in a synchronized and homogeneous cell population.
Considering how transcription initiating at the nmt1 promoter might repair the SSB at mat1, we first reasoned that the RNA polymerase II machinery would run into the SSB, inducing a direct repair event. This idea is supported by the fast disappearance of the break upon induction of transcription (Fig. 2). Transcription-induced repair of the break at mat1 also partially depends on Rad13, the homolog of the nucleotide excision repair factor, Rad2, and XPG in S. cerevisiae and mammals, a nuclease necessary for the incision of UV and oxidative damages during transcription-coupled repair (A. Holmes, unpublished results). However, transcription through the cis-acting elements essential for MT switching, which are located between the promoter and SSB, could disrupt the chromatin structure and/or binding of certain factors required for establishing and/or maintaining the imprint, thus allowing the isolation of stable virgin cell populations.
At the time transcription is turned on, the mat1 locus contains P or M information in equal proportions. Therefore, we first assumed that transcription would capture and freeze the existing allele at mat1, giving rise to similar proportions of Pu or Mu virgin cells. It was intriguing to us, however, that we were able to isolate Mu cells much more frequently than Pu cells. This bias is not due to the death of P cells, since viability and growth were not perturbed during the initial waves of transcription (data not shown). A similar bias (M > P) has been observed from studies focused on the mechanisms of silencing at the donor loci, mat2-P and mat3-M, and on donor preference (16, 46).
Our work raises the possibility that transcription within the mat1-distal region alters the proposed intrachromosomal folding, bridging the silent loci with the active mat1 locus, revealing a default folding pattern between mat1-P and mat3-M. This indicates that cis-acting elements within the mat1-distal sequence (200 bp) are actively involved in donor choice processes to counteract this default pathway. Our favorite candidate for this is the Sap1 protein, shown to be required for chromosomal organization (14). In S. cerevisiae, donor preference is controlled, in part, by a recombination enhancer element unlinked to the MAT locus (reviewed in reference 19). Alternatively, the bias producing more Mu cells could be due to mat1-M repair using the complementary strand rather than the mat2-P donor, and mat1-P repair using the mat3-M donor, rather than the complementary strand.
Although transcription efficiently and stably removes the SSB at mat1, some colonies retain a lightly sectored, streaky switching phenotype (Fig. 1C, middle panel). These streaks reveal a boundary between M and P cells, formed early within the growing colony. Importantly, S. pombe cells spend 70% of their cell cycle in G2, which means that two different sister chromatid-containing mat1 loci normally coexist (Mu and Ms, Ms and Pu, Pu and Ps, and Ps and Mu). Thus, transcription occurring in MM- or PP-containing cells should give rise to homogeneously white colonies, whereas heterogeneous cells containing mat1 sister loci MP or PM should yield streaky colonies. The donor preference bias we observe towards M upon transcription could create streaky sectors during the growth of the colony, and this is consistent with the observed data (Fig. 1C, middle panel).
Kinetics of SSB formation and MPS1.
An important finding from this work is that transcription destabilizes the MPS1 site (Fig. 4B). It is possible that the ectopic nmt1 promoter continues to fire after it is replicated and destabilizes the replication fork stalled there, located only 200 to 300 bp downstream of the start of transcription. Interestingly, this is the first example in eukaryotes in which transcription has been shown to disrupt a stalled replication structure in vivo. This may be similar to studies of Escherichia coli and Bacillus subtilis, in which transcription was also shown to disrupt termination of replication at Ter sites and to disrupt the activities of DnaB and PriA helicases (10), and of yeast, in which transcription disrupted CEN and ARS activities in vivo (38, 41). One possible function of replication fork barrier sites, cited often in the literature, might be to avoid the collision between transcription and replication machineries, as was recently demonstrated (8, 45). This should not normally occur at the MPS1 pause site, since the mat1 transcripts (Mc and Pi) are terminated before the SSB and pause site, and during ectopic nmt1 expression, transcription and replication progress in the same direction (Fig. 2A).
Previous attempts to directly determine the timing of SSB formation were impossible, due to the omnipresence of SSB-containing cells at mat1. However, previous work inferred that the SSB formed during S phase (4). Starting with a clonal population of cells with a single MT and no imprint or SSB at mat1, we show for the first time that the SSB is made during S phase. Furthermore, the replication pause and SSB are formed simultaneously on the replication intermediate, and the SSB is maintained or protected after the fork passes mat1. These results, together with the strand specificity of SSB formation and the polarity of replication, strongly suggest that the imprint is made on the nascent lagging-strand DNA. The appearance of the SSB on the replicating intermediate molecules 10 min before its appearance on the linear DNA indicates that the half-life of the MPS1 pause is within 10 min (Fig. 4B and C), about one-half of the entire S phase.
Swi1 binding at mat1 is enriched in replicating Mu cells.
Enrichment of Swi1 and Tof1 with chromatin indicates that these factors form a complex with elongating and stalled replication forks in fission and budding yeast, respectively (22, 37). At the G2/M transition when transcription is turned off and the cells are released into the cell cycle, the quantity of Swi1 associated with mat1 is close to background levels. The initial accumulation of Swi1 coincides with what appears to be early mat1 replication (26), the accumulation of replication forks at MPS1 (11), and the appearance of the SSB (Fig. 4). The overall kinetics of Swi1 accumulation in this study (during 60 min) extend beyond the estimated half-life of the MPS1 pause, suggesting that the role of Swi1 at MPS1 is not restricted to pause formation and may also participate in the initial protection and/or maintenance of the break (Fig. 7). One could imagine that the imprint is a bound protein dislodged by transcription, marking the DNA prior to replication, pausing, and cleavage at mat1. However, repression of transcription as late as G1 still allows wild-type levels of SSB formation during the following S phase (data not shown). Similarly, in asynchronous growth, the MPS1 pause and SSB are formed within 10 min of transcriptional repression (Fig. 4). Altogether, this indicates that the de novo imprint is made in G1, 10 min before replication of mat1. Alternatively, the DNA sequence information of the imprint might be detected by the moving replication machinery and transformed into a pause and SSB in a Swi1-dependent manner.
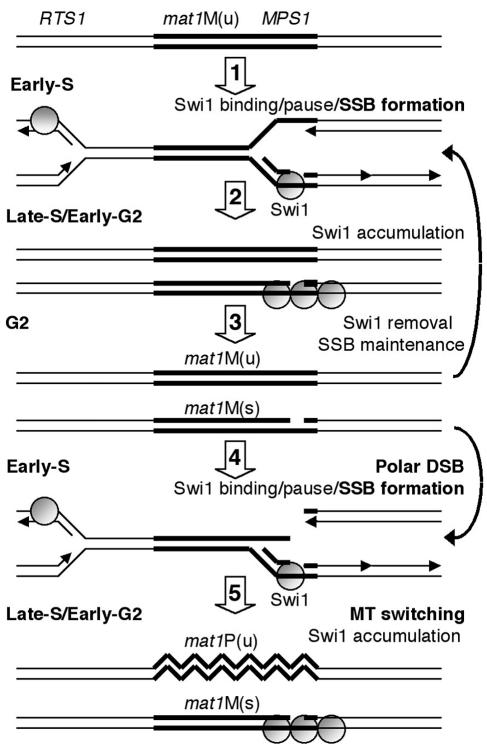
FIG. 7.
Working model for imprinting and MT switching. (Step 1) During DNA replication, two forks approaching the virgin mat1 [mat1-M (u)] locus from opposing directions are blocked in a Swi1-dependent manner at RTS1 and MPS1. (Step 2) Simultaneously, the imprint or SSB appears on the stalled, nascent lagging strand at MPS1, stimulating accumulation of Swi1 until early G2. For simplicity, Swi1 is depicted as associating with the lagging strand template and because swi1Δ is synthetically lethal with a swi7 mutation, encoding for the lagging-strand DNA polymerase. (Step 3) During G2, two different sister chromatids are produced, mat1-M (u) and mat1-M (s), without and with the SSB at mat1, respectively. The removal of Swi1 will allow the full process to restart. (Step 4) Due to the stability of the SSB at the mat1-M(s) allele until the subsequent S phase, the leading-strand polymerase will form a blunt-ended recombination intermediate, triggering MT switching. (Step 5) During G2, one sister has repaired the DSB and switched to mat1-P (u), whereas the other unswitched sister contains a newly formed SSB.
In our system, we cannot rule out the possibility that Swi1 binding is also important during gene conversion (Fig. 7). If Swi1 is able to form different complexes, one as a normal component of the elongating replication fork and another as part of a pausing checkpoint complex, what kind of pause complex might be formed at mat1, upon break formation or its subsequent conversion to a DSB after one generation? Recent work indicates the formation of a chicken foot structure at mat1, in the absence of the donors (47). Swi1 interacts with Cds1 (Rad53 in yeast and Chk2 in humans) and Rad3 (Mec1 in yeast and ATM/ATR in humans), but Cds1 and Rad3 appear not to be necessary for MT switching, as measured by iodine staining (37). Perhaps these factors do play a subtle role not revealed in their assay (37), and/or other unidentified checkpoint mediators are necessary at mat1, which interact with Swi1.
S. pombe MT switching is a two-step developmental process.
The overall process of MT switching in S. pombe is a two-step process, involving two rounds of DNA replication, in which the first generates a mark or imprint on one sister chromatid, leaving behind a stable nick on the DNA. This imprinting event determines the fate of future progeny one generation in advance. During the second round of replication, MT switching is triggered when the fork encounters the SSB, creating a polar, double-stranded end, and a new SSB is made on the other strand. As recently proposed, lagging-strand reinitiation (47) may allow formation of this newly formed imprint or SSB (Fig. 7). The SSB is formed during lagging-strand synthesis and/or processing (Fig. 7, steps 2 and 4), and gene conversion is initiated by the leading-strand machinery (step 5). The H1 box is also present at the donor loci, and the relatively short homology (59 bp or less) allows invasion of one donor locus, inducing MT switching (5). This type of recombination does not lead to crossovers and proceeds by a conservative mode of replication in which both mat1 DNA strands are neosynthesized during one round of replication and 25% of the mat1-containing molecules are labeled (2). The PCR and pedigree experiments demonstrate that switching from M to P occurs after only two generations (one-in-four rule) and that the newly switched allele is functional soon after gene conversion (Fig. 5 and 6). Such features are reminiscent of break-induced replication, a process involved in DSB repair and telomere-length maintenance, in the absence of telomerase (21, 29).
The connection between replication and recombination pathways in eukaryotic systems is well established with many players already identified, although their respective molecular roles in recombination are still unclear. The inducible MT switching system in S. pombe provides two unique advantages. The first is its dependence on DNA replication, and the second is its site-specificity, which provides a basis for further molecular and cellular studies of processes required for genomic stability.
Acknowledgments
We thank Guy-Franck Richard for critical reading of the manuscript and Michele Gilbert for technical assistance.
This work was funded, in part, by the Association pour la Recherche sur le Cancer (ARC) and the Human Frontiers Science Program. A.M.H. was supported by the EMBO long-term and Fondation pour la Recherche Medicale fellowships, and A.K. was supported by the Ministère de l'Education National de la Recherche et de la Technologie (MENRT) and Ligue contre le Cancer and Pasteur-Weismann fellowships.
REFERENCES
- 1.Arcangioli, B. 1998. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 17:4503-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcangioli, B. 2000. Fate of mat1 DNA strands during mating-type switching in fission yeast. EMBO Rep. 1:145-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcangioli, B., and A. J. Klar. 1991. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 10:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arcangioli, B., and G. Thon. 2004. Mating-type cassettes: structure, switching and silencing, p. 129-147. In R. Egel (ed.), The molecular biology of Schizosaccharomyces pombe. Springer Verlag, Berlin, Germany.
- 5.Arcangioli, B., and R. de Lahondes. 2000. Fission yeast switches mating type by a replication-recombination coupled process. EMBO J. 19:1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahler, J., J. Q., Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 7.Beach, D. H. 1983. Cell type switching by DNA transposition in fission yeast. Nature 305:682-688. [Google Scholar]
- 8.Brewer, B. J. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53:679-686. [DOI] [PubMed] [Google Scholar]
- 9.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 10.Bussiere, D. E., and D. Bastia. 1999. Termination of DNA replication of bacterial and plasmid chromosomes. Mol. Microbiol. 31:1611-1618. [DOI] [PubMed] [Google Scholar]
- 11.Dalgaard, J. Z., and A. J. S. Klar. 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400:181-184. [DOI] [PubMed] [Google Scholar]
- 12.Dalgaard, J. Z., and A. J. S. Klar. 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102:745-751. [DOI] [PubMed] [Google Scholar]
- 13.Dalgaard, J. Z., and A. J. S. Klar. 2001. Does S. pombe exploit the intrinsic asymmetry of DNA synthesis to imprint daughter cells for mating-type switching? Trends Genet. 17:153-157. [DOI] [PubMed] [Google Scholar]
- 14.de Lahondes, R., V. Ribes, and B. Arcangioli. 2003. Fission yeast Sap1 protein is essential for chromosome stability. Eukaryot. Cell 2:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egel, R., and H. Gutz. 1981. Gene activation by copy transposition in mating-type switching of a homothallic fission yeast. Curr. Genet. 3:5-12. [DOI] [PubMed] [Google Scholar]
- 16.Egel, R., D. Beach, and A. J. S. Klar. 1984. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc. Natl. Acad. Sci. USA 81:3481-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egel, R., and B. Eie. 1987. Cell lineage asymmetry in Schizosaccharomyces pombe: unilateral transmission of a high-frequency state for mating-type switching in diploid pedigree. Curr. Genet. 12:429-433. [Google Scholar]
- 18.Foss, E. J. 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 20.Hanawalt, P. C. 2002. Subpathways of nucleotide excision repair and their regulation. Oncogene 21:8949-8956. [DOI] [PubMed] [Google Scholar]
- 21.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078-1083. [DOI] [PubMed] [Google Scholar]
- 23.Kaykov, A., and B. Arcangioli. 2004. A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Current Biol. 14:1924-1928. [DOI] [PubMed] [Google Scholar]
- 24.Kaykov, A., A. M. Holmes, and B. Arcangioli. 2004. Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J. 23:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, M., J. Burke, M. Smith, A. J. S. Klar, and D. Beach. 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S. M., D. D. Dubey, and J. A. Huberman. 2003. Early-replicatiing heterochromatin. Genes Dev. 1:330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klar, A. J. S. 1987. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature 326:466-470. [DOI] [PubMed] [Google Scholar]
- 28.Klar, A. J. S. 1990. The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J. 9:1407-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Page, F., E. E. Kwoh, A. Avrutskaya, A. Gentil, S. A. Leadon, A. Sarasin, and P. K. Cooper. 2000. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101:159-171. [DOI] [PubMed] [Google Scholar]
- 31.Leupold, U. 1955. Methodishes zur Genetik von Schizosaccharomyces pombe. Schweiz. Z. Pathol. Bakteriol. 18:1141-1146. [PubMed] [Google Scholar]
- 32.Maundrell, K. 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265:10857-10864. [PubMed] [Google Scholar]
- 33.Meluh, P. B., and J. R. Broach. 1999. Immunological analysis of yeast chromatin. Methods Enzymol. 304:414-430. [DOI] [PubMed] [Google Scholar]
- 34.Miyata, H., and M. Miyata. 1981. Mode of conjugation in homothallic cells of Schizosaccharomyces pombe. J. Gen. Appl. Microbiol. 27:365-371. [Google Scholar]
- 35.Moreno, S., A. J. S. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, O., and R. Egel. 1989. Mapping the double-strand breaks at the mating-type locus in fission yeast by genomic sequencing. EMBO J. 8:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noguchi, E., C. Noguchi, L. L. Du, and P. Russell. 2003. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 23:7861-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panzeri, L., I. Groth-Clausen, J. Shepard, A. Stotz, and P. Philippse. 1984. Centromeric DNA in yeast. Chromosomes Today 8:46-58. [Google Scholar]
- 39.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 40.Singh, J., and A. J. S. Klar. 1993. DNA polymerase-alpha is essential for mating-type switching in fission yeast. Nature 361:271-273. [DOI] [PubMed] [Google Scholar]
- 41.Snyder, M., R. J. Sapolsky, and R. W. Davis. 1988. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2184-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strathern, J. N., A. J. Klar, J. B. Hicks, J. A. Abraham, J. M. Ivy, K. A. Nasmyth, and McGill, C. 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31:183-192. [DOI] [PubMed] [Google Scholar]
- 43.Svejstrup, J. Q. 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 3:21-29. [DOI] [PubMed] [Google Scholar]
- 44.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi, Y., T. Horiuchi, and T. Kobayashi. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thon, G., and A. J. Klar. 1993. Directionality of fission yeast mating-type interconversion is controlled by the location of the donor loci. Genetics 134:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vengrova, S., and J. Z. Dalgaard. 2004. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 18:794-804. [DOI] [PMC free article] [PubMed] [Google Scholar]