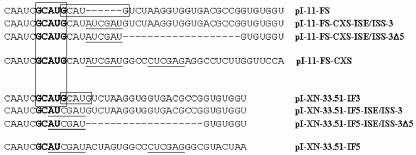Abstract
Mutually exclusive splicing of fibroblast growth factor receptor 2 (FGFR2) exons IIIb and IIIc yields two receptor isoforms, FGFR2-IIIb and -IIIc, with distinctly different ligand binding properties. Several RNA cis elements in the intron (intron 8) separating these exons have been described that are required for splicing regulation. Using a heterologous splicing reporter, we have identified a new regulatory element in this intron that confers cell-type-specific inclusion of an unrelated exon that mirrors its ability to promote cell-type-specific inclusion of exon IIIb. This element promoted inclusion of exon IIIb while at the same time silencing exon IIIc inclusion in cells expressing FGFR2-IIIb; hence, we have termed this element ISE/ISS-3 (for “intronic splicing enhancer-intronic splicing silencer 3”). Silencing of exon IIIc splicing by ISE/ISS-3 was shown to require a branch point sequence (BPS) using G as the primary branch nucleotide. Replacing a consensus BPS with A as the primary branch nucleotide resulted in constitutive splicing of exon IIIc. Our results suggest that the branch point sequence constitutes an important component that can contribute to the efficiency of exon definition of alternatively spliced cassette exons. Noncanonical branch points may thus facilitate cell-type-specific silencing of regulated exons by flanking cis elements.
Alternative splicing is an important mechanism by which the same gene transcript is differentially processed in separate tissues or during different developmental stages to yield proteins with distinct functions. Although it has been estimated that over half of all human genes generate more than one mRNA due to alternative splicing, molecular mechanisms that explain how cell-type-specific splicing regulation occurs in mammals are limited (7). The most frequently described systems of alternative splicing are those in which internal exons are either included or skipped in the mature mRNAs; such exons are commonly referred to as cassette exons. Consensus sequences at the 5′ splice site, the 3′ splice site and associated polypyrimidine tract, and the branch point sequence (BPS) upstream of the 3′ splice site are required elements for the splicing of all cassette exons. The degree to which these sequences match the consensus sequences is one factor that influences whether an exon is included or skipped. In addition, the proximity of the 5′ and 3′ splice sites to one another is a major determinant of splicing efficiency. Internal exons longer than 300 nucleotides (nt) are generally not efficiently spliced, suggesting that exon definition occurs through coordinated interactions between factors recruited to both ends of an exon (5). The selection of a cassette exon for splicing is further determined by RNA cis elements generally referred to as exonic or intronic splicing enhancers (ESEs or ISEs) and exonic or intronic splicing silencers (ESSs or ISSs). For the most part, it appears that these cis elements function by binding to splicing regulatory proteins that stimulate or inhibit exon definition. Most alternatively spliced cassette exons contain more than one such regulatory cis element in the exon and/or in the flanking introns, and as such, combinatorial models of splicing regulation have been proposed whereby inclusion or skipping of an exon is determined by the net activity of several proteins associated with these elements (7, 46). Studies of numerous alternatively spliced transcripts support such models of combinatorial control, but the means by which tissue-specific differences in splicing are achieved remains unclear (7, 46). It is possible that cell-type-specific differences in splicing result from the net balance of positive and negative regulatory elements due to differences in the relative expression levels of a number of ubiquitously expressed factors that bind them. Alternatively, expression of a single cell-type-specific factor may switch the splicing pattern of a regulated transcript. A particularly robust example of regulation by a mammalian tissue-specific splicing factor is that seen with the neural-specific Nova proteins, Nova-1 and Nova-2, that are expressed exclusively in distinct subsets of neurons and that may be sufficient to achieve brain-specific alternative splicing of several transcripts (52). It is likely that the splicing outcome for many alternatively spliced mammalian transcripts results from the combinatorial effects of cell-type-specific factors as well as differential expression of other regulatory factors.
Mutually exclusive cassette exons present an additional level of complexity, because mechanisms of splicing regulation must coordinate the cell-type-specific inclusion of one exon with exclusion of another exon(s). A highly cell-type-specific example of such a mechanism occurs during splicing of mutually exclusive exon IIIb or IIIc of fibroblast growth factor receptor 2 (FGFR2). These exons encode the second half of the third immunoglobulin-like domain in the extracellular portion of the protein and yield receptors (FGFR2-IIIb or FGFR2-IIIc) with distinctly different ligand binding preferences. FGFR2-IIIb is expressed in epithelia whereas FGFR2-IIIc is mesenchymal, and exclusive expression of either receptor in the proper cell type is critical for organ development and maintenance of normal intercellular communication (14). Several studies using FGFR2 minigenes have characterized regulatory cis elements within or flanking exons IIIb or IIIc and are shown schematically in Fig. 1. RNA binding proteins that influence FGFR2 splicing through binding these elements have been described previously and include polypyrimidine tract binding protein (PTB; binds upstream intronic splicing silencer [UISS], downstream intronic splicing silencer (DISS), and exon IIIc ESSs), TIA-1 (binds ISE-1), and hnRNPA1 (binds the exon IIIb ESS) (11, 16, 17, 29, 53). ISE-2 and ISAR base pair with one another to form a stem structure, and this structure has been shown to play a role both in activation of exon IIIb splicing and silencing of exon IIIc in cells expressing FGFR2-IIIb (4, 15, 33). Despite these findings, the means by which cell-type-specific regulation occurs remains a mystery. On the one hand, the regulatory factors identified thus far are present in cells expressing FGFR2-IIIb or FGFR2-IIIc and, thus, it is not clear how they differentially affect splicing in either cell type. In addition, several of the cis elements can be shown to influence splicing in both cell types depending on the context of other FGFR2 RNA sequences. For example, UISS silences exon IIIb inclusion in both cell types (11). However, in cells expressing FGFR2-IIIb, this silencing is opposed by elements in the downstream intron (including ISE-2 and ISAR) that promote exon IIIb inclusion. In such cells, when UISS is deleted these downstream elements are no longer required for exon IIIb inclusion. When UISS is deleted in cells that express FGFR2-IIIc, they then splice exon IIIb efficiently. Further, exon IIIb silencing by UISS appears to also require the cooperation of DISS elements in the downstream intron as well (53). In this case, if the role of other elements in the FGFR2 transcript were not considered, the ability of UISS could have been interpreted to be that of a cell-type-specific repressor of exon IIIb inclusion in cells expressing FGFR2-IIIc.
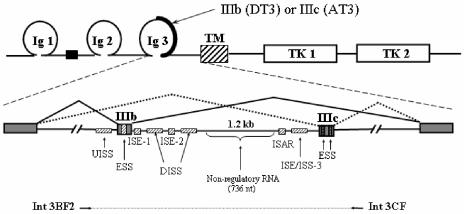
FIG. 1.
Schematic representation of alternatively spliced variants of FGFR2 and RNA cis elements shown to regulate splicing. At the top is a protein domain map showing the region encoded by exons IIIb or IIIc. At the bottom is a map of the pre-mRNA with mutually exclusive pathways leading to production of FGFR2-IIIb in DT3 cells or FGFR2-IIIc in AT3 cells. TM, transmembrane domain; TK, tyrosine kinase domains. Shaded boxes represent exons, and solid lines represent introns. Hatched boxes represent intronic and exonic cis elements (enhancers and silencers). Note that human sequences highly similar to ISE-1, ISE-2, and ISAR have been referred to as IAS1, IAS2, and IAS3, respectively, and can be considered to be functionally equivalent. The horizontal arrows and dashed line underneath the pre-mRNA represent the FGFR2 genomic sequence that was amplified by PCR and positioned in the intron of pI-11 to generate the full-length FGFR2 minigenes as described in Results.
In order to independently investigate the functions of cis elements required for expression of FGFR2-IIIb, we elected to study candidate elements in the context of an otherwise completely heterologous minigene. We used the previously described rat prostate cancer cell lines, DT3 and AT3, that exclusively express FGFR2-IIIb and FGFR2-IIIc, respectively. The minigene contained a single heterologous internal exon, and we tested the ability of sequences located in the intron between exons IIIb and IIIc (intron 8; see Fig. 1) to exhibit DT3-cell-type-specific splicing enhancement of this exon. These studies identified a new element, ISE/ISS-3, that is located downstream of ISAR and which was required to activate splicing in DT3 cells, but it displayed no function on the splicing pathway in AT3 cells. Most importantly, we were able to determine that ISE/ISS-3 displays cell-type-specific splicing regulation in the absence of other previously described FGFR2 regulatory cis elements. In addition to functioning in activation of exon IIIb inclusion, this element also represses splicing to the downstream exon IIIc, potentially coupling these activities that are both needed for mutually exclusive splicing. To better understand the mechanism leading to exon IIIc silencing in DT3 cells, we performed branch point mapping. We determined that the branch point sequence upstream of exon IIIc specified use of G as the primary branch nucleotide in the first step of splicing. This suboptimal branch point sequence was necessary in order for ISE/ISS-3 to repress exon IIIc. Introducing an optimized branch point resulted in nearly complete inclusion of exon IIIc in DT3 cells. We also noted that, compared to an optimized branch point, the natural exon IIIc branch point was able to complete the first step of splicing with only slightly delayed kinetics in vitro. In contrast, the second step of splicing was severely impaired. Our results further highlight the importance of branch point sequences during exon definition of a regulated cassette exon. These results also raise the interesting possibility that regulated silencing of FGFR2 exon IIIc could occur during the second catalytic step of splicing.
MATERIALS AND METHODS
Plasmid construction.
To construct pI-XN-33.51 we used PCR to amplify RTB33.51 (kindly provided by T. Cooper) using primers TNT-Xba-F (GGCCTCTAGAGTCGAAGTCCTCACCTGGTG) and RTB33.51-PNB-R (TTAATTAAGCGGCCGCCCTGTACAAACTACCTGTCAGACATCT) followed by insertion of the PCR product between the XbaI and NotI sites of pI-11(-H3)-PL. The latter plasmid was similar to the previously described pI-11 (10), except that the HindIII site in the multicloning site of PCDNA3 was eliminated (by digestion, blunt fill-in, and religation) and the sequences between the XbaI and XhoI sites of the intron were replaced with the sequence 5′-GGCCGTCGACGCTAGCGCGGCCGCGATATCATCGATACTAGTGGCC-3′. FGFR2 intron 8 fragments 1, 3, 5, 6, and 8 (IF1, -3, -5, -6, and -8) were obtained by PCR from the previously described pI-11-FS-ΔBclI/NdeI (10), and IF2, -4, and -7 were obtained by PCR using template previously described as pI-11-FS-ΔBclI/NsiI (10). The resulting fragments were obtained using the following primer pairs: IF1 and IF2, Intron 2F-Not (5′-GCGGCCGCCAACGTTTTTGTGTTTGTGT-3′) and Intron 2-R2 (5′-CCGGCTCGAGGGCTAGACATAGGAATGATT-3′); IF3 and IF4, Intron-2F-Not and Stu/Cla-R (5′-CCGGAGGCCTATCGATGTTCCCAGCAGGTCCAGGGG-3′); IF5, Intron-2F-Not and Nsi/Cla-R (5′-CCGGATGCATATCGATGCGATTGAACACATGGAAAA-3′); IF6 and IF7, IAS2-F2 (5′-GCGGCCGCTGGCCATGGAAAAATGCCCA3′) and Stu/Cla-R; IF8, IAS2-F2 and Nsi/Cla-R. The β-globin (BG) fragment (Fig. 2A) was obtained by PCR of the second intron of the human β-globin gene with β-Globin-F (5′-CCGGGCGGCCGCTATATTAATGCCTTAACAT-3′) and β-Globin-R (5′-CCGCATCGATGATTGTAGCTGCTATTAGCA-3′) primers. These sequences were cloned between NotI and ClaI sites in pI-XN-33.51 plasmid to obtain the constructs shown in Fig. 2A. The pI-11-FS adenoviral splicing construct was made as previously described (10). To create pI-11-FS-CXS-No ISE/ISS3, the sequence between NsiI and StuI in the parent pI-11-FS plasmid was replaced with the sequence 5′-ATCGATGGCCCTCGAG-3′ containing ClaI and XhoI sites. In this construct, an XhoI site downstream of exon IIIc was eliminated to facilitate cloning between the ClaI and XhoI sites upstream of exon IIIc. Minigenes with ISE/ISE-3 fragment deletions were obtained by PCR of pI-11-FS with the primers shown in Fig. 3B and cloning the sequences in the ClaI/XhoI sites of pI-11-FS-CXS or pI-XN-33.51-IF5, respectively. The BG sequence (Fig. 4B and C) was obtained by PCR of the human β-globin gene with the following primer pair: BG105-F (CATCGATGTCTATGGGACCCTTGATGT) and BG105-R (CTCGAGCAGACAAAGGGTAAGATTTG). The mutation shown in Fig. 3B (ISE/ISS-3 Mut) was introduced by PCR-based site-directed mutagenesis into a minigene containing the ISE/ISS-3Δ5 fragment. pI-11-FS-IIIB Mut and pI-11-FS-IIIC Mut series plasmids were made in two consecutive steps with the QuikChange kit (Stratagene) to replace the 5′ and 3′ splice sites in the pI-11-FS-CXS minigene as shown in Fig. 4A. The ISE/ISS-3Δ5, ISE/ISS-3 Mut, and BG fragments were then cloned in the ClaI/XhoI sites of the respective minigenes to obtain the minigenes shown in Fig. 4. The construct for in vitro splicing of exon IIIc (as shown in Fig. 5A) was made by PCR of pI-11-FS with primers Nde/Not-F (CCGGCATATGGCGGCCGCCAAACAAATTCAAAGAGAAC) and IIIc-5′ss-R (CATCGATACTTACCTGGCATCCTCAAAAGTTACATT) and cloning of the resulting product into the NotI/ClaI sites of pI-11(-H3)-PL, pI-11-FS-IIIc-BP-Mut1, pI-11-FS-IIIc-yeast branch point (YBP), and pI-11-FS-IIIc-Mam. BP minigenes were also made with the QuikChange kit (Stratagene). The same branch point modifications were introduced into the related plasmids for in vitro splicing by PCR amplification of pI-11-FS-BP-Mut1 and pI-11-FS-YBP, respectively, with Nde/Not-F and IIIc-5′ss-R primers and cloning the product of amplification in NotI/ClaI sites of pI-11(-H3)-PL. All plasmid constructs were prepared with QIAGEN Midi kits. Sequences of all the minigenes described were confirmed by sequencing at the University of Pennsylvania sequencing facility.
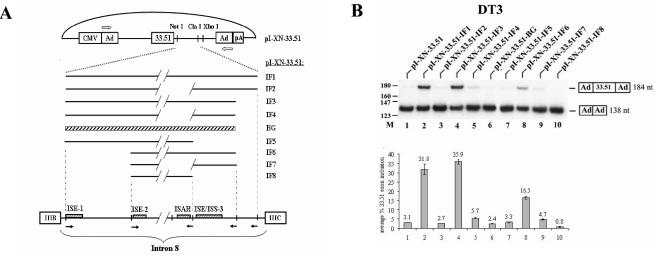
FIG. 2.
Intron 8 sequences from FGFR2 can activate splicing of an upstream heterologous troponin I (TNI) exon in DT3 cells but not in AT3 cells. (A) Schematic presentation of pI-XN-33.51 series plasmids and intron 8 elements from FGFR2 that are positioned downstream of exon 33.51. Ad, adenoviral exons; CMV, cytomegalovirus exons. Solid lines represent intronic fragments (IF) that were inserted downstream of the TNI exon 33.51. Diagonal lines indicate deleted sequence, and the cross-hatched box represents a β-globin intron sequence. Outlined arrows indicate primers used for RT-PCR analysis. At the bottom is a representation of intron 8 to illustrate the derivation of the intron fragments. Open boxes indicate the exons, and solid arrows indicate positions of PCR primers used to generate the intron fragments. (B) Intron 8 sequences from FGFR2 can activate splicing of a heterologous exon in DT3 cells. At the top is shown a representation of a single experiment using RT-PCR of RNA from DT3 cells stably transfected with these minigenes, and at the bottom is shown graphical results representing the average of at least three independent experiments (this same format applies to most subsequent transfection results). RT-PCR products with inclusion or skipping of the cTNI exon are indicated at the right. M, molecular weight markers.
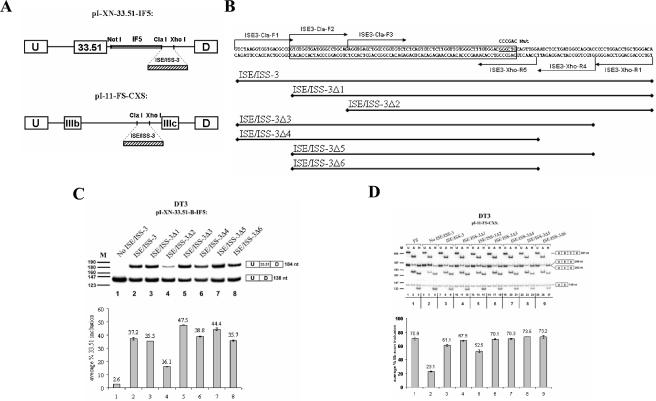
FIG. 3.
An 85-nt sequence, ISE/ISS-3Δ6, is sufficient to restore splicing regulation by sequences downstream of ISAR. (A) Schematic representation of the minigene constructs used to study the influence of ISE/ISS-3 deletions and mutations on splicing of exon 33.51 splicing in the heterologous minigenes (top) or on exon IIIb and IIIc splicing in the full-length FGFR2 minigenes (bottom). (B) Sequence of the ISE/ISS-3 RNA cis element. Deletions from the 5′ or 3′ end were obtained by PCR of the corresponding DNA with primer positions indicated by arrows. The resulting DNA sequences were inserted between ClaI and XhoI sites in the pI-XN-33.51-IF5 and pI-11-FS-CXS plasmids. The boxed sequence indicates the minimal 78-nt sequence that is sufficient for full ISE/ISS-3 function. Underlined is a sequence that, when mutated to the sequence shown above (Mut), results in loss of exon 33.51 inclusion and a switch from exon IIIb to IIIc in full-length minigenes. (C) Results of RT-PCR of RNA prepared from DT3 cells stably transfected by the minigenes containing the truncated sequences downstream of the heterologous cTNI exon 33.51. (D) Results of RT-PCR and restriction analysis to determine the percentage of exon IIIb inclusion using full-length FGFR2 minigenes in DT3 cells. When undigested (U) RT-PCR products are analyzed, three bands are observed: a 431-bp product corresponding to a spliced product containing both exon IIIb and IIIc (double inclusion), a 138-bp band corresponding to spliced products in which both exon IIIb and IIIc are skipped, and 286- or 283-bp products corresponding to products containing either exon IIIb or exon IIIc (single inclusion), respectively, that cannot be easily distinguished from each other by size. To distinguish exon IIIb from exon IIIc, the RT-PCR products are digested with AvaI (A) or HincII (H), which recognize restriction sites present only in exon IIIb- or exon IIIc-containing products, respectively. We calculated the percentage of the single-inclusion products that contain exon IIIb versus exon IIIc, and the results are presented graphically at the bottom. U, undigested RT-PCR products; A, products digested with AvaI; H, products digested with HincII. M, pBR322/MspI molecular weight markers. At the right-hand side, spliced products represented by each band are shown. U and D indicate the upstream and downstream adenoviral exons; B and C, exons IIIb and IIIc, respectively.
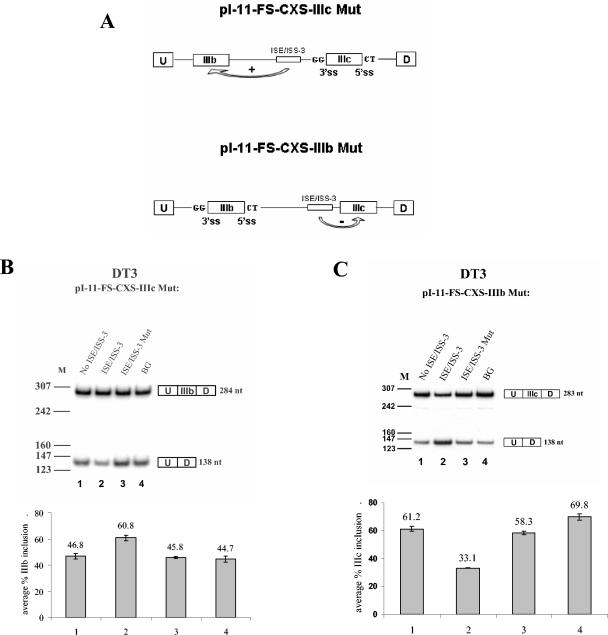
FIG. 4.
ISE/ISS-3 enhances splicing of exon IIIb and represses splicing of exon IIIc. (A) Schematic representation of the constructs showing splice site mutations in which GG and CT replace the AG and GU dinucleotides, respectively. (B) Results of RT-PCR for pI-11-FS-CXS-IIIc Mut minigene series stable transfected in DT3 cells. Results in which no sequence is inserted to replace ISE/ISS-3, ISE/ISS-3, ISE/ISS-3 Mut, or the unrelated BG sequence are shown. (C) Similar results showing the effect of ISE/ISS-3 on exon IIIc splicing using the pI-11-FS-CXS-IIIb Mut minigenes. ss, splice site; U, D, and M, as defined in the legend to Fig. 3.
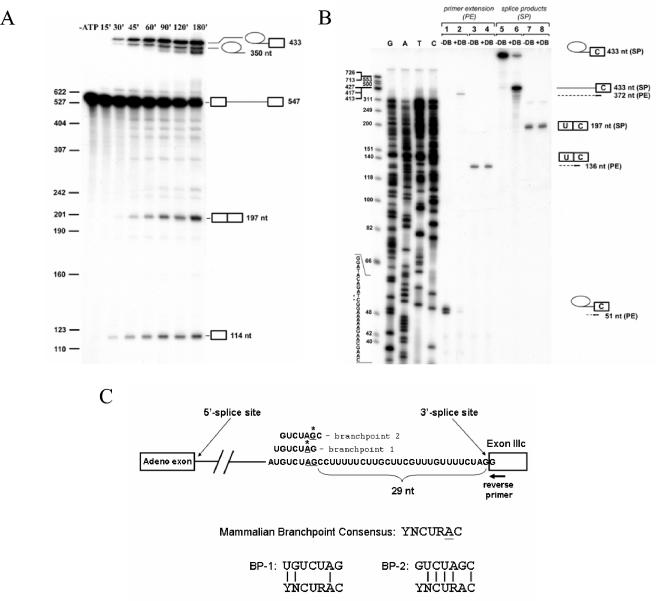
FIG. 5.
Branch point mapping for exon IIIc. (A) A time course of in vitro splicing of a pre-mRNA containing an upstream adenoviral exon followed by FGFR2 intron 8 sequences and exon IIIc. From top to bottom, icons at the right indicate the lariat intermediate, lariat product, pre-mRNA, spliced product, and free 5′ exon. (B) Branch point determination by primer extension. At the far right-hand side, icons present splicing products. Arrows indicate the primer used for sequencing and primer extension, and dotted lines show products of the primer extension. Samples in lines 1 to 4 were primer extended (PE) before loading the gel; samples in lines 5 to 8 represent gel-purified products from the spliced reaction. +DB, debranched; −DB, not debranched. At far left are ΦX174/HinfI molecular weight markers followed by a sequencing ladder obtained using the same primer that was used for primer extension. Sequence of the minus strand and the positions at which primer extension is blocked are indicated by stars. (C) Schematic presentation of branch point sequences identified. On the bottom, the degree of matching of the branch point sequences to the degenerate mammalian BPS consensus is shown. Adeno, adenovirus.
Transfections, RNA purification, and reverse transcription-PCR (RT-PCR) analysis.
Transfection of DT3 and AT3 cells and analysis of minigene splicing were performed as previously described (59), with the exception that only 25 cycles of denaturation and an annealing temperature of 60°C were used during PCR. After correction of molar equivalents for differences in sizes, quantification of data was performed with the use of a Molecular Dynamics PhosphorImager.
Extract preparation and in vitro splicing assays.
HeLa nuclear extract and S100 cytoplasmic extract from KATO III type cells were prepared as described previously (18). Capped 32P-labeled pre-mRNA substrates were made by in vitro transcription of ClaI-digested plasmids using T7 RNA polymerase (Ambion). Transcriptions were performed in a 25-μl volume containing 1× T7 RNA polymerase buffer (Ambion), 5 mM dATP and dCTP, 250 μM UTP, 3.2 mM 5′,7-methyl guanosine nucleotide [m7G(5′)ppp(5′)G], 400 μM GTP, 2.67 × 10−8 mmol of [α-32P]UTP (3000 mCi/mmol, 10 mCi/ml), 2 μg of plasmid, and 40 U of T7 RNA polymerase. Full-length transcripts were gel purified by denaturing polyacrylamide gel electrophoresis. In vitro splicing was performed with 8.0 μl of HeLa nuclear extract in 25-μl reaction mixtures containing 21 fmol (100,000 counts per million [cpm]) of pre-mRNA in the presence of 2.8 mM ATP, 14 mM creatine phosphate (Sigma), 4.5 mM MgCl2, and 85 mM KCl. The mixtures were incubated at 30°C for 15 to 180 min followed by addition of 125 μl of stop buffer (100 mM Tris-HCl [pH 7.5], 10 mM EDTA-Na2 [pH 8.0], 1% sodium dodecyl sulfate [SDS], 150 mM NaCl, 300 mM sodium acetate [pH 5.2]). After phenol-chloroform extraction and ethanol precipitation, the pellet was resuspended in loading buffer (95% formamide, 0.09% bromphenol blue, 0.09% xylene cyanol FF) and loaded on denaturing polyacrylamide gels, followed by autoradiography and/or PhosphorImager analysis.
Branch point identification.
Debranching reactions with S100 extract were performed as previously described (43), except that S100 was prepared from KATO III cells and RNA splicing products were all gel purified on denaturing gels before debranching. Ten picomoles of EX-3CPE-R primer (CCGTGGTGTTAACACCGGCGGC) was labeled with 50 pmol of [γ-32P]ATP in the presence of 20 U of T4 polynucleotide kinase (New England Biolabs) and 1× polynucleotide kinase buffer and was incubated for 45 min at 37°C. Unincorporated nucleotides were removed using MicroSpin G-25 Sephadex Columns (Amersham Pharmacia). Ten femtomoles (approximately 50,000 cpm) of the labeled primer was annealed in a volume of 20 μl with hybridization buffer (150 mM KCl, 10 mM Tris-HCl [pH 8.3], 1.0 mM EDTA) and 1 to 3 fmol of splicing product at 65°C for 1 h and slow cooling (20 to 30 min) to room temperature. To each tube containing RNA and the primer, 30 μl of a primer extension reaction mix (30 mM Tris-Cl [pH 8.3], 15 mM MgCl2, 8.3 mM dithiothreitol, 6.75 μg of actinomycin D, 0.22 mM each deoxynucleotide triphosphate, 5 U of avian myeloblastosis virus reverse transcriptase) was added followed by incubation for 1 h at 42°C. Samples were treated with 20 μg of RNase A, incubated for 15 min at 37°C, extracted using phenol-chloroform, ethanol precipitated, and resuspended in loading buffer. Sequencing was performed using a Sequenase version 2.0 DNA Sequencing kit and the EX-3CPE-R primer.
RESULTS
FGFR2 intron 8 sequence elements downstream of ISAR are required to activate cell-type-specific splicing of a heterologous exon.
In order to better understand the mechanism that results in cell-type-specific splicing of exon IIIb in DT3 cells, we chose to focus on the role of intron 8 RNA cis elements. To study the functions of these elements in isolation from other sequences in the endogenous FGFR2 transcript, we elected to use a heterologous minigene lacking any FGFR2 sequences other than the intron 8 elements themselves. We developed a heterologous minigene derived from RTB33.51 as originally described by Ryan and Cooper (44). RTB33.51 consists of a 46-nucleotide skeletal troponin I (sTNI) exon (henceforth the 33.51 exon) flanked by cardiac troponin T (cTNT) intron sequences. We inserted the 33.51 exon together with the upstream 99 nt of cTNT intron sequence into the intron of a previously described minigene, pI-11, that contains two adenoviral exons and a single intron (10). The resulting minigene, pI-XN-33.51 (Fig. 2A), also contains 6 nucleotides downstream of exon 33.51 that are components of the 5′ splice site consensus sequence. DT3 and AT3 cells were stably transfected with pI-XN-33.51, RNA was harvested, and levels of exon 33.51 inclusion versus skipping were analyzed by RT-PCR. In both cell types, we noted very inefficient splicing of exon 33.51; only about 3% of spliced RNAs included this exon in either cell type (Fig. 2B, lane 1 and data not shown). A series of intron 8 fragments were cloned downstream of exon 33.51, and the levels of exon 33.51 inclusion from these minigenes were determined (Fig. 2A). A deletion of 736 nucleotides between ISE-2 and ISAR had no effect on FGFR2 splicing regulation, and therefore all intron fragments contained this deletion (indicated by hash marks in Fig. 2A) (10). The largest sequence tested, IF1, began with the sixth nucleotide in intron 8 and ended 26 nucleotides upstream of the 3′ splice site of exon IIIc. This intron fragment included three previously described cis elements (ISE-1, ISE-2, and ISAR) involved in exon IIIb splicing enhancement, as well as additional sequences downstream of ISAR. In DT3 cells, IF1 robustly activated splicing of exon 33.51 (Fig. 2B, lane 2). However, in AT3 cells, no increase in exon 33.51 inclusion was observed with this or any other intron 8 fragments tested (data not shown). When ISAR was deleted in IF2, exon 33.51 splicing was no longer seen in DT3 cells, recapitulating the reduction in exon IIIb splicing when ISAR was deleted from FGFR2 minigenes (10). IF3, containing a deletion of an additional 49 nucleotides at the 3′ end (compared to IF1), was also able to activate exon 33.51 splicing to a level equivalent to that seen with IF1 (Fig. 2B, lane 4). However, an intronic sequence of the same size as IF3 derived from the second intron of human β-globin was not capable of activating exon 33.51 splicing (Fig. 2B, lane 6). Thus, a 514-nt sequence (IF3), which contains all of the intron 8 cis elements (ISE-1, ISE-2, and ISAR) previously implicated in FGFR2 splicing regulation, is necessary and sufficient to confer robust, cell-type-specific splicing activation to an unrelated exon. Deletion of ISAR to yield IF4 again negated the ability of this intron fragment to activate exon 33.51 splicing (Fig. 2B, lane 5). Further shortening from the 3′ end of intron 8 to a point 11 nucleotides downstream of ISAR also resulted in a loss of 33.51 exon inclusion (Fig. 2B, lane 7). This result demonstrated that, in this heterologous context, ISE-1, ISE-2, and ISAR are not sufficient to activate cell-type-specific activation of an upstream exon. This suggested that an additional regulatory cis element was located downstream of ISAR that plays an important role in splicing regulation. As will be discussed subsequently, this element also plays a role in silencing splicing of a downstream exon, and as such we will henceforth refer to this element as ISE/ISS-3 (intronic splicing enhancer/intronic splicing silencer-3).
Cell-type-specific ISE/ISS-3-mediated enhancement of splicing by ISE/ISS-3 does not require a specific contribution by other intron 8 splicing enhancers.
ISE-1 is a uridine-rich sequence element located immediately downstream of the exon IIIb 5′ splice site. Intron fragment 6 (IF6) contains ISE-2, ISAR, and ISE/ISS-3, but all sequences upstream of ISE-2, including ISE-1, have been deleted. This intronic sequence is also capable of mediating cell-type-specific inclusion of exon 33.51, albeit at a level approximately half that seen with IF1 or IF3 (Fig. 2B, lane 8). However, it can again be seen that further elimination of either ISAR or ISE/ISS-3 results in loss of exon 33.51 splicing (Fig. 2B, lanes 9 and 10). Although deletion of ISAR also resulted in loss of exon 33.51 splicing, when ISAR and ISE-2 are both deleted, together with the sequences between them, exon 33.51 splicing in DT3 cells is restored (data not shown). These results suggest that formation of a stem structure between ISE-2 and ISAR may be required either to approximate ISE/ISS-3 to the upstream exon or to counter the effect of a DISS between them (4, 33). Regardless, it is evident that ISE/ISS-3 can carry out cell-type-specific enhancement of splicing without a specific contribution by ISE-1, ISE-2, or ISAR. While ISE-1 enhanced exon 33.51 inclusion when present together with ISE-2, ISAR, and ISE/ISS-3, it was not sufficient to activate exon 33.51 splicing at all in the context of IF5. This is likely due to sequences downstream from ISE-1 that function as silencers counteracting the ability of ISE-1 to activate splicing of an upstream exon. Consistent with this, a downstream intronic splicing silencer (DISS) has also been described between ISE-1 and ISE-2 that represses exon IIIb splicing (53). When we inserted only the 51-nt rat ISE-1 sequence (of which 38 nt are uridines) downstream of exon 33.51, we observed over 90% exon 33.51 inclusion in both DT3 and AT3 cells (data not shown). Thus, ISE-1 can function to activate splicing, but this effect is not cell type specific and can be prevented by downstream silencer sequences. Similar observations were made with a similar sequence in the human FGFR2 transcript, IAS-1 (16). In the case of the rat transcript, it would appear that the ability of sequences downstream of ISE-1 to function as splicing silencers is also not cell type specific, because these elements can prevent ISE-1 from activating splicing in both cell types as well (Fig. 2B, lane 7, and data not shown).
Deletion analysis defines the minimal sequence downstream of ISAR that constitutes a functional splicing regulatory element.
The length of the intronic sequence (containing ISE/ISS-3) downstream of ISAR that is present in IF3 but deleted in IF5 is 144 nt (Fig. 3B, top). To further define the ISE/ISS-3 cis element, we performed deletion analysis in both the heterologous minigene context as well as in the context of FGFR2 minigenes containing exons IIIb and IIIc (Fig. 3A). We introduced progressive 19- to 20-nt deletions from the 5′ and/or 3′ end of this region and determined the effects of these deletions on exon 33.51 inclusion or exon IIIb inclusion in each minigene context. In the heterologous minigenes, these deletions were tested by inserting the sequences containing the deletions downstream of the intron fragment (IF5) that was, by itself, unable to activate exon 33.51 inclusion (Fig. 3A). The shortest element shown here that preserved the same level of exon 33.51 inclusion in DT3 cells, ISE/ISS-3Δ6, was 85 nt. Any further deletion from the 5′ end resulted in a substantial decrease in exon 33.51 inclusion (Fig. 3C, lane 4, and data not shown). An additional 7 nucleotides could be eliminated from the 3′ end of ISE/ISS-3Δ6 without a reduction in 33.51 inclusion, but further truncation led to a significant decrease in 33.51 inclusion (data not shown). Thus, a 78-nt sequence appeared to constitute the minimal ISE/ISS-3 element (sequence boxed in Fig. 3B). To similarly assess the effects of the deletions on exon IIIb inclusion, we used a minigene, pI-11-FS-CXS, that is a derivative of the previously described FGFR2 minigene, pI-11-FS (10, 11). This minigene contains both exons IIIb and IIIc and differs from the parent construct only by containing restriction sites introduced to facilitate testing of the 144-nt sequence and deletions thereof (Fig. 3A). These minigenes will henceforth we referred to as full-length FGFR2 minigenes. To analyze spliced mRNAs from the full-length FGFR2 minigenes, we used a previously validated RT-PCR assay to determine the percentage of exon IIIb, as opposed to exon IIIc, inclusion (10, 11, 33). It can be seen that with pI-11-FS, over 70% of these spliced products contain exon IIIb in DT3 cells (Fig. 3D, lanes 1 to 3). When pI-11-FS-CXS-No ISE/ISS-3 (in which the 144-nt sequence is absent) was transfected in DT3 cells, only 23% of the spliced products contained exon IIIb with a switch towards predominant exon IIIc inclusion (Fig. 3D, lanes 4 to 6). Insertion of ISE/ISS-3Δ6 was sufficient to restore restored predominant exon IIIb inclusion (Fig. 3D, lanes 25 to 27).
To further investigate sequences in ISE/ISS-3 involved in splicing regulation, we also sequentially mutated blocks of six nucleotides in ISE/ISS-3. As an additional control, we also tested an unrelated, size-matched sequence from the second intron of human β-globin (BG). Introduction of the BG sequence led to a reduction in exon 33.51 inclusion in the heterologous minigenes as well as a switch from exon IIIb to exon IIIc in the full-length minigenes, confirming that specific sequences of ISE/ISS-3 were required for its function (data not shown). We also identified several mutations that similarly caused both a reduction in exon 33.51 inclusion and a switch from exon IIIb to exon IIIc splicing, one of which is shown in Fig. 3B (data not shown).
ISE/ISS-3 functions as an ISE of exon IIIb and an ISS of exon IIIc.
The heterologous minigenes clearly indicated that ISE/ISS-3 functions as a splicing enhancer of an upstream exon. However, deletion of ISE/ISS-3 in the context of the full-length FGFR2 minigenes resulted in a decrease in single-inclusion products containing exon IIIb and an increase in exon IIIc inclusion. This suggested that this element functions as a splicing enhancer of the upstream exon IIIb and a silencer of downstream exon IIIc. To more definitively demonstrate that ISE/ISS-3 activates exon IIIb splicing and represses exon IIIc splicing, we generated the minigenes shown schematically in Fig. 4A. In pI-11-FS-CXS-IIIc Mut, we mutated both the 3′ and 5′ splice sites of exon IIIc, eliminating the option of exon IIIc splicing. We then evaluated the ability of ISE/ISS-3 to activate exon IIIb splicing in DT3 cells. Because exon IIIc cannot be utilized, we observe only two predominant spliced RNA products with these minigenes: those that include exon IIIb and those in which exon IIIb is skipped and the flanking exons are directly ligated. When we transfected a minigene in which ISE/ISS-3 was deleted in DT3 cells, we noted that most products did not include exon IIIb (Fig. 4B, lane 1). However, when we inserted the wild-type ISE/ISS-3 sequence, exon IIIb inclusion was greater than 60% (Fig. 5B, lane 2). Insertion of the ISE/ISS-3 fragment containing a functional 6-nt mutation (Mut; see Fig. 4B) or the size-matched β-globin intron sequence (BG) each returned the level of inclusion to that observed when the element was deleted (Fig. 3B, lanes 3 and 4). It should be noted that because we are determining exon IIIb inclusion versus skipping, results shown here cannot be directly compared to those in which we compare exon IIIb inclusion to exon IIIc inclusion (for example, see Fig. 3D). Nonetheless, while the magnitude of the effect of deleting or mutating ISE/ISS-3 on exon IIIb inclusion appears less in this case, there is no question that its ability to activate exon IIIb inclusion can occur independent of any role in silencing exon IIIc inclusion.
In pI-11-FS-CXS-IIIb Mut, we mutated the 3′ and 5′ splice sites of exon IIIb in order to test the role of ISE/ISS-3 in exon IIIc splicing independent from regulation of exon IIIb. When the ISE/ISS-3Δ5 element was present, exon IIIc inclusion was quite low compared to the level of skipping. However, deletion of ISE/ISS-3, mutation (Mut), or replacement with the BG sequence led to an approximately twofold increase in the percentage of exon IIIc inclusion in DT3 cells (Fig. 4C). Therefore, these data confirm that ISE/ISS-3 also silences exon IIIc splicing. Furthermore, we have also shown that ISE/ISS-3 is able to repress exon IIIc even when all upstream exon 8 sequences are deleted (data not shown). It is also noteworthy that the ISE/ISS-3 mutation tested here clearly impairs both DT3-specific activation of exon IIIb splicing and repression of exon IIIc splicing. The series of mutations described previously also showed a good correlation between the effects of the mutations on activation of exon 33.51 and a switch from exon IIIb to exon IIIc inclusion in full-length FGFR2 minigenes. Collectively these results suggest that ISE/ISS-3 functions as a single element with dual roles in splicing.
Branch point sequences used for exon IIIc splicing are suboptimal matches to the consensus BPS.
To further investigate the mechanism by which ISE/ISS-3 silences exon IIIc splicing, we mapped the branch point used during exon IIIc splicing in vitro. We generated a radiolabeled pre-mRNA as a runoff transcript containing the same upstream adenoviral exon used in the transfected minigenes as well as the last 282 nt of intron 8 (including ISAR and ISE/ISS-3) and the first 71 nt of exon IIIc. Splicing was carried out in HeLa cell nuclear extracts, and products of the splicing reaction were analyzed by denaturing gel electrophoresis. A time course of the splicing reaction is shown in Fig. 5A with splice products indicated schematically at the right. We noted that only a small fraction of pre-mRNAs that completed the first step of splicing also underwent the second step of splicing, even after prolonged incubation. This phenomenon appeared to be substrate specific and not due to any general deficiency in second-step activity in these extracts, because other pre-mRNA substrates demonstrated efficient second-step activity with the same extracts (data not shown; also see below). To confirm the identification of the splice products, the bands corresponding to the predicted lariat intermediate and lariat product were gel purified and treated with a debranching extract. When the predicted lariat intermediate was debranched, the resulting linear RNA migrated with mobility consistent with the 433-nt length expected for the intron and 3′ exon (Fig. 5B, lane 6). When the product migrating with a slightly faster mobility than the lariat intermediate was similarly analyzed, the resulting product migrated with a mobility consistent with a linear 350-nt molecule that corresponds to the expected size of the linear intron (data not shown). We saw no change in the mobility of either the predicted mRNA or the 5′ exon upon debranching, which likewise migrated as predicted based on nucleotide length (Fig. 5B and data not shown). The identity of the predicted spliced mRNA was confirmed by RT-PCR and sequencing.
To identify the branch point nucleotide, we performed primer extension from gel-purified lariat intermediates with a primer complementary to exon IIIc in combination with a sequencing reaction containing the plasmid encoding the pre-mRNA substrate. Because reverse transcriptase is arrested at the branch nucleotides, we were able to determine that the extended products stopped at a position corresponding to use of either an A or G as the branch nucleotide as indicated in Fig. 5C. To confirm that these primer extension products represent a block in transcription due to a branch structure and not to unrelated polymerase pausing or transcriptional termination, we also performed the same primer extension with a molar equivalent of the same lariat intermediate after debranching. This resulted in continuation of the reverse transcription to a point corresponding to the predicted 5′ end of the intron (Fig. 5B, lane 2). As further evidence that the reduction in primer extension products from the lariat intermediate after debranching was not due to RNA degradation in the debranching extract, it can be seen that the amount of RNA degradation seen with both the lariat intermediate and spliced mRNA is minimal. Furthermore, debranching results in no reduction in the amount of primer extension product seen with the spliced mRNA when the reactions are run in parallel (Fig. 5B, lanes 3 and 4).
In contrast to the highly conserved yeast branch point consensus sequence, UACUAAC, a more degenerate consensus sequence, YNCURAC, is generally sufficient for splicing in mammals (the underline indicates the branch nucleotide). Although the vast majority of mammalian branch point sequences utilize adenosine as the branch nucleotide, use of other nucleotides has been observed in rare cases (34). If we compare the sequences flanking the branch point A (BP-1) to the consensus sequence, we note an extremely poor match to the consensus when the A is used to align the other position (Fig. 5C). In contrast, alignment relative to the G as the branch nucleotide (BP-2) reveals a complete match to the consensus at the −3, −2, −1, and +1 positions, positions that are the most highly conserved. Thus, although G is least frequently used as the branch nucleotide, the other flanking nucleotides are perfect matches to the most important sequences in the mammalian branch point consensus sequence.
The first and second steps of splicing occur more efficiently in vitro when a consensus branch point sequence replaces the wild-type branch point sequences upstream of exon IIIc.
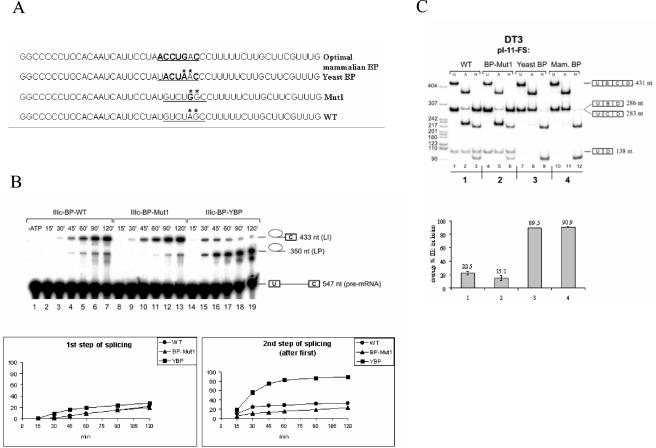
The observations made above suggested that both of the identified branch points were very weak (due to a poor match to the consensus or to use of G as the branch nucleotide) and were likely to be poorly recognized during branch point identification during spliceosome assembly and splicing catalysis. This suggested that, although exon IIIc is efficiently spliced in AT3 cells, the ability of splicing regulatory factors to silence exon IIIc splicing in DT3 cells may be facilitated by these poor branch point sequences. To investigate this further, we tested the effects of inserting sequences in place of the wild-type sequences that were a more optimal match to the branch point consensus on exon IIIc splicing in vitro and in vivo (Fig. 6A). In one case we replaced the wild-type sequence with the yeast branch point consensus sequence UACUAA, which has previously been shown to be an optimal BPS for mammalian splicing (42, 61). We also tested another sequence, ACCUGAC (optimal mammalian BP), which was shown to be the most optimal branch point sequence that could rescue splicing of an otherwise inefficiently spliced human lecithin:cholesterol acyltransferase (LCAT) intron (31). In addition, we also changed the branch point A to G (Mut1). Pre-mRNAs containing the yeast BPS and the A-to-G mutation (Mut1) were spliced in vitro in HeLa nuclear extracts, and splice products were evaluated (Fig. 6B). We noted little difference in splicing efficiency of the first step of splicing using the Mut1 branch point compared to that of the wild-type branch point. There was, however, a slightly higher rate of the second step of splicing using the wild-type branch point relative to Mut1. However, pre-mRNAs containing the yeast BPS displayed greater splicing efficiency of both steps of splicing compared to that of the wild-type or Mut1 branch point. In the case of the first step, this proceeded with slightly faster kinetics with the yeast branch point (note that there was some lariat intermediate already present after 15 min of incubation, in contrast to the wild type or Mut1). Even so, the percentage of pre-mRNAs completing at least the first step of splicing was not appreciably different after 2 h of incubation (28% for the yeast BPS versus 22 and 20% for wild type and Mut1, respectively). Much more dramatic than the difference in first-step activity, we noted that after 120 min of incubation the majority of lariat intermediates (90%) were converted to lariat products when the yeast branch point was used (Fig. 6B). This contrasted with the results seen with pre-mRNAs with the wild-type or Mut1 BPS, where only 31 or 23% of pre-mRNAs completing the first step subsequently completed the second step, respectively. This indicated that, at least in vitro, the second step of splicing was impaired to a greater degree by the weak branch points upstream of exon IIIc than was the first step of splicing.
FIG. 6.
Replacement of the weak exon IIIc branch point sequence with consensus branch points rescues the second step of splicing in vitro and leads to constitutive inclusion of exon IIIc in DT3 cells in vivo. (A) Sequences showing alterations in the branch point region upstream of exon IIIc. (B) A time course of in vitro splicing reactions with pre-mRNAs containing the wild type (WT), A-to-G-mutated branch point (Mut1), and yeast branch point sequence in HeLa nuclear extract. Above each gel the duration of incubation in HeLa cell nuclear extracts is indicated. At the right, icons indicate (from top to bottom) lariat intermediate, lariat product, and pre-mRNA. The lower graphs show the efficiency of the first and second steps of splicing of exon IIIc. Calculation of the first-step efficiency represents the percentage of pre-mRNA processed to lariat intermediate (LI) or lariat product (LP) (LI + LP/LI + LP + pre-mRNA). The efficiency of the second step (after step 1) represents the percentage of products undergoing the first step that also complete the second step (LI/LI + LP). (C) Results of RT-PCR of RNA prepared from DT3 cells stably transfected by the minigenes containing the indicated branch point sequences. Lane and band designations are the same as those described in the legend for Fig. 3D. In the graph, the percentage of exon IIIc inclusion in single-inclusion products is indicated compared to that of exon IIIb inclusion.
Exon IIIc repression in DT3 cells is dependent on poor branch point sequences.
Although the weak branch points upstream of exon IIIc display reduced splicing efficiency in vitro in HeLa nuclear extracts, this limitation does not prevent highly efficient exon IIIc inclusion in vivo in AT3 cells. In fact, when the same full-length rat FGFR2 minigene, pI-11-FS, is transfected in HeLa cells, spliced products also contain almost exclusively exon IIIc and not exon IIIb (data not shown). In order to test our hypothesis that exon IIIc silencing in DT3 cells is facilitated by these weak branch point sequences, we also determined the effects of changing the wild-type branch point sequence on splicing in vivo. All three changes to the wild-type branch point sequence were introduced in the context of the full-length pI-11-FS minigene. Analysis of the spliced products from stably transfected DT3 and AT3 cells was carried out as described previously. As expected, in AT3 cells the already highly efficient exon IIIc inclusion was unaffected by the two improved branch point sequences (data not shown). The mutation of branch point A similarly showed no change in efficient exon IIIc splicing, suggesting that the branch point G is most likely used in vivo as well. Results from DT3 cells, however, showed that replacing either of the two more optimal branch point sequences resulted in a nearly complete elimination of any exon IIIc silencing (Fig. 6C). When we analyzed single-inclusion products (U-B-D or U-C-D), approximately 90% of products contained exon IIIc, in contrast to results with the wild-type BPS in which products predominantly contained exon IIIB. However, because improving the BPS upstream of exon IIIc was not predicted to impair exon IIIb splicing, we also evaluated products in which both exon IIIb and exon IIIc (U-B-C-D) are included. Notably, we see a significant increase in this product compared to that of single-inclusion splice products and compared to that of the products using the wild-type BPS. This indicated that loss of exon IIIc repression did not significantly impair exon IIIb splicing, but most products containing exon IIIb were now represented in the double-inclusion products, and fewer single-inclusion products containing only exon IIIb were obtained because of the dramatic increase in exon IIIc splicing efficiency. The mutation in branch point A may have led to a slightly decreased level of exon IIIc, but this result did not appear significant. Therefore, it is clear that exon IIIc splicing cannot be silenced in vivo in the absence of the suboptimal branch points identified in the in vitro experiments.
DISCUSSION
Previous studies of FGFR2 are consistent with combinatorial models of splicing regulation, in that several cis elements that influence exon IIIb or IIIc inclusion collectively contribute to the outcome in different cell types. We have identified a novel element, ISE/ISS-3, that exhibits cell-type-specific activation of an upstream exon and can do so independently of ISE-1, ISE-2, or ISAR, although optimal splicing enhancement occurs when all of these elements are present. We also showed that this element plays a role both in activation of the upstream exon IIIb and in silencing of exon IIIc, providing a means of integrating these functions. ISE/ISS-3 consists of a high GU content, suggesting a possible involvement of RNA binding proteins that bind to GU-rich sequences. For example, the CELF (CUG-binding protein and ETR-3-like factors; also referred to as Bruno-like) family of RNA binding proteins regulate splicing and preferentially bind to UG- or UR-rich sequences (12, 27, 49, 50, 58). Similarly, TLS (for “translocated in liposarcoma”) has been implicated in splicing regulation and also binds GU-rich sequences (30). Experiments are under way to explore the potential role of these, or other, splicing regulatory proteins that interact with the ISE/ISS-3 element to regulate FGFR2 splicing.
It was recently shown that two overlapping GCAUG motifs also located downstream of ISAR are also involved in cell-type-specific activation of exon IIIb splicing in DT3 cells (4). One or both of these GCAUG elements were disrupted when we generated minigenes to study ISE/ISS-3. The resulting sequences of the junctions between the GCAUG elements and the downstream sequences are shown in Fig. 7. For the full-length FGFR2 minigenes, insertion of a ClaI site resulted in disruption of the downstream GCAUG, but the 5′ GCAUG element was preserved. However, when the ISE/ISS-3 sequences are not also present, exon IIIb inclusion is highly reduced (compare lanes 1 to 3 and 4 to 6 in Fig. 3D). Introduction of ISE/ISS-3 restores exon IIIb inclusion to levels similar to those achieved by the pI-11-FS minigene containing both GCAUG sequences (Fig. 3D). In the heterologous minigenes, pI-XN-33.51-IF5 does not contain either GCAUG element and exon 33.51 is skipped. However, insertion of ISE/ISS-3 alone restores exon 33.51 inclusion (compare lane 4 in Fig. 2B with lane 2 in 3C). Thus, our studies clearly distinguish that ISE/ISS-3 is a separate and important regulatory element.
FIG. 7.
Diagram demonstrating the sequences present between ISAR and ISE/ISS-3. GCAUG elements are boxed where present, with the unaltered wild-type (WT) sequence junction for pI-11-FS and pI-XN-33.51-IF3 showing two overlapping copies. Underlined sequences represent those encoded by ClaI or XhoI sites inserted to facilitate study of ISE/ISS-3. pI-11-FS-CXS and pI-XN-33.51-IF5 indicate the sequences in which no ISE/ISS-3 element was inserted into the ClaI and XhoI sites.
We mapped the branch points upstream of exon IIIc and identified a highly suboptimal branch point sequence and/or branch nucleotide. 5′ and 3′ splice site sequences associated with alternatively spliced exons have generally been observed to be poorer matches to the consensus sequence than those flanking constitutive exons (25, 48). The role of branch point sequences in alternative splicing is less frequently described, largely reflecting the need to experimentally define them. Compilations of mammalian branch point sequences identified a degenerate consensus of YNCURAC (or the looser YNYURAY) that defines the branch point (the underline indicates the branch nucleotide), although many of the branch point sequences in these compilations have not been confirmed experimentally (26, 45). In the majority of experimentally verified mammalian branch points, adenosine is used as the branch nucleotide and a U at the −2 position relative to the branch site is the next most conserved position in the consensus (34). In general, it appears that the most conserved sequences that comprise the branch point sequence are the three nucleotides preceding the branch nucleotide and the single nucleotide 3′ from it, with CURAC thus forming the minimal mammalian branch point consensus (45). If we analyze the exon IIIc branch point sequence by using the branched A to align the other nucleotides, none of these other nucleotides is a match to the minimal consensus (Fig. 6C). Conversely, alignment of the exon IIIc branch point sequence by using the branched G reveals a complete match at each position. Therefore, although G is the least preferred branch nucleotide, this is compensated for by an optimal match to the consensus at the flanking positions. Use of A as a secondary branch site in this case likely reflects its position within the consensus, because the nucleotide immediately preceding the primary branch nucleotide is also used for branching with varying frequency (39). We are not aware of any other example in which G has been shown to be a major branch site in a cellular gene transcript. The only case in which a G has previously been directly demonstrated to be a primary site of branching was in a herpes simplex virus latency-associated transcript (57). While the precise frequency of G as the primary branch nucleotide is unknown, previous biochemical identification of branched nucleotides from nuclear poly(A) RNA suggest that G is the least preferred nucleotide for this role in splicing (54). Infrequently, U and C have been demonstrated to be used as the branch nucleotides, and in these cases the flanking nucleotides are also a better match to the consensus sequence than most branch point sequences (3, 13, 23, 56). These observations suggest that use of non-adenosine branch sites requires a higher degree of complementary binding to U2 to enforce their use (39).
The role of the branch point sequence during spliceosome assembly commences with binding of mBBP/SF1 during formation of the ATP-independent E complex in mammals (1). In the first ATP-dependent step in spliceosome assembly, U2 base pairs with the branch point sequence and the branch nucleotide becomes bulged, which is proposed to position its 2-OH for nucleophilic attack on the 5′ splice site (34, 36, 39, 55, 60). The ability of more degenerate mammalian branch points to be recognized in E complex and to be base paired with U2 during early steps in spliceosome assembly implicates the function of other factors in stabilizing these interactions, including U2 auxiliary factor (U2AF), UAP56, and the multimeric splicing factors SF3a and SF3b (6, 19, 22, 37, 41). In mammals, recognition of the branch point sequence and polypyrimidine tract appears to be coupled such that a strong polypyrimidine tract (i.e., one of longer length and/or with greater uridine content) can compensate for a weak branch point sequence and vice versa (9, 32). In addition, U1 is also a prerequisite for productive recognition of both the polypyrimidine tract and the branch point sequence. While this role of U1 is traditionally considered as occurring across introns, models of exon definition suggest that U1 binding to the downstream 5′ splice site across the exon may also promote use of the upstream 3′ splice site (5, 8, 46).
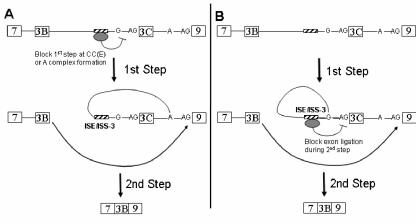
A number of examples have been described in which weak branch point sequences contribute to mechanisms of alternative splicing, mostly of viral pre-mRNAs (21, 35, 47, 51, 59). Fewer cases have been described in which weak branch points play a role in alternative splicing of cellular pre-mRNAs. The branch point upstream of the alternative 3′ splice site of exon 4 in the human calcitonin-calcitonin gene-related peptide (CT/CGRP) gene pre-mRNA contains a noncanonical U as the primary branch nucleotide, and substitution of adenosine for the uridine results in constitutive splicing to the associated 3′ splice site (2, 3, 13, 56). In the mouse and rat CT/CGRP gene, the same consensus sequence is observed in the same location upstream of exon 4 but with C as the predicted branch nucleotide (56). The identification of a weak branch point associated with exon IIIc and use of G as a primary branch nucleotide raises interesting questions regarding the mechanisms by which splicing of this exon is regulated. Compared to use of adenosine in the context of an optimal BPS, use of G as the branch nucleotide upstream of exon IIIc results in greater impairment in the second step of splicing than in the first step. Previous studies have similarly demonstrated that when branch site A residues in the context of otherwise consensus branch point sequences are replaced with C, G, or U, the first step of splicing can proceed with any of these alternative nucleotides (20, 24, 40). When C is the alternative branch site, the second step is nearly as efficient as when A is the branch nucleotide, whereas the second step shows nearly a complete block with G as the branch nucleotide and U shows an intermediate inhibition. Such studies suggest that the branch structure is involved in structural rearrangements that occur in the catalytic core between the first and second step or during the second step of splicing (38). This low efficiency of the second step of splicing using G as the branch nucleotide likely contributes to the rarity with which it has been identified as the branch nucleotide. Consistent with the suggestion that most cases of spicing regulation involve alterations in formation of early spliceosomal complexes (7), it is possible that regulatory factors in DT3 cells silence exon IIIc splicing by interfering with branch point recognition during E complex formation or by preventing U2 base pairing in complex A prior to the first step of splicing (Fig. 8A). However, given our results showing that the second step of splicing shows the greatest impairment using the weak wild-type branch site relative to a consensus BPS, it is also possible that silencing of exon IIIc can be effected during the second step of splicing (Fig. 8B). In fact, studies of splicing regulation of the Drosophila melanogaster sex-lethal (SXL) pre-mRNA have set a precedent for regulated splicing silencing during the second step of splicing (28).
FIG. 8.
Schematic illustrating two models in which the noncanonical branch point facilitates inhibition of exon IIIc inclusion in DT3 cells. In model A, a factor(s) associated with ISE/ISS-3 (and, potentially, other factors) is able to prevent the first step of splicing by preventing recognition of the exon IIIc weak branch point sequence during commitment complex (CC or E complex) formation or A complex assembly. As a result, the branch point associated with exon 9 forms a lariat structure with the 5′ end of intron 8 and exon IIIc is sequestered in the lariat intermediate. Ligation of exon IIIb (3B) to exon 9 ensues in the second step. In model B, the first step of splicing can occur using the weak exon IIIc branch nucleotide(s) to generate the lariat intermediate shown. However, ISE/ISS-3-associated factors (and possibly others as well) are able to prevent the second step of splicing using the 3′ splice site of exon IIIc. Thus, ligation of exon IIIb to exon 9 occurs instead. Note that only the branch G nucleotide upstream of exon IIIc is shown for clarity. Also omitted are other intron 8 elements that may also contribute to exon IIIc silencing and/or exon IIIb activation.
Further insight into the mechanism by which ISE/ISS-3 can function to enhance exon IIIb splicing and repress exon IIIc splicing will require identification of regulatory factors that carry out this role. Optimal activation of exon IIIb splicing clearly also requires a contribution by upstream ISEs, and the net effect of these sequences likely includes countering the activity of adjacent ISSs as well as direct recruitment of splicing factors to the exon. In the context studied here, the ability of ISE/ISS-3 to silence exon IIIc splicing clearly depends on the weak BPS, because this silencing is eliminated by a consensus BPS. Thus, we suggest that factors that bind this element may interfere with branch point recognition or block the inefficient second step of splicing that is a consequence of the weak BPS. However, a more systematic study of ISE/ISS-3-mediated silencing will be needed to establish whether or not it can only repress splicing in the context of a weak BPS. Nonetheless, it is clear that noncanonical branch point sequences can render cassette exons amenable to regulation, and it will be of interest to determine whether they are a common feature of alternatively spliced exons.
Acknowledgments
We thank Gilbert Cote and Zhi-Ming Zheng for critical review of the manuscript. We also thank Tom Cooper for providing plasmids containing troponin minigenes.
This work was supported by Public Health Service grant CA093769, Department of Defense grant PC 991539, and start-up funds from the University of Pennsylvania School of Medicine.
REFERENCES
- 1.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403-412. [DOI] [PubMed] [Google Scholar]
- 2.Adema, G. J., R. A. Bovenberg, H. S. Jansz, and P. D. Baas. 1988. Unusual branch point selection involved in splicing of the alternatively processed calcitonin/CGRP-I pre-mRNA. Nucleic Acids Res. 16:9513-9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adema, G. J., K. L. van Hulst, and P. D. Baas. 1990. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-l pre-mRNA. Nucleic Acids Res. 18:5365-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baraniak, A. P., E. L. Lasda, E. J. Wagner, and M. A. Garcia-Blanco. 2003. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 23:9327-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411-2414. [DOI] [PubMed] [Google Scholar]
- 6.Berglund, J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branch point region recognition. Genes Dev. 12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 8.Boukis, L. A., N. Liu, S. Furuyama, and J. P. Bruzik. 2004. Ser/Arg-rich Protein-mediated communication between U1 and U2 small nuclear ribonucleoprotein particles. J. Biol. Chem. 279:29647-29653. [DOI] [PubMed] [Google Scholar]
- 9.Buvoli, M., S. A. Mayer, and J. G. Patton. 1997. Functional crosstalk between exon enhancers, polypyrimidine tracts and branch point sequences. EMBO J. 16:7174-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carstens, R. P., W. L. McKeehan, and M. A. Garcia-Blanco. 1998. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol. Cell. Biol. 18:2205-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carstens, R. P., E. J. Wagner, and M. A. Garcia-Blanco. 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7388-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlet, B. N., P. Logan, G. Singh, and T. A. Cooper. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9:649-658. [DOI] [PubMed] [Google Scholar]
- 13.Cote, G. J., I. N. Nguyen, C. J. Lips, S. M. Berget, and R. F. Gagel. 1991. Validation of an in vitro RNA processing system for CT/CGRP precursor mRNA. Nucleic Acids Res. 19:3601-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Moerlooze, L., B. Spencer-Dene, J. Revest, M. Hajihosseini, I. Rosewell, and C. Dickson. 2000. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127:483-492. [DOI] [PubMed] [Google Scholar]
- 15.Del Gatto, F., A. Plet, M. C. Gesnel, C. Fort, and R. Breathnach. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Gatto-Konczak, F., C. F. Bourgeois, C. Le Guiner, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Gatto-Konczak, F., M. Olive, M. C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branch point interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 20.Freyer, G. A., J. Arenas, K. K. Perkins, H. M. Furneaux, L. Pick, B. Young, R. J. Roberts, and J. Hurwitz. 1987. In vitro formation of a lariat structure containing a G2′-5′G linkage. J. Biol. Chem. 262:4267-4273. [PubMed] [Google Scholar]
- 21.Fu, X. D., R. A. Katz, A. M. Skalka, and T. Maniatis. 1991. The role of branch point and 3′-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 5:211-220. [DOI] [PubMed] [Google Scholar]
- 22.Gozani, O., R. Feld, and R. Reed. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 23.Hartmuth, K., and A. Barta. 1988. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol. Cell. Biol. 8:2011-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornig, H., M. Aebi, and C. Weissmann. 1986. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature 324:589-591. [DOI] [PubMed] [Google Scholar]
- 25.Itoh, H., T. Washio, and M. Tomita. 2004. Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes. RNA 10:1005-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller, E. B., and W. A. Noon. 1984. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc. Natl. Acad. Sci. USA 81:7417-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladd, A. N., N. Charlet, and T. A. Cooper. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lallena, M. J., K. J. Chalmers, S. Llamazares, A. I. Lamond, and J. Valcarcel. 2002. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109:285-296. [DOI] [PubMed] [Google Scholar]
- 29.Le Guiner, C., A. Plet, D. Galiana, M. C. Gesnel, F. Del Gatto-Konczak, and R. Breathnach. 2001. Polypyrimidine tract-binding protein represses splicing of a fibroblast growth factor receptor-2 gene alternative exon through exon sequences. J. Biol. Chem. 276:43677-43687. [DOI] [PubMed] [Google Scholar]
- 30.Lerga, A., M. Hallier, L. Delva, C. Orvain, I. Gallais, J. Marie, and F. Moreau-Gachelin. 2001. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 276:6807-6816. [DOI] [PubMed] [Google Scholar]
- 31.Li, M., and P. H. Pritchard. 2000. Characterization of the effects of mutations in the putative branch point sequence of intron 4 on the splicing within the human lecithin:cholesterol acyltransferase gene. J. Biol. Chem. 275:18079-18084. [DOI] [PubMed] [Google Scholar]
- 32.Lund, M., T. O. Tange, H. Dyhr-Mikkelsen, J. Hansen, and J. Kjems. 2000. Characterization of human RNA splice signals by iterative functional selection of splice sites. RNA 6:528-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muh, S. J., R. H. Hovhannisyan, and R. P. Carstens. 2002. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2). J. Biol. Chem. 277:50143-50154. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, K. K., and M. R. Green. 1989. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 3:1562-1571. [DOI] [PubMed] [Google Scholar]
- 35.Noble, J. C., C. Prives, and J. L. Manley. 1988. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 2:1460-1475. [DOI] [PubMed] [Google Scholar]
- 36.Pascolo, E., and B. Seraphin. 1997. The branch point residue is recognized during commitment complex formation before being bulged out of the U2 snRNA-pre-mRNA duplex. Mol. Cell. Biol. 17:3469-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peled-Zehavi, H., J. A. Berglund, M. Rosbash, and A. D. Frankel. 2001. Recognition of RNA branch point sequences by the KH domain of splicing factor 1 (mammalian branch point binding protein) in a splicing factor complex. Mol. Cell. Biol. 21:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Query, C. C., and M. M. Konarska. 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell 14:343-354. [DOI] [PubMed] [Google Scholar]
- 39.Query, C. C., M. J. Moore, and P. A. Sharp. 1994. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 8:587-597. [DOI] [PubMed] [Google Scholar]
- 40.Query, C. C., S. A. Strobel, and P. A. Sharp. 1996. Three recognition events at the branch-site adenine. EMBO J. 15:1392-1402. [PMC free article] [PubMed] [Google Scholar]
- 41.Rain, J. C., Z. Rafi, Z. Rhani, P. Legrain, and A. Kramer. 1998. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA 4:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed, R., and T. Maniatis. 1988. The role of the mammalian branch point sequence in pre-mRNA splicing. Genes Dev. 2:1268-1276. [DOI] [PubMed] [Google Scholar]
- 43.Ruskin, B., and M. R. Green. 1990. RNA lariat debranching enzyme as tool for analyzing RNA structure. Methods Enzymol. 181:180-188. [DOI] [PubMed] [Google Scholar]
- 44.Ryan, K. J., and T. A. Cooper. 1996. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol. Cell. Biol. 16:4014-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senapathy, P., M. B. Shapiro, and N. L. Harris. 1990. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 183:252-278. [DOI] [PubMed] [Google Scholar]
- 46.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 47.Staffa, A., and A. Cochrane. 1994. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 68:3071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamm, S., J. Zhu, K. Nakai, P. Stoilov, O. Stoss, and M. Q. Zhang. 2000. An alternative-exon database and its statistical analysis. DNA Cell Biol. 19:739-756. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki, H., Y. Jin, H. Otani, K. Yasuda, and K. Inoue. 2002. Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells 7:133-141. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi, N., N. Sasagawa, K. Suzuki, and S. Ishiura. 2000. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem. Biophys. Res. Commun. 277:518-523. [DOI] [PubMed] [Google Scholar]
- 51.Tange, T. O., C. K. Damgaard, S. Guth, J. Valcarcel, and J. Kjems. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 20:5748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ule, J., K. B. Jensen, M. Ruggiu, A. Mele, A. Ule, and R. B. Darnell. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302:1212-1215. [DOI] [PubMed] [Google Scholar]
- 53.Wagner, E. J., and M. A. Garcia-Blanco. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10:943-949. [DOI] [PubMed] [Google Scholar]
- 54.Wallace, J. C., and M. Edmonds. 1983. Polyadenylylated nuclear RNA contains branches. Proc. Natl. Acad. Sci. USA 80:950-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, J., and J. L. Manley. 1989. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 3:1553-1561. [DOI] [PubMed] [Google Scholar]
- 56.Yeakley, J. M., F. Hedjran, J. P. Morfin, N. Merillat, M. G. Rosenfeld, and R. B. Emeson. 1993. Control of calcitonin/calcitonin gene-related peptide pre-mRNA processing by constitutive intron and exon elements. Mol. Cell. Biol. 13:5999-6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zabolotny, J. M., C. Krummenacher, and N. W. Fraser. 1997. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J. Virol. 71:4199-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, W., H. Liu, K. Han, and P. J. Grabowski. 2002. Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR. RNA 8:671-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng, Z. M., P. He, and C. C. Baker. 1996. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J. Virol. 70:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhuang, Y., and A. M. Weiner. 1989. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 3:1545-1552. [DOI] [PubMed] [Google Scholar]
- 61.Zhuang, Y. A., A. M. Goldstein, and A. M. Weiner. 1989. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc. Natl. Acad. Sci. USA 86:2752-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]