Abstract
Introduction
Chronic obstructive pulmonary disease (COPD) is a significant cause of morbidity in persons living with HIV (PLWH) and HIV appears to uniquely cause COPD, independent of smoking. The mechanisms by which HIV leads to COPD are not clear. The objective of this study was to identify metabolomic biomarkers and potential mechanistic pathways of HIV-associated COPD (HIV-COPD).
Methods
We performed case–control metabolite profiling via mass spectrometry in plasma from 38 individuals with HIV-COPD (cases), comparing to matched controls with/without HIV and with/without COPD. Untargeted metabolites of interest were identified with liquid chromatography with mass spectrometry (LC-MS/mass spectrometry (MS)), and targeted metabolomics for tryptophan (Trp) and kynurenine (Kyn) were measured by selective reaction monitoring (SRM) with LC-MS/MS. We used mixed-effects models to compare metabolite concentrations in cases compared with controls while controlling for relevant biological variables.
Results
We identified 1689 analytes associated with HIV-COPD at a false discovery rate (FDR) of 10%. In PLWH, we identified 263 analytes (10% FDR) between those with and without COPD. LC MS/MS identified Trp and 17 lipids, including sphingolipids and diacylglycerol. After adjusting for relevant covariates, the Kyn/Trp ratio measured by SRM was significantly higher in PLWH (p=0.022), but was not associated with COPD status (p=0.95).
Conclusions
There is a unique metabolite profile in HIV-COPD that includes sphingolipids. Trp metabolism is increased in HIV, but does not appear to independently contribute to HIV-COPD.
Trial registration numbers
Keywords: COPD ÀÜ Mechanisms, Immunodeficiency, COPD Pathology
Key messages.
HIV-associated chronic obstructive pulmonary disease (HIV-COPD) has a unique plasma metabolomic profile that includes sphingolipids and fatty acids.
Tryptophan catabolism is increased in PLWH, but does not correlate with COPD status.
Additional studies are needed to determine how such metabolic pathways contribute to HIV-COPD and whether therapeutic interventions to alter sphingolipid expression can reduce the risk of COPD in persons living with HIV.
Introduction
Combination antiretroviral therapy (ART) has markedly improved the survival of individuals living with HIV-1. However, improved survival has led to a higher prevalence of several chronic illnesses including chronic obstructive pulmonary disease (COPD), which affects an estimated 3–23% of persons living with HIV (PLWH).1–8 Although the prevalence of smoking is high in PLWH, observational studies suggest that HIV infection is an independent risk factor for accelerated lung function decline and subsequent COPD.1 2 5 In the era prior to the common use of highly active ART (HAART), pulmonary obstruction was mostly associated with advanced HIV/AIDS and frequent pulmonary infections.9 In the HAART era, COPD persists as a frequent comorbidity in PLWH even in the absence of AIDS or frequent pulmonary infections.1–5 8 10–14 It is highly likely that several factors are involved in HIV-associated COPD (HIV-COPD) and many investigators have implicated the HIV virus itself. Currently, the underlying mechanisms of HIV-COPD remain unclear and there is no biomarker to identify which patients with HIV will be at a higher risk for the subsequent development of COPD. For this study, we identified individuals in the Strategic Timing of Antiretroviral Treatment (START) study with HIV-COPD prior to the diagnosis of AIDS or initiation of HAART, offering a unique opportunity to characterise HIV-COPD.15
Recent developments in metabolomics and metabolic profiling have enabled the measurements of thousands of metabolites simultaneously in biological samples leading to new discoveries of biomarkers of disease.16 Metabolomics, the study of small molecules, is a field that complements genomics and proteomics to provide a snapshot into the physiology of human disease. As such, global, untargeted metabolomic profiling is a method for analysing complex diseases to understand the underlying pathways involved in disease pathogenesis and to identify biomarker candidates for disease. In this study, the primary aim was to perform large-scale untargeted analysis with mass spectrometry of plasma to identify metabolites associated with HIV-COPD. Our results revealed that HIV-COPD has a unique metabolomic signature and among the metabolome are sphingolipids.
Methods
We performed a cross-sectional, matched case–control study using plasma samples from two large cohort studies.
Study population
Cases were HIV positive and were selected from a pulmonary substudy of the START trial.15 17 START enrolled HIV-positive, ART-naive persons with a CD4+ count >500 cells/mm3. From this cohort, we selected 38 participants with HIV-COPD (defined as forced expiratory volume in 1 s (FEV1)/forced vital capacity (FVC)< lower limits of normal and FEV1<80% predicted at baseline pulmonary function testing, table 1) for metabolomic profiling. HIV-positive controls consisted of 40 individuals with normal lung function (defined as FEV1/FVC>lower limits of normal and FEV1>80% predicted) matched on age, sex, region and smoking status. HIV-negative controls were identified in the Genetic Epidemiology of Chronic Obstructive Pulmonary Disease (COPDGene) study, an observational study that enrolled current or former smokers. COPDGene controls were matched on lung function, age, sex and race. These consisted of 17 individuals without COPD and 20 individuals with COPD (table 1). This study was approved by the University of Minnesota Institutional Review Board.
Table 1.
Characteristics of cases and controls
| HIV(±) COPD(±) (n=38) | HIV(±) COPD(−) (n=40) | HIV(−) COPD(±) (n=20) | HIV(−) COPD(−) (n=17) | |
|---|---|---|---|---|
| Age, years | 38.97±9.21* | 38.93±7.78* | 48.18±3.67* | 55.91±5.48 |
| Sex | 27M/11F | 29M/11F | 18M/2F | 15M/2F |
| FEV1pp | 70.95±11.6* | 94.90±15.6* | 73.08±12.8* | 109.6±13.2 |
| FEV1/FVC | 0.663±0.0704* | 0.815±0.0476 | 0.618±0.065* | 0.818±0.0341 |
| HIV RNA log10, copies/mL | 3.95±1.04 | 3.86±0.994 | NA | NA |
| CD4+ T-cell count, cells/mm3 | 707.03±198.89 | 716.26±182.84 | NA | NA |
| CD8+ T-cell count, cells/mm3 | 1129.68±596.74 | 1142.86±912.3 | NA | NA |
| CD4/CD8 ratio | 0.793±0.395 | 0.936±0.722 | NA | NA |
| Smoking status† | 16/3/19 | 16/4/20 | 16/4/0 | 0/0/17 |
| Race‡ | 15/22/1 | 16/22/2 | 15/5/0 | 15/2/0 |
| Region§ | 6/13/17/2 | 8/13/17/2 | 20/0/0/0 | 17/0/0/0 |
*Significant from COPD(−) HIV(−) using Student's t-test.
†Smoking status (current/former/never).
‡Race (Caucasian/black/Latino).
§Region (North America/Europe/Africa/South America).
COPD, chronic obstructive pulmonary disease; FEV1pp, forced expiratory volume in 1 s, per cent predicted; FVC, forced vital capacity; NA, not available.
Solvents and reagents
The acetonitrile (ACN) used in this study was of high pressure liquid chromatography (HPLC) grade and purchased from Fisher Scientific (Pittsburg, Pennsylvania, USA). HPLC grade methanol (MeOH) was acquired from Sigma-Aldrich (St Louis, Missouri, USA) and analytical grade formic acid from Fluka (Sigma, St Louis, Missouri, USA). The mass spectrometer lock-spray solution, leucine encephaline, was purchased from Waters (Milford, Massachusetts, USA). Ultra high purity bottled water was procured from Invitrogen (Grand Island, New York, USA).
Sample preparation:
Aliquots of 100 µL of plasma had a heavy standard of 3 µL of 100 µM of kynurenine (Kyn)-D6 and 3 µL of 1 mM of tryptophan (Trp)-C1113 (Cambridge Isotope Laboratories, Tewksbury, Massachusetts, USA) added in prior to any preparation. The aliquots of plasma were then mixed with 400 µL of ice-cold solvent (100% MeOH), vortexed and placed on ice for 10 min. Samples were then centrifuged at 13 000×g for 10 min at 4°C, and the supernatant was removed and transferred into a clean low-retention phial. This step was repeated once. Samples were concentrated using a vacuum centrifuge to ∼50 µL. Formic acid was used to acidify the plasma samples which were added to the starting buffer used in ultraperformance liquid chromatography (5% ACN, 95% water, 0.1% formic acid) to ∼100 µL.
MS analysis
For analyte profiling and quantitation, 10 µL of the undiluted sample was injected into Thermo Q-exactive LC-MS (ThermoFisher Scientific, Marietta, Ohio, USA) with Acquity ultra performance liquid chromatography (UPLC) C18 2.1×100 mm, 1.7 µm column (Waters, Milford, Massachusetts, USA) at 40˚C. All of the samples were subjected to a gradient going from buffer A to B over 15 min then flushing for 5 min. Buffer A consisted of 99.9% water with 0.1% formic acid, while buffer B was 99.9% ACN with 0.1% formic acid. To avoid batch effects, all samples were run in one continuous MS run. For quality control, internal standards consisted of blank samples spiked with four metabolites (phenylalanine-D6, hippuric acid-D5, Kyn-D6 and Trp-C11) and were placed at random intervals in the MS run. For quantitation, the individual samples were spiked with the same four metabolites for normalisation of the peak intensities. For metabolite identification, two samples from each group (±COPD, ±HIV) were run on the Thermo Q-exactive LC-MS in MS/MS data-independent acquisition mode with a 25 min of gradient A to B followed by 5 min of flushing.
MS data processing
The .RAW files from the Q-Exactive were converted into mzXML using MSconvert (http://proteowizard.sourceforge.net/tools.shtml), and processed using the XCMS algorithm as implemented in the xcms package for R using the recommended pipeline (the symmetric family was used for retention time (RT) correction). Data from each sample were normalised to the mean of the heavy standards we introduced for Trp (mass/charge (m/z)=215.1231 and RT=1.841226) and Kyn (m/z=216.1318 and RT=1.941770), so that on the log scale the means of these two analytes were zero for all individuals.
LC-MS/MS analysis for analyte identification
The .RAW files from Q-exactive LC-MS/MS were analysed using Thermo Xcalibur Qual Browser for fragmentation data of the analytes of interest. These analytes were matched to fragmentation spectra based on m/z and RT matching between LC-MS and LC-MS/MS. The fragmentation data for all of these analytes were compared and matched to metabolites via Metlin's MS/MS metabolite database within a 12 ppm difference.
LC-MS/MS selective reaction monitoring analysis of Trp and Kyn
Diluted samples (20 µL, diluted 1:1000 for Trp and 1:100 for Kyn) were subjected to injection using an Agilent autosampler LC-MS/MS with an analytical Waters Symmetry C18, 3.5 µm column connected to the 5500 iontrap (Sciex, Framingham, Massachusetts, USA) fitted with a turbo V electrospray source. The samples were subjected to a linear gradient of 2% ACN, 0.1% formic acid to 98% ACN 0.1% formic acid for 10 min at a column flow rate of 250 µL/min. Transitions monitored are in online supplementary table 1S. The data were analysed using MultiQuant (Applied Biosystems, Foster City, California, USA) providing the peak area for the transitions. A standard curve was constructed using concentration ratios of Trp/Trp and Kyn /Kyn from fentomole to nanomole in 20 µL.
bmjresp-2017-000180supp_table1.pdf (34.7KB, pdf)
Statistics
To test for differences in metabolite concentrations, mixed-effects models were used that included fixed effects for HIV status, COPD status, the interaction between HIV and COPD status, FEV1 per cent predicted, race and smoking status and a random effect for match group. All matched groups were matched on age and sex. For analysis of individual metabolites, a logarithmic transformation was employed prior to model fitting, whereas the component of the targeted analysis which focused on the ratio of Kyn to Trp did not employ such a transformation. Otherwise, the same approach was employed for the analysis of the metabolites identified in the untargeted analysis as for the targeted analysis, except that we report an unadjusted p value for the targeted analysis, whereas we use the q-value approach to control the false discovery rate (FDR) at 10% for the untargeted analysis. V.3.2.3 of the statistical software package R was used for all statistical analyses.
Analyte bioinformatics
MS/MS spectra .Raw files were observed using Thermo Xcalibur Qual Browser. MS/MS spectra for each analyte of interest were matched to metabolite spectra through the Metlin metabolite database from Scripps (https://metlin.scripps.edu/index.php). A degree of restraint, within 10% of the RT, was used when matching to feasible options based on RT of the analyte. Analytes illuted in the first 30s and after 15 min were eliminated.
Results
Study participant characteristics
Cases consisted of 38 individuals with HIV-COPD (table 1). HIV(+) controls were 40 individuals with HIV, but without COPD, matched on age, sex, smoking status, region and race. Baseline plasma samples were drawn prior to ART initiation or any AIDS-defining illness. All had CD4+ T-cell counts >500 cells/mm3 and there were no significant differences in plasma HIV RNA, CD4+ and CD8+ T-cell counts between HIV-positive cases and controls. The HIV(−) controls were significantly older than the HIV-positive participants.
Metabolomics analyte profiles
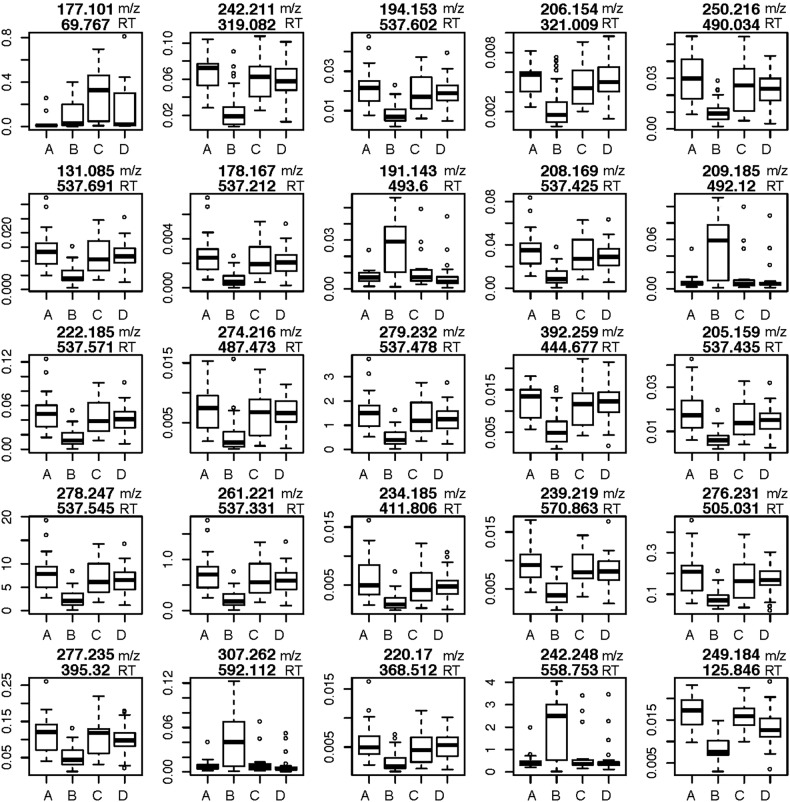
MS analysis detected 1689 plasma analyte signals, defined as a discreet m/z and RT, that correlated with a yet unknown metabolite. Since there is no universally accepted methodology to quantify analytes detected by mass spectrometry, we chose relative abundance as the sum of all peak intensities detected by the mass spectrometer that associated with the given analyte. Using a mixed-effects model with fixed effects for COPD status, HIV status, their interaction, FEV1 per cent predicted, race, sex and smoking status, we identified 880 analytes at 10% FDR that were attributable to the main effect of HIV status, that is, differences due to HIV averaging over COPD status; we identified no analytes that differed at 10% FDR for the main effect of COPD. Tests of the statistical interaction between HIV status and COPD status identified 263 analytes that displayed differences of interest (ie, a q-value <0.1). Of these 263 analytes, 232 (88%) also had significant main effects for HIV status. On examination of the estimated regression coefficients for HIV status and the interaction between HIV and COPD status of the 263 analytes, it was found that differences between HIV(+) and HIV(−) participants are, on average, smaller in COPD(+) participants compared with COPD(−) participants. This is reflected in figure 1 that displays the raw data for 25 analytes with the lowest q-values that met our FDR criterion for the interaction term. After eliminating isotopes and contaminants, 172 of the original 263 analytes differed based on HIV and COPD status and therefore qualified for further analysis described next.
Figure 1.
The top 25 analytes by relevant q-value differentially expressed in HIV with COPD. The top number is m/z and the bottom number is RT. Y-axis is intensity. (A) HIV− COPD−, (B) HIV+ COPD−, (C) HIV− COPD+, (D) HIV+ COPD+. COPD, chronic obstructive pulmonary disease; m/z, mass/charge; RT, retention time.
Metabolite identification
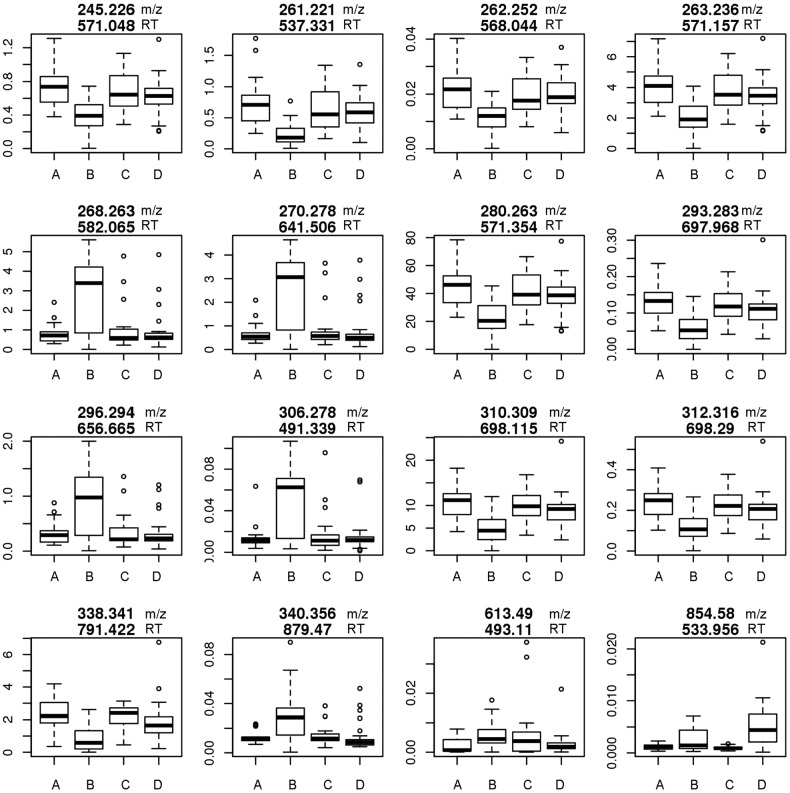
To identify some of the significant analytes of interest, we performed LC-MS/MS targeting the 172 analytes we identified that differed in HIV-COPD. We were able to attribute 17 of these analyte spectra to lipids, including sphingolipids, diacylglycerol and fatty acids (table 2, figure 2, online supplementary table 2S). Ceramide (m/z 560.50948) concentrations were higher in HIV-COPD compared with +HIV/normal lung function, but did not quite meet our FDR threshold (q-value=0.11). Diacylglycerol was lower in HIV-COPD compared with +HIV/normal lung function and similar to those without HIV. We identified three lipids as fatty acids and one as a phospholipid. Generally, the fatty acids were higher in the HIV-COPD persons compared with +HIV/normal lung function, whereas the different sphingolipids demonstrated both increased and decreased concentrations.
Table 2.
Metabolites identified by LC-MS/MS
| m/z | RT (s) | Adduct | Accurate mass | Formula | Tentative identification | q-Value |
|---|---|---|---|---|---|---|
| 245.22577 | 571.05 | M+H-H2O | 262.2297 | C18H30O | Fatty acid | 0.035141 |
| 261.22061 | 537.33 | M+H-H2O | 278.2246 | C16H32O2 | Fatty acid | 0.002931 |
| 262.25198 | 568.04 | M+H-H2O | 279.2562 | C18H33NO | Linoleamide | 0.031476 |
| 263.23627 | 571.16 | M+H | 262.2297 | C18H32O2 | Fatty acid | 0.023163 |
| 268.26277 | 582.06 | M+H-H2O | 268.2640 | C17H35NO2 | Sphingolipid | 0.014423 |
| 270.27842 | 641.51 | M+H-H2O | 287.2824 | C17H37NO2 | Sphingolipid | 0.010165 |
| 280.26257 | 571.35 | M+H-H2O | 297.2668 | C18H33NO | Sphingolipid | 0.046177 |
| 293.28267 | 697.97 | M+Na | 270.2923 | C18H38O | Octadecanol | 0.024386 |
| 296.29396 | 656.67 | M+H-H2O | 313.2981 | C19H39NO2 | Sphingolipid | 0.021558 |
| 306.27791 | 491.34 | M+Na | 283.2875 | C18H37NO | Stearamide | 0.002926 |
| 310.30938 | 698.11 | M+H-H2O | 327.3137 | C20H41NO2 | Sphingolipid | 0.021558 |
| 312.31609 | 698.29 | M+H-H2O | 329.3197 | C20H43NO2 | Sphingolipid | 0.021623 |
| 338.34061 | 791.42 | M+H | 337.3345 | C22H43NO | Docosenamide | 0.005362 |
| 340.35618 | 879.47 | M+H | 339.3501 | C22H45NO | Docosenamide | 0.027448 |
| 560.50948 | 568.79 | M+H-H2O | 577.5070 | C36H67NO4 | Ceramide | 0.113237 |
| 613.49004 | 493.11 | M+H | 612.4754 | C39H65O5 | Diacylglycerol | 0.014779 |
| 854.57988 | 533.96 | M+Na | 831.5989 | C45H86NO10P | Phospholipid | 0.006038 |
m/z, Mass to charge ratio of metabolite; RT, retention time (s) in mass spectrometry column.
Figure 2.
Lipids differentially expressed in HIV. The top number is m/z and the bottom number is RT. Y-axis is intensity. (A) HIV− COPD−, (B) HIV+ COPD−, (C) HIV− COPD+, (D) HIV+ COPD+. COPD, chronic obstructive pulmonary disease; m/z, mass/charge; RT, retention time.
bmjresp-2017-000180supp_table2.pdf (105.7KB, pdf)
Targeted Trp and Kyn concentrations
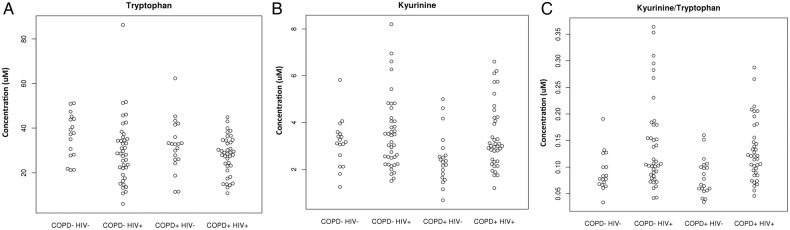
One of the analytes differentially expressed in HIV+ participants was consistent with the essential amino acid Trp (m/z 205.097). Since Trp catabolism has been associated with both HIV infection and COPD,18–20 and standards are readily available, we sought to quantify Trp and its major metabolite, Kyn (m/z 209.094) by performing targeted selective reaction monitoring (SRM; figure 3). Indoleamine-2,3-dioxygenase (IDO) is the main inducible and rate-limiting enzyme involved in Trp catabolism, and IDO activity is expressed as the Kyn/Trp ratio. We found that Trp was lower in HIV(+) individuals compared with HIV(−) participants as previously reported,21–23 although this was not significant after adjustment for relevant covariates (figure 3A, p=0.0681). Trp was also lower in individuals with COPD, but this was not statistically significant (figure 3A, p=0.39). Kyn was higher in HIV-positive participants but was not statistically significant after adjustment for relevant covariates (figure 3B, p=0.5586), and was lower in COPD participants, albeit not significantly (figure 3B, p=0.1888). As a reflection of IDO activity, the Kyn/Trp ratio was significantly higher in HIV (figure 3C, p=0.022), but there was no difference due to COPD status (p=0.95).
Figure 3.
(A) Plasma tryptophan concentrations. (B) Plasma kynurenine concentrations. (C) IDO activity as reflected by kynurenine/tryptophan ratio. COPD, chronic obstructive pulmonary disease; IDO, indoleamine-2,3-dioxygenase.
Discussion
A major gap in knowledge is the limited understanding of why PLWH are more susceptible to developing COPD independent of smoking status.1 2 5 10 Currently, there is no biomarker to identify risk or lend insight into the mechanisms of developing rapid decline in lung function and subsequent COPD in PLWH. The pulmonary substudy of START offers a unique opportunity to identify biomarkers of HIV-COPD prior to the development of an AIDS-defining illness. This is important since pulmonary infection itself can lead to a loss of lung function.24 25 In this exploratory study using an untargeted metabolomic approach, we identified 172 unique plasma analytes that associated with HIV-COPD compared with participants with HIV without COPD after controlling for multiple confounders such as smoking status, race and sex.
We performed LC-MS/MS to identify metabolites of interest and found that 17 of these metabolites were lipids. Among these lipids were diacylglycerol and several sphingolipids. Sphingolipids serve as a structural component of the plasma membrane lipid bilayer and also participate in cell recognition and signalling. In individuals with COPD, increased concentrations of sphingolipids have been measured in sputum. Telenga et al26 identified 168 sphingolipids and 36 phosphatidylethanolamine lipids in the sputum that were significantly higher in smokers with COPD compared with smokers without COPD. These lipids correlated with reduced lung function, sputum inflammation and smoking history. In our study, we found a complex expression of sphingolipids with both elevated and decreased plasma concentrations of various sphingolipids. This complex plasma expression of sphingolipids was observed by Bowler et al, where they used both targeted and untargeted metabolomic platforms that identified 23 sphingolipids common to both platforms in COPD. Some of these lipids correlated to COPD phenotype with five associating with emphysema and seven associating with COPD exacerbations.26 27 In addition, sphingolipid expression was complex depending on lipid type and COPD phenotype. Our data support these findings in individuals with HIV, as we found that sphingolipids concentrations were altered in the setting of HIV-COPD when controlling for smoking status.
Among the lipids, we also identified ceramide. Although ceramide concentrations were higher in HIV-COPD, in this relatively small exploratory study, the difference did not quite meet statistical significance (q-value=0.11). Ceramide consists of a sphingolipid backbone (sphingosine) plus a fatty acid. It has been implicated in the pathogenesis of COPD. Petrache et al28 found that ceramide acts as a second messenger and a crucial mediator of apoptosis and thus alveolar destruction in a murine model of emphysema. In human studies, elevated concentrations of ceramide have been found in sputum and lung tissue.26 28 29 Bowler et al27 further characterised plasma lipids in various COPD phenotypes and found that ceramides had a negative association with emphysema as measured by quantitative high-resolution CT (HRCT); however, trihexosylceramides had a positive association with frequent exacerbators. We also found an elevation of diacylglycerol, a protein kinase C activator that cooperates with ceramide in inducing apoptosis,30 but we did not have additional studies, such as HRCT, for further phenotyping.
Lipid dysregulation is a known complication of AIDS and is a well-known side effect of protease inhibitors. Chetwynd et al31 found that ART resulted in reductions in certain urinary metabolites, including sphingamines. Our study is significant as all of the START participants were ART-naïve and prior to the onset of AIDS. Therefore, the differences in lipid profiles between those with and without COPD cannot be attributed to ART. In addition, lipid abnormalities have been linked to markers of inflammation in HIV compared with healthy controls; however, all of the participants were on HAART and it is unclear whether the lipid abnormalities are due to HIV infection versus ART.32 Pertinent to our study, Scarpelini et al33 identified metabolites that associated with rapid progression of HIV infection and these included sphingomyelin metabolism. It is possible that lipid dysregulation in HIV infection is a marker of worsened disease, including the development of COPD.
In addition to lipids, we identified an analyte consistent with Trp that trended towards an association with HIV-COPD in our metabolite profiling. Trp and its main catabolic enzyme, IDO, have been associated with both COPD and HIV infection.18 20 Therefore, we used targeted SRM to measure Trp and its major metabolite, Kyn, to determine if the Trp catabolism is a biomarker for HIV-COPD. Consistent with previous reports, we found decreased Trp and increased Kyn concentrations in individuals with HIV. As a reflection of IDO activity, the Kyn/Trp ratio was significantly elevated in the setting of HIV, but not in COPD participants. Although not statistically significant, we did find that Kyn concentrations were higher than one would predict in those with both COPD and HIV (data not shown). One possibility is that the effects of HIV on IDO activation are much larger than those of COPD, therefore requiring a larger cohort to determine if IDO activation plays a role in HIV-COPD.
There are several limitations to this study. While HIV-positive controls were matched on age, sex, race, region and smoking status to our cases, our non-HIV controls were difficult to match on age and race as our HIV-COPD cases were much younger and over half were black. In this study, we found that metabolites associated with HIV-COPD were not associated with COPD in HIV-negative controls. This may be due to confounders, such as age and race; however, COPD in HIV-negative individuals is most likely a very different phenotype and disease. Therefore, diagnosis and treatment may need to be very different for HIV-COPD and these metabolites have the potential to identify those with HIV who are at risk of developing COPD. This study was relatively small and may have been underpowered to determine all significant biomarkers and metabolomics pathways of HIV-COPD. Future studies would benefit from larger sample sizes and longitudinal study designs such as that recently completed and now possible in the START Pulmonary Substudy.
Conclusion
HIV-COPD has a unique plasma metabolomic profile that includes sphingolipids and fatty acids. Additional studies are needed to determine how such metabolic pathways contribute to HIV-COPD and whether therapeutic interventions to alter sphingolipid expression can reduce the risk of COPD in PLWH.
Acknowledgments
The authors wish to thank the START participants and investigators. A full list of START investigators can be found in N Engl J Med 2015;373:795–807 and a full list of START Pulmonary Substudy investigators can be found in Lancet Respir Med 2016;4(12):980–989.
Footnotes
Contributors: SHo, TJG, CR, SHa, BAW, BJS, KMK and CHW provided substantial contributions to the conception and design of the work, and the acquisition, analysis and interpretation of data. SHo, CR, SHa, BAW, BJS, KMK and CHW were involved in drafting the work or revising it critically for important intellectual content. SHo, TJG, CR, SHa, BAW, BJS, KMK and CHW were involved in final approval of the version published. SHo, TJG, CR, SHa, BAW, BJS, KMK and CHW were involved in the agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding: The project described was supported by the University of Minnesota Developmental Center for AIDS Research (Research Award). START and its Pulmonary Substudy were funded by the National Heart, Lung and Blood Institute (USA, R01 HL096453), the National Institute of Allergy and Infectious Diseases (USA, UM1AI068641 and UM1AI120197), Agence Nationale de Recherches sur le SIDA et les Hipatites Virales (France), National Health and Medical Research Council (Australia), National Research Foundation (Denmark), Bundes ministerium f|r Bildung und Forschung (Germany), European AIDS Treatment Network, Medical Research Council (UK), National Institute for Health Research, National Health Service (UK), and the University of Minnesota. Antiretroviral drugs were donated to the START central drug repository by AbbVie, Bristol-Myers Squibb, Gilead Sciences, GlaxoSmithKline/ViiV Healthcare, Janssen Scientific Affairs and Merck. COPDGene was supported by the National Heart, Lung and Blood Institute (USA, R01HL089897 and R01HL089856) and the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, Sunovion, and GlaxoSmithKline.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Department of Veterans Affairs, the US Government, other funding agencies or any of the authors' affiliated institutions or organisations.
Competing interests: None declared.
Ethics approval: University of Minnesota Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1.Drummond MB, Merlo CA, Astemborski J et al. . The effect of HIV infection on longitudinal lung function decline among injection drug users: a prospective cohort. AIDS 2013;27:1303–11. doi:10.1097/QAD.0b013e32835e395d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.George MP, Kannass M, Huang L et al. . Respiratory symptoms and airway obstruction in HIV-infected subjects in the HAART era. PLoS ONE 2009;4:e6328 doi:10.1371/journal.pone.0006328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gingo MR, George MP, Kessinger CJ et al. . Pulmonary function abnormalities in HIV-infected patients during the current antiretroviral therapy era. Am J Respir Crit Care Med 2010;182:790–6. doi:10.1164/rccm.200912-1858OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirani A, Cavallazzi R, Vasu T et al. . Prevalence of obstructive lung disease in HIV population: a cross sectional study. Respir Med 2011;105:1655–61. doi:10.1016/j.rmed.2011.05.009 [DOI] [PubMed] [Google Scholar]
- 5.Crothers K, Huang L, Goulet JL et al. . HIV infection and risk for incident pulmonary diseases in the combination antiretroviral therapy era. Am J Respir Crit Care Med 2011;183:388–95. doi:10.1164/rccm.201006-0836OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui Q, Carruthers S, McIvor A et al. . Effect of smoking on lung function, respiratory symptoms and respiratory diseases amongst HIV-positive subjects: a cross-sectional study. AIDS Res Ther 2010;7:6 doi:10.1186/1742-6405-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madeddu G, Fois AG, Calia GM et al. . Chronic obstructive pulmonary disease: an emerging comorbidity in HIV-infected patients in the HAART era? Infection 2013;41:347–53. doi:10.1007/s15010-012-0330-x [DOI] [PubMed] [Google Scholar]
- 8.Kristoffersen US, Lebech AM, Mortensen J et al. . Changes in lung function of HIV-infected patients: a 4.5-year follow-up study. Clin Physiol Funct Imaging 2012;32:288–95. doi:10.1111/j.1475-097X.2012.01124.x [DOI] [PubMed] [Google Scholar]
- 9.Raynaud C, Roche N, Chouaid C. Interactions between HIV infection and chronic obstructive pulmonary disease: Clinical and epidemiological aspects. Respir Res 2011;12:117 doi:10.1186/1465-9921-12-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crothers K, Butt AA, Gibert CL et al. . Increased COPD among HIV-positive compared to HIV-negative veterans. Chest 2006;130:1326–33. doi:10.1378/chest.130.5.1326 [DOI] [PubMed] [Google Scholar]
- 11.Diaz PT, King MA, Pacht ER et al. . Increased susceptibility to pulmonary emphysema among HIV-seropositive smokers. Ann Intern Med 2000;132:369–72. doi:10.7326/0003-4819-132-5-200003070-00006 [DOI] [PubMed] [Google Scholar]
- 12.Drummond MB, Kirk GD, McCormack MC et al. . HIV and COPD: impact of risk behaviors and diseases on quality of life. Qual Life Res 2010;19:1295–302. doi:10.1007/s11136-010-9701-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drummond MB, Kirk GD, Ricketts EP et al. . Cross sectional analysis of respiratory symptoms in an injection drug user cohort: the impact of obstructive lung disease and HIV. BMC Pulm Med 2010;10:27 doi:10.1186/1471-2466-10-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzpatrick ME, Singh V, Bertolet M et al. . Relationships of pulmonary function, inflammation, and T-cell activation and senescence in an HIV-infected cohort. AIDS 2014;28:2505–15. doi:10.1097/QAD.0000000000000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunisaki KM, Niewoehner DE, Collins G et al. . Pulmonary effects of immediate versus deferred antiretroviral therapy in HIV-positive individuals: a nested substudy within the multicentre, international, randomised, controlled Strategic Timing of Antiretroviral Treatment (START) trial. Lancet Respir Med 2016;4:980–9. doi:10.1016/S2213-2600(16)30319-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suhre K, Shin SY, Petersen AK et al. . Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011;477:54–60. doi:10.1038/nature10354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunisaki KM, Niewoehner DE, Collins G et al. . Pulmonary function in an international sample of HIV-positive, treatment-naive adults with CD4 counts >500 cells/muL: a substudy of the INSIGHT Strategic Timing of AntiRetroviral Treatment (START) trial. HIV Med 2015;16(Suppl 1):119–28. doi:10.1111/hiv.12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Favre D, Mold J, Hunt PW et al. . Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med 2010;2:32ra36 doi:10.1126/scitranslmed.3000632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maneechotesuwan K, Kasetsinsombat K, Wongkajornsilp A et al. . Decreased indoleamine 2,3-dioxygenase activity and IL-10/IL-17A ratio in patients with COPD. Thorax 2013;68:330–7. doi:10.1136/thoraxjnl-2012-202127 [DOI] [PubMed] [Google Scholar]
- 20.Ubhi BK, Riley JH, Shaw PA et al. . Metabolic profiling detects biomarkers of protein degradation in COPD patients. Eur Respir J 2012;40:345–55. doi:10.1183/09031936.00112411 [DOI] [PubMed] [Google Scholar]
- 21.Fu X, Lawson MA, Kelley KW et al. . HIV-1 Tat activates indoleamine 2,3 dioxygenase in murine organotypic hippocampal slice cultures in a p38 mitogen-activated protein kinase-dependent manner. J Neuroinflamm 2011;8:88 doi:10.1186/1742-2094-8-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huengsberg M, Winer JB, Gompels M et al. . Serum kynurenine-to-tryptophan ratio increases with progressive disease in HIV-infected patients. Clin Chem 1998;44:858–62. [PubMed] [Google Scholar]
- 23.Samikkannu T, Saiyed ZM, Rao KV et al. . Differential regulation of indoleamine-2,3-dioxygenase (IDO) by HIV type 1 clade B and C Tat protein. AIDS Res Hum Retroviruses 2009;25:329–35. doi:10.1089/aid.2008.0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanner RE, Anthonisen NR, Connett JE et al. . Lower respiratory illnesses promote FEV(1) decline in current smokers but not ex-smokers with mild chronic obstructive pulmonary disease: results from the lung health study. Am J Respir Crit Care Med 2001;164:358–64. doi:10.1164/ajrccm.164.3.2010017 [DOI] [PubMed] [Google Scholar]
- 25.Morris AM, Huang L, Bacchetti P et al. . Permanent declines in pulmonary function following pneumonia in human immunodeficiency virus-infected persons. The Pulmonary Complications of HIV Infection Study Group. Am J Respir Critl Care Med 2000;162:612–16. doi:10.1164/ajrccm.162.2.9912058 [DOI] [PubMed] [Google Scholar]
- 26.Telenga ED, Hoffmann RF, Ruben tK et al. . Untargeted lipidomic analysis in chronic obstructive pulmonary disease. Uncovering sphingolipids. Am J Respir Crit Care Med 2014;190:155–64. doi:10.1164/rccm.201312-2210OC [DOI] [PubMed] [Google Scholar]
- 27.Bowler RP, Jacobson S, Cruickshank C et al. . Plasma sphingolipids associated with chronic obstructive pulmonary disease phenotypes. Am J Respir Crit Care Med 2015;191:275–84. doi:10.1164/rccm.201410-1771OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrache I, Natarajan V, Zhen L et al. . Ceramide upregulation causes pulmonary cell apoptosis and emphysema-like disease in mice. Nat Med 2005;11:491–8. doi:10.1038/nm1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scarpa MC, Baraldo S, Marian E et al. . Ceramide expression and cell homeostasis in chronic obstructive pulmonary disease. Respiration 2013;85:342–9. doi:10.1159/000341185 [DOI] [PubMed] [Google Scholar]
- 30.Denis U, Lecomte M, Paget C et al. . Advanced glycation end-products induce apoptosis of bovine retinal pericytes in culture: involvement of diacylglycerol/ceramide production and oxidative stress induction. Free Radic Biol Med 2002;33:236–47. doi:10.1016/S0891-5849(02)00879-1 [DOI] [PubMed] [Google Scholar]
- 31.Chetwynd AJ, Samarawickrama A, Vera JH et al. . Nanoflow-Nanospray mass spectrometry metabolomics reveals disruption of the urinary metabolite profiles of HIV-Positive patients on combination antiretroviral therapy. J Acquir Immune Defic Syndr 2017;74:e45–53. doi:10.1097/QAI.0000000000001159 [DOI] [PubMed] [Google Scholar]
- 32.Cassol E, Misra V, Holman A et al. . Plasma metabolomics identifies lipid abnormalities linked to markers of inflammation, microbial translocation, and hepatic function in HIV patients receiving protease inhibitors. BMC Infect Dis 2013;13:203 doi:10.1186/1471-2334-13-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scarpelini B, Zanoni M, Sucupira MC et al. . Plasma etabolomics biosignature according to HIV stage of infection, pace of disease progression, viremia level and immunological response to treatment. PLoS ONE 2016;11:e0161920 doi:10.1371/journal.pone.0161920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjresp-2017-000180supp_table1.pdf (34.7KB, pdf)
bmjresp-2017-000180supp_table2.pdf (105.7KB, pdf)