Abstract
The concentrations and functions of many eukaryotic proteins are regulated by the ubiquitin pathway, which consists of ubiquitin activation (E1), conjugation (E2), and ligation (E3). Cullins are a family of evolutionarily conserved proteins that assemble by far the largest family of E3 ligase complexes. Cullins, via a conserved C-terminal domain, bind with the RING finger protein Roc1 to recruit the catalytic function of E2. Via a distinct N-terminal domain, individual cullins bind to a protein motif present in multiple proteins to recruit specific substrates. Cullin 3 (Cul3), but not other cullins, binds directly with BTB domains to constitute a potentially large number of BTB-CUL3-ROC1 E3 ubiquitin ligases. Here we report that the human BTB-Kelch protein Keap1, a negative regulator of the antioxidative transcription factor Nrf2, binds to CUL3 and Nrf2 via its BTB and Kelch domains, respectively. The KEAP1-CUL3-ROC1 complex promoted NRF2 ubiquitination in vitro and knocking down Keap1 or CUL3 by short interfering RNA resulted in NRF2 protein accumulation in vivo. We suggest that Keap1 negatively regulates Nrf2 function in part by targeting Nrf2 for ubiquitination by the CUL3-ROC1 ligase and subsequent degradation by the proteasome. Blocking NRF2 degradation in cells expressing both KEAP1 and NRF2 by either inhibiting the proteasome activity or knocking down Cul3, resulted in NRF2 accumulation in the cytoplasm. These results may reconcile previously observed cytoplasmic sequestration of NRF2 by KEAP1 and suggest a possible regulatory step between KEAP1-NRF2 binding and NRF2 degradation.
Covalent conjugation of proteins by ubiquitin or ubiquitin-like modifiers usually involves a cascade of three enzymatic activities for activating (E1), conjugating (E2), and ligating (E3) ubiquitin or ubiquitin-like modifiers to a substrate. The E3 ubiquitin ligases contain two distinct functions: catalyzing isopeptide bond formation and recruiting the substrate (15, 16, 22). Two major families of E3 ligases have been described; the HECT domain family that is defined by its homology to E6-associated protein carboxyl terminus (named HECT for homology to E6-associated protein carboxyl terminus) and the RING family that contains either an intrinsic RING finger domain or an associated RING finger protein subunit essential for ubiquitin ligase activity (6, 35, 48).
Of several hundred RING finger proteins, ROC1 (named ROC for RING of cullins; also known as Rbx1, Hrt1, and SAG1) is uniquely linked with the ubiquitination of a potentially large number of substrates (8, 20, 34, 37, 39, 41). Unlike most other RING finger proteins, ROC1 (108 residues) is a small protein with the RING finger taking up 60% of the coding region. Extensive mutational analyses have demonstrated the requirement of the integrity of the RING finger for ubiquitin ligase activity (5, 20, 33, 34). Purified recombinant ROC1 and ROC2 or their RING finger alone, like that of APC11 (14, 26), are capable of activating E2-UbcH5 to synthesize polyubiquitin chains in the presence of E1, and removal of N-terminal sequences flanking the RING domain severely reduced ROC1-cullin 1 (CUL1) binding, but not the catalytic function of ROC1 (11). These results suggest that RING-E2 constitutes the catalytic core of the ubiquitin ligase and that the cullins assemble productive E3 ligases by bringing the RING-E2 catalytic core and substrates together.
The cullins are a family of evolutionarily conserved proteins, which contains three related genes in budding yeast and fission yeast and six related genes in worms, fruit flies, and humans (23, 27). In addition, three additional proteins, APC2 in all eukaryotes (47, 49), and PARC and CUL7 in mammals (7, 32), contain significant sequence homology to cullins over a ∼180-amino-acid region involved in binding with ROC1 or APC11. A remarkable aspect of the cullins is that each individual cullin can assemble into multiple distinct E3 ligases by interacting with a protein motif present in multiple proteins.
To recruit specific substrates to CUL1-dependent ligases, the cell utilizes an adaptor protein, SKP1. This adaptor binds simultaneously to an N-terminal domain found in CUL1/Cdc53p, but not other cullins (29), and to a conserved 40-residue protein motif known as an F-box, which was first recognized in a study of cyclin F and two additional SKP1-binding proteins. F-box proteins contain additional variable protein-protein interaction modules and recruit various substrates, often phosphorylated, to the CUL1-ROC1 catalytic core (2, 9, 38, 51).
To recruit specific substrates to CUL2- and possibly CUL5-dependent ligases, a different adaptor is used. A heterodimeric adaptor complex containing elongins B and C binds simultaneously to an analogous N-terminal domain in CUL2 and a conserved 40-residue protein motif, the SOCS box (named SOCS for suppressor of cytokine signaling), which was initially identified in the suppressor of cytokine signaling family of proteins. The SOCS proteins, via their additional protein-protein interaction modules, target various substrates differently to the CUL2- or CUL5-ROC1 catalytic cores (19, 21, 40, 50). Omitting the adaptor, CUL3 utilizes its N-terminal domain to bind to a conserved 100-residue protein motif known as a BTB domain (named BTB for Drosophila broad-complex C, Tramtrack, and Bric-a-brac) which was first identified in the Drosophila broad-complex C (BR-C), Tramtrack (Ttk), and Bric-a-brac (Bab) proteins (53). BTB proteins, via additional protein-protein interaction domains, then target potentially different substrates to the CUL3-ROC1 catalytic core (10, 13, 36, 46). The presence of multiple substrate specificity factors—mammals express more than 60 F-box, 40 SOCS, and 200 BTB proteins—suggests that cullins may form by far the largest family of E3 ligases and control the ubiquitination of a potentially large number of substrates. The ability of each cullin-dependent ligase to target the ubiquitination of a large number of substrates may explain various physiological functions linked with individual cullins, such as cell cycle regulation, cell growth control, tumor suppression, and organism development.
In response to oxidative stress and electrophiles, cells express different genes encoding antioxidative and phase II detoxification enzymes. Induction of these genes is regulated largely at the level of transcription, mediated by the antioxidant response element (ARE). The transcriptional activator Nrf2 is a key regulator that interacts with the ARE and is negatively regulated in nonstressed cells by a 75-kDa Kelch-BTB protein, Keap1 (named Keap1 for Kelch-like ECH-associated protein 1) (31). Targeted deletion of the mouse Keap1 gene resulted in a constitutive accumulation of Nrf2 protein in the nucleus, activation of Nrf2 target genes, and postnatal lethality (in <21 days) that could be rescued by deleting the Nrf2 gene also (43). These results establish Keap1 as a major regulator of Nrf2 and Nrf2 as a major downstream target of Keap1. The molecular mechanism by which Keap1 negatively regulates Nrf2 function remains incompletely understood. Two different views—sequestering NRF2 in the cytoplasm and promoting NRF2 degradation—are currently being pursued (17, 28). In this paper, we tested the idea that Keap1 may exert its inhibitory activity toward Nrf2 by targeting it for ubiquitination by the CUL3-ROC1 ligase and subsequent proteasomal degradation.
MATERIALS AND METHODS
Plasmids, antibodies, and chemicals.
Plasmids expressing wild-type human CUL3 and CUL3 in which the N-terminal 41 residues had been deleted and human ROC1 were described previously (10, 33). Human Nrf2 and Keap1 cDNA were cloned by PCR amplification from a HeLa cell cDNA library and verified by DNA sequencing. Mutations were introduced by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing. Antibodies to CUL3 (10), ROC1 (34), hemagglutinin (HA) (12CA5; Boehringer-Mannheim), Myc (9E10; NeoMarker), T7 (Novagen), FLAG (M2; Sigma), NRF2 (C-20 and H-300; Santa Cruz), KEAP1 (E-20; Santa Cruz), actin (C-11; Santa Cruz), and glutathione S-transferase (GST) (B-14; Santa Cruz) were previously described or purchased commercially. MG132 (Peptides International, Louisville, Ky.), and cycloheximide (Sigma) were purchased commercially.
Cell culture, transfection, and immunoprecipitation.
HeLa and 293T cells used in this study were cultured in Dulbecco's modified Eagle medium (Gibco-BRL) supplemented with 10% fetal bovine serum (Sigma) in a 37°C incubator with 5% CO2. Cell transfection was performed by using the FuGENE 6 (Roche) transfection reagent according to the manufacturer's instructions (for HeLa cells) or by using calcium phosphate buffer (for 293T cells). Except where indicated, cells were lysed with Nonidet P-40 (NP-40) lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 50 mM NaF, 0.5% NP-40, 1 mM Na3VO4, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 25 mg of leupeptin per liter, 25 mg of aprotinin per liter, 150 mg of benzamidine per liter, 10 mg of trypsin inhibitor per liter). Procedures for immunoprecipitation and immunoblotting have been described previously (11, 12).
Fluorescence microscopic analyses.
Full-length Nrf2 and Keap1 cDNA were cloned into pEGFP-C1 and DsRed2-C1 (Clontech), respectively. Transiently expressed green fluorescent protein (GFP)-NRF2 and DsRed-KEAP1 in live cells were analyzed with an Olympus IX70 microscope fitted with appropriate fluorescence filters.
GST fusion protein pull-down assay.
Full-length human Keap1 and Nrf2 cDNA were cloned into pGEX and pET-His vectors, respectively. The six-histidine (His6)-tagged fusion proteins were purified with Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) after induction with 0.4 mM isopropyl-1-β-d-galactopyranoside (IPTG) for 4 h in exponentially growing Escherichia coli BL21(DE3) cells cultured at 30°C. GST or GST fusion proteins were purified with glutathione agarose (Sigma) after induction with 0.4 mM IPTG for 24 h in exponentially growing E. coli XL1-Blue cells cultured at 20°C. One microgram of purified His6-tagged fusion proteins were incubated with 1 μg of purified GST or Keap1-GST fusion proteins immobilized on the glutathione agarose in buffer A, which contains 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 120 mM NaCl, 10% glycerol, 0.5% NP-40, 1 mM DTT, 0.5 mM PMSF, and 1 mg of bovine serum albumin. After 1 h incubation at room temperature, the glutathione agarose beads were washed with buffer A three times and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting with anti-GST, anti-NRF2, or anti-CUL3 antibodies.
Ubiquitin ligase activity assay.
The procedures for the ubiquitin ligase activity assay were performed as previously described (11, 12). Briefly, 293T cells were cotransfected with plasmids expressing Myc-tagged CUL3, T7-tagged KEAP1, and HA-tagged ROC1. Twenty-four hours after transfection, cells were lysed and immunoprecipitated with an anti-Myc antibody. Myc-CUL3 immunocomplexes immobilized on protein A-agarose beads were incubated with 1 μg of purified His-FLAG-tagged Nrf2 in a ubiquitin ligation reaction mixture (final volume, 30 μl) containing 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 1 μg of His6-tagged ubiquitin, 60 ng of E1 (Boston Biochem), and 300 ng of E2-UbcH5c at 37°C for 1 h. The reaction was terminated by boiling for 5 min in a SDS sample buffer containing 0.1 M DTT, and the proteins were resolved by SDS-PAGE, followed by immunoblotting with an anti-FLAG antibody.
RNA interference.
The desired 19- or 27-bp stem-loop RNAs were expressed from the pHTPsiRNA vector (a gift from Bill Reed [University of North Caroling at Chapel Hill]) driven by the human H1 gene promoter. The vector also contains a simian virus 40 promoter that drives transcription of a puromycin N-acetyltransferase gene conferring puromycin resistance. Vectors expressing the hairpin RNAs were transfected into 293T cells, and cells were selected with 1 μg of puromycin per ml for 3 days before lysis. The target sequences were 5′-GUCGUAGACAGAGGCGCAA-3′ (for Cul3) and 5′-CAUGAACGGUGCUGUCAUGUACCAGAU-3′ (for Keap1)
RESULTS
NRF2 is ubiquitinated, has a short half-life, and is degraded by a proteasomal pathway.
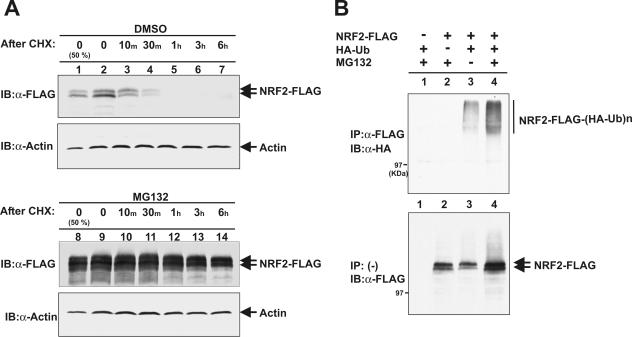
Two different models have been proposed to account for Keap1-mediated inhibition of Nrf2 function; KEAP1 binds to and sequesters NRF2 in the cytoplasm (17), and KEAP1 promotes NRF2 ubiquitination and subsequent degradation by a pathway yet to be identified (18, 28). The discovery that the BTB domain binds to CUL3 and may function to recruit substrate proteins to the CUL3-ROC1 ligase promoted us to test the idea that KEAP1 protein may target NRF2 for ubiquitination by the CUL3-ROC1 ligase. We first determined the half-life and sensitivity to proteasome-mediated degradation of ectopically expressed NRF2 protein. FLAG-tagged NRF2 was ectopically expressed in 293T cells and as endogenously expressed NRF2, exhibited two bands (see Fig. 5 below). The nature of these two forms is not known at present. The half-lives of both forms of ectopically expressed FLAG-tagged NRF2, as determined by cycloheximide-mediated pulse-chase experiments, were less than 30 min (Fig. 1A, top panel, lanes 1 and 4). Inhibition of the 26S proteasome by the addition of a proteasome inhibitor, MG132, effectively stabilized NRF2 (Fig. 1A, bottom panel). An in vivo ubiquitination assay demonstrated that ectopically expressed NRF2-FLAG protein is efficiently polyubiquitinated and that inhibition of 26S proteasome by MG132 accumulated both unmodified NRF2 as well as polyubiquitinated NRF2 (Fig. 1B). These results demonstrated that NRF2 is a short-lived protein and is degraded, in large part, by the 26S proteasome through a yet-to-be identified ubiquitin-dependent pathway. Our results support the view that controlling the stability of NRF2 contributes to, if not is primarily responsible for, negative regulation of Nrf2 (28).
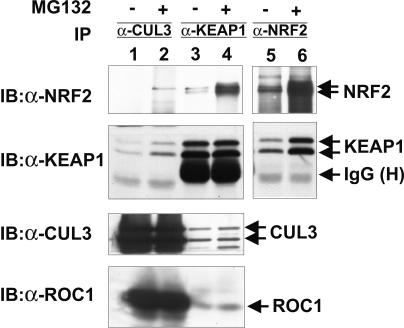
FIG. 5.
Endogenous CUL3, KEAP1, and NRF2 form a complex. 293T cells were treated with MG132 or DMSO (−) for 4 h prior to lysis. Cell lysates were immunoprecipitated (IP) with antibodies to CUL3 (α-CUL3), KEAP1 (α-KEAP1), or NRF2 (α-NRF2). Washed immunocomplexes were resolved by SDS-PAGE, followed by immunoblotting (IB) with the indicated antibodies. IgG (H), immunoglobulin G (heavy chain).
FIG. 1.
NRF2 is rapidly ubiquitinated and degraded by proteasomes. (A) 293T cells were transfected with the plasmid expressing FLAG-tagged NRF2 (NRF2-FLAG). Twenty-four hours after transfection, cells were treated first with MG132 (25 μM) or dimethyl sulfoxide (DMSO) for 5 h and then with cycloheximide (CHX) (75 μg/ml) for periods of time ranging from 10 min to 6 h. Cells were lysed, and 50 μg (lane 1) or 100 μg (lanes 2 to 7) of total lysates were resolved by SDS-PAGE, followed by immunoblotting (IB) with anti-FLAG (α-FLAG) or antiactin (α-Actin) antibodies. (B) 293T cells were transfected with the indicated plasmids expressing FLAG-tagged NRF2 and HA-tagged ubiquitin (HA-Ub). Twenty hours after transfection, cells were treated with MG132 (25 μM) for 2 h prior to cell lysis. Cells were lysed in a SDS lysis buffer and boiled for 15 min. Lysates were then diluted with NP-40 lysis buffer and immunoprecipitated (IP) with anti-FLAG antibody. The washed immunoprecipitates and 50 μg of lysates were resolved by SDS-PAGE, followed by immunoblotting with anti-HA and anti-FLAG antibodies, respectively.
KEAP1 promotes NRF2 degradation.
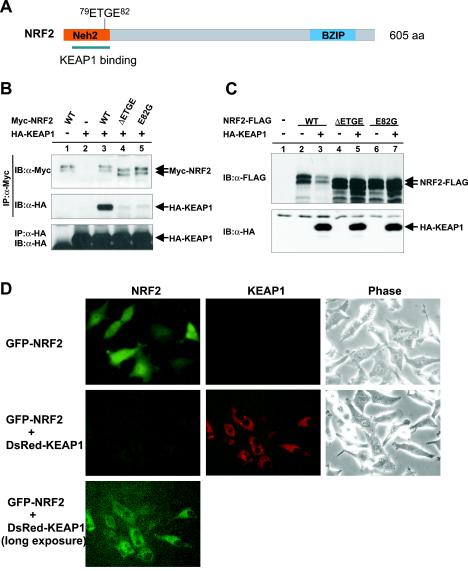
To determine how KEAP1 regulates the stability and subcellular localization of NRF2, we coexpressed these two proteins and determined the effect of KEAP1 on NRF2 protein stability. We first confirmed the KEAP1-NRF2 association by coupled immunoprecipitation and Western blotting (IP-Western). As shown in Fig. 2B, when these two proteins were coexpressed, a KEAP1-NRF2 complex was readily detected in cells treated with proteasome inhibitor MG132 but barely detected in untreated cells (data not shown). This observation is consistent with the hypothesis that KEAP1-NRF2 association may represent an intermediate complex that is rapidly degraded or dissociated in a 26S proteasome-dependent manner. Confirming the results of a previous report (25), deletion of four amino acids (ΔETGE) or a substitutive mutation at residue 82 (E82G) both substantially reduced the binding of NRF2 with KEAP1 (Fig. 2A and B). Coexpression with wild-type KEAP1, but not either mutant of KEAP1, considerably reduced the steady-state level of NRF2 (Fig. 2C).
FIG. 2.
KEAP1 promotes NRF2 degradation. (A) Schematic illustration of NRF2 protein and mutations characterized in this study. BZIP, basic region leucine zipper; aa, amino acids. (B) 293T cells were cotransfected with the plasmids expressing HA-tagged KEAP1 (HA-KEAP1) and Myc-tagged NRF2 (Myc-NRF2) (wild-type [WT] or mutant NRF2). Twenty hours after transfection, cells were treated with MG132 (25 μM) for 4 h prior to lysis, and the NRF2-KEAP1 association was examined by IP-Western. IB, immunoblotting; α-Myc, anti-Myc antibody; α-HA, anti-HA antibody. (C) HeLa cells were cotransfected with the plasmids expressing HA-tagged KEAP1 and FLAG-tagged wild-type or mutant NRF2. Twenty-four hours after transfection, cells were lysed, and lysates (0.1 mg) were resolved by SDS-PAGE, followed by immunoblotting (IB) with anti-FLAG (α-FLAG) or anti-HA antibodies. (D) GFP-tagged NRF2 was transfected to HeLa cells either alone or with the plasmid expressing DsRed-tagged KEAP1. Twenty-four hours after transfection, the levels of expression of GFP-NRF2 and DsRed-KEAP1 were examined by fluorescence or phase-contrast microscopy.
To determine how KEAP1 may affect the subcellular localization of NRF2, we fused NRF2 with GFP and KEAP1 with DsRed and examined the expression and localization of both proteins in live cells by fluorescence microscopy. GFP-NRF2, when singly expressed, localized predominantly in the nucleus and also exhibited visible cytoplasmic staining (Fig. 2D). KEAP1-positive cells, on the other hand, exhibited a clear fluorescence pattern of nuclear exclusion, indicating that KEAP1 was localized mostly in the cytoplasm. When coexpressed with KEAP1, the GFP signals were substantially reduced, consistent with a role of KEAP1 in promoting NRF2 degradation. Only after a long exposure could we detect GFP signals, localized mostly in the cytoplasm of a fraction of KEAP1-expressing cells (Fig. 2D). Treatment of cells with MG132 significantly increased the GFP signals in KEAP1-expressing cells, mostly localized in the cytoplasm (data not shown; discussed below also). These results suggest that, in HeLa cells, KEAP1-NRF2 interaction leads primarily to NRF2 protein degradation, not cytoplasmic sequestration, that the cytoplasm is most likely where KEAP1 promotes NRF2 degradation, and that when KEAP1-mediated NRF2 degradation is inhibited, NRF2 is accumulated or sequestered in the cytoplasm.
Via its BTB domain, KEAP1 binds to the N-terminal region of CUL3.
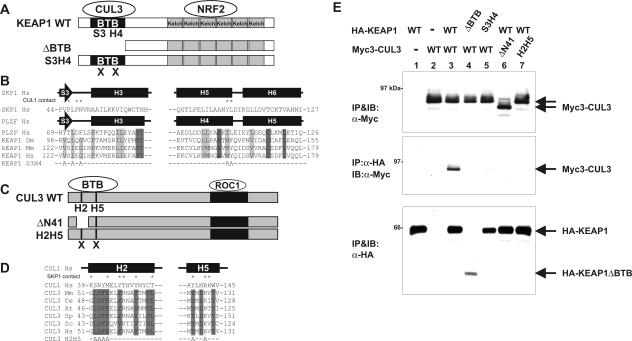
It has previously been demonstrated that CUL3, but not other cullins, utilizing a N-terminal sequence analogous to the SKP1-binding region in CUL1, binds to the BTB domain, which is present in multiple proteins (10, 13, 36, 46). To directly test the possibility that KEAP1, a BTB-Kelch protein (Fig. 3A), may interact with CUL3, we coexpressed both proteins and examined their association by IP-Western. A KEAP1-CUL3 association was readily detected (Fig. 3E, lane 3). Deletion of the BTB domain from KEAP1 abolished its binding with CUL3 (Fig. 3E, lane 4), demonstrating that the BTB domain is required for KEAP1-CUL3 binding. Crystallographic structure analysis revealed that the BTB domain of human promyelocytic leukemia zinc finger (PLZF) protein and SKP1 adopt similar three-dimensional structures (1, 51). We introduced a triple alanine substitution mutation, V123A, I125A, and G127A, into residues in the third β-sheet and a double mutation, M161A and Y162A, in the fourth α-helix of KEAP1 (S3H4) (Fig. 3B), which correspond to the residues in SKP1 that contact with CUL1. The S3H4 mutant completely lost its ability to bind with CUL3 (Fig. 3E, lane 5), further supporting the conclusion that the BTB domain of KEAP1 mediates the binding with CUL3.
FIG. 3.
BTB domain in Keap1 binds to the N terminus of CUL3. (A) Schematic illustration of wild-type (WT) and mutant Keap1. (B) Conserved α/β-structure in BTB/POZ fold of SKP1, PLZF, and Keap1. Identical and conserved residues are indicated by dark gray and light gray shading, respectively. Residues in the SKP1 making contact with CUL1 are marked by asterisks. The mutated residues in the β-sheet S3 and α-helix H4 (KEAP1S3H4 mutant) are shown at the bottom. (C) Schematic figures of wild-type CUL3, ΔN41 (CUL3 in which the N-terminal 41 residues were deleted), and helix 2 helix 5 mutant (H2H5) CUL3. (D) Sequence comparison of helix 2 (H2) and helix 5 (H5) of CUL1 and CUL3. Residues in the H2 and H5 helices of CUL1 that make contact with SKP1 are marked by asterisks. Residues conserved in CUL3 are marked by shading. The residues mutated in the CUL3H2H5 mutant are shown at the bottom. (E) 293T cells were cotransfected with the indicated plasmids expressing Myc-tagged CUL3 (Myc3-CUL3) or HA-tagged KEAP1 (HA-KEAP1) (wild-type [WT] or mutant CUL3 or KEAP1). Twenty-four hours after transfection, cells were lysed, and the CUL3-KEAP1 association was examined by IP-Western. IB, immunoblotting; α-Myc, anti-Myc antibody.
Two hydrophobic helical surfaces in the N-terminal regions of CUL1, H2, and H5, pack with hydrophobic and polar residues from SKP1 to form a large interface (Fig. 3C). The N-terminal regions of other cullins form similar H2 and H5 helices, which contain residues that are invariably conserved in orthologues but are different in paralogues (51). This suggests that other cullins could use analogous protein-binding sites in their N-terminal regions to associate with different adaptors, thereby conferring different specificities. Deletion of the N-terminal 41 residues from CUL3 (from Trp34 to Tyr74 [CUL3ΔN41]) removed the entire H2 helix and abolished CUL3's binding with KEAP1 (Fig. 3E, lane 6). To further confirm the binding of KEAP1 to the N-terminal region in CUL3, we introduced a quaternary alanine substitution mutation in the H2 (52LSFE55) helix and a double mutation in the H5 (Y125 and R128) helix (CUL3H2H5m) (Fig. 3D). The CUL3H2H5m mutant, like the CUL3ΔN41 mutant, completely abolished CUL3's association with KEAP1. As a result of both observations together, we conclude that KEAP1 and CUL3 bind to each other via the BTB domain in KEAP1 and the N-terminal sequence in CUL3, respectively.
KEAP1 binds to both NRF2 and CUL3 directly.
Detection of KEAP1-NRF2 and KEAP1-CUL3 associations in vivo in cultured cells led us to determine whether KEAP1 binds directly to these two proteins or requires an additional factor(s). We expressed recombinant GST-KEAP1, His-NRF2, and His-CUL3N197 and purified these proteins from bacteria. After in vitro incubation, GST-KEAP1 protein was precipitated with glutathione agarose beads, the bands were resolved by SDS-PAGE, and binding with NRF2 and CUL3 was analyzed by direct immunoblotting. GST-KEAP1, but not control GST protein, binds to His-NRF2 (Fig. 4A), demonstrating that KEAP1-NRF2 association is not dependent on an additional factor(s) and that KEAP1-NRF2 interaction can occur independently of CUL3. Phosphorylation of NRF2 by several kinases (protein kinase C [PKC], mitogen-activated protein kinase [MAPK]/ERK, phosphatidylinositol 3-kinase [PI3K], and JNK) have been implicated in regulating NRF2-KEAP1 association or NRF2 stability (3, 4, 30), but the physiological significance and molecular consequence of this phosphorylation are yet to be determined. Our results indicate that NRF2 phosphorylation, if involved in the regulation of its association with KEAP1, may influence the efficiency of, but is not required for, NRF2-KEAP1 association.
FIG. 4.
Keap1 binds to Nrf2 and CUL3 directly. (A) His-tagged NRF2 (His-NRF2) fusion protein was purified from bacteria and mixed with bacterially purified GST or GST-KEAP1 fusion proteins immobilized on the glutathione agarose beads. NRF2-KEAP1 binding was examined by immunoblotting (IB). α-NRF2, anti-NRF2 antibody; α-GST, anti-GST antibody. (B) His-CUL3N197 fusion protein was purified from bacteria and mixed with bacterially purified GST or GST-KEAP1 fusion proteins immobilized on the glutathione agarose beads. CUL3N197-KEAP1 binding was examined by immunoblotting (IB). α-CUL3, anti-CUL3 antibody.
After in vitro incubation, recombinant GST-KEAP1, but not control GST protein, binds to His-CUL3N197 (Fig. 4B). This result demonstrated that KEAP1 can bind directly with CUL3 and that the N-terminal 197 residues of CUL3 are sufficient for binding. We note that the efficiency of this in vitro binding appeared to be lower than that of in vivo KEAP1-CUL3 association (Fig. 3; also see Fig. 5 below). We speculate that either additional sequences or a modification of KEAP1 may influence its association with CUL3.
In vivo NRF2-KEAP1-CUL3-ROC1 complex.
The association of KEAP1 with NRF2 and CUL3 led us to determine whether endogenous CUL3 associates with KEAP1 and NRF2 in vivo. A two-component CUL3-KEAP1 complex can be readily detected by reciprocal IP-Western assay using both CUL3 and KEAP1 antibodies (Fig. 5). Under the same assay conditions, we could also readily detect NRF2 in the KEAP1 immunocomplex, but we could barely detect it in the CUL3 immunocomplex (longer exposure not shown). Reasoning that the NRF2 substrate-CUL3 ligase interaction may be transient and thus difficult to trap, we treated cells with MG132 to block NRF2 degradation and examined the NRF2-CUL3 association. Inhibition of proteasome activity resulted in an increase in the level of KEAP1-NRF2 and readily allowed detection of NRF2 in the CUL3 immunocomplex (Fig. 5). These results are consistent with the interpretation that a quaternary NRF2-KEAP1-CUL3-ROC1 complex exists in vivo and that the NRF2-CUL3 interaction is sensitive to proteasome activity.
KEAP1-CUL3-ROC1 complex ubiquitinates NRF2 in vitro.
We next examined the in vitro ubiquitination of NRF2 by the KEAP1-CUL3-ROC1 ligase complex. Anti-myc immunocomplexes were precipitated from either control, nontransfected cells or from cells cotransfected with Myc-CUL3, T7-KEAP1, and HA-ROC1 and incubated in vitro with purified FLAG-NRF2 in the presence or absence of exogenously added ubiquitin. High-molecular-weight smears characteristic of polyubiquitination were readily detected by an anti-FLAG antibody after incubation of FLAG-NRF2 with the myc immunocomplex derived from triply transfected cells (Fig. 6B, lane 4), but not with an empty myc immunocomplex (lane 3). Omitting either FLAG-NRF2 or ubiquitin abolished the polyubiquitin smears, demonstrating that the detected polyubiquitin chain was formed on exogenously added NRF2 and was synthesized in vitro after the addition of ubiquitin, excluding possible contamination by other proteins present in the myc complexes. To confirm the nature of polyubiquitinated product to be NRF2, we reblotted the same membrane with an antibody detecting NRF2 and observed virtually the same result (Fig. 6B, lower panel).
FIG. 6.
KEAP1-CUL3-ROC1 complex ubiquitinates NRF2 in vitro. (A) KEAP1-CUL3-ROC1 complex. 293T cells were cotransfected with the plasmids expressing the indicated proteins, Myc-tagged CUL3 (Myc-CUL3), T7-tagged KEAP1 (T7-KEAP1), and HA-tagged ROC1 (HA-ROC1). Twenty-four hours after transfection, cells were lysed and immunoprecipitated (IP) with anti-Myc antibody (α-Myc). Total lysates (lane 1 and 2) and immunoprecipitates (lanes 3 and 4) were resolved by SDS-PAGE, followed by immunoblotting (IB) with the indicated antibodies. (B) In vitro ubiquitination of NRF2. KEAP1-CUL3-ROC1 complexes were prepared from triply transfected cells by immunoprecipitation using myc antibody and used as the source of E3 ligase. Bacterially expressed and purified FLAG-tagged NRF2 was incubated with KEAP1-CUL3-ROC1 complex, E1, E2, ubiquitin (Ub), and ATP, and reaction mixtures were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
NRF2 accumulated if Keap1 or Cul3 was silenced.
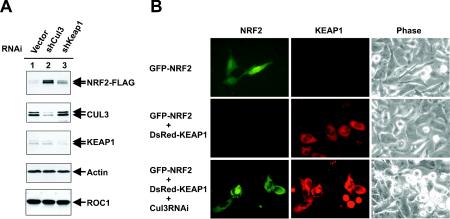
To obtain in vivo evidence supporting KEAP1-CUL3-mediated NRF2 ubiquitination, we silenced expression of either the Keap1 or Cul3 gene by RNA interference and determined the steady-state level of NRF2 protein by direct immunoblotting. Transfection of cells by plasmids expressing short interfering RNA targeting either Keap1 or Cul3 achieved estimated 60 and 75% reduction of both proteins, respectively, but had no significant effect on the levels of other proteins (Fig. 7A). A substantial accumulation of NRF2 protein was seen when Cul3 or Keap1 was silenced, with the silencing of Cul3 causing a higher level of accumulated NRF2 than that of Keap1 (Fig. 7A).
FIG. 7.
NRF2 accumulated when Cul3 or Keap1 was silenced. (A) 293T cells were cotransfected with the plasmid expressing FLAG-tagged NRF2 and the indicated short hairpin RNA plasmids (shCul3 and shKeap1). Transfected cells were selected by puromycin (1 μg/ml). Ninety-six hours after transfection, selected cells were lysed, and 50-μg samples of total lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. RNAi, RNA interference. (B) The expression vector for GFP-NRF2 alone or for GFP-NRF2 and DsRed-Keap1 was cotransfected with CUL3 short hairpin RNA plasmid or control short hairpin RNA plasmid to HeLa cells. After puromycin selection, the expression and localization of GFP-NRF2 and DsRed-KEAP1 were examined by fluorescence or phase-contrast microscopy.
To further confirm the function of Cul3 in promoting NRF2 degradation, we determined the level and subcellular localization of GFP-NRF2 in response to Cul3 silencing. Ectopically expressed GFP-NRF2, as expected, localized predominantly in the nucleus with visible cytoplasmic staining (Fig. 7B). Coexpression of DsRed-Keap1 nearly completely abolished the GFP signals. Silencing of Cul3 in cells coexpressing both GFP-NRF2 and DsRed-KEAP1 accumulated NRF2, indicating that Keap1-promoted NRF2 degradation requires the function of CUL3. Notably, NRF2 that accumulated after Cul3 silencing localized mostly in the cytoplasm (Fig. 7B). These results are consistent with the interpretation that when KEAP1 is coexpressed with NRF2 but inhibited from degrading NRF2, NRF2 accumulates in the cytoplasm.
DISCUSSION
In this study, we demonstrate that Keap1 negatively regulates the function of Nrf2, at least in part, by targeting NRF2 protein to the CUL3-ROC1 ligase for ubiquitination and subsequent proteasomal degradation. Four lines of evidence support this conclusion. First, NRF2 is a short-lived protein with a half-life of between 10 to 30 min, and KEAP1 promotes NRF2 degradation (18, 28, 30) (Fig. 1 and 2). KEAP1-mediated NRF2 degradation appears to be direct and requires binding with NRF2. Mutations in NRF2 disrupting its binding with KEAP1 also abolished the ability of KEAP1 to promote NRF2 degradation and inhibit the function of NRF2 (28) (Fig. 2). Second, endogenous NRF2, KEAP1 and CUL3 proteins can form a ternary complex in a proteasome-sensitive manner (Fig. 5). Third, a KEAP1-CUL3-ROC1 immunocomplex can cause efficient polyubiquitination in vitro (Fig. 6). Last, knocking down either Cul3 or Keap1 by RNA interference resulted in an accumulation of NRF2 (Fig. 7). These studies established an important and direct role of Cul3 in the regulation of Nrf2, and by extension, in mediating the cellular response to oxidative stress.
During the preparation of this report, Kobayashi and colleagues reported a similar finding that Keap1 functions as an adaptor for a Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2 (24). Several conclusions they reached—that Keap1-mediated Nrf2 degradation requires direct binding between the two proteins, that Keap1 associates with Cul3, and that overexpression of Keap1 or a combination of Cul3 and ROC1 promoted Nrf2 ubiquitination in vivo—are in agreement with our findings. Additionally, we have demonstrated that the KEAP1-CUL3-ROC1 immunocomplex caused polyubiquitination of NRF2 in vitro and knocking down either Keap1 or Cul3 accumulated NRF2 protein in vivo. Together with three previously identified substrates of CUL3-ROC1: the catalytic subunit of Caenorhabditis elegans katanin MEI-1 targeted by the BTB protein MEL-26 (10, 36), the human Rho GTPase, RhoBTB2 (45), and Arabidopsis 1-aminocyclopropane-1-carboxylic acid synthase (ACS) targeted by BTB protein ETO1 (44), these findings support the view that CUL3, via its binding with multiple BTB domains, may function in multiple, and potentially a large number of, distinct ubiquitin ligases that are involved in multiple cellular processes.
There is one critical discrepancy between our study and that of Kobayashi et al. (24). While we demonstrated that the BTB domain of KEAP1 mediates the binding of KEAP1 with CUL3, Kobayashi et al. reported that deletion of a sequence outside the BTB domain, referred to as the intervening region or IVR, but not deletion of the BTB domain, completely abolished the association between KEAP1 and CUL3 (24). Given the reports that the BTB domains derived from more than a dozen otherwise distinct proteins have been shown to bind with CUL3 (10, 13, 36, 44, 45, 46), the conclusion that the IVR, but not the BTB, in KEAP1 mediates the binding with CUL3 is surprising. The exact cause of the discrepancy is not clear. KEAP1 has been reported to form a homodimer (42, 52). The discrepancy could be reconciled if ectopically expressed KEAP1ΔBTB mutant retained its ability to bind with endogenous wild-type KEAP1 and was coprecipitated with CUL3 indirectly, rather than directly binding to CUL3. Another possibility is that the ΔBTB mutants used in two studies are different; while we removed residues 1 to 179, their ΔBTB mutant deleted residues 61 to 179. We have not determined whether the remaining 60 N-terminal residues may have contributed to the weak binding they observed. However, we also noticed that in their study, deletion of the BTB domain significantly reduced the binding of KEAP1 with CUL3 and impaired the ability of KEAP1 to promote NRF2 degradation. Both findings are consistent with the idea that the BTB domain binds to and is essential for the function of CUL3.
The mechanism underlying the contribution of the IVR to KEAP1-mediated NRF2 degradation remains to be determined. Although it is clear from the study of Kobayashi et al. (24) that deletion of this internal sequence situated between the N-terminal BTB domain and the C-terminal NRF2-binding Kelch repeats disrupted the binding of KEAP1 with CUL3, how the IVR contributes to CUL3 binding—whether by directly binding to CUL3 or by contributing to the overall conformation of KEAP1 protein—is not known. Previous studies have demonstrated that for those proteins tested, the BTB domain is both required and sufficient for binding with CUL3. A contribution by the sequence flanking the BTB domain to the binding with CUL3 could, in theory, provide further regulation of the BTB-CUL3 association. This interesting possibility remains to be investigated.
Two models have been proposed to explain the molecular basis for the negative regulation of Nrf2 function by Keap1: cytoplasmic sequestration and degradation of Nrf2. Our studies shed some light that may reconcile these two seemingly incompatible models. We observed that when KEAP1-mediated NRF2 degradation was inhibited, either by the inhibition of proteasome activity (Fig. 2D) or by silencing of Cul3 (Fig. 7B), NRF2 was accumulated in the presence of KEAP1, and accumulated NRF2 localized predominantly in the cytoplasm. These observations suggest the possibility that a regulatory step between KEAP1-NRF2 association and CUL3-dependent NRF2 degradation may exist in vivo. While the existence of such a regulatory event and the signal triggering the degradation of KEAP1-bound NRF2 are not known, it is conceivable that such a regulatory step would allow cells to maintain a certain level of NRF2, in a KEAP1-associated form, and the ability to rapidly respond to oxidative condition. The challenge remains to elucidate the signal(s) controlling KEAP1-NRF2 complex formation and/or subsequent NRF2 ubiquitination by the CUL3-ROC1 ligase.
Acknowledgments
We thank Joe He for technical assistance, Chad McCall for reading the manuscript, and other members of the Xiong lab for helpful discussions throughout this work.
This study was supported in part by an NIH grant (GM067113) to Y.X.
REFERENCES
- 1.Ahmad, K. F., C. K. Engel, and G. G. Prive. 1998. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 95:12123-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 3.Balogun, E., M. Hoque, P. Gong, E. Killeen, C. J. Green, R. Foresti, J. Alam, and R. Motterlini. 2003. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem. J. 371:887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom, D. A., and A. K. Jaiswal. 2003. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 278:44675-44682. [DOI] [PubMed] [Google Scholar]
- 5.Chen, A., K. Wu, S. Y. Fuchs, P. Tan, C. Gomez, and Z. Q. Pan. 2000. The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J. Biol. Chem. 275:15432-15439. [DOI] [PubMed] [Google Scholar]
- 6.Deshaies, R. J. 1999. SCF and cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 7.Dias, D. C., G. Dolios, R. Wang, and Z. Q. Pan. 2002. CUL7: a DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex. Proc. Natl. Acad. Sci. USA 99:16601-16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan, H., Y. Wang, M. Aviram, M. Swaroop, J. A. Loo, J. Bian, Y. Tian, T. Mueller, C. L. Bisgaier, and Y. Sun. 1999. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol. Cell. Biol. 19:3145-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman, R. M. R., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 10.Furukawa, M., Y. J. He, C. Borchers, and Y. Xiong. 2003. Targeting of protein ubiquitination by BTB-cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5:1001-1007. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa, M., T. Ohta, and Y. Xiong. 2002. Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J. Biol. Chem. 277:15758-15765. [DOI] [PubMed] [Google Scholar]
- 12.Furukawa, M., Z. Yanping, J. McCarville, T. Ohta, and Y. Xiong. 2000. The C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20:8185-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geyer, R., S. Wee, S. Anderson, J. Yates, and D. A. Wolf. 2003. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12:783-790. [DOI] [PubMed] [Google Scholar]
- 14.Gmachl, M., C. Gieffers, A. V. Podtelejnikov, M. Mann, and J.-M. Peters. 2000. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 97:8973-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 16.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 17.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, T. O'Connor, and M. Yamamoto. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379-391. [DOI] [PubMed] [Google Scholar]
- 19.Kamura, T., D. Burian, Q. Yan, S. L. Schmidt, W. S. Lane, E. Querido, P. E. Branton, A. Shilatifard, R. C. Conaway, and J. W. Conaway. 2001. Muf1, a novel elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276:29748-29753. [DOI] [PubMed] [Google Scholar]
- 20.Kamura, T., M. N. Conrad, Q. Yan, R. C. Conaway, and J. W. Conaway. 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, Jr., R. C. Conaway, and J. W. Conaway. 1998. The elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King, R. W., R. J. Deshaies, J.-M. Peters, and M. W. Kirschner. 1996. How proteolysis drives the cell cycle. Science 274:1652-1659. [DOI] [PubMed] [Google Scholar]
- 23.Kipreos, E. T., L. E. Lander, J. P. Wing, W.-W. He, and E. M. Hedgecock. 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85:829-839. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, M., K. Itoh, T. Suzuki, H. Osanai, K. Nishikawa, Y. Katoh, Y. Takagi, and M. Yamamoto. 2002. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7:807-820. [DOI] [PubMed] [Google Scholar]
- 26.Leverson, J. D., C. A. P. Joazeriro, A. M. Page, H.-K. Huang, P. Hieter, and T. Hunter. 2000. The APC11 RING-H2 mediates E2-dependent ubiquitination. Mol. Biol. Cell 11:2315-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathias, N., S. J. Johnson, M. Winey, A. E. M. Adams, L. Goetsch, J. R. Pringle, B. Byers, and M. G. Gobel. 1996. Cdc53p acts in concert with cdc4p and cdc34p to control the G1-to-S phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 16:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 29.Michel, J. J., and Y. Xiong. 1998. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 9:435-449. [PubMed] [Google Scholar]
- 30.Nguyen, T., P. J. Sherratt, H. C. Huang, C. S. Yang, and C. B. Pickett. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26S proteasome. J. Biol. Chem. 278:4536-4541. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen, T., P. J. Sherratt, and C. B. Pickett. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43:233-260. [DOI] [PubMed] [Google Scholar]
- 32.Nikolaev, A. Y., M. Li, N. Puskas, J. Qin, and W. Gu. 2003. ParcA cytoplasmic anchor for p53. Cell 112:29-40. [DOI] [PubMed] [Google Scholar]
- 33.Ohta, T., J. Michel, and Y. Xiong. 1999. Association with cullin partners protects ROC proteins from proteasome-dependent degradation. Oncogene 18:6758-6766. [DOI] [PubMed] [Google Scholar]
- 34.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 35.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 36.Pintard, L., J. H. Willis, A. Willems, J. L. Johnson, M. Srayko, T. Kurz, S. Glaser, P. E. Mains, M. Tyers, B. Bowerman, and M. Peter. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425:311-316. [DOI] [PubMed] [Google Scholar]
- 37.Seol, J. H., R. M. R. Feldman, W. Zachariae, A. Shevchenko, C. C. Correll, S. Lyapina, Y. Chi, M. Galova, J. Claypool, S. Sandmeyer, K. Nasmyth, and R. J. Deshaies. 1999. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13:1614-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skowyra, D., K. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 39.Skowyra, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 40.Stebbins, C. E., W. G. Kaelin, Jr., and N. P. Pavletich. 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284:455-461. [DOI] [PubMed] [Google Scholar]
- 41.Tan, P., S. Y. Fuches, A. Angus, K. Wu, C. Gomez, Z. Ronai, and Z.-Q. Pan. 1999. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell 3:527-533. [DOI] [PubMed] [Google Scholar]
- 42.Wakabayashi, N., A. T. Dinkova-Kostova, W. D. Holtzclaw, M. I. Kang, A. Kobayashi, M. Yamamoto, T. W. Kensler, and P. Talalay. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 101:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakabayashi, N., K. Itoh, J. Wakabayashi, H. Motohashi, S. Noda, S. Takahashi, S. Imakado, T. Kotsuji, F. Otsuka, D. R. Roop, T. Harada, J. D. Engel, and M. Yamamoto. 2003. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 35:238-245. [DOI] [PubMed] [Google Scholar]
- 44.Wang, K. L., H. Yoshida, C. Lurin, and J. R. Ecker. 2004. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428:945-950. [DOI] [PubMed] [Google Scholar]
- 45.Wilkins, A., Q. Ping, and C. L. Carpenter. 2004. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 18:856-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, L., Y. Wei, J. Reboul, P. Vaglio, T. H. Shin, M. Vidal, S. J. Elledge, and J. W. Harper. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425:316-321. [DOI] [PubMed] [Google Scholar]
- 47.Yu, H., J.-M. Peters, R. W. King, A. M. Page, P. Hieter, and M. W. Kirschner. 1998. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science 279:1219-1222. [DOI] [PubMed] [Google Scholar]
- 48.Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039-2058. [DOI] [PubMed] [Google Scholar]
- 49.Zachariae, W., A. Shevchenko, P. D. Andrews, R. Ciosk, M. Galova, M. J. R. Stark, M. Mann, and K. Nasmyth. 1998. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science 279:1216-1219. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, J. G., A. Farley, S. E. Nicholson, T. A. Willson, L. M. Zugaro, R. J. Simpson, R. L. Moritz, D. Cary, R. Richardson, G. Hausmann, B. J. Kile, S. B. Kent, W. S. Alexander, D. Metcalf, D. J. Hilton, N. A. Nicola, and M. Baca. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. USA 96:2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703-709. [DOI] [PubMed] [Google Scholar]
- 52.Zipper, L. M., and R. T. Mulcahy. 2002. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 277:36544-36552. [DOI] [PubMed] [Google Scholar]
- 53.Zollman, S., D. Godt, G. G. Prive, J. L. Couderc, and F. A. Laski. 1994. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 91:10717-10721. [DOI] [PMC free article] [PubMed] [Google Scholar]