Abstract
The story of how preColumbian civilizations developed goes hand-in-hand with the process of plant domestication by Mesoamerican inhabitants. Here, we present the almost complete sequence of a mitochondrial genome and a partial chloroplast genome from an archaeological maize sample collected at the Valley of Tehuacán, México. Accelerator mass spectrometry dated the maize sample to be 5,040–5,300 years before present (95% probability). Phylogenetic analysis of the mitochondrial genome shows that the archaeological sample branches basal to the other Zea mays genomes, as expected. However, this analysis also indicates that fertile genotype NB is closely related to the archaeological maize sample and evolved before cytoplasmic male sterility genotypes (CMS-S, CMS-T, and CMS-C), thus contradicting previous phylogenetic analysis of mitochondrial genomes from maize. We show that maximum-likelihood infers a tree where CMS genotypes branch at the base of the tree when including sites that have a relative fast rate of evolution thus suggesting long-branch attraction. We also show that Bayesian analysis infer a topology where NB and the archaeological maize sample are at the base of the tree even when including faster sites. We therefore suggest that previous trees suffered from long-branch attraction. We also show that the phylogenetic analysis of the ancient chloroplast is congruent with genotype NB to be more closely related to the archaeological maize sample. As shown here, the inclusion of ancient genomes on phylogenetic trees greatly improves our understanding of the domestication process of maize, one of the most important crops worldwide.
Keywords: long branch attraction, plant domestication, mitochondria, chloroplast
Introduction
Genetic evidence indicates that maize diverged ∼9,000 years ago from its wild ancestor the teosinte Zea mays subsp. parviglumis, somewhere near the Balsas river (Matsuoka et al. 2002; Piperno et al. 2009; van Heerwaarden et al. 2011). Its domestication boosted the development of preColumbian civilizations. Nowadays maize is one of the most important crops. Extant populations of maize in Mexico and worldwide show high genetic and phenotypic diversity (Arteaga et al. 2016; Hufford et al. 2012).
Although nuclear genetic diversity of maize is relatively well studied, there is much less information regarding the genetic diversity of organellar genomes. As of today, only five complete mitochondrial genomes from Zea mays subsp. mays and one from Zea mays subsp. parviglumis (hereafter parviglumis) have been sequenced and are publicly available. These genomes are from genotypes: NA, NB, CMS-S, CMS-T, and CMS-C (Allen et al. 2007; Clifton et al. 2004). Genotypes NA and NB are from fertile commercial maize, whereas CMS varieties are from cytoplasmic male sterility groups. Regarding maize chloroplasts, six genomes were sequenced recently (Bosacchi et al. 2015). These correspond to the same genotypes for which there are complete mitochondrial sequences, plus a plastid genome from the B73 inbred line. This last reference line has its nuclear genome completely sequenced (Schnable et al. 2009). In addition, there is a report of a complete chloroplast genome from 1995 (Maier et al 1995). However, recent analysis showed this genome to have several sequencing errors (Bosacchi et al. 2015).
Phylogenetic analysis of mitochondrial genomes showed NB genotype to be more closely related to parviglumis than to NA (Darracq et al. 2010). These phylogenies also indicate that CMS genotypes branch earlier relative to fertile genotypes NA, NB, and parviglumis. This has been interpreted as evidence that organelles differentiated before domestication of maize (Darracq et al. 2010).
In addition, phylogenetic analysis of available chloroplast showed a congruent evolutionary history to that of the mitochondria (Bosacchi et al. 2015). This is a clear indication that both organelles have been cotransmitted via the maternal line. In maize, mitochondria and chloroplasts are transmitted to seed via the maternal parent (Conde et al. 1979).
Here we report the almost complete genome sequence of a mitochondrial genome from a ∼5,000 years old maize sample. We also report a partial chloroplast genome from the same maize sample. We show that as expected, ancient mtDNA branched earlier than other sequenced genotypes from maize. We also show that this ancient maize had organelles closely related to NB genotype.
Materials and Methods
Archaeological Maize Sample
Under the guidance of Instituto Nacional de Antrolopología e Historia (INAH), remains of archaeobotanical maize specimens were collected at San Marcos cave (Valley of Tehuacán, México), under controlled conditions to avoid contamination. DNA sequences reported here belong to sample SM10 that was dated by using accelerator mass spectrometry (Vallebueno-Estrada et al. 2016). DNA extraction was performed in the Ancient DNA laboratory of UGA Langebio, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, using a modification of the protocol developed by Cappellini et al (2010). Reads were sequenced by using Solid 5,500 (average read length 73 bp), Illumina Hi-Seq (average read length 100 bp) and Illumina Next-Seq (average read length 100 bp). Reads from Solid were transform to fastq. All reads were treated as single end. Although nuclear DNA was sequenced with SOLID and Illumina, the organellar genomes were assembled only using Illumina sequences. A detailed report of archaeological maize samples, extraction and sequencing of DNA has been published elsewhere (Vallebueno-Estrada et al. 2016).
Assembly of Organellar Contigs from Ancient DNA
The reads were filtered using the bbsplit tool from the BBMap software suite (BBMap—Bushnell B.—https://sourceforge.net/projects/bbmap/; last accessd March 16, 2017). In the case of the mitochondria, the reference genomes used for the splitting of the reads were employed in the following order: Zea mays (AY506529), Zea mays ssp. parviglumis (DQ645539), Zea luxurians (DQ645537), Zea perennis (DQ645538), Rickettsia rickettsii (CP000766.3), Rickettsia typhi (AE017197.1), and Saccharomyces cerevisiae (NC_001224.1). All reference genomes were mitochondrial, except, of course, the ones from the rickettsias. The bbsplit tool allocated the reads to a specific genome due to sequence similarity. In case of a draw, the input order of the genomes set the priority order of allocation. The Rickettsia rickettsii, Rickettsia typhi, and Saccharomyces cerevisiae genomes were used to filter out contaminating sequences that could alter the assembly results due to the nature of the biological sample (ancient DNA). All the reads that matched the Zea mitogenomes were assembled using the Velvet genomic assembler (Zerbino 2010); using default settings and a kmer length of 31. The quality of the contigs was analyzed using the QUAST software (Gurevich et al. 2013).
In the case of the chloroplast, the reads were filtered using the bbsplit tool from the BBMap software suite. The reference chloroplast genomes used for the splitting of the reads were employed in the following order: Zea mays B73 (AY928077), Zea mays A188 (KF241980) and Zea mays B37 (KP966114). The bbsplit tool allocated the reads to a specific genome due to sequence similarity. In case of a draw, the input order of the genomes set the priority order of allocation. All the reads that matched the chloroplast genomes were assembled using the Velvet genomic assembler; using default settings and a kmer length of 31. The quality of the contigs was analyzed using the QUAST software.
The distribution of contig sizes for mitochondrial and chloroplast ancient DNA is shown in supplementary material figure S1, Supplemental Material online. In supplementary material figure S2, Supplemental Material online, we show the distribution of alignment sizes of these contigs to maize mitochondria (AY506529) and chloroplast (KP966114) genomes. The organellar genomes are deposited in GenBank under accession numbers: KY018916 (mitochondria) and KY018917 (chloroplast). Filtered reads from ancient mtDNA and ptDNA are available upon request.
Identification of SNPs in Ancient DNA
The obtained contigs from ancient mitochondrial and chloroplast DNA were aligned separately to the mitochondrial and chloroplast genomes shown in table 1 by using the MUGSY software (Angiuoli and Salzberg 2011). We then refined the MUGSY alignments with MUSCLE software (Edgar 2004). The distribution of alignment sizes (regions in organellar DNA continuously covered by aligned contigs) for mitochondrial and chloroplast ancient DNA is shown in supplementary material figure S2, Supplemental Material online.
Table 1.
Organellar Genomes Used in This Study
| Spp. | Cytotype | ptDNA Accession No. | ptDNA Reference | mtDNA Accession No. | Reference |
|---|---|---|---|---|---|
| Z. mays sub. mays (ancient) | — | KY018917 | This study | KY018916 | This study |
| Z. mays sub. mays (B37N) | NB | KP966114 | Bosacchi et al. (2015) | AY506529 | Clifton et al. (2004) |
| Z. mays sub. mays (B73) | NB | KF241981 | Bosacchi et al. (2015) | ||
| Z. mays sub. mays (B73) | NB | AY928077 | Unpublished | ||
| Z. mays sub. mays (A188) | NA | KF241980 | Bosacchi et al. (2015) | DQ490952 | Allen et al. (2007) |
| Z. mays sub. parviglumis | DQ645539 | Darracq et al. (2010) | |||
| Z. mays sub. mays (B37C) | CMS-C | KP966115 | Bosacchi et al. (2015) | DQ645536 | Allen et al. (2007) |
| Z. mays sub. mays (B37S) | CMS-S | KP966116 | Bosacchi et al. (2015) | DQ490951 | Allen et al. (2007) |
| Z. mays sub. mays (B37T) | CMS-T | KP966117 | Bosacchi et al. 2015 | DQ490953 | Allen et al. 2007 |
| Z. perennis | DQ645538 | Darracq et al. (2010) | |||
| Z. luxurians | DQ645537 | Darracq et al. (2010) |
Next, we reconstructed phylogenetic trees for mitochondrial as well as chloroplast alignments and inferred ancestral sequences for each node on the trees by Maximum-likelihood as implemented in MEGA6 (Tamura et al. 2013). By using Perl scripts, we identified all gap-free variable sites in the alignment. And then for each gap-free variable site, we retrieved the nucleotide in the immediate ancestor for each sequence and recorded the kind of nucleotide substitution. The total number of kinds of substitutions (i.e., A⇊T; A⇊C; A⇊G; etc.) between organellar sequences (including ancient DNA) and their immediate ancestral sequences is shown in supplementary material figures S4 and S13, Supplemental Material online. Next, we eliminated from the alignment all sites in which the nucleotide from ancient DNA (A, T, C, G) was not represented among any of the other modern organellar sequences (see table 1) and repeated the above procedure. By this, we guarantee that each SNP in ancient DNA is supported by at least one of the other organellar genomes. The total number of kinds of substitutions between organellar sequences (including ancient DNA) and their immediate ancestral sequences, after the filter, is shown in supplementary material figures S6 and S15, Supplemental Material online. We also counted the number of filtered SNPs shared between ancient DNA and the other organellar genomes (see supplementary material tables S1–S4, Supplemental Material online). Filtered SNPs and its coordinates in relation to mitochondrial genome AY506529 and chloroplast genome KP966114 are shown in supplementary tables S22 and S23, Supplemental Material online.
We also identified reliable SNPs by using mapDamage2.0 (Jónsson et al. 2013). MapDamage2.0 requires a reference genome to identify reliable SNPs. Therefore, we aligned reads from ancient mtDNA identified with BBsplit to sequenced mitogenomes from table 1 and then identified all trusted SNPs. Polymorphism in ancient DNA of more than one nucleotide were eliminated.
Phylogenetic Analysis
For all phylogenetic analysis we used the alignments with filtered SNPs (see the above section). Mitochondrial and chloroplast Bayes trees from figures 3 and 4 were reconstructed with MrBayes by using GTR model of evolution plus Gamma distribution and a proportion of invariant sites (Ronquist et al. 2012). Chains of the mcmc were run until the standard deviation of split frequencies was below 0.01 and the potential scale reduction factor was close to 1.0 for most parameters. Trees were visualized with Figtree (http://tree.bio.ed.ac.uk/software/figtree/; last accessed March 16, 2017).
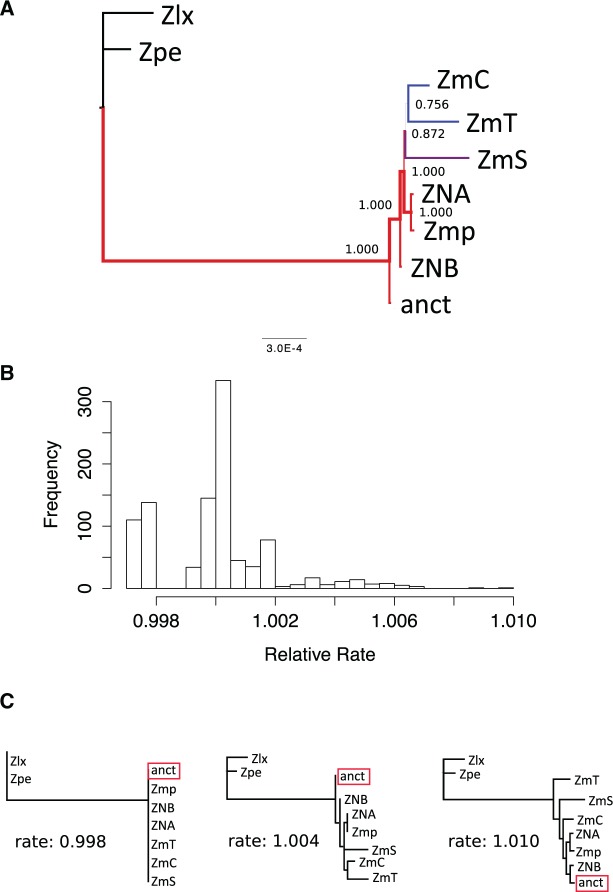
Fig. 3.—
Phylogenetic analysis of maize mitochondrial genomes. (a) Bayesian phylogenetic tree reconstructed from all sites of the alignment. The color of the branches corresponds to posterior probabilities support; (b) frequency of sites evolving at different relative rates along the alignment of mitochondrial genomes; The site rates are scaled such that the average evolutionary rate across all sites is 1. Hence most sites have this relative rate of evolution. Sites showing values larger than 1 are evolving at rates faster than average and sites showing values smaller than 1 are evolving at rates slower than average. (c) Maximum-likelihood (ML) trees using progressively faster sites (i.e., the left-most tree is reconstructed with sites evolving at a relative rate of at most 0.998 and right-most tree is reconstructed with all sites in the alignment). Note that the topology of the ML tree reconstructed from sites evolving at a relative rate of at most 1.004 has a topology identical to that of the Bayesian tree. Abbreviates: anct, ancient; NB, Z. mays NB (AY506529); NA, Z. mays NA (DQ490952); Zmp, Z. parviglumis (DQ645539); ZmC, Z. mays CMS-C (DQ645536); ZmT, Z. mays CMS-T (DQ490953); ZmS, Z. mays CMS-S (DQ490951); Zpe, Z. perennis (DQ645538); Zlx, Z. luxurians (DQ645537).
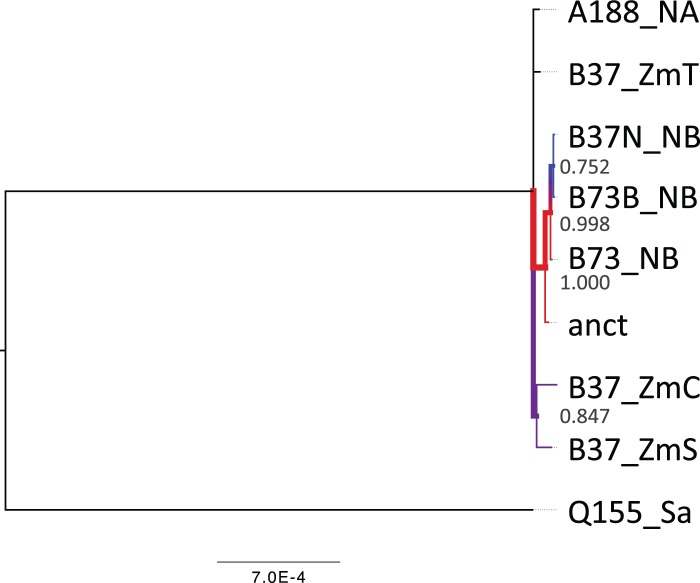
Fig. 4.—
Bayesian phylogenetic tree of maize chloroplast genomes. The colors of the branches correspond to posterior probabilities support. Abbreviates: anct, ancient; B73_NB, Z. mays NB (AY928077); B73B_NB, Z. mays NB (KF241981); B37N_NB, Z. mays NB (KP966114); A188_NA, Z. mays NA (KF241980); B37_ZmC, Z. mays CMS-C (KP966115); B37_ZmT, Z. mays CMS-T (KP966117); B37_ZmS, Z. mays CMS-S (KP966116); Q155_Sa, Saccharum hybrid (KU214867).
Mitochondrial Maximum-likelihood trees from figure 3 and supplementary material, Supplemental Material online, were reconstructed by using PhyML 3 (Guindon et al., n.d.). For Maximum-likelihood trees, we first identified the best model of evolution by using JModelTest (Posada 2008). To infer the relative rates of evolution of sites in the alignment in figure 3, we used MEGA 6 (Tamura et al. 2013). By using Perl scripts, we partitioned the alignment of mitochondrial sequences according to a maximum relative rate of evolution allowed. These maximum allowed relative rates were: 0.998, 1.000, 1.002, 1.004, 1.006, and 1.010. The last category contained the full alignment. For each partition, we reconstructed four trees. Two Maximum-likelihood and two Bayesian trees. The two trees reconstructed by each method differ by having or not having the ancient DNA sequences. The results of these analyses are shown in supplementary material, Supplemental Material online, together with all trees in newick format.
Molecular Clock Analysis
To determine whether the sequences are suitable for time tree reconstruction by Bayesian methods, we used TempEst (Rambaut et al. 2016). Results are shown in supplementary material, Supplemental Material online. Molecular clock analysis was done by using BEAST 2 (Bouckaert et al. 2014). XML files for BEAST analysis were prepared by using BEAUti. Results of BEAST were inspected by using Tracer. Trees were obtained by TreeAnnotator and visualized with FigTree. All the above software was retrieved from “Molecular evolution and phylogenetics and epidemiology” website (http://tree.bio.ed.ac.uk; last accessed March 16, 2017).
To test whether a strict or a lognormal relaxed molecular clock better fits the data, we examined the distribution of the 95% highest posterior density (HPDs) of the likelihoods of the two models in Tracer. As a rule of thumb, if the distributions do not overlap, then it is safe to assume that the one with the highest likelihood performs better. Therefore, we used a relaxed lognormal clock for all the following analysis. Results are shown in supplementary material, Supplemental Material online.
To assess whether the data has influence on the posterior, we prepared three files with BEAUti differing in their prior of the age of the root of the tree. To do so, we changed the beta parameter from the gamma distribution describing the age of the root. Used beta values and their associated times of divergence are shown in table 2. The three XML files for BEAST analysis are provided in supplementary material, Supplemental Material online. To compare the three models, we sampled from the prior and compared the results under Tracer. The rationale is straightforward, if the posterior mirrors changes on the prior, then data has no influence on the posterior. Results of this analysis are shown in supplementary material, Supplemental Material online. Also, the three models were compared by the Akaike’s information criterion through MCMC (AICM) as implemented in Tracer (Baele et al. 2012). Although the best model according to AICM was the one in which the beta parameter equals 10, we conclude that making an inference based on it is not reliable because of the lack of strong influence of the data on the posterior.
Table 2.
Estimates of the Age of the Root are Strongly Influenced by Priors
| Beta | Prior of the Age of the Roota | Posterior of the Age of the Rootb | Age of the Root When Sampling from Priorb | Clock Ratec | AICM |
|---|---|---|---|---|---|
| 10 | 7,500 (7,310–7,710) | 7,496 (7,302–7,699) | 7,424 (7,350–7,639) | 1.38E-8 (9.12E-9–1.86E-8) | 910951.55 ± 0.016 |
| 20 | 8,490 (8,130–8,910) | 8,487 (8,107–8,870) | 8,424 (8,338–8,658) | 1.14E-8 (7.45E-9–1.53E-8) | 910952.01 ± 0.011 |
| 30 | 9,490 (8,940–10,000) | 9,472 (8,916–10,081) | 11,050 (9,273–11,440) | 9.75E-9 (6.51E-9–1.32E-8) | 910952.12 ± 0.021 |
Note.—We show the effect in the posterior of using different values in the beta parameter of the gamma distribution for the age of the root prior.
Median and quantiles 2.5% to 97.5.
Median of the distribution and the 95% HPDs.
Mean number of substitutions per site per year and the 95% HPDs.
Results
Ancient DNA was recovered from a ∼5,000 (5,040–5,300) year old maize sample from San Marcos cave in Tehuacán, México (SM10; Vallebueno-Estrada et al. 2016). Ancient DNA was sequenced with both SOLiD and Illumina platforms. Obtained reads were filtered to separate sequences of nuclear origin from those of ancient mitochondrial and chloroplasts genomes. These filtered sequences were assembled into contigs. We obtained larger contigs for ancient mtDNA than for ancient ptDNA (see supplementary material figs. S1 and S2, Supplemental Material online). While in mtDNA ∼47% of contigs have a size of 300 or more nucleotides, in ptDNA only ∼42% of contigs have a size of only 200 or more nucleotides. Assembled contigs were aligned with MUGSY and MUSCLE to reference mitochondria and chloroplast genomes (table 1).
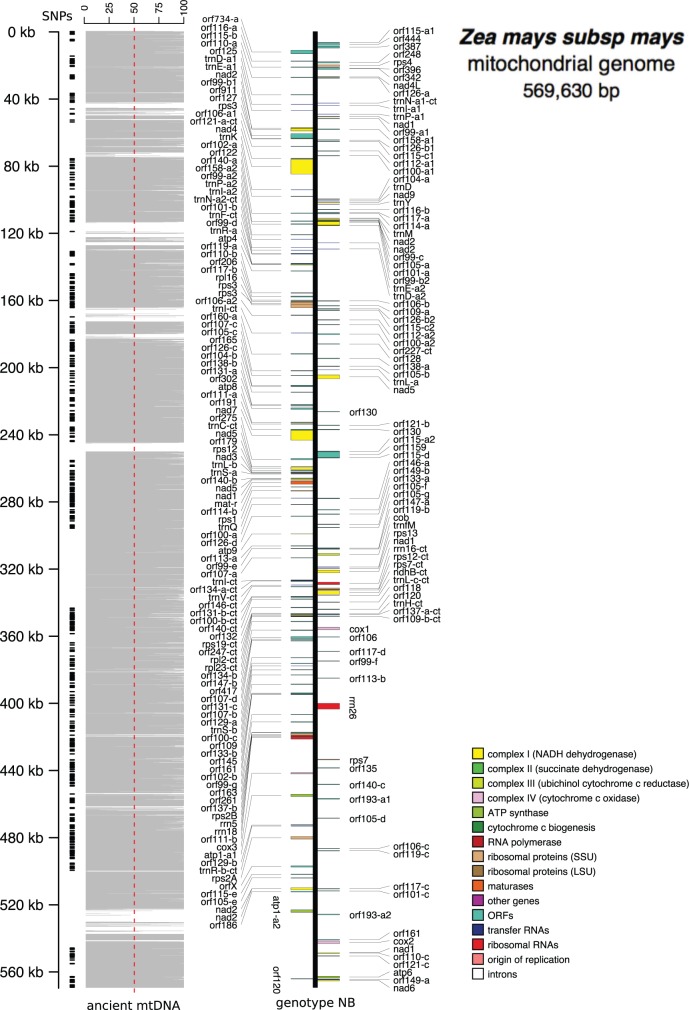
In figure 1, we show a linear representation of the mitochondrial genome genotype NB indicating regions covered by ancient DNA. Each gray bar represents the percentage of covered nucleotides by ancient DNA to mitochondrial genome NB along nonoverlapping windows of 100 bp. As shown, ancient DNA covers ∼89% of NB mitochondrial maize genome.
Fig. 1.—
Ancient Zea mays mitochondrial genome. Each horizontal gray bar denotes the percentage of coverage by ancient DNA to maize mitochondrial genome NB (Genbank: AY506529) along nonoverlapping windows of 100 bp. Approximately 89% of mitochondrial genome is covered by ancient DNA. We also show the position of ∼1,000 validated SNPs. Linear representation of mitochondrial genome was draw with OrganellarGenomeDraw (Lohse et al. 2013).
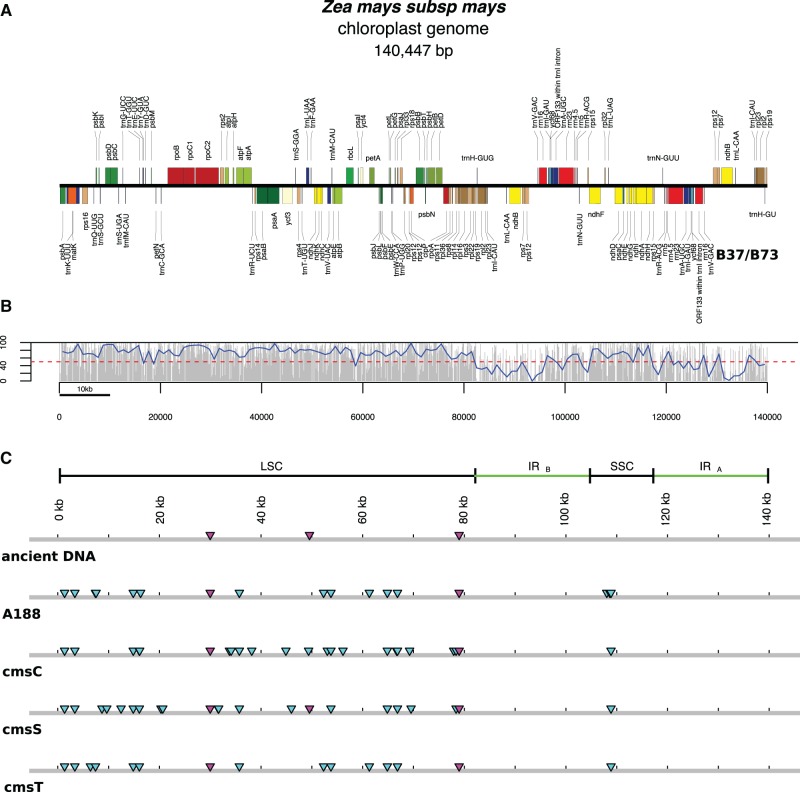
In figure 2, we show a linear representation of chloroplast from NB genotype along regions covered by ancient DNA. Ancient DNA covers ∼64% of plastid maize genome. Each gray bar represents the percentage of covered nucleotides by ancient DNA to the chloroplast genome NB along nonoverlapping windows of 100 bp. As shown, the percentage of coverage is much less than in the case of the mitochondria. In particular, there are two regions where coverage is very low. These regions correspond approximately to IRA and IRB elements. To investigate why IRA and IRB elements show low coverage, we assembled the chloroplast genome with Spades (Bankevich et al. 2012). By this, we increased coverage. However, IRA and as small fraction of IRB continued unmapped by reads (see supplementary material fig. S22, Supplemental Material online).
Fig. 2.—
Ancient Zea mays chloroplast genome. (a) Linear representation of maize chloroplast genome NB (Genbank: KP966114). (b) Each vertical gray bar denotes the percentage of coverage by ancient DNA to maize chloroplast genome NB along nonoverlapping windows of 100 bp. Approximately 64% of chloroplast genome is covered by ancient DNA. (c) Representation of the SNPs found in sequenced maize chloroplast genomes in relation to B37/B73; cyan triangles indicate SNPs not present in ancient DNA; magenta triangles indicate SNPs in ancient DNA shared with chloroplast genomes other than B37/B73; LSC, large-single-copy region; SSC, small-single copy region; IRA and IRB are inverted repeats A and B. Based on figure 1 from Bosacchi et al. (2015). Linear representation of chloroplast genome was draw with OrganellarGenomeDraw (Lohse et al. 2013).
SNP Identification in Ancient DNA
Ancient DNA decays because of a variety of environmental factors. When this degraded DNA is amplified by PCR, sequences with incorrect bases may be obtained. The most frequent errors in these amplified sequences are transitions (A→G)/(T→ C) and (C→T)/(G→A)(Hansen et al. 2001). These errors are called miscoding lesions type I and type II respectively. In particular, the second type of lesions is more common in nuclear and mitochondrial DNA (Binladen et al. 2006). Because of this, it is very important to have a way to distinguish true polymorphisms from miscoding lesions when studying ancient DNA samples (Mateiu and Rannala 2007).
Here, we took a straightforward approach to identify reliable SNPs in ancient DNA. We considered a SNP in ancient DNA to be real only if this SNP was also found among sequenced organellar maize and teosinte genomes available in table 1. This approach has the disadvantage that SNPs found only in ancient DNA will not be identified. In addition, miscoding lesions coincident with SNPs in extant genomes will be misidentified as reliable SNPs.
In the case of mitochondria, this approach allowed us to identify ∼1000 SNPs. This approach also reduced the number of (C→T)/(G→A) transitions found between ancient DNA and its immediate ancestor on a phylogenetic tree (see supplementary material figs. S3–S6, Supplemental Material online). We found that genotype NB shares the largest number of SNPs with ancient DNA, followed by NA, parviglumis and then the CMS genotypes. And, as expected, Z. perennis and Z. luxurians share the smallest number of SNPs with ancient DNA (see supplementary material tables S1 and S2, Supplemental Material online).
In the case of the chloroplast, the number of SNPs assigned to ancient DNA is much smaller (see supplementary material tables S3 and S4, Supplemental Material online). It is known that in maize, chloroplast DNA evolves much slower than mitochondria, which parallels several protists (Bosacchi et al. 2015; Smith 2015). As well as in the mitochondria, ancient chloroplast DNA shares the largest number of SNPs with genotype NB, followed by NA and then by the CMS genotypes (see supplementary material tables S3 and S4, Supplemental Material online).
The complete list of trusted SNPs is shown in supplementary material table S22, Supplemental Material online. Each SNP is presented with its homologous nucleotide from mitochondrial (AY506529) and chloroplast (KP966114) genomes. By using these genomes as templates, we also provide scaffolds of ancient organellar genomes in FASTA format (supplementary material, Supplemental Material online).
Phylogenetic Analysis of Ancient Z. mays Organelles
As expected for an archaeological maize sample, Bayesian phylogenetic analysis indicates that ancient DNA branches at the base of extant maize mitochondria, with Z. luxurians and Z. perennis as outgroups (fig. 3a). Unexpectedly, genotype NB also branches from a basal position relative to the other genotypes. This result is contradictory with previous published phylogenies of mtDNA from maize in which CMS genotypes are basal to NA, NB, and parviglumis (Darracq et al. 2010).
One possible explanation of the differences in topologies between both studies is the occurrence of long-branch attraction (LBA). LBA tends to occur when in the correct tree short internal branches separate two or more large external branches (Yang and Rannala 2012). Then, an incorrect topology may be inferred in which long branches are grouped together.
Previous phylogenies of mtDNA were reconstructed with maximum-likelihood, distance and parsimony methods (Darracq et al. 2010). These analyses were based on concatenated protein coding gene sequences (maximum likelihood and distance methods) and on the number of genome rearrangements between mitochondrial genomes (distance and maximum parsimony methods).
To investigate whether LBA is causing the differences in the topology of the trees, we partitioned the sites in the multiple alignment based on their relative rate of evolution (fig. 3b). The rate of evolution was estimated by using the GTR + G model of evolution with four site categories. The site rates are scaled such that the average evolutionary rate across all sites is 1. Then, sites showing values >1 are evolving at rates faster than average and sites showing values <1 are evolving at rates slower than average. The analysis was performed with MEGA6 (Tamura et al. 2013). We then reconstructed phylogenies starting with slowly evolving sites and progressively reconstructed phylogenies by adding faster evolving sites to the multiple alignment.
As shown in figure 3c, maximum-likelihood recovers the same topology as the Bayesian analysis when using sites evolving at an intermediate rate (from relative rate 1.002–1.006). Only when using all sites (relative rate up to 1.010), Maximum-Likelihood infers a topology where CMS genotypes are basal to fertile genotypes. Therefore, we conclude that the position of CMS genotypes in the base of the Zea mays subtree is due to LBA. Although sites evolving up to a relative rate of 1.010 are only ∼1% faster than slower sites and they represent a small proportion of all variable sites (fig. 3c), their inclusion in the phylogenetic analysis is enough to attract the long branches of CMS genotypes toward the base of the tree in maximum-likelohood analysis.
It is worth mentioning that MrBayes inferred trees where ancient DNA and NB genotype were at the base of the Zea mays subtrees even when including fastest sites (relative rate up to 1.010). In supplementary material figure S7, Supplemental Material online, we show the complete list of trees reconstructed from all relative rate partitions of the alignment. In supplementary material figure S8, Supplemental Material online, we show the Bayesian and Maximum-Likelihood trees inferred by using all sites having a maximum relative rate up to 1.006 along with their support values.
To exclude the possibility that the topology of figure 3a is the result of artifactual SNPs, we repeated the above analysis excluding ancient DNA. In this case, we found that as soon as internal branches become larger than 0 (corresponding to relative rate 1.002), maximum likelihood reconstructs a phylogeny where CMS-T genotype occupies the basal position among Zea mays species (see supplementary material fig. S9, Supplemental Material online). However, this is not the case in Bayesian analysis. In Bayesian analysis, genotype NB occupies the basal position when using sites evolving at a relative rate up to 1.002. For relative rates larger that 1.002, Bayesian analysis also reconstructs a tree where CMS genotypes branch at the base of Zea mays species. This shows that when lacking ancient DNA sequences, the effects of LBA are stronger. However, Bayesian analysis still recovers a tree where NB branches at the base of the maize subtree when using sites evolving at a relative rate up to 1.002. It is well known that the inclusion of more sequences corrects LBA artifacts (Yang and Rannala 2012).
As previously mentioned, our approach to identify reliable SNPs will fail to identify true SNPs from ancient DNA not present among sequenced mitochondrial genomes (mitogenomes). At the same time, we expect that a proportion of the SNPs identified by our method should be a consequence of DNA decay. Because such errors in SNP identification could mislead our phylogenetic analysis, we tested the robustness of our SNP identification inference by using mapDamage2.0 to filter out spurious SNPs from ancient DNA. By using this method, we identified 634 SNPs in ancient mtDNA. The pattern of base changes for these SNPs shows a reduced pattern of DNA damage (see supplementary material fig. S10, Supplemental Material online). The Bayesian phylogenetic tree reconstructed from such SNPs shows that ancient mtDNA branches at the base of Z. mays together with genotype NB (see supplementary material fig. S11, Supplemental Material online), supporting the basal position of NB genotype relative to the other varieties of maize. Although in this case the values of posterior probabilities are lower than those obtained by our Bayesian phylogeny, perhaps due to a smaller number of SNPs.
We also reconstructed the phylogeny of the chloroplast by using Bayesian analysis. In this case, paralleling the phylogeny of the mitochondria, ancient DNA branches with genotype NB (fig. 4). As well as in the case of the mitochondria, we followed the same procedure to identify reliable SNPs (see supplementary material figs. S12–S15, Supplemental Material online) However, there is little phylogenetic information in chloroplast sequences to resolve other nodes in the tree. Also, by using the sequence assembled with Spades, we obtain a phylogeny were ancient chloroplast branches with genotype NB (see supplementary material fig. S23, Supplemental Material online).
Evolution of Organelles on a Time Frame
Our sample of ancient DNA is dated to be ∼5,000 year old (95% of probability between 5,040 and 5,300 years). This offers the possibility to estimate the coalescence time of Z. mays organellar genomes studied here. Nowadays, several methods have been developed to calculate divergence times when there is moderate rate variation among lineages (Ho and Duchêne 2014). In particular, Bayesian methods estimate divergence times by incorporating prior information. These priors come in form of statistical distributions representing our knowledge (or our uncertainty) of the evolutionary process that generated the gene sequences.
As previously mentioned, maize originated from teosinte ∼9,188 years ago (95% confidence limits are between 5,689 and 13,093) (Matsuoka et al. 2002). This data is consistent with phytholits from the Xihuatoxtla shelter indicating that maize remains were present in grinding tools dating to 8,700 before present (Piperno et al. 2009). It is also consistent with archaeological estimates suggesting that the origin of crop domestication in Mexico occurred less than 10,000 years ago (Smith 1999).
To estimate divergence times from sequences sampled at different times, it is good practice to evaluate first whether the sequences in question accumulated measurable amounts of evolutionary change between them (Rambaut et al. 2016). This is done by making a regression between the sampling time of the sequences versus the root-to-tip genetic distance between the sequences. If the slope of the regression turns out to be positive, this is an indication that the sequences have accumulated measurable amounts of evolutionary change and are suitable for divergence time estimation using Bayesian methods.
As shown in supplementary material figure S16, Supplemental Material online, this is indeed the case for the phylogenetic tree composed of mitochondrial genomes NA, NB, parviglumis, and the ancient DNA sequences. Notice that because of their long branches, we excluded genomes from CMS genotypes. Nevertheless, there is large branch rate variation among studied genomes as can be appreciated by the scattering of dots in the regression. This simple analysis suggests that mitochondrial genomes coalesce ∼13,290 years ago. However, this result must be taken only as a gross approximation because of the scattering of the data.
To better estimate the time of coalescence of mitochondrial genomes, we performed a Bayesian analysis. We first compared the performance of the strict versus the relaxed lognormal clock models under a Bayesian framework. We found that the relaxed lognormal model performs better. As shown in supplementary material figure S17, Supplemental Material online, the distributions containing 95% of the highest posterior densities (HPDs) of the likelihood between the two models do not overlap. This is an indication that the model with the highest likelihood performs better (Drummond and Bouckaert 2015). This is consistent with previous analysis showing that the molecular clock hypothesis was rejected for mitochondrial genomes (Darracq et al. 2010). In fact, there is a lot of rate variation among branches as indicated by the standard deviation (∼0.91) of the uncorrelated lognormal relaxed clock model (supplementary material fig. S18, Supplemental Material online). Values larger than 1.0 indicate that standard deviation in branch rates is greater than the mean rate (Drummond et al. 2007).
When estimating divergence times under a Bayesian framework, it is extremely important to evaluate the sensitivity of the estimated parameters to changes in the priors (Drummond and Bouckaert 2015). The idea is to determine whether there is enough information in the sequences to inform the posterior. Otherwise, the posterior will be only a reflection of the prior. Accordingly, we analyzed the sensitivity of the estimated age of the root to changes in three different priors. We proceeded as follows: We used a gamma distribution with an alpha parameter of 100 as a prior for the age of the root (the alpha parameter controls the shape of the curve of the gamma distribution and large values of the alpha parameters create normal-like curves). Then, we changed the beta parameter of the gamma distribution to three different values: 10, 20, and 30 (table 2; supplementary material fig. S19, Supplemental Material online). Larger values of the beta parameter flatten the gamma distribution and move the median towards older time divergences. As a result of this tuning of the beta parameter, we ended up with three priors for the age of the root: 7,500, 8,490, and 9,490 years ago (table 2). For all cases, we used a minimum time of divergence of 6,500 years.
Of the three models, the best evaluated by Akaike’s information criterion through MCMC (AICM) was the one using a value of 10 for the beta parameter. The rate estimated by using this value for the beta parameter (1.38E-8) is faster than the rate estimated by noncoding regions in the mitochondria of Arabidopsis thaliana 9.6E-10 (Christensen 2013).
However, we were not able to infer an estimate of the time of divergence of mitochondrial genomes. This is because we found that the results of the analysis were strongly influenced by the priors (table 2 and supplementary material fig. S20, Supplemental Material online). Only in the case when the parameter beta is equal to 30, the estimated age of coalescence when sampling from the prior is larger (11,050) than the estimated age informed by the sequences (9,472). Nevertheless, the 95% HPD partially overlap.
Therefore, we only provide a time inference based on a prior for the origin of maize (9,190 years ago with 95% of the distribution falling between 8,640 and 9,820 years). The resulting time tree is shown in supplementary material figure S21, Supplemental Material online. This inference must be taken with caution because the time of coalescence of mitochondrial genomes studied here can be different from the divergence of maize from teosinte ∼9,000 years ago. Clearly, we need more genome sequences to make an inference on the time of coalescence of mitochondrial maize genomes.
Discussion
The rise of paleogenetics more than 30 years ago has opened a new way to conduct research related to the study of fast evolutionary changes, the genetics of climate adaptation, domestication, the spread of pathogens, etc. (Hofreiter et al. 2015). It is now possible to obtain more genetic information from small and damaged DNA samples than before, allowing the reconstruction of virtually complete genomes (from organellar and prokaryotic to eukaryotic genomes) for population and phylogenetic studies involving extant and ancient species.
DNA sequences from the archaeological maize sample reported here allowed us to redraw the phylogeny of maize mitochondria. It showed that CMS genotypes evolved after NB fully fertile genotype.
That previous analysis failed to identify the LBA artifact is not surprising. The internal branches separating Z. mays mitochondrial genomes are too short when compared with tip branches from CMS genotypes and to the large branch connecting Z. mays to Z. luxurians and Z. perennis. These situations easily led to LBA artifact (Misof et al. 2012). Elimination of fast evolving sites as well as addition of more sequences are well known techniques to correct for LBA (Jeffroy et al. 2006; Yang and Rannala 2012).
Organellar genomes reported here also provided partial support for coinheritance of mitochondria and chloroplasts via the maternal line (Bosacchi et al. 2015). On the chloroplast phylogeny, only two nodes are resolved with a posterior probability > 0.8 within the Z. mays subtree. These are the nodes grouping ancient chloroplast with genotype NB (posterior probability of 1.00); and the node grouping CMS-S and CMS-C genotypes (posterior probability of 0.85). Of them, only the first one is congruent with the mitochondrial tree. However, chloroplast tree is reconstructed from very few SNPs. Therefore, is very likely that as more chloroplast genomes are sequenced, the phylogeny will change, likely supporting the coinheritance of both organelles via the maternal line.
The rate of evolution of the maize mitochondrial genome is larger than that of the chloroplast (Bosacchi et al. 2015). The opposite pattern is found in most land plants (Drouin et al. 2008). However, few other lineages of plants have larger rates of evolution in their mitochondria than in their chloroplasts. These include Geraniaceae, Plantago, and Silene (Cho et al. 2004; Mower et al. 2007; Parkinson et al. 2005; Sloan et al. 2012). What is becoming clear is that rates of evolution in mitochondria vary more than in chloroplasts (Smith and Keeling 2015). Failures in DNA repair mechanisms have been suggested to explain these differences in rate variation among mitochondria from different plant lineages (Christensen 2013).
The close association between genotype NA and parviglumis is somehow unexpected. Because maize originally diverged from parviglumis, one would expect the teosinte to branch prior other maize mitochondrial genomes. However, the phylogenetic tree suggests a much more recent origin. According to our preliminary molecular clock analysis, the branch connecting parviglumis to NA is no older than ∼3,300 years. A parsimonious scenario to explain the proximity of NA to parviglumis is to consider that both genotypes were already present in the population of teosinte that originated maize. It is also possible that the small sample of available mitochondrial genomes for phylogenomic analysis hinders our ability to detect recent cases of introgression.
Domestication of maize is one of the key processes in the development of preColumbian civilizations. Although the evolutionary genomics of nuclear DNA is relatively well understood, that of the organelles is just beginning to be addressed. Genetic evidence suggest that maize diverged in a single event near the Balsas river ∼9,000 years ago (Matsuoka et al. 2002). The genetic diversity of extant organellar genomes can or cannot predate this point of divergence. Unfortunately, our molecular clock analysis failed to provide a time divergence of mitochondrial genomes. Sequencing of more and selected mitochondrial genomes can help to overcome this and provide a better time estimation for its coalescence. A better understanding of maize mitochondrial evolution can shed light on the population size at the time of divergence and during domestication.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Acknowledgments
We thank Mtra. Diana Rossetti for useful comments to the manuscript and to three anonymous reviewers. This research was supported by a grant from CONACyT (CB256826) and the Instituto Nacional de Antropología e Historia (INAH), through the CINVESTAV-INAH collaboration.
Literature Cited
- Allen JO, et al. 2007. Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177(2):1173–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli SV, Salzberg SL. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27(3):334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga MC, et al. 2016. Genomic variation in recently collected maize landraces from Mexico. Genomics Data 7:38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, et al. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 29(9):2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binladen J, et al. 2006. Assessing the fidelity of ancient DNA sequences amplified from nuclear genes. Genetics 172(2):733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosacchi M, Gurdon C, Maliga P. 2015. Plastid genotyping reveals the uniformity of cytoplasmic male sterile-T maize cytoplasms. Plant Physiol. 169(3):2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, et al. 2014. BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput Biol. 10(4):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellini E, et al. 2010. A multidisciplinary study of archaeological grape seeds. Naturwissenschaften 97(2):205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Mower JP, Qiu Y-L, Palmer JD. 2004. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci U S A. 101(51):17741–17746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC. 2013. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol Evol. 5(6):1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton SW, et al. 2004. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 136(3):3486–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde MF, Pring DR, Levings CS., III 1979. Maternal inheritance of organelle DNA’s in Zea mays–Zea perennis reciprocal crosses. J Heredity 70:2–4. [Google Scholar]
- Darracq A, Varré J-S, Touzet P. 2010. A scenario of mitochondrial genome evolution in maize based on rearrangement events. BMC Genomics 11:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 49(3):827–831. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Bouckaert RR. 2015. Bayesian evolutionary analysis with BEAST. Cambridge University Press.
- Drummond AJ, Ho SYW, Rawlence N, Rambaut A. (2007). A Rough Guide to BEAST 1. 4. Com/P/Beast-, p. 1–41. Available from: http://www.molecularevolution.org/molevolfiles/beast/BEAST14_MANUAL-7-6-07.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Willerslev E, Wiuf C, Mourier T, Arctander P. 2001. Statistical evidence for miscoding lesions in ancient DNA templates. Mol Biol Evol. 18(2):262–265. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Duchêne S. 2014. Molecular-clock methods for estimating evolutionary rates and timescales. Mol Ecol 23(24):5947–5965. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, et al. 2015. The future of ancient DNA: technical advances and conceptual shifts. BioEssays 37(3):284–293. [DOI] [PubMed] [Google Scholar]
- Hufford MB, et al. 2012. Comparative population genomics of maize domestication and improvement. Nat Genet. 44(7):808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. 2006. Phylogenomics: the beginning of incongruence?. Trends Genet. 22(4):225–231. [DOI] [PubMed] [Google Scholar]
- Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29(13):1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. (2013). OrganellarGenomeDRAW—A Suite of Tools for Generating Physical Maps of Plastid and Mitochondrial Genomes and Visualizing Expression Data Sets, p. 1–7. Available from: http://doi.org/10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed]
- Maier RM, Neckermann K, lgloi GL, Kössel H. 1995. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 251:614–628. [DOI] [PubMed] [Google Scholar]
- Mateiu LM, Rannala BH. (2007). Bayesian Inference of Errors in Ancient DNA Caused by Postmortem Degradation. Available from: http://doi.org/10.1093/molbev/msn095. [DOI] [PubMed]
- Matsuoka Y, et al. 2002. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci U S A. 99(9):6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, Mayer C, Wa J, Ku P. 2012. Long branch effects distort maximum likelihood phylogenies in simulations despite selection of the correct model. PLoS One 7(5):31–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. 2007. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 7:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson CL, et al. 2005. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol. 5:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno DR, Ranere AJ, Holst I, Iriarte J, Dickau R. 2009. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc Natl Acad Sci U S A. 106(13):5019–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Lam TT, Carvalho LM, Oliver G. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115. [DOI] [PubMed] [Google Scholar]
- Sloan DB, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10(1): e1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BD. 1999. The emergence of agriculture “Scientific American” library. New York and Oxford: W H Freeman & Co. 231 pp. [Google Scholar]
- Smith DR. 2015. Mutation rates in plastid genomes: they are lower than you might think. Genome Biol Evol. 7(5):1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ. 2015. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 112(33):201422049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallebueno-Estrada M, et al. 2016. The earliest maize from San Marcos Tehuacán is a partial domesticate with genomic evidence of inbreeding. Proc Natl Acad Sci U S A. 113(49):14151–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heerwaarden J, et al. 2011. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci U S A. 108(3):1088–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. 2012. Molecular phylogenetics: principles and practice. Nat Rev Genet. 13(5): 303–314. [DOI] [PubMed] [Google Scholar]
- Zerbino DR. (2010). Using the velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics, 1–13. doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed]