Abstract
The Ccr4-Not complex is a conserved global regulator of gene expression, which serves as a regulatory platform that senses and/or transmits nutrient and stress signals to various downstream effectors. Presumed effectors of this complex in yeast are TFIID, a general transcription factor that associates with the core promoter, and Msn2, a key transcription factor that regulates expression of stress-responsive element (STRE)-controlled genes. Here we show that the constitutively high level of STRE-driven expression in ccr4-not mutants results from two independent effects. Accordingly, loss of Ccr4-Not function causes a dramatic Msn2-independent redistribution of TFIID on promoters with a particular bias for STRE-controlled over ribosomal protein gene promoters. In parallel, loss of Ccr4-Not complex function results in an alteration of the posttranslational modification status of Msn2, which depends on the type 1 protein phosphatase Glc7 and its newly identified subunit Bud14. Tests of epistasis as well as transcriptional analyses of Bud14-dependent transcription support a model in which the Ccr4-Not complex prevents activation of Msn2 via inhibition of the Bud14/Glc7 module in exponentially growing cells. Thus, increased activity of STRE genes in ccr4-not mutants may result from both altered general distribution of TFIID and unscheduled activation of Msn2.
All living organisms are proficient at adapting within a defined physiological range to changing environmental conditions. Such adaptation processes are inherently coupled to changes in the expression of functional proteins which—particularly in eukaryotic cells—are based on regulation of different processes, such as transcription initiation, mRNA stability, translation, or posttranslational protein modification. Interestingly, the Ccr4-Not complex, which is conserved from yeast to human, acts on most of these processes to control the appropriate expression of functional proteins and may therefore play a critical role in adaptive responses to environmental challenges. The complex exists in at least two distinguishable forms of 1.2 and 2 MDa and harbors nine core subunits, which include Ccr4, Caf1, Caf40, Caf130, and Not1-5 (reviewed in references 11, 14, and 17). While the smaller complex may contain solely the core subunits, the larger form is likely to be associated with various additional proteins that are involved in transcription initiation, such as the SAGA-complex subunit Ada2 (4), the RNA polymerase II (Pol II) holoenzyme subunits Srb9, Srb10, and Srb11 (32), and the TFIID-complex subunits TBP, Taf13, and Taf1 (16, 28, 29, 44). The larger complex may further include proteins that are involved in mRNA degradation, such as the putative RNA helicase Dhh1, which resides in the decapping complex (35). Interestingly, both core subunits Ccr4 and Caf1 are also known to control initial steps in mRNA degradation as key components of the yeast deadenylase complex (8, 15, 52, 53). Finally, the Ccr4-Not complex has also been found to interact directly with the cell cycle-regulated Dbf2 protein kinase (33) as well as with the E2 ubiquitin-conjugating enzymes Ubc4 and Ubc5, which—by analogy to the situation in mammalian cells—are thought to interact with the putative E3 ubiquitin ligase Not4 (1).
The NOT genes were originally isolated in a selection for mutants that cause an increase in transcription of the HIS3 gene (12, 13, 41). The not mutants displayed core promoter-specific defects which, together with the reported interaction between specific Not proteins and TFIID subunits, indicated that the NOT gene products may be involved in control of TFIID function. In line with this suggestion, it was recently found that Not5 not only associates with promoter DNA in a Taf1-dependent manner but also controls appropriate Taf1-DNA association, particularly during adaptation to nutrient-limiting conditions (16). In parallel, the Ccr4-Not complex may exert an additional control over transcription initiation by directly or indirectly inhibiting the function of the zinc finger transcription factor Msn2 (30), which is known to control expression from the stress response element (STRE) in response to environmental signals (20, 23, 38, 45). While in principle it is possible that the Ccr4-Not complex may simply regulate the presence of TFIID at Msn2-regulated promoters, an alternative model suggests that the Ccr4-Not complex, possibly in response to high protein kinase A (PKA) levels under conditions of nutrient abundance, inhibits Msn2 function via direct or indirect posttranslational modification (30). Notably, in this context, both subcellular localization and STRE-binding activity of Msn2 are regulated by phosphorylation and dephosphorylation processes that are likely to involve different protein kinases and yet-unknown protein phosphatases (9, 21, 23, 24, 26).
It has been proposed that the Ccr4-Not complex may regulate mRNA levels of Msn2-controlled genes, such as HSP12, via more than one mechanism. Accordingly, Ccr4 negatively regulates HSP12 mRNA stability (8), while the Not5 subunit of the Ccr4-Not complex controls the recruitment of Taf1 to the HSP12 core promoter (16). Moreover, since posttranslational modification of Msn2 appeared different in those mutants of the Ccr4-Not complex in which Msn2-dependent transcription was increased, the Ccr4-Not complex may also directly or indirectly regulate the activity status of Msn2 (30). Here we study in more detail how the Ccr4-Not complex controls transcription of Msn2-dependent genes. We show that the complex acts independently on TFIID to control its promoter-specific distribution and on Msn2 to control its posttranslational modification, possibly via a newly identified Bud14/Glc7 protein phosphatase module. Thus, the Ccr4-Not complex regulates STRE-dependent transcription via at least two different mechanisms, namely, modification of TFIID distribution and modification of Msn2 activity.
MATERIALS AND METHODS
Yeast strains, media, and general methods.
The Saccharomyces cerevisiae strains used in this work are listed in Table 1. Strains KT1960 and KT1961 are congenic to KT1112. Strains KT1703 and KT1708 carrying integrated glc7-132 and glc7-127 alleles, respectively (for details, see reference 54), and KT1705 and KT1706 carrying the integrated glc7-133 allele were kindly provided by K. Tatchell. Mating of MY2052 with MY4 and subsequent sporulation of the resulting diploid yielded MY2050, MY2051, and MY2053. PCR-based gene deletions (bud14Δ::kanMX2 transformed into W303-1A, SGY446, and PD6517 to create PE27, PE15, and PE6; bud14Δ::kanMX6 transformed into MY1 to create MY2904; not2Δ::kanMX6 transformed into MY2 to create MY2182; and msn2Δ::TRP1 transformed into MY2 to create MY3496) and tagging of chromosomal genes (BUD14-myc13-kanMX6 transformed into KT1961 to create IP19 and MSN2-myc13-kanMX6 transformed into MY1 and MY2050 to obtain MY3590 and MY3591, respectively) were done as described previously (34). Mating of MY2904 with MY2051, KT1705 with MY2053, and MY3496 with MY2050 and subsequent sporulation of the resulting diploid strains yielded the segregants MY2995 and MY2998, MY3317, and MY3498, respectively. Mating of MY2998 with MY3591 and subsequent sporulation of the resulting diploids yielded the segregants MY3633 and MY3634. The linearized, NcoI-cut integrative vector pCTT1-18/7x (see below) was transformed into strains MY2050, MY2998, MY2995, KT1705, and MY3317 to construct MY2596, MY3234, MY3235, MY3362, and MY3363, respectively. Strains were grown at 30°C (except where noted) in standard rich yeast extract-peptone-dextrose (YPD) medium with 2% glucose (unless otherwise stated) or in synthetic defined media lacking specific amino acids as described previously (46). Yeast transformations, manipulation of Escherichia coli, and the preparation of bacterial growth media were performed as described previously (2).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| MY1 | MATaura3-52 trp1 leu2Δ::PET56 gcn4 gal2 | 13 |
| MY2 | MATα ura3-52 trp1 leu2Δ::PET56 gcn4 gal2 | 36 |
| MY4 | MATα ura3-52 trp1 leu2Δ::PET56 gcn4 gal2 his3::TRP1 | 36 |
| MY508 | MY1 not3Δ::URA3 | 13 |
| MY1719 | MY2 not5Δ::LEU2 | 41 |
| MY1728 | MY1 ccr4Δ::URA3 | 36 |
| MY1729 | MY1 caf1Δ::LEU2 | 36 |
| MY2050 | MY1 not4Δ::LEU2 his3Δ::TRP1 | This study |
| MY2051 | MY2 not4Δ::LEU2 his3Δ::TRP1 | This study |
| MY2052 | MY1 not4Δ::LEU2 | 30 |
| MY2053 | MY2 not4Δ::LEU2 | This study |
| MY2182 | MY2 not2Δ::kanMX6 | This study |
| MY2489 | MY2 his3Δ::TRP1 ura3Δ::STRE-lacZ-URA3 | 30 |
| MY2596 | MY2050 ura3Δ::STRE-lacZ-URA3 | This study |
| MY2904 | MY1 bud14Δ::kanMX6 | This study |
| MY2995 | MY2 his3Δ::TRP1 not4Δ::LEU2 bud14Δ::kanMX6 | This study |
| MY2998 | MY2 bud14Δ::kanMX6 | This study |
| MY3317 | MATaura3-52 his3 glc7-133 not4Δ::LEU2 | This study |
| MY3234 | MY2998 ura3Δ::STRE-lacZ-URA3 | This study |
| MY3235 | MY2995 ura3Δ::STRE-lacZ-URA3 | This study |
| MY3362 | KT1705 ura3Δ::STRE-lacZ-URA3 | This study |
| MY3363 | MY3317 ura3Δ::STRE-lacZ-URA3 | This study |
| MY3496 | MY2 msn2Δ::TRP1 | This study |
| MY3498 | MY1 his3Δ::TRP1 msn2Δ::TRP1 not4Δ::LEU2 | This study |
| MY3590 | MY1 MSN2-myc13::kanMX6 | This study |
| MY3591 | MY2050 MSN2-myc13::kanMX6 | This study |
| MY3633 | MY1 not4Δ::LEU2 his3Δ::TRP1 bud14Δ::kanMX6 MSN2-myc13::kanMX6 | This study |
| MY3634 | MY1 his3Δ::TRP1 bud14Δ::kanMX6 MSN2-myc13::kanMX6 | This study |
| PD6517 | MATα ade8 leu2 trp1 cdc35-10 | 18 |
| PE6 | PD6517 bud14Δ::kanMX2 | This study |
| SGY446 | MATα tpk1Δ::ADE8 tpk2-63(Ts) tpk3::TRP1 ura3-52 his3 leu2-3,11 trp1 ade8 | 48 |
| PE15 | SGY446 bud14Δ::kanMX2 | This study |
| W303-1A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 51 |
| PEY78 | W303-1A msn2-Δ3::HIS3 msn4-1::TRP1 | 38 |
| PE27 | W303-1A bud14Δ::kanMX2 | This study |
| EGY48 | MATahis3 trp1 ura3 LEU2::pLexAop6-LEU2 | 55 |
| KT1112 | MATaura3 leu2 his3 | 50 |
| KT1960 | MATα ura3-52 leu2 his3 trp1 | K. Tatchell |
| KT1961 | MATaura3-52 leu2 his3 trp1 | K. Tatchell |
| KT1703 | KT1960 glc7-132 | K. Tatchell |
| KT1705 | KT1961 glc7-133 | K. Tatchell |
| KT1706 | KT1960 glc7-133 | K. Tatchell |
| KT1708 | KT1960 glc7-127 | K. Tatchell |
| IP19 | KT1961 BUD14-myc13::kanMX6 | This study |
Plasmid construction.
Plasmids are listed in Table 2. To fuse the various genes to the LexA DNA-binding domain coding sequences in plasmid pEG202 and/or to the activation domain coding sequences in plasmid pJG4-5, yeast glc7-133, GLC7, BUD14, REG1, REG2, REF2, and GIP2 full-length coding sequences were amplified by PCR using Pfx Platinum polymerase (Invitrogen) and genomic wild-type DNA as template DNA (except for glc7-133, where genomic DNA from strain KT1706 was used). Appropriate restriction sites were introduced with the primers. The PCR products were cloned at the EcoRI-XhoI sites of pJG4-5 (using BUD14 and REG1 to yield pCDV472 and pCDV690, respectively), the NcoI-XhoI sites of pJG4-5 (using REG2, REF2, and GIP2 to yield pCDV697, pCDV688, and pCDV692, respectively), or the NcoI-XhoI sites of pEG202 (using GLC7 and glc7-133 to yield pCDV471 and pIP760, respectively). All constructs have the original start and stop codons of the fused genes. Plasmids pEG202-MSB2 and pJG4-5-MSB2 were described previously (47). To express GLC7 under the control of the ADH1 promoter, restriction sites were introduced immediately upstream (HindIII) and 616 nucleotides downstream (SacI) of the Glc7 coding sequence by PCR using wild-type genomic DNA (of strain KT1960) as template. The resulting amplicon was ligated into HindIII-SacI-digested YCpADH1 (43), thus creating pFD668. To obtain hemagglutinin (HA) epitope-tagged versions of Glc7, Glc7-127, Glc7-132, and Glc7-133, the corresponding full-length coding regions were amplified by PCR using genomic DNA of strains KT1960, KT1708, KT1703, and KT1706, respectively, as template. Restriction sites were introduced immediately upstream of the ATG codon (PstI) and 151 nucleotides downstream of the stop codons of the GLC7, glc7-127, glc7-132, and glc7-133 coding regions (HindIII). The PCR products were cloned at the PstI-HindIII sites of pAR498, which is YCplac22 containing an EcoRI-HindIII GAL1-HA2 fragment isolated from pAS24, to yield pIP607 (GAL1-HA2-GLC7), pIP765 (GAL1-HA2-glc7-127), pIP766 (GAL1-HA2-glc7-132), and pIP767 (GAL1-HA2-glc7-133). The STRE-lacZ reporter plasmid (pCTT1-18/7x) has been described previously (37).
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pEG202 | 2μm HIS3 ADH1-LexA-DBD | 55 |
| pEG202-MSB2 | pEG202 ADH1-LexA-DBD-MSB2 | 47 |
| pSH18-34 | 2μm URA3 LexAop(8)-lacZ | 25 |
| pJG4-5 | 2μm TRP1 GAL1-AD | 25 |
| pJG4-5-MSB2 | pJG4-5 GAL1-AD-MSB2 | 47 |
| YCplac22 | CEN TRP1 | 22 |
| pAS24 | CEN LEU2 GAL1-HA2 | 5 |
| YCpADH1 | CEN LEU2 ADH1-promoter | 43 |
| pCTT1-18/7x | URA3 STRE(7X)-lacZ | 37 |
| pFD668 | CEN LEU2 ADH1-GLC7 | This study |
| pCDV471 | pEG202 ADH1-LexA-DBD-GLC7 | This study |
| pIP760 | pEG202 ADH1-LexA-DBD-glc7-133 | This study |
| pCDV472 | pJG4-5 GAL1-AD-BUD14 | This study |
| pCDV690 | pJG4-5 GAL1-AD-REG1 | This study |
| pCDV697 | pJG4-5 GAL1-AD-REG2 | This study |
| pCDV688 | pJG4-5 GAL1-AD-REF2 | This study |
| pCDV692 | pJG4-5 GAL1-AD-GIP2 | This study |
| pAR498 | YCplac22 GAL1-HA2 | This study |
| pIP607 | YCplac22 GAL1-HA2-GLC7 | This study |
| pIP765 | YCplac22 GAL1-HA2-glc7-127 | This study |
| pIP766 | YCplac22 GAL1-HA2-glc7-132 | This study |
| pIP767 | YCplac22 GAL1-HA2-glc7-133 | This study |
2μm indicates high-copy-number plasmids, and CEN indicates low-copy-number plasmids. ADH1 and GAL1 designate only the promoter regions of the corresponding genes.
IP and immunoblotting.
To perform coprecipitation experiments with Bud14 and wild-type or mutant Glc7 proteins, strain IP19 expressing genomically tagged Bud14-myc13 was transformed with a control plasmid (pAR498) and with pIP607, pIP765, pIP766, and pIP767, which express HA2-GLC7, HA2-glc7-127, HA2-glc7-132, and HA2-glc7-133, respectively, under the GAL1 promoter. Overnight cultures grown at 30°C in synthetic defined medium with 2% (wt/vol) raffinose were diluted to an optical density at 600 nm (OD600) of 0.4 in the same medium and grown for an additional 2 h. Subsequently, 4% (wt/vol) galactose was added to induce GAL1-driven expression of the various Glc7 and Bud14 proteins during an additional 4 h at 30°C. Cells were then harvested by centrifugation and resuspended in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 0.1 M NaCl, 1 mM EDTA, 1% NP-40, one tablet of Complete Protease Inhibitor Cocktail [Roche Diagnostics GmbH]). Following addition of an equal volume of acid-washed glass beads, cells were broken by subjecting them to four 30-s cycles in a cell disruptor (FastPrep FP120). The extracts were clarified three times by centrifugation for 10 min at 4°C in a microfuge (17,000 rpm). HA2-tagged Glc7 and Glc7 variants were purified from clarified extracts with a protein G-agarose immunoprecipitation (IP) kit (Roche Diagnostics GmbH) following the manufacturer's instructions using monoclonal mouse anti-HA antibodies (HA.11 and immunoglobulin G1; Covance). Protein G-agarose beads carrying immunoprecipitates were resuspended in 50 μl of 2× sample buffer (62.5 mM Tris [pH 6.8], 25% glycerol, 2% sodium dodecyl sulfate [SDS], 0.01% bromphenol blue, 5% β-mercaptoethanol), boiled for 5 min, and electrophoresed on 10% polyacrylamide-SDS gels. After electrophoresis, proteins were transferred to nitrocellulose for immunoblotting and subsequent detection (ECL system; Amersham) of coprecipitated myc-tagged Bud14 using monoclonal mouse anti-myc antibodies (Santa Cruz Biotechnology, Inc.). Immunoblot analyses of total lysates were carried out as described previously (42). For analyses of Msn2 electrophoretic mobility, proteins (of the equivalent of an OD600 of 1) of exponentially growing cells were extracted by the post-alkaline extraction method, separated on 7% polyacrylamide-SDS gels, transferred to nitrocellulose, and probed with anti-Msn2 antibodies (kind gift from E. Estruch).
Two-hybrid analyses.
Quantitative β-galactosidase assays were performed as described previously (2) with reporter plasmid pSH18-34. For the assays (see Table 5), we used strain EGY48 that had been cotransformed with a pEG202-based plasmid and a pJG4-5-based plasmid.
TABLE 5.
Two-hybrid interactions between Glc7, Glc7-133, and various regulatory subunitsa
| AD fusion gene | β-Galactosidase activity (Miller units) with indicated DBD fusion geneb
|
||
|---|---|---|---|
| GLC7 | glc7-133 | MSB2 | |
| BUD14 | 2,098 | 19 | 23 |
| REG1 | 365 | 77 | 1 |
| REG2 | 445 | 98 | 5 |
| REF2 | 120 | 55 | 5 |
| GIP2 | 1,785 | 851 | 1 |
| MSB2 | 12 | 2 | 14 |
Two-hybrid assays were performed as described in Materials and Methods. AD, activation domain; DBD, DNA-binding domain.
β-Galactosidase activities were measured in three independent isolates of each strain after growth for 16 h at 30°C in minimal medium containing 1% raffinose and 2% galactose. The average activities (in Miller units) are shown. Values that are at least 10-fold higher than those for each of the corresponding controls (pJG4-5-MSB2 and pEG202-MSB2) are shown in boldface.
mRNA preparation and synthesis of cDNA.
Yeast strains were grown at 30°C in YPD medium. Overnight cultures of 5 ml were diluted to an OD600 of 0.2 and maintained in exponential growth phase (OD600 < 1.0) for a period of 12 h by repeated dilution in fresh YPD medium to ensure complete depletion of stationary phase-specific transcripts. At this point exponential-phase samples were harvested. Subsequently, the cultures were grown until glucose was exhausted in the medium, and diauxic shift samples were harvested 30 min after glucose exhaustion. Total RNA was then extracted using the RNApure kit (GeneHunter Corporation) according to the manufacturer's instructions. Radiolabeled cDNA probes were generated from 1 μg of total RNA by reverse transcription of mRNA using Superscript II (Invitrogen), an oligo(dT) primer (10- to 20-mer mixture; Research Genetics), and [33P]dCTP. Labeled probes were purified by passage through Bio-Spin 6 Chromatography Columns (Bio-Rad) and denatured for 5 min at 95°C.
GeneFilter hybridization and data analysis.
Yeast Index GeneFilters (Research Genetics-Invitrogen) were hybridized with the labeled probes according to the manufacturer's protocol. The filters were scanned by use of a PhosphorImager (Fuji BAS-1000) to obtain digital images. Images produced by MacBas (Fuji) were converted to TIFFs and imported into the Pathways version 4.0 software (Research Genetics) for subsequent normalization against all data points and quantification of spot intensities. The average ratio was calculated from log2 expression ratios during the exponential phase of growth relative to the diauxic-shift transition from two independent experiments using either wild-type or mutant strains. Noninterpretable spots were manually flagged and excluded. A selection from the remaining spots was made to include only those open reading frames (ORFs) for which the discrepancy between the two independent experiments was less than 2.5-fold. Of the selected 3,466 ORFs, those with an average ratio in wild-type cells of at least 2.0 were analyzed for Bud14 and Msn2/Msn4 dependency. To this end, we calculated fold decrease values by dividing the average ratio in wild-type strains by the average ratio in bud14Δ and msn2 msn4 mutant strains for any given ORF. Descriptions of gene products were derived from the Saccharomyces Genome Database and/or the Comprehensive Yeast Genome Database (MIPS). Original data are available upon request.
Miscellaneous.
Glucose concentrations were determined by the glucose oxidase method (Roche Diagnostics, GmbH). DNA sequences were obtained using the BigDye primer cycle sequencing kit and an ABI 301 automated sequencer (Applied Biosystems) according to the manufacturer's instructions. Protein concentrations were measured by use of the Bio-Rad protein assays according to the manufacturer's instructions using bovine serum albumin as a standard. For β-galactosidase assays and analyses of mRNA levels, cells were grown exponentially in rich medium at 30°C to an OD600 between 0.8 and 1.2. Protein extracts (50 μg) were then tested for β-galactosidase activity as previously described (30). For analysis of mRNA levels, total cellular RNA was extracted by the hot acid phenol method, and lacZ transcript levels were measured by S1 analysis using a specific oligonucleotide as previously described (12). Chromatin IP (ChIP) and quantitative real-time PCR were also performed as previously described (16). Polyclonal antibodies against Taf3, Taf8, Taf9, Taf11, and Taf12 were raised in rabbits following purification of the corresponding recombinant proteins expressed in E. coli from pET15b-derived plasmids (Elevage Scientifique des Dombes). Oligonucleotide sequences for the specific promoter DNAs measured in this study are available upon request.
RESULTS
The Ccr4-Not complex controls differential TFIID promoter association.
We have previously shown that cross-linking of Taf1 (expressed from a centromeric plasmid) to the HSP12 promoter was increased in not5Δ cells (16). To investigate this effect in more detail, we decided to perform a series of ChIP experiments again using wild-type and not5Δ cells yet with different TFIID subunits and a larger set of promoters. The cross-linking of TBP and of the TFIID-specific Taf1 and Taf8 proteins (expressed from their own promoters) to the promoters of HSP12, HSP26, HSP104, ADH1, RPS8A, RPS9B, and BAT1 in unstressed and heat-shocked (10 min at 39°C) wild-type and not5Δ cells is shown in Table 3. In unstressed wild-type cells, we found that TBP, Taf1, and Taf8 cross-linking to the promoters of the highly expressed ADH1 or the RPS8A and RPS9B ribosomal protein genes was high compared to the observed cross-linking to the promoter of the weakly expressed HSP12 gene. TBP and Taf proteins (Tafs) are therefore differentially distributed across different promoters (e.g., the amount of Taf8 cross-linked to the RPS9B promoter is about 50-fold higher than the amount cross-linked to the HSP12 promoter) (Table 3), and their distribution pattern correlates well with the expression pattern from the corresponding promoters. When wild-type cells were subjected to a brief heat shock, cross-linking of TBP and Tafs strongly increased on highly expressed heat shock gene promoters (HSP12, HSP26, and HSP104) and generally decreased on repressed ribosomal protein gene promoters (RPS8A and RPS9B) (Table 3). Accordingly, in wild-type cells subjected to heat stress, the distribution pattern of TBP and Tafs also correlated well with the expression pattern from the corresponding promoters.
TABLE 3.
Cross-linking of the TFIID subunits TBP, Taf1, and Taf8 to various promoters in wild-type and not5Δ cellsa
| Pro- tein | Incuba- tion temp (°C) | Strain | Relative amount of DNA of:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| HSP12 | HSP26 | HSP104 | ADH1 | RPS8A | RPS9B | BAT1 | |||
| TBP | 30 | WT | 0.36 | 0.43 | 0.48 | 1.97 | 0.83 | 2.45 | 0.76 |
| not5Δ | 1.18 | 0.83 | 1.12 | 2.27 | 0.97 | 1.82 | 0.94 | ||
| 39 | WT | 6.10 | 11.80 | 16.40 | 6.37 | 1.35 | 1.17 | 1.10 | |
| not5Δ | 18.50 | 25.30 | 21.90 | 8.52 | 1.09 | 1.72 | 0.68 | ||
| Taf1 | 30 | WT | 0.36 | 0.38 | 0.43 | 0.74 | 1.20 | 3.23 | 0.52 |
| not5Δ | 0.62 | 0.46 | 0.61 | 0.89 | 0.93 | 2.31 | 0.41 | ||
| 39 | WT | 0.95 | 1.33 | 1.42 | 1.04 | 0.53 | 0.49 | 0.45 | |
| not5Δ | 2.02 | 3.09 | 2.40 | 1.77 | 0.73 | 0.80 | 0.45 | ||
| Taf8 | 30 | WT | 0.34 | 0.33 | 1.17 | 2.82 | 5.37 | 17.10 | 1.75 |
| not5Δ | 0.62 | 0.37 | 0.48 | 0.69 | 0.99 | 2.27 | 0.42 | ||
| 39 | WT | 3.47 | 4.29 | 5.01 | 2.75 | 1.14 | 1.11 | 1.18 | |
| not5Δ | 4.11 | 5.41 | 4.47 | 2.88 | 1.12 | 1.15 | 0.63 | ||
Wild-type (WT) (MY1) and not5Δ (MY1719) cells were grown to exponential phase and cross-linked following incubation for 10 min at 30 or 39°C. The indicated proteins (i.e., TBP, Taf1, and Taf8) were immunoprecipitated from total cell extracts, and the amount of DNA of each of the indicated promoters in the immunoprecipitates was quantified by real-time PCR and expressed relative to the amount of DNA in the corresponding total extracts (in arbitrary units). One representative data set (of a total of three) (unpublished data) is shown.
Loss of Not5 caused a dramatic change in the distribution of TBP and Tafs on various promoters in unstressed cells (Table 3). In general, the differential distribution of TBP, Taf1, and Taf8 was significantly reduced in not5Δ mutant cells (e.g., while the largest difference in cross-linking of Taf8 is 50-fold in wild-type cells [17.1 and 0.34 for the RPS9B and HSP12 promoters, respectively], this value is reduced to 6-fold in not5Δ cells [2.27 and 0.37 for the RPS9B and HSP26 promoters, respectively]). In particular, Taf1 and Taf8 cross-linking appeared to increase on the HSP12 promoter and to decrease on the RPS8A/9B promoters in not5Δ cells. Since we obtained similar results for additional TFIID-specific Tafs (Taf3 and Taf11) and Tafs that are shared between TFIID and SAGA (Taf9 and Taf12) (data not shown), our data indicate that Not5 is involved in recruitment and/or stabilization of TFIID on ribosomal protein gene promoters and in detraction and/or destabilization of TFIID on promoters of stress genes in unstressed cells. This assumption is further supported by additional experiments (data available on request), where the amount of TBP and Tafs cross-linked to various promoters was normalized to the corresponding amount cross-linked to the HSP12 promoter. Accordingly, loss of Not5 consistently caused a significant increase of the amount of TBP, Taf1, and Taf8 cross-linked to the HSP12 promoter relative to that cross-linked to the RPS9B promoter. It is worth mentioning that the heat shock-induced changes in distribution of Taf8 (and other TFIID subunits) (data not shown) across promoters remained largely unaffected by the loss of Not5 (Table 3) (unpublished data), indicating that Not5 exerts its control over TFIID distribution mainly under nonstress conditions.
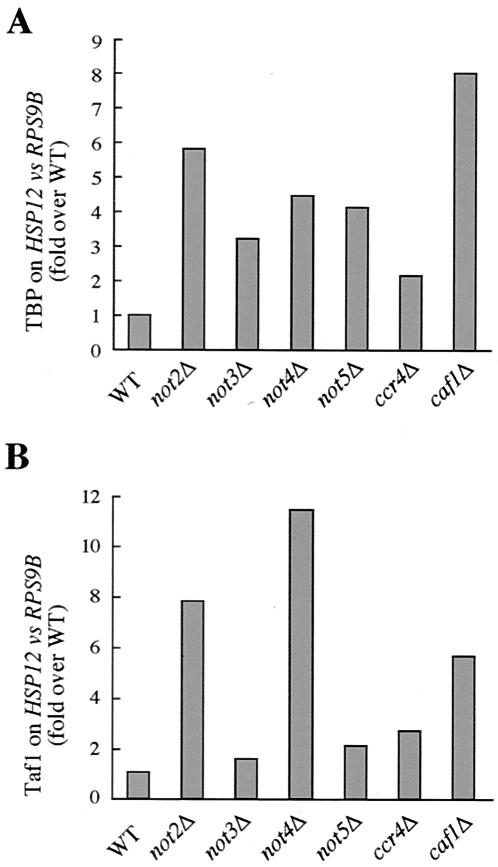
Finally, as with the loss of Not5, loss of various additional Ccr4-Not complex subunits (i.e., Not2, Not3, Not4, Ccr4, or Caf1) significantly increased the amount of both TBP (Fig. 1A) and Taf1 (Fig. 1B) that was cross-linked to the HSP12 promoter relative to that cross-linked to the RPS9B promoter (similar results were obtained when using an alternative pair of an STRE-controlled gene [HSP26] and a ribosomal protein gene [RPS8A]) (data not shown). The control over differential distribution of TFIID across promoters appears therefore to reflect a general function of the Ccr4-Not complex.
FIG. 1.
Distribution of TBP (A) and Taf1 (B) on HSP12 and RPS9B promoters in wild-type (WT) and various ccr4-not mutant cells. Wild-type (MY1), not2Δ (MY2182), not3Δ (MY508), not4Δ (MY2052), not5Δ (MY1719), ccr4Δ (MY1728), and caf1Δ (MY1729) strains were grown to exponential phase at 30°C. Following chromatin IP from whole cell extracts using either anti-TBP or anti-Taf1 antibodies, the levels of HSP12 and RPS9B promoter DNA in the immunoprecipitates relative to their levels in whole cell extracts were determined via reverse transcription-PCR. The values represent ratios of the levels of HSP12 to RPS9B promoter DNA precipitated with either anti-TBP or anti-Taf1 antibodies. Each value has been brought arbitrarily to the wild-type value (WT = 1). Results from one representative experiment are shown (similar results were obtained three times).
The Ccr4-Not complex controls TFIID distribution independently of Msn2.
To address the question of whether the redistribution of TFIID in ccr4-not mutants may result from constitutive activation of Msn2, we analyzed the amount of Taf8 that was cross-linked to the HSP12 promoter relative to that cross-linked to the RPS9B promoter in wild-type, not4Δ, msn2Δ, and not4Δ msn2Δ cells before and after heat shock (10 min at 39°C). Cross-linking of Taf8 to the HSP12 promoter relative to the RPS9B promoter was similarly increased in both exponentially growing not4Δ and not4Δ msn2Δ cells when compared to wild-type cells (Table 4), indicating that the changes in Taf8-promoter association following loss of Not4 do not require the presence of Msn2 (in unstressed cells). Under heat shock conditions, loss of Msn2 significantly reduced cross-linking of Taf8 to the HSP12 promoter relative to the RPS9B promoter in both wild-type and not4Δ cells (Table 4). This effect was mainly due to a decrease in cross-linking of Taf8 to HSP12 following heat shock (data not shown). Thus, while Msn2 is involved in recruitment of Taf8 to the HSP12 promoter under heat-shock conditions, it is dispensable for the redistribution of TFIID following inactivation of the Ccr4-Not complex in unstressed cells.
TABLE 4.
Relative cross-linking of Taf8 to the HSP12 and RPS9B promoters in unstressed and heat-shocked wild-type and various mutant cellsa
| Strain | Relative HSP12/RPS9B association ratio at:
|
|
|---|---|---|
| 30°C | 39°C for 10 min | |
| Wild type | 1.0 | 1.0 |
| not4Δ | 2.5 | 0.8 |
| msn2Δ | 0.8 | 0.2 |
| not4Δ msn2Δ | 2.5 | 0.1 |
| bud14Δ | 0.7 | 2.2 |
| not4Δ bud14Δ | 2.9 | 1.0 |
Wild-type (MY3590), not4Δ (MY3591), msn2Δ (MY3496), not4Δ msn2Δ (MY3498), bud14Δ (MY3634), and not4Δ bud14Δ (MY3633) cells were grown to exponential phase at 30°C and treated for 10 min at 39°C. The relative association of Taf8 to the promoters of HSP12 and RPS9B was evaluated from the average of three independent ChIP experiments. The ratio HSP12/RPS9B before (30°C) and after (10 min at 39°C) heat shock was brought arbitrarily to 1.0 in wild-type cells, and the other ratios are expressed relative to the wild-type ratio.
Activation of Msn2-dependent transcription in ccr4-not mutants requires normal protein phosphatase type I function.
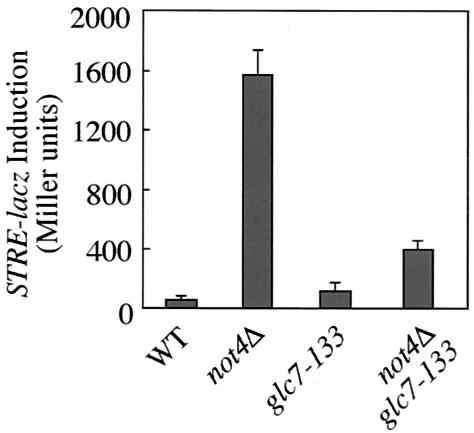
While the altered distribution of TFIID in not mutants appears to be independent of the presence of Msn2, the constitutively high expression of Msn2-controlled genes in these mutants strongly depends on the presence of Msn2 (30). Since we observed earlier that the loss of Ccr4-Not function resulted in aberrant posttranslational modification of Msn2, we considered the possibility that the Ccr4-Not complex may—via control of the activity of a protein phosphatase or kinase—regulate the capacity of Msn2 to activate target genes. In this context, we discovered that loss of Not4, similar to defined mutations in the essential type I protein phosphatase Glc7 (54), resulted in synthetic lethality when combined with loss of the two partially redundant Ppz1 and Ppz2 protein phosphatases (data not shown). One interpretation of these genetic data is that Not4 and Glc7 act in a common pathway that (e.g., due to a role in repression of Msn2-controlled, growth-inhibitory genes) becomes essential for cellular fitness in the absence of Ppz1 and Ppz2. To study both the epistatic relationship between not4Δ and glc7 and the potential contribution of Glc7 in Msn2-dependent transcription, we determined the effect of glc7 alleles on STRE-dependent gene expression in wild-type and not4Δ cells. As shown in Fig. 2, we found that the glucose-derepressed, recessive glc7-133 allele (3) dramatically reduced the constitutive STRE-dependent transcription in not4Δ mutant cells while having little impact on STRE-dependent transcription in unstressed wild-type cells. These data show that normal Glc7 function is required for the increased STRE-controlled gene expression in ccr4-not mutants and suggest that Glc7 may act directly or indirectly downstream of the Ccr4-Not complex to regulate Msn2 function. Intriguingly, in this context, we isolated the catalytic protein phosphatase domain of Ppz1, which is highly conserved between Ppz1, Ppz2, and Glc7 (54), in a two-hybrid screen for Not1 interactors (data not shown). Thus, even though Glc7 and the Ccr4-Not complex may independently converge on Msn2 function, an attractive model posits that Glc7 may act as an effector of the Ccr4-Not complex.
FIG. 2.
Constitutive activation of STRE-dependent transcription in not4Δ mutant cells depends partially on the function of the Glc7 phosphatase. Proteins were extracted from wild-type (MY2489), not4Δ (MY2596), glc7-133 (MY3362), and not4Δ glc7-133 (MY3363) cells growing exponentially (OD600 of 0.8 to 1.2) at 30°C on rich medium. Protein extracts (50 μg of proteins in total) were tested for β-galactosidase activity (indicated in Miller units).
Bud14, a new Glc7-binding protein, is involved in control of Msn2 function by the Ccr4-Not complex.
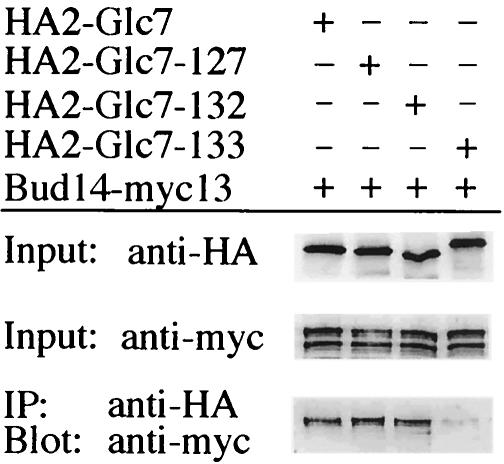
Glc7 itself has little substrate specificity, and it has been proposed that its specificity is dictated by different regulatory subunits that target the catalytic subunit to its site of action and/or regulate its substrate specificity (49). Consequently, we sought to isolate the corresponding Glc7 subunit(s) that specifies Glc7 activity in its presumed effector function for the Ccr4-Not complex. To this end, we performed a screen that was based on the assumption that specific activation of Glc7 due to overproduction of the appropriate regulatory subunit may mimic downregulation of the Ccr4-Not complex. Since SSA3 is one of the genes that is most strongly induced in exponentially growing ccr4-not complex mutants (30), we screened for genes that, when overexpressed from a 2μm plasmid, resulted in enhanced transcription of an SSA3-lacZ reporter gene (6). One of the genes isolated in this screen (i.e., BUD14) encodes a 78.3-kDa protein containing a potential SH3 domain and a V/IXF motif (at positions 377 to 379) that generally serves as a recognition site for the type 1 protein phosphatase Glc7 (10, 19). Both two-hybrid and co-IP assays confirmed that Bud14 specifically associates with Glc7 (Fig. 3; Table 5). Interestingly, the protein encoded by the glc7-133 allele, unlike two other Glc7 mutant proteins (i.e., Glc7-127 and Glc7-132) (3), was seriously defective for Bud14 binding (Fig. 3). Furthermore, while Glc7-133 was almost entirely defective for Bud14 binding (the Glc7-133-Bud14 interaction was reduced by more than 99% compared to the corresponding Glc7-Bud14 value), it was only partially defective for binding of several other known subunits, including Reg1, Reg2, Ref2, and Gip2 (Table 5) (for a review, see reference 49). The Glc7-Bud14 interaction is therefore particularly sensitive to the mutations encoded by the glc7-133 allele, which was able to suppress a ccr4-not mutant phenotype (see above).
FIG. 3.
Interaction between Bud14 and Glc7. Strain IP19 expressing genomically tagged Bud14-myc13 was transformed with a control plasmid (pAR498) and with pIP607, pIP765, pIP766, and pIP767, which express HA2-GLC7, HA2-glc7-127, HA2-glc7-132, and HA2-glc7-133, respectively, under the GAL1 promoter. Cell lysates (Input) and immunoprecipitates (IP) were subjected to PAGE, and immunoblots were probed using anti-HA or anti-myc antibodies. Note that the faster-migrating second band revealed in the anti-myc input blot may represent a degradation product of Bud14-myc13, which appears to poorly bind Glc7.
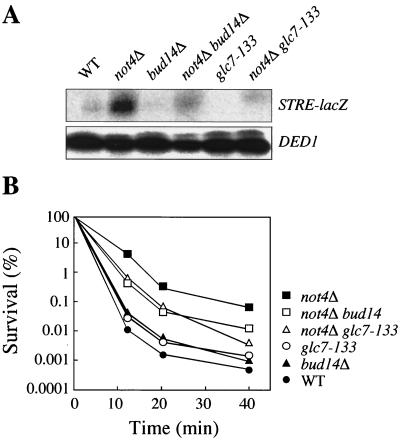
To determine whether the newly identified Bud14/Glc7 module might positively regulate Msn2-dependent transcription downstream of the Ccr4-Not complex, we deleted the nonessential BUD14 gene in both wild-type and not4Δ mutant cells and assayed STRE-dependent transcription in these strains. Interestingly, loss of Bud14—like introduction of the Bud14-binding-deficient glc7-133 allele—significantly reduced the observed derepression of STRE-dependent transcription in the not4Δ mutant (Fig. 4A) while having little impact on STRE-dependent transcription in unstressed wild-type cells. Moreover, the intrinsically high level of thermotolerance observed in not4Δ cells, which depends on Msn2 (data not shown), was significantly reduced following loss of Bud14 or introduction of glc7-133 (Fig. 4B). Together, these results formally place Bud14/Glc7 downstream of or in parallel to Not4 and support the assumption that Bud14/Glc7 and the Ccr4-Not complex have a common role in the regulation of STRE-dependent transcription.
FIG. 4.
Genetic interaction between NOT4 and GLC7/BUD14. (A) Constitutive activation of STRE-dependent transcription in not4Δ cells depends on the function of the Glc7/Bud14 module. Total RNA was extracted from wild-type (WT) (MY2489), not4Δ (MY2596), bud14Δ (MY3234), not4Δ bud14Δ (MY3235), glc7-133 (MY3362), and not4Δ glc7-133 (MY3363) cells growing exponentially (OD600 of 0.8 to 1.2) at 30°C on rich medium. LacZ transcript levels were measured via S1 analysis using DED1 as a control. (B) Mutations in GLC7 and BUD14 suppress the constitutive thermotolerance in not4Δ cells. The thermotolerance of exponentially growing cells (same strains as shown in panel A) was measured as survival following incubation at 50°C for the times indicated.
Bud14 and Glc7 control Msn2 posttranslational modification.
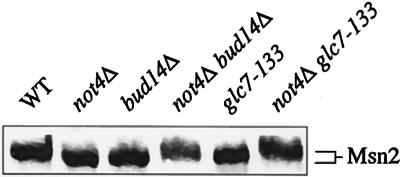
In an attempt to define whether the Bud14/Glc7 module is implicated in posttranslational modification of Msn2, we next analyzed the Msn2 isoform pattern in wild-type and various mutant cells. As shown in Fig. 5, Msn2 exhibited a higher electrophoretic mobility in not4Δ than wild-type cell extracts. Msn2 mobility was also altered in bud14Δ and glc7-133 cell extracts, supporting the notion that the Bud14/Glc7 phosphatase module is required for appropriate Msn2 modification in unstressed wild-type cells. Importantly, the high electrophoretic mobility of Msn2 in not4Δ cell extracts appears to be reversed by loss of Bud14 or introduction of the glc7-133 allele. Thus, the Bud14/Glc7 module (directly or indirectly) affects Msn2 posttranslational modifications in both wild-type and not4Δ cells. Notably, the fact that Msn2 electrophoretic mobility is similarly high in cells exhibiting strong (not4Δ) or weak (bud14Δ and glc7-133) STRE-driven transcription indicates that Msn2 electrophoretic mobility per se is not indicative of Msn2 activity. Since similarly migrating isoforms of Msn2 may even represent differentially phosphorylated proteins, resolution of this issue will ultimately depend on identification of the residues that are responsive to the Ccr4-Not complex, to Bud14/Glc7, and to the various protein kinases that have been implicated in Msn2 phosphorylation (9, 21, 23, 24, 26). Nevertheless, our data allow us to conclude at present that both the Ccr4-Not complex and the Glc7/Bud14 module, which is required for the enhanced Msn2-driven transcription following loss of Ccr4-Not function, control the posttranslational modification of Msn2.
FIG. 5.
Altered posttranslational modifications of Msn2 in not4Δ mutant cells depend partially on the function of the Glc7/Bud14 module. Proteins (of the equivalent of an OD600 of 1) of exponentially growing cells were extracted by the post-alkaline lysis method, separated on SDS-7% PAGE, transferred to nitrocellulose, and probed with anti-Msn2 antibodies. WT, wild type.
Neither the Ccr4-Not complex nor Bud14 contributes to Msn2 DNA association.
To answer the question of whether the Ccr4-Not complex and the Bud14/Glc7 module may impinge on the same Msn2 control mechanism, we studied both nuclear localization and promoter affinity of Msn2 in wild-type cells and corresponding mutant cells. We found that Msn2 was normally localized (i.e., cytoplasmic in exponentially growing cells) in various notΔ, bud14Δ, and glc7-133 mutants (data not shown). In addition, not4Δ, bud14Δ, and not4Δ bud14Δ cells exhibited a similar level of Msn2 cross-linked to the STRE region of the HSP26 promoter in both exponentially growing and heat-shocked wild-type cells (Table 6). Thus, neither a change in subcellular localization of Msn2 nor altered STRE-promoter affinity is sufficient to explain the dramatic increase in STRE-dependent transcription observed in exponentially growing not mutants or the suppression of this phenotype following loss of Bud14.
TABLE 6.
Cross-linking of Msn2 to HSP26 STRE in unstressed and heat-shocked wild-type, not4Δ, bud14Δ, and not4Δ bud14Δ mutant cellsa
| Strain | Relative amount of HSP26 STRE promoter DNA at:
|
|
|---|---|---|
| 30°C | 39°C for 10 min | |
| Wild type | 1.3 | 3.1 |
| not4Δ | 1.1 | 2.7 |
| bud14Δ | 1.0 | 6.4 |
| not4Δ bud14Δ | 1.6 | 5.0 |
Wild-type (MY3590), not4Δ (MY3591), bud14Δ (MY3634), and not4Δ bud14Δ (MY3633) cells were grown to exponential phase at 30°C and treated for 10 min at 39°C. Msn2 was immunoprecipitated by use of a myc tag using monoclonal anti-myc antibodies. The levels of HSP26 STRE promoter DNA in the immunoprecipitates of unstressed (30°C) or heat-shocked (10 min at 39°C) cells are expressed relative to their corresponding presence in the total cell extract.
Since our results indicated that the Ccr4-Not complex controls Msn2 activity and TFIID distribution via two independent pathways, we also tested whether Bud14 may, in addition to playing a role in Msn2 activation, be involved in TFIID distribution. As shown in Table 4, cross-linking of Taf8 to the HSP12 promoter relative to the RPS9B promoter was increased in not4Δ cells independently of the presence or absence of Bud14, indicating that Bud14, like Msn2, is not involved in the redistribution of TFIID in not4Δ mutant cells. Finally, Bud14 also was not required for the increased recruitment of TFIID to heat shock promoters following heat shock (Table 4), which is in line with our observation that loss of Bud14 or introduction of the glc7-133 allele did not significantly alter the induction of STRE-controlled genes following a 10-min heat shock at 39° (data not shown).
The Ccr4-Not complex may prevent Msn2 activation by negative control of the Bud14/Glc7 module under high-PKA conditions.
We have previously shown that the Ccr4-Not complex may function as an effector of the PKA pathway that contributes to downregulation of Msn2-dependent transcription of growth-inhibitory genes under conditions of high PKA (30, 42, 48). Based on the results shown above, the Ccr4-Not complex may perform this function at least in part via inhibition of the Glc7/Bud14 module, which—particularly following release from repression by the Ccr4-Not complex (for instance, under conditions of low PKA at the diauxic shift)—may positively regulate Msn2. In line with this model, we found that the dosage-dependent effect of Bud14 on SSA3-lacZ transcription was mainly apparent in cells that have entered the diauxic shift (data not shown), which temporally coincides with the time of Ccr4-Not complex downregulation (30). Furthermore, whole-genome array analysis confirmed a positive role of Bud14 in regulation of Msn2-controlled genes at the diauxic transition. Accordingly, we found that of 375 genes that were induced in wild-type cells at the diauxic transition, a large fraction (54.6% or 205 genes) required Msn2/Msn4 for induction, which is in accordance with previously published data (7). Importantly, 57 (27.8%) of these Msn2/Msn4-dependent genes also required Bud14 for induction, and application of a less stringent cutoff value for Bud14-dependent genes resulted in an almost complete overlap of the Bud14- and Msn2/Msn4-dependent gene sets. This is further illustrated in Table 7, which shows the entire set of genes that were most strongly dependent on Bud14 for induced expression at the diauxic shift (i.e., at least 2.5-fold reduced in bud14Δ cells compared to wild-type cells), including the corresponding values for fold decrease in bud14Δ and msn2 msn4 mutant cells. The fact that the defect of bud14Δ mutant cells for induction of Msn2/Msn4-dependent genes at the diauxic transition was on average much lower (2.1-fold decrease) than the corresponding defect of msn2 msn4 mutant cells (3.3-fold decrease) indicates the presence of additional (possibly redundant) regulatory mechanisms, which allow nutrient limitation-induced activation of Msn2/Msn4-dependent transcription in the absence of Bud14. Our observation that loss of Bud14 did not affect the expression levels of STRE-controlled genes in later postdiauxic growth phases (data not shown) supports the idea that the main role of Bud14 in regulation of transcription is confined to the diauxic transition phase.
TABLE 7.
Bud14- and Msn2/Msn4-dependent gene induction at the diauxic transition
| ORF | Gene | Description | Fold decrease ina:
|
|
|---|---|---|---|---|
| bud14Δ cells | msn2 msn4 cells | |||
| YOR135C | IDH2 | Oxidoreductase | 4.7 | 3.5 |
| YGR101W | Involved in energy generation | 4.3 | 2.4 | |
| YEL070W | Mannitol-1-phosphate 5-dehydrogenase | 4.1 | 4.2 | |
| YML087C | Molecular function unknown | 4.1 | 3.9 | |
| YGR110W | Molecular function unknown | 3.9 | 3.8 | |
| YLR116W | Branch point bridging protein | 3.8 | 2.7 | |
| YLR070C | Putative sugar dehydrogenases | 3.2 | 2.7 | |
| YER096W | Sporulation-specific protein | 3.1 | 1.8 | |
| YHR211W | Flocculation-specific protein | 3.1 | 1.8 | |
| YDR497C | ITR1 | Major myo-inositol permease | 3.0 | 1.6 |
| YML120C | NDII | NADH-ubiquinone-6 oxidoreductase | 2.9 | 1.9 |
| YKR052C | MRS4 | Mitochondrial membrane transporter | 2.9 | 4.1 |
| YBR280C | Molecular function unknown | 2.8 | 4.6 | |
| YBL075C | SSA3 | Heat shock protein of HSP70 family | 2.8 | 4.8 |
| YNL125C | Molecular function unknown | 2.8 | 4.3 | |
| YJL225C | ATP-dependent DNA helicase | 2.8 | 2.4 | |
| YPR026W | ATH1 | Acid trehalase | 2.7 | 2.3 |
| YDL078C | MDH3 | Malate dehydrogenase | 2.7 | 2.0 |
| YCLX05C | Molecular function unknown | 2.7 | 1.7 | |
| YKL146W | Neutral amino acid transporter | 2.6 | 3.2 | |
| YIR043C | Molecular function unknown | 2.6 | 1.8 | |
| YOR005C | DNL4 | DNA ligase | 2.6 | 2.2 |
| YHR033W | Putative glutamate 5-kinase | 2.6 | 1.1 | |
| YLR178C | TFS1 | Cdc25-dependent nutrient response regulator | 2.6 | 3.5 |
| YER060W-A | FCY22 | Purine/cytosine permease | 2.6 | 3.3 |
| YJL082W | Molecular function unknown | 2.5 | 2.0 | |
| YHR006W | STP2 | Transcription factor | 2.5 | 2.9 |
| YBR139W | Protease | 2.5 | 3.7 | |
Genes with a fold decrease in bud14Δ (PE27) cells of >2.5 were selected from the pool of genes that were at least two-fold induced (average ratio, > 2) in wild-type cells at the diauxic transition. The corresponding fold decrease in msn2 msn4 mutant (PEY78) cells is also shown.
To determine whether Bud14, as expected for an activator of Msn2, also modulates PKA-dependent growth, we then studied whether loss of Bud14 could suppress growth defects that are associated with attenuated PKA activity. To this end, a temperature-sensitive PKA mutant strain with only one functional tpk2(Ts) gene was transformed with the bud14Δ::kanMX2 cassette. Even though loss of Bud14 did not restore growth of the tpk2(Ts) cells at the nonpermissive temperature, it increased the growth rate of the tpk2(Ts) cells at a semipermissive temperature (31°C) from 0.107 ± 0.010 h−1 (SGY446) to 0.287 ± 0.023 h−1 (PE15). Loss of Bud14 therefore at least partially relieves dependence on PKA function. Overproduction of Bud14, in contrast, strongly inhibited the growth at 31°C of tpk2(Ts) cells (similar results were obtained using temperature-sensitive Ras GTP exchange factor and adenylate cyclase mutant strains harboring cdc25(Ts) and cdc35(Ts) mutations, respectively) (data not shown). Together, these results show that Bud14, possibly via Glc7, antagonizes PKA-mediated cell proliferation control. This is also in line with a previous report in which overexpression of GLC7 was found to prevent growth of ras1 ras2(Ts) mutant cells (39) and our own observation that Glc7 overproduction (using an ADH1-GLC7 construct, pFD688) strongly reduced the growth rate at 36°C of cdc35(Ts) (PD6517) cells (i.e., from 0.0730 ± 0.003 h−1 to 0.0014 ± 0.003 h−1), while it only slightly reduced the growth rate of cdc35(Ts) bud14Δ (PE6) cells (i.e., from 0.083 ± 0.004 h−1 to 0.068 ± 0.003 h−1). Thus, Glc7 antagonizes PKA-mediated growth at least in part through Bud14. Taken together, our combined genetic and molecular experiments support a model in which PKA-dependent repression of Msn2 is mediated at least in part by the Ccr4-Not complex, possibly through control of the Glc7/Bud14 module.
DISCUSSION
In this work, we have studied the association of TFIID subunits across different promoters in wild-type cells or cells with deletions of various subunits of the Ccr4-Not complex. In accordance with our previous study (16), which was focused on the analysis of Taf1, we show here that loss of Ccr4-Not complex subunits dramatically changes the overall promoter distribution pattern of several TFIID subunits. Accordingly, in unstressed exponentially growing wild-type cells, we found a high level of differential promoter cross-linking of Tafs (27, 31, 40), with a particular bias for promoters of ribosomal protein genes, whose expression is strong and Taf dependent, and against promoters of heat shock genes, whose expression is weak and Taf independent under these conditions. In Ccr4-Not complex mutant cells, in contrast, we found a generally low level of differential promoter cross-linking of Tafs, with a particular bias against promoters of ribosomal protein genes. Thus, the Ccr4-Not complex functions to ensure that the differential promoter association of Tafs remains high and appears to particularly stabilize ribosomal protein gene promoter association of TFIID under nonstress conditions (Fig. 6). Since we have previously demonstrated that the dramatically increased expression of STRE-controlled genes in ccr4-not mutants was dependent on the presence of Msn2, we reasoned that the TFIID promoter distribution defect in ccr4-not mutants may result indirectly from constitutive activation of Msn2. Interestingly, however, the observed changes in TFIID promoter distribution due to mutations in the Ccr4-Not complex appeared similar in the presence or absence of Msn2, indicating that the Ccr4-Not complex controls TFIID promoter association independently of Msn2.
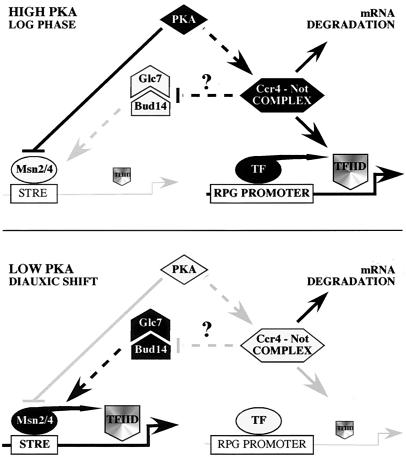
FIG. 6.
Model for the function of the Ccr4-Not complex. (Top) Cells growing exponentially (high-PKA conditions). The level of TFIID on STRE-controlled promoters is low compared to the level on RPG promoters. In parallel, STRE gene expression is low, and ribosomal protein gene (RPG) expression is high. The Ccr4-Not complex positively regulates TFIID presence on RPG promoters and may negatively control the Bud14/Glc7 module. TF, presumed transcription factor(s). (Bottom) Cells entering the diauxic shift (low-PKA conditions). The level of TFIID on STRE-controlled promoters is increased compared to the level on RPG promoters. In parallel, STRE gene expression increases, and RPG expression decreases. The Glc7/Bud14 module is relieved from repression and may activate STRE genes following posttranslational modification of Msn2. Note that this activation may actually occur before both the exhaustion of glucose and the subsequent transfer of the major portion of Msn2 to the nucleus. Arrows and bars denote positive and negative interactions, respectively. Grey and black bars and symbols denote low and high activity, respectively.
While in principle it is possible that Msn2 activation results from the altered TFIID distribution in ccr4-not mutant cells, we also considered the possibility that the Ccr4-Not complex may activate Msn2 via a mechanism that is separate from its function in TFIID promoter distribution. Definition of such an independent mechanism requires the identification of signaling molecules that are necessary for proper STRE-dependent transcription without affecting the general distribution of TFIID across promoters. In this context, we isolated a new type I protein phosphatase module in yeast (i.e., Bud14/Glc7) that fulfilled both of these criteria (i.e., it is required for proper STRE-dependent transcription at the diauxic shift and does not appear to affect TFIID promoter association). Intriguingly, we found that both the dramatic, constitutive Msn2/STRE-dependent transcription and the constitutive thermotolerance of various ccr4-not mutants are strongly dependent on the presence of Bud14 or a wild-type Glc7 protein. Moreover, mutations in the Ccr4-Not complex caused Msn2 posttranslational modifications that appeared to be reverted by loss of Bud14 or a Glc7-133 mutant protein. The simplest model that emerges from our genetic analyses is therefore that the Ccr4-Not complex may regulate the activity of Msn2 via a Glc7/Bud14-dependent, TFIID-independent mechanism (Fig. 6). It must be noted however, that while our results do not exclude additional, more complex models (particularly involving further intermediary steps or nonlinear interactions), our simple linear model provides an initial framework which predicts a variety of biochemical interactions that can be tested in the future.
How does the Ccr4-Not complex impinge on Msn2 activation? During heat shock, Msn2 activation appears to be accomplished through both nuclear accumulation and enhanced STRE binding (24, 26). We found that the Ccr4-Not complex and Bud14/Glc7 regulate neither nucleocytoplasmic localization of Msn2 nor its recruitment to STRE-controlled promoters. From these observations, we infer that the mechanism of Msn2 activation following both inactivation of the Ccr4-Not complex and activation of the Bud14/Glc7 complex differs significantly from the one observed following heat stress. (This is also in line with our findings that (i) enhanced Taf8 cross-linking to the HSP12 promoter is independent of Msn2 following loss of Not4 yet strongly dependent on Msn2 following heat stress and (ii) STRE-driven gene expression is strongly dependent on Bud14 following loss of Not4 yet independent of Bud14 following heat stress). One possibility is that the Ccr4-Not complex primarily impinges on the ability of Msn2 to communicate with the general transcription machinery, particularly in response to the availability of nutrients. Strikingly, Msn2 has previously been shown to interact with Srb10, a component of the RNA Pol II holoenzyme (9), suggesting that Msn2 may communicate with the SRB-Mediator complex. Thus, a likely scenario that is based on our present and previously published data (16, 30) is that the Ccr4-Not complex and Bud14/Glc7, rather than controlling the subcellular localization or STRE binding of Msn2, may control the ability of Msn2 to activate Pol II-dependent transcription following nutrient limitation. Accordingly, in exponentially growing (high-PKA) cells, the Ccr4-Not complex may serve to prevent activation of Msn2 (possibly via inactivation of Bud14/Glc7), while inactivation of the Ccr4-Not complex during the diauxic shift (low PKA) may allow activation of Msn2 (possibly via Bud14/Glc7) (Fig. 6). This model is also in line with two additional observations, namely, that Bud14 increases in abundance (>10-fold) and—like Msn2—accumulates in the nuclei of cells entering the diauxic shift (data not shown). In summary, the Ccr4-Not complex may, as proposed previously, function as an effector of the PKA pathway that contributes, via inactivation of Bud14/Glc7, to downregulation of Msn2-dependent transcription of growth-inhibitory genes in cells growing on glucose (30, 42, 48). Elucidation of the precise nature of the biochemical interactions between the proteins in this proposed separate effector pathway is warranted to provide further insight into how the Ccr4-Not complex contributes to the control of yeast cell growth.
Acknowledgments
We thank K. Tatchell, E. Estruch, and S. Garrett for reagents necessary to carry out this work and N. Bürckert for technical assistance. We are grateful to A. Wiemken and T. Boller for their support during the initial phase of this project, which was carried out at the Botanical Institute of the University of Basel.
This work was supported by grants of the Katholieke Universiteit Leuven and the Fund for Scientific Research of Flanders to J.W. and grants of the Swiss National Science Foundation to M.A.C. (3100-059199) and C.D.V. (631-62731.00).
REFERENCES
- 1.Albert, T. K., H. Hanzawa, Y. I. A. Legtenberg, M. J. de Ruwe, F. A. J. van den Heuvel, M. A. Collart, R. Boelens, and H. T. M. Timmers. 2002. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 21:355-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Baker, S. H., D. L. Frederick, A. Bloecher, and K. Tatchell. 1997. Alanine-scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics 145:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, J. D., M. Benson, Peter M. Howley, and K. Struhl. 1998. Association of distinct yeast Not2 functional domains with components of Gcn5 histone acetylase and Ccr4 transcriptional regulatory complexes. EMBO J. 17:6714-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickle, M., P.-A. Delley, A. Schmidt, and M. N. Hall. 1998. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 17:2235-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boorstein, W., and E. Craig. 1990. Regulation of a yeast HSP70 gene by a cAMP responsive transcriptional control element. EMBO J. 9:2543-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie, and M. Jacquet. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., Y.-C. Chiang, and C. L. Denis. 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21:1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cicchetti, P., B. J. Mayer, G. Thiel, and D. Baltimore. 1992. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science 257:803-806. [DOI] [PubMed] [Google Scholar]
- 11.Collart, M. A. 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313:1-16. [DOI] [PubMed] [Google Scholar]
- 12.Collart, M. A., and K. Struhl. 1993. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 12:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collart, M. A., and K. Struhl. 1994. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8:525-537. [DOI] [PubMed] [Google Scholar]
- 14.Collart, M. A., and H. T. Timmers. 2004. The eukaryotic ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 77:289-322. [DOI] [PubMed] [Google Scholar]
- 15.Daugeron, M.-C., F. Mauxion, and B. Séraphin. 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deluen, C., N. James, L. Maillet, M. Molinete, G. Theiler, M. Lemaire, N. Paquet, and M. A. Collart. 2002. The Ccr4-Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Mol. Cell. Biol. 22:6735-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denis, C. L., and J. Chen. 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73:221-250. [DOI] [PubMed] [Google Scholar]
- 18.dos Passos, J. B., M. Vanhalewyn, R. L. Brandao, I. M. Castro, J. R. Nicoli, and J. M. Thevelein. 1992. Glucose-induced activation of plasma membrane H+-ATPase in mutants of the yeast Saccharomyces cerevisiae affected in cAMP metabolism, cAMP-dependent protein phosphorylation and the initiation of glycolysis. Biochim. Biophys. Acta 1136:57-67. [DOI] [PubMed] [Google Scholar]
- 19.Egloff, M.-P., D. F. Johnson, G. Moorhead, P. T. W. Cohen, P. Cohen, and D. Barford. 1997. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16:1876-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estruch, F., and M. Carlson. 1993. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garreau, H., R. N. Hasan, G. Renault, F. Estruch, E. Boy-Marcotte, and M. Jacquet. 2000. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 146:2113-2120. [DOI] [PubMed] [Google Scholar]
- 22.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 23.Görner, W., E. Durchschlag, M. T. Martínez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schüller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Görner, W., E. Durchschlag, J. Wolf, E. L. Brown, G. Ammerer, H. Ruis, and C. Schüller. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 26.Hirata, Y., T. Andoh, T. Asahara, and A. Kikuchi. 2003. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol. Biol. Cell 14:302-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuras, L., P. Kosa, M. Mencía, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 28.Lee, T. I., J. J. Wyrick, S. S. Koh, E. G. Jennings, E. L. Gadbois, and R. A. Young. 1998. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 18:4455-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemaire, M., and M. A. Collart. 2000. The TATA-binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4-Not complex and physically interacts with the Not5 subunit. J. Biol. Chem. 275:26925-26934. [DOI] [PubMed] [Google Scholar]
- 30.Lenssen, E., U. Oberholzer, J. Labarre, C. De Virgilio, and M. A. Collart. 2002. Saccharomyces cerevisiae Ccr4-Not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol. 43:1023-1037. [DOI] [PubMed] [Google Scholar]
- 31.Li, X.-Y., S. R. Bhaumik, X. Zhu, L. Li, W.-C. Shen, B. L. Dixit, and M. R. Green. 2002. Selective recruitment of TAFs by yeast upstream activating sequences: implications for eukaryotic promoter structure. Curr. Biol. 12:1240-1244. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H.-Y., Y.-C. Chiang, J. Pan, J. Chen, C. Salvadore, D. C. Audino, V. Badarinarayana, V. Palaniswamy, B. Anderson, and C. L. Denis. 2001. Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4-NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J. Biol. Chem. 276:7541-7548. [DOI] [PubMed] [Google Scholar]
- 33.Liu, H.-Y., J. H. Toyn, Y.-C. Chiang, M. P. Draper, L. H. Johnston, and C. L. Denis. 1997. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 16:5289-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longtine, M. S., A. McKenzie III, D. J. DeMarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 35.Maillet, L., and M. A. Collart. 2002. Interaction between Not1p, a component of the Ccr4-Not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 277:2835-2842. [DOI] [PubMed] [Google Scholar]
- 36.Maillet, L., C. Tu, Y. K. Hong, E. O. Shuster, and M. A. Collart. 2000. The essential function of Not1 lies within the Ccr4-Not complex. J. Mol. Biol. 303:131-143. [DOI] [PubMed] [Google Scholar]
- 37.Marchler, G., C. Schüller, G. Adam, and H. Ruis. 1993. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 12:1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martínez-Pastor, M. T., G. Marchler, C. Schüller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuura, A., and Y. Anraku. 1994. Genetic interaction between the Ras-cAMP pathway and the Dis2s1/Glc7 protein phosphatase in Saccharomyces cerevisiae. Mol. Gen. Genet. 242:257-262. [DOI] [PubMed] [Google Scholar]
- 40.Mencía, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 41.Oberholzer, U., and M. A. Collart. 1998. Characterization of NOT5 that encodes a new component of the Not protein complex. Gene 207:61-69. [DOI] [PubMed] [Google Scholar]
- 42.Pedruzzi, I., N. Bürckert, P. Egger, and C. De Virgilio. 2000. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 19:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinders, A., N. Bürckert, T. Boller, A. Wiemken, and C. De Virgilio. 1998. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 12:2943-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 47.Simon, M.-N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376:702-705. [DOI] [PubMed] [Google Scholar]
- 48.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark, M. J. R. 1996. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast 12:1647-1675. [DOI] [PubMed] [Google Scholar]
- 50.Stuart, J. S., D. L. Frederick, C. M. Varner, and K. Tatchell. 1994. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol. Cell. Biol. 14:896-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 52.Tucker, M., R. R. Staples, M. A. Valencia-Sanchez, D. Muhlrad, and R. Parker. 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 54.Venturi, G. M., A. Bloecher, T. Williams-Hart, and K. Tatchell. 2000. Genetic interactions between GLC7, PPZ1 and PPZ2 in Saccharomyces cerevisiae. Genetics 155:69-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zervos, A. S., J. Gyuris, and R. Brent. 1993. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72:223-232. [DOI] [PubMed] [Google Scholar]