Abstract
The luxR homolog aviR in Agrobacterium vitis strain F2/5 was recently shown to be associated with induction of a hypersensitive response (HR) on tobacco and necrosis on grape plants, indicating that the responses are regulated by quorum sensing. We now report a second luxR homolog, avhR, whose disruption (mutant M1320) results in HR-negative and reduced grape necrosis phenotypes. The deduced AvhR protein has characteristic autoinducer binding and DNA binding domains and is unique among reported functional LuxR homologs in having substitutions at highly conserved Asp70, Trp57, and Trp85 residues, which are predicted to play important roles in autoinducer binding in TraR. M1320 was fully complemented with cloned avhR. The same array of N-acylhomoserine lactones (AHL) from F2/5, M1320, and complemented M1320 were observed; however, the signal strength from extracts of 6-day-old M1320 cultures was stronger than that of F2/5. Cultures of F2/5 amended with AHL extracts from overnight and 6-day cultures of F2/5 and M1320 were not affected in ability to cause HR or necrosis. A region of about 14 kb flanking avhR was sequenced and compared with homologous regions of A. tumefaciens C58 and Sinorhizobium meliloti Rm1021 genomes. Gene order and homology are conserved between the species. A site-directed mutation in a putative gene that resides downstream of avhR and that has homology to genes belonging to the ATP-binding cassette transporter family did not affect HR or necrosis phenotypes. It was determined that avhR and aviR are expressed independently and that neither regulates the expression of a clpA homolog in F2/5.
Quorum-sensing gene regulation is a means of cell-cell communication in diverse species of gram-negative bacteria and is characterized by the presence of specific proteins that function as transcriptional regulators (LuxR homologs) and N-acylhomoserine lactone (AHL) synthases (LuxI homologs) (29). Several important physiological functions, such as symbiosis, conjugation, and virulence, have been shown to be regulated by quorum sensing. Amino acid sequences of members of the LuxR protein family have relatively low identity (18 to 25%), and only five consensus residues have been identified in currently published functional proteins. However, all possess a characteristic amino terminus autoinducer binding domain and a carboxy terminus helix-turn-helix DNA binding domain (29). Specific amino acid residues that are highly conserved and are associated with AHL and DNA binding have been identified in TraR (28, 31).
There are currently several LuxR/LuxI homologs that have been identified; however the functions of relatively few have been identified. Notable examples are LuxR in Vibrio fischeri (regulation of bioluminescence) (8), TraR/TraI in Agrobacterium tumefaciens (regulation of Ti plasmid conjugal transfer) (9), and SinR/SinI and associated ExpR in Sinorhizobium meliloti (regulation of symbiotically active EPS II) (19, 22).
Agrobacterium vitis causes crown gall, a serious disease of grape plants, which may result in poor growth and death of vines (1). In addition to causing tumors, A. vitis induces a tissue-specific necrosis on its grape host and a hypersensitivity-like response (HR) on nonhost plants, such as tobacco (13). To investigate mechanisms associated with necrosis and HR, A. vitis strain F2/5 Tn5 mutants were selected based on altered necrosis and HR phenotypes. One mutant, M1154, which is completely necrosis and HR negative, was found to have the Tn5 insertion in a luxR homolog that was named aviR (32). It was determined that M1154 produces fewer AHLs than wild-type F2/5. Complemented M1154 regained full ability to cause grape necrosis and the HR, indicating that the phenotypes are regulated by a quorum-sensing system. In this paper we identify a second luxR homolog, avhR, in F2/5, which is expressed independently of aviR and which is essential for induction of the HR on tobacco and for full expression of necrosis on grape plants.
MATERIALS AND METHODS
Culture of bacteria.
A. vitis strain F2/5 was propagated on potato dextrose agar (PDA) or in potato dextrose broth (PDB) (Difco Laboratories, Detroit, Mich.) at 28°C. Tn5 mutants M1154 and M1320 were cultured on PDA or PDB amended with kanamycin (50 μg/ml) at 28°C. Escherichia coli was grown on Luria-Bertani (LB) medium with appropriate antibiotics at 37°C. Assays for necrosis on grape shoot explants and for HR on tobacco leaves were done as previously described (13).
Characterization of the Tn5-flanking sequences.
It was previously shown that the F2/5 mutant M1320 carries a single Tn5 insertion (13). Genomic DNA of M1320 was isolated and digested with EcoRI, ligated to plasmid vector pBluescript SK(−), and transformed into E. coli strain JM109 competent cells as previously described (13). Transformants containing inserts that included Tn5 were selected on LB plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 50 μg/ml), kanamycin (50 μg/ml), and carbenicillin (100 μg/ml). The clones were characterized by restriction analysis and sequencing.
Complementation of M1320.
Based on the DNA sequence flanking the Tn5 insertion in M1320, primers M1320-F and M1320-R were designed (Table 1). They amplified a 1.2-kb DNA fragment from F2/5 genomic DNA that was subsequently used to probe an F2/5 cosmid library by colony hybridization (13). Clone CPM201 was transformed into E. coli S17-1 and transferred to M1320 by conjugation. The complementation of HR and grape necrosis phenotypes by M1320(pCPM201) was determined.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| Primers for amplification of probe to screen cosmid clones | |
| M1320-F | TTGGCTGTAACTGCCGGAATG |
| M1320-R | TTGAGAGCCATGGGGACCAGC |
| Primers for amplifying sequences used in site-directed mutagenesis | |
| avhR-F1 | ACATGAGCTCGCGAATTGAGCGGCTATAAGAC |
| avhR-R1 | TCGATCTAGAGACCACATGAATATCGAAAACAAC |
| avhR-Int | AAGAGAACAAGGCTTTTTATC |
| ORF8-F | TACTGAGCTCACTGGTTGCGGCGGTGGAA |
| ORF8-R | ACGATCTAGAAGGCGGCAGGGACTTAGC |
| ORF8-Int | CACCCGCAAGCTGGATACC |
| PVIK165-P | TACTCATCTTTGTTTCCTCC |
| Primers for amplification of avhR | |
| avhR-F2 | GTAGAATTCATAAGGCTTTTTATCGATGGGTGACGATCTGGT |
| avhR-R2 | TCTCCTGCAGCTATTTGATCAGGCTCAACCGTAG |
| Primers for RT-PCR | |
| aviR-F | TACGGCCTAGTGCGCAGTCCCAAAC |
| aviR-R | TCAGCTGATCAGGCCGAGACGA |
| avhR-F3 | GATATGGCGTCCCTGCTGTCGTC |
| avhR-R3 | CGGGTGGCGCGGTTGAGATAATG |
| clpA-F | GCTGCCAAGCGAAGTGATCCACA |
| clpA-R | TCCAGCAAAAGTCGAAGCGTTACAG |
Site-directed mutagenesis.
Analysis of DNA sequence flanking the Tn5 insertion revealed that two putative genes that could have been disrupted by the transposon, i.e., a luxR homolog that we designate avhR and a downstream open reading frame (ORF8) that has homology with genes belonging to the ATP-binding cassette (ABC) transport family (14). Therefore, site-directed mutant versions of both putative genes were generated by mating F2/5 with E. coli strain S17-1/λpir carrying suicide plasmid pVIK165, into which a PCR-amplified fragment of the ORF to be disrupted was cloned (16). Gene disruption occurs following a single homologous recombination event whereby the entire plasmid inserts into a portion of the target gene. Primer pairs into which XbaI and SacI restriction sites were engineered included avhR-F1 and avhR-R1 and ORF8-F and ORF8-R (Table 1). They amplified 259- and 353-bp internal fragments that include nucleotides (nt) 5 to 264 from avhR and 75 to 427 from ORF8, respectively. The PCR products were amplified from F2/5 genomic DNA, purified, digested and then cloned into pVIK165. Mutants generated following conjugation with F2/5 were selected on AB minimal medium (4) amended with kanamycin (50 μg/ml). Mutations were verified by PCR with primers derived from sequences of the F2/5 chromosome that flank the insertion site (avhR-Int and ORF8-Int) and from within the PVIK165 vector (PVIK-P) (Table 1). The mutants were also confirmed by the expression of gfp by UV microscopy. Mutants were tested for their ability to cause the HR and grape necrosis.
Cloning avhR and complementation.
Primers avhR-F2 and avhR-R2 containing EcoRI and PstI restriction sites, respectively, were designed to amplify avhR from F2/5 genomic DNA (Table 1). A 768-bp PCR product was amplified, digested with EcoRI and PstI, and ligated to broad-host-range vector pPZP201 (11) to make complemented M1320(pGHavhR). The ligated DNA was transformed into DH5α competent cells, and positive colonies were selected on LB agar containing spectinomycin (100 μg/ml). The consensus clone in the recombinant plasmid was verified by restriction digestion and sequencing. It was then transformed into competent cells of S17-1 by heat shock transformation and transferred to M1320 by conjugation. Transformants were selected on AB minimal medium containing spectinomycin (400 μg/ml) and kanamycin (100 μg/ml) and verified by PCR. It was necessary to do selection on 400-μg/ml spectinomycin as nontransformed M1320 was able to grow on media amended with 300 μg/ml but not 400 μg/ml. The complemented M1320 clones were screened for their ability to cause the tobacco HR and grape necrosis.
Detection of AHLs.
AHLs were extracted from F2/5, M1320, and complemented M1320 cells harvested from PDA or PDA amended with appropriate antibiotics from overnight and 6-day-old cultures grown at 28°C, as previously described (32). Bacteria from four agar plates were harvested and suspended in a volume of 20 ml of acidified ethyl acetate-acetonitrile mixture (50:50). The first extraction was shaken overnight, and the second was shaken for 1 h at 150 rpm. The extracts were centrifuged at 10,000 rpm for 10 min (Sorvall rotor SA300), after which the supernatant was dried by rotary evaporation and then under a stream of N2. Extractions were made from whole cells because we previously reported that F2/5 produces long-chain AHLs and because it has been indicated that the permeability of such AHLs through bacterial membranes may be limited and may require active transport mechanisms (21). AHL profiles were determined by reverse-phase thin-layer chromatography of plates overlaid with a cell suspension of biosensor strain NTL4(pZLR4) (3). Two other sensors, including one that employs a T7 expression system to overproduce TraR in Agrobacterium (33) and a Chromobacterium violaceum mutant CV026 (20), were also used to compare AHLs in extracts from F2/5 and M1320 and complemented M1320.
Because the signal strength of AHLs from the M1320 6-day culture is noticeably greater than that of AHLs from F2/5, we wished to determine if extracts from M1320 contain concentrations or derivatives of AHLs that inhibit the regulation of the F2/5-induced HR. AHL extract volumes of 2 and 20 μl from the overnight and 6-day M1320 and F2/5 cultures were added to tubes, dried in a fume hood to evaporate the solvent, and then were mixed with F2/5 cells (from an overnight plate) suspended to an optical density at 600 nm (OD600) of 1.5 in sterile distilled water and incubated for 1 to 2 h. The culture was then infiltrated into tobacco leaves for HR induction. Tubes of PDB medium amended with AHL extracts were also inoculated with lower concentrations of F2/5 (OD600 ≈ 0.5) and shaken overnight at 28°C until they reached an OD600 of approximately 1.5, and the cultures were tested for their ability to cause the HR. Controls consisted of non-AHL-amended F2/5 cultures and AHL-amended media without bacteria.
Sequence analysis of cosmid CPM201.
An approximately 14-kb region of clone CPM201 that carries avhR was sequenced by a strategy used previously in our laboratory (32). In general, cosmid DNA was digested with EcoRI and subcloned into plasmid vector pUC18. Subclones were selected based on insert size and were sequenced from both ends. Clones with large inserts were digested with restriction enzyme HindIII or BamHI and subcloned into vector pUC18. If there were no suitable restriction sites for subcloning, a primer walking strategy, employing primers designed from sequenced ends of fragments, was used. The order of each fragment within CPM201 was determined by identification of overlapping sequences. DNA sequence homology searches were performed with the Basic Local Alignment Search Tool (BLAST) algorithm provided by the National Center for Biotechnology Information. Analysis of DNA and deduced protein sequences was carried out with DNASTAR software.
RT-PCR.
Expression of aviR, avhR, and a putative HR-associated gene identified in F2/5 mutant M852 (13) was determined by reverse transcriptase PCR (RT-PCR). The M852 ORF has homology with clpA, which encodes one of two ATPase subunits of the ClpAP protease of E. coli (23). Total RNAs were isolated from overnight PDB cultures (OD600 = 1.4 to 1.5, corresponding to 3 × 109 to 5 × 109 CFU/ml) of F2/5, M1154, M1320, and complemented M1154 and M1320 with the TRI-Reagent kit (Molecular Research Center, Cincinnati, Ohio) by following the manufacturer's protocol. RNA samples were treated with RNase-free DNase (Promega, Madison, Wis.) to prevent DNA contamination, and then PCRs were run with the treated RNA samples as templates to verify the absence of contaminating DNA. RT-PCR was conducted with the Superscript One-Step RT-PCR system plus Platinum Taq DNA polymerase (Invitrogen, Carlsbad, Calif.) by following the manufacturer's protocol. Products were separated by electrophoresis on 1.0% agarose in 0.5× Tris-borate-EDTA buffer. Primers for determining expression of aviR (aviR-F and aviR-R), avhR (avhR-F3 and avhR-R3), and clpA (clpA-F and clpA-R) are shown in Table 1. All assays were repeated at least once.
Nucleotide sequence accession number.
The sequence of CPM201 has been deposited in GenBank under accession number AY519466.
RESULTS
avhR is a luxR homolog that is associated with HR and necrosis.
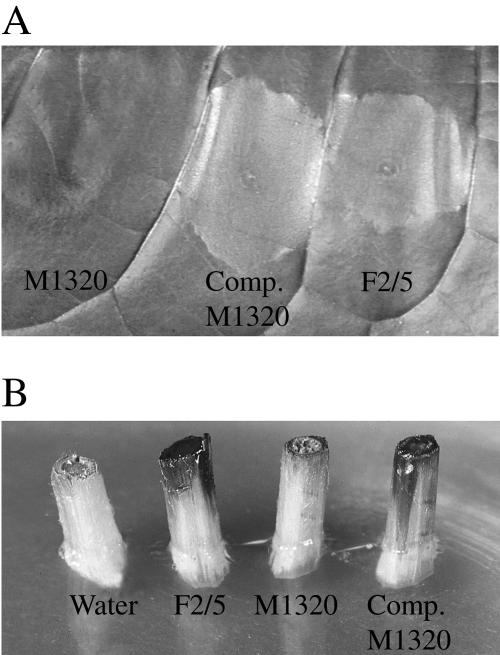
The F2/5 mutant M1320 is HR negative and causes less necrosis on grape shoot explants than F2/5 (Fig. 1). Sequence analysis of the EcoRI fragment carrying the Tn5 insertion revealed disruption of a putative gene designated avhR (for A. vitis hypersensitive response), which encodes a predicted 230-amino-acid protein with a molecular mass of 25.8 kDa. The putative start codon of avhR is GTG, and a likely ribosomal binding site, GGAA, is located 6 nt upstream from the start codon. The deduced AvhR protein has consensus autoinducer binding and helix-turn-helix DNA binding domains that are characteristic of the LuxR family of transcriptional regulators. The Tn5 insertion in avhR occurred near the helix-turn-helix domain, toward the 3′ end of the gene (Fig. 2). The amino acid sequence deduced from avhR is most similar to those of AGR-c-1279 (50.6%) from A. tumefaciens C58 and SMc00878 (47.2%) and SMc00877 (43.0%) from S. meliloti Rm1021, all which have identity with the LuxR family but whose functions have not been determined. Similarities to AviR (23.4%) and to other LuxR homologs are compared in Fig. 2.
FIG. 1.
Comparison of tobacco HR (A) and necrosis of grape shoot explants (B) induced by F2/5, M1320, and complemented M1320.
FIG. 2.
Alignment of the protein sequence deduced from avhR with those of selected members of the LuxR family. Proteins whose sequences have similarities to the amino acid sequence deduced from avhR include AviR (23.4%) from A. vitis (accession number AF521015), LuxR (19.0%) from V. fischeri (accession number M 96844), TraR (17.3%) from A. tumefaciens (accession number L08596), SinR (22.6%) from S. meliloti Rm1021 (accession number NC003062), EsaR (17.4%) from Erwinia stewartii (accession number L32184), RhiR (22.6%) from Rhizobium leguminosarum (accession number M98835), RhlR (20.4%) from P. aeruginosa (accession number AE 004768), AGR-c-1279 (50.6%) from A. tumefaciens C58 (accession number NC003062), and SMc00877 (47.2%) and SMc00878 (43.0%) from S. meliloti Rm1021 (accession number NC003047). Arrows, insertion sites in AvhR (arrow A, site-directed mutant; arrow B, Tn5 mutant); *, “strictly conserved” amino acid residues; ▵, substitutions at strictly conserved amino acid residues; #, substitutions at conserved residues that are involved in pheromone binding.
The amino acid sequence of the product of avhR has four of the five residues (Tyr61, Gly113, Glu178, and Gly188) that are absolutely conserved among known functional members of the LuxR family. Asp70, however, which contributes to pheromone binding by forming a hydrogen bond with the imino group, is replaced by Ser. Other highly conserved amino acids that are predicted to play important roles in pheromone binding of LuxR proteins have also been identified (28, 31). It is particularly interesting that two of these highly conserved residues, Trp57 and Trp85, are also replaced in AvhR (Fig. 2).
Detection of AHLs.
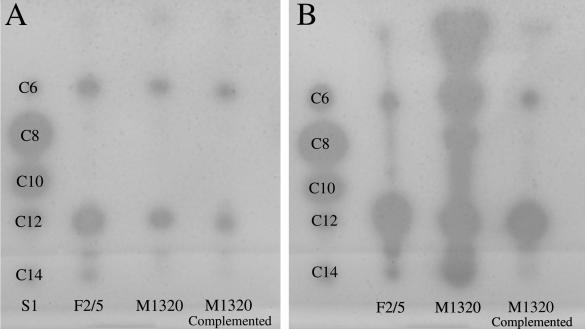
There were no observed differences between the arrays of AHL signals from extracts of F2/5 and M1320, as determined with the NTL4(pZL4) biosensor (Fig. 3). Only two predominant signals were observed from overnight cultures, whereas at least five were observed from 6-day cultures. The signal strength from the 6-day M1320 extracts, however, was much stronger than that from extracts of F2/5 or the complemented mutant. This was especially clear for signals that migrated with AHLs having fatty acid side chains with 10 or fewer carbons. Analysis of extracts using the sensors CV026 and T7 also revealed no differences between AHL patterns of F2/5 and M1320 (not shown). More specifically, only signals that migrated with C8 and C6 AHL standards were observed with the CV026 sensor. For the T7 sensor the AHL pattern looked identical to that for the NTL4 sensor except that signals were noticeably stronger. When different amounts of extracts from the overnight and 6-day cultures of F2/5 or M1320 were added to F2/5 or to M1320 cultures at different times, they did not alter the ability of the bacteria to induce the HR on tobacco.
FIG. 3.
Profiles of AHLs from F2/5, M1320, and complemented M1320 from extracts of overnight (A) and 6-day (B) cultures. Volumes of 10 μl from overnight cultures and 5 μl from 6-day cultures of F2/5, M1320, and complemented M1320 were spotted and separated on C18 reverse-phase thin-layer chromatography plates developed with methanol-water (70:30) and were visualized following overlay with Agrobacterium sensor strain NTL4(pZLR4). Extracts were compared to a mixture of unsubstituted AHLs that were run as standards (S1).
Site-directed mutagenesis.
ORF8 lies 94 nt downstream of avhR, contains 1,248 nt, and encodes a predicted 415-amino-acid protein that shows identity to the nitrate transporter component (encoded by nrtA) from Mesorhizobium loti (49.3%), ABC transporter AGR-c-1281p from A. tumefaciens (39.2%), and nitrate transporter protein SMb21114, encoded by the megaplasmid pSymB of. S. meliloti (35.6%). The ORF8 site-directed mutant was positive for the tobacco HR and for grape necrosis and thus did not differ from F2/5 with regard to these phenotypes (not shown). Therefore, the disrupted gene in M1320 affecting HR is avhR.
The avhR site-directed mutant (mutant M1320B) resulted from insertion of pVIK165 in a region that would be in proximity of the putative AHL binding site of the gene (Fig. 2). M1320B responded identically to M1320 when inoculated into grape and tobacco plants, i.e., it was HR negative and caused reduced necrosis.
Complementation of M1320.
M1320 was first complemented by transforming it with cosmid CPM201 by conjugation. The ability to induce the HR on tobacco and necrosis on grape plants was fully restored in complemented M1320 (Fig. 1).
To further verify that avhR is necessary for the HR and full induction of necrosis on grape plants, the gene was cloned into expression vector pPZP201 and was transferred to mutant M1320 by conjugation. Again, the complemented M1320 was fully restored in ability to cause HR on tobacco and grape necrosis.
Analysis of avhR flanking sequence.
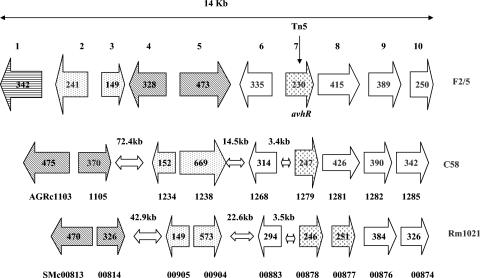
A 14-kb fragment of CPM201 that includes avhR contains 10 (including 2 incomplete) ORFs. The overall GC content of the fragment is identical to that of the A. tumefaciens strain C58 genome (58%) (30). Sequence comparison revealed that all of the proteins deduced from the CPM201 sequence have corresponding homologs in A. tumefaciens and S. meliloti except for that deduced from the partial ORF1, which is similar to a hypothetical protein of Burkholderia fungorum and Ralstonia metallidurans (Table 2; Fig. 4). Sequence comparisons show that ORFs in CPM201 are homologs of genes located on the circular chromosomes of A. tumefaciens and S. meliloti except for ORF8, which is similar to the ORF encoding SMb21114, which is located on the megaplasmid pSym of S. meliloti. The relative order of ORF6 to -10 is identical for all three bacterial species except that Rm1021 has a second luxR-related ORF (encoding SMc00877) that is immediately downstream of the ORF encoding SMc00878 (Fig. 4). In addition, ORF2, -3, -4, and -5 in A. vitis exist in different relative locations and orientations as compared to the corresponding ORFs in C58 and Rm1021, and there are considerably greater distances between some of the ORFs in these strains (Fig. 4).
TABLE 2.
Homologs from F2/5 that were identified within a 14-kb region of cosmid CPM201
| F2/5 ORF (size of product [aag]) | Homolog of product (size [aa]) | Putative product | Identity (%) |
|---|---|---|---|
| ORF1a (>342) | Bcep2846b (700) | Hypothetical protein | 39.8 |
| Ruet3075c (659) | Hypothetical protein | 40.4 | |
| ORF2 (241) | AGR-c-1238d (669) | Hypothetical protein encoded by rluB | 81.3 |
| SMc00904e (573) | Hypothetical protein | 79.3 | |
| ORF3 (149) | AGR-c-1234 (152) | Deaminase | 69.3 |
| SMc00905 (149) | Deaminase | 69.8 | |
| ORF4 (328) | AGR-c-1105 (370) | Hypothetical protein | 72.9 |
| SMc00814 (326) | Signal peptide | 61.3 | |
| ORF5 (473) | AGR-c-1103 (475) | MFSh permease | 70.0 |
| SMc00813 (470) | Transmembrane protein | 68.7 | |
| ORF6 (335) | AGR-c-1268 (314) | Calcium binding protein | 56.4 |
| SMc00883 (294) | Hypothetical protein | 56.1 | |
| ORF7 (230) | AGR-c-1279 (247) | Transcriptional regulator | 50.6 |
| SMc00878 (246) | Transcriptional regulator | 47.2 | |
| SMc0877 (251) | Transcriptional regulator | 43.0 | |
| ORF8 (415) | AGR-c-1281 (426) | ABC transporter | 39.2 |
| SMb21114f (441) | Nitrate transporter | 35.6 | |
| ORF9 (389) | AGR-c-1282 (390) | Multidrug-related protein | 68.9 |
| SMc00876 (384) | ATP binding protein | 70.6 | |
| ORF10a (>250) | AGR-c-1285 (384) | Magnesium and cobalt transporter | 53.6 |
| SMc00874 (326) | Magnesium and cobalt transporter | 56.8 |
FIG. 4.
Comparison of ORF organization within the 14-kb region in CPM201 that includes avhR to corresponding regions in A. tumefaciens C58 and S. meliloti Rm1021. The length of each arrow represents the relative ORF size and indicates the direction of transcription. ORF numbers for C58 and Rm1021 are given below arrows, and amino acid numbers are in boldface within arrows. Related gene clusters are indicated by arrow background patterns.
RT-PCR.
The expression of aviR, avhR, and the clpA homolog in F2/5 and mutants M1154 and M1320 and their complemented derivatives was determined by RT-PCR (Fig. 5). As expected, avhR is expressed in F2/5 and in complemented M1320; it is also expressed in the aviR mutant M1154. Expression of aviR was also detected in F2/5 and in M1320. Therefore aviR and avhR appear to be expressed independently and not in a hierarchical manner. The clpA homolog was expressed in F2/5 and in all mutants, thereby serving as a positive control and revealing that the gene is not regulated by aviR or avhR.
FIG. 5.
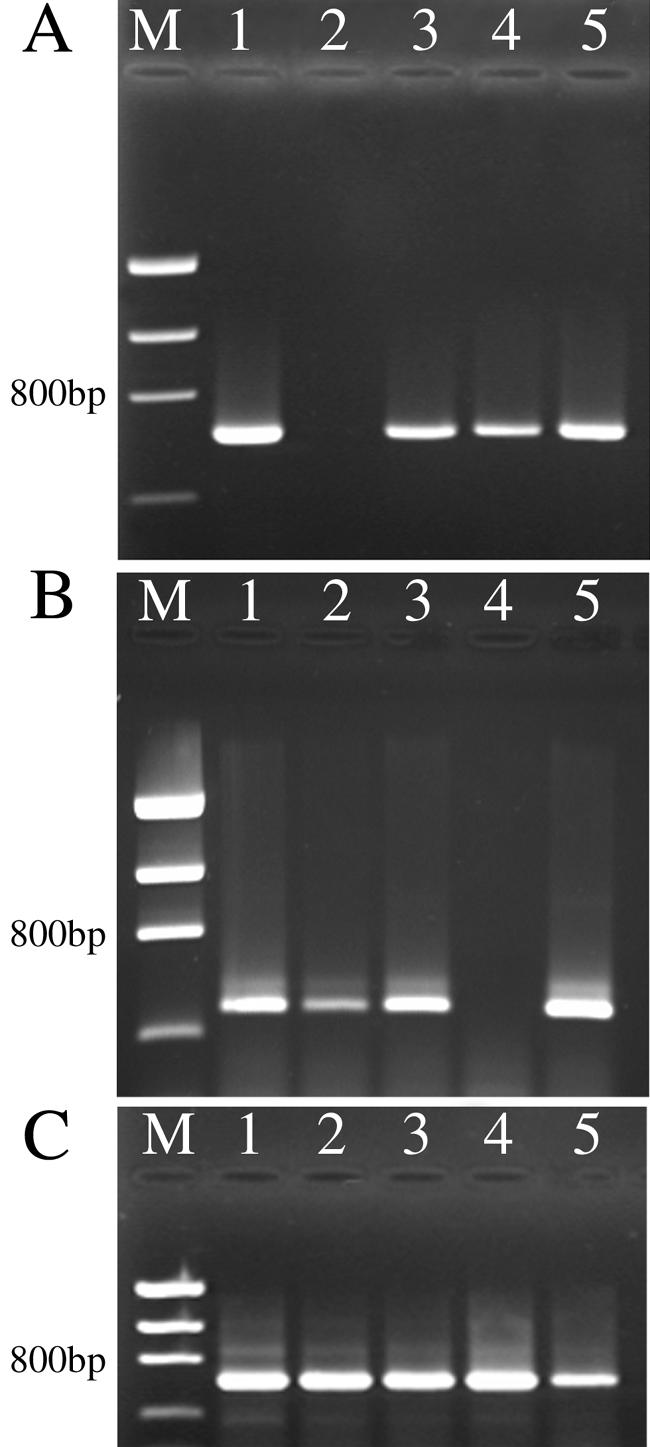
Expression of aviR (A), avhR (B), and the clpA homolog (C) in F2/5, M1154 (aviR mutant), M1320 (avhR mutant), and complemented M1154 and M1320, as determined by RT-PCR. Lane 1, F2/5; lane 2, M1154; lane 3, complemented M1154; lane 4, M1320; lane 5, complemented M1320; lane M, low-molecular-size DNA ladder.
DISCUSSION
The HR is a form of programmed cell death in plants that is characterized by a rapid localized plant cell death that is associated with resistance against disease. It is induced by several species of gram-negative bacteria on nonhost plants and often involves the delivery of specific avirulence (Avr) proteins to the plant via a type III secretion system (TTSS) (2). The TTSS is encoded by a set of hrp and hrc (for hypersensitivity response and pathogenicity and hrp conserved, respectively) genes that are also essential for induction of the HR. Such genes are often grouped in the bacterial genome on pathogenicity islands and may encode effector proteins that are delivered into the plant cell via the TTSS pilus (15). Subsequent interaction of the effectors with plant receptors, generally encoded by plant resistance genes, can lead to disease (in a compatible host) or induction of the HR (incompatible response). Although it has been reported that A. tumefaciens induces responses in maize that resemble an HR (12) and that S. meliloti induces an oxidative burst leading to localized cell death during the early infection process of alfalfa (24), A. vitis is thus far the only member of the Rhizobiaceae that has been shown to cause a rapid collapse and cell death of infiltrated tobacco leaves (13). The A. vitis-induced response resembles HRs caused by other plant-associated bacteria in that it is dependent on bacterial-cell concentration, is affected by inhibitors of plant metabolism, and results in collapse of infiltrated leaf tissue within 24 h. The nature of the HR elicitor of A. vitis and the means by which it is delivered to the plant remain to be determined.
The TTSS has been identified in several members of the Rhizobiaceae in previous studies; these include recent reports of nodulation-associated outer membrane proteins being secreted via a TTSS in Rhizobium sp. strain NGR234 (18) and the association of extracellular nodulation proteins of Sinorhizobium fredii USDA257 with a TTSS pilus (17). In contrast the TTSS is absent from A. tumefaciens C58 (30), which employs a type IV system for transfer of its transferred DNA and effector proteins to plant cells (6). It remains to be determined whether type III or type IV systems are present in A. vitis and play roles in the delivery of effectors involved in the HR and grape necrosis. Thus far, however, none of the A. vitis HR and necrosis mutants that we have examined result from disruption of genes that are known to be associated with these transport systems.
The HR on tobacco that is induced by A. vitis is the first example where the response is associated with quorum sensing. We previously reported that aviR is essential for expression of the HR and grape necrosis and that the aviR mutant M1154 has a significantly altered AHL profile, indicating its likely involvement in regulation of at least one luxI homolog that has not yet been identified (32). In contrast, avhR is necessary for the HR but when disrupted only partially affects grape necrosis. The avhR mutation does not appear to affect the AHL profile, suggesting that the putative AvhR protein may not regulate expression of a cognate luxI or other genes encoding AHL synthases in the F2/5 genome. However, such a conclusion cannot be drawn from biosensor assays alone, and the issue is currently being investigated by biochemical characterization of AHLs that are produced by the different F2/5 derivatives. It is also interesting that AHL extracts from 6-day-old M1320 cultures produce a much stronger signal on the thin-layer chromatography biosensor plates. The reason for this response is unknown but could indicate that avhR suppresses expression of AHL synthases or genes that are associated with specific substrates. When AHLs extracted from F2/5 and M1320 were added to cultures of F2/5, they did not affect its ability to induce the HR, indicating that the apparently increased AHL concentration that is produced by M1320 does not negatively affect the regulation of the response. Further research to identify specific genes that are regulated by avhR is needed to clarify which factors are responsible for increased AHL production by M1320.
The fact that there are at least two LuxR homologs in A. vitis that are involved in regulation of the HR and grape necrosis indicates the presence of a complex quorum-sensing network. Several other bacteria also employ multiple quorum-sensing systems, including some that are known to function in a hierarchical manner to regulate diverse phenotypes (27). For example, in Pseudomonas aeruginosa the LasIR and RhlIR systems act in series for expression of the different virulence factors that are produced during different stages of infection. LasI/LasR initiates the infection process by inducing the transcription of genes that encode virulence factors as well as by activating expression of rhlR. RhlI/RhlR further activates genes controlled by LasI/LasR and a second class of specific genes. In addition to the well-studied TraR/TraI system of A. tumefaciens, which regulates conjugal transfer of the Ti plasmid, other and more complicated quorum-sensing systems have been discovered in several other members of the Rhizobiaceae (10). For example, in S. meliloti ExpR is regulated by the SinR/SinI system and together they are involved in the production of symbiotically active exopolysaccharide EPS II (19, 22). We are particularly interested in comparisons between the S. meliloti Rm1021 genome and F2/5 because regions that we have sequenced thus far show a high level of similarity with regard to gene arrangement (including the presence of luxR homologs at positions corresponding to aviR and avhR). For example, AviR has an unusually high similarity (>56%) to ExpR and AvhR has relatively high similarity to SMc00877 and SMc00878. Further investigations are under way to determine whether AviR also regulates exopolysaccharide production and whether SinI and SinR homologs are present in A. vitis.
Two mutations made in avhR resulted in loss of HR and reduced grape necrosis. For M1320 the Tn5 insertion results in the deletion of 65 amino acids from the C terminus of AvhR, thereby predictably disrupting the helix-turn-helix domain of the protein. It has been reported for LuxR that this domain plays a role in activating transcription of the lux operon (25, 26) and that, when more than 40 amino acids are deleted from its C terminus, transcription of the operon and autoregulation of luxR do not occur (5). For the site-directed mutant M1320B, the insertion of pVIK165 occurs within the region that would constitute the putative autoinducer binding domain of the protein.
Known functional LuxR homologs only have about 20% amino acid sequence identity; however they all possess five conserved residues (29). The recent reports elucidating the crystalline structure of TraR have led to the identification of several amino acid residues that are involved in pheromone binding, dimerization, and DNA binding (28, 31). The putative AvhR protein is the first member of the LuxR family that has a substitution at Asp70, which together with highly conserved Trp57 (also replaced in AvhR) is located in the AHL binding pocket of TraR and stabilizes the pheromone through the formation of hydrogen bonds. The highly conserved Trp85, which contributes to AHL binding through van der Waals contact, is also replaced in AvhR. Binding of pheromone in several, but not all, LuxR regulators is thought to promote multimerization. This has been specifically demonstrated for the TraR protein, which requires binding of its cognate AHL, N-(3-oxo-octanoyl)-l-homoserine lactone, for proper protein folding, dimerization into its active form, and protection against proteolysis (34, 35). Therefore, a critical question is whether these seemingly important substitutions in AvhR affect binding of a cognate AHL. At this time an associated luxI homolog has not been identified for avhR to allow identification of the possible cognate AHL. Other possibilities are that AvhR binds a signal molecule that is not an AHL (29) and that the function of AvhR is pheromone independent (7). Our group is pursuing this research to further elucidate the function of avhR and to understand its role in regulation of the HR and necrosis.
Acknowledgments
This research was funded by NRI Competitive Grants Program/USDA award number 2002-35319-12582.
We thank S. Winans (Cornell) for vector pVIK165 and the T7 biosensor, S. Farrand (University of Illinois) for biosensor NTL4(pZLR4), and A. Eberhard (Cornell) for providing AHL standards.
REFERENCES
- 1.Burr, T. J., and L. Otten. 1999. Crown gall of grape: biology and disease management. Annu. Rev. Phytopathol. 37:53-80. [DOI] [PubMed] [Google Scholar]
- 2.Buttner, D., and U. Bonas. 2002. Getting across—bacterial type III effector proteins on their way to the plant cell. EMBO 21:5313-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha, C., Y. Gao, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 4.Chilton, M., T. C. Currier, S. K. Farrand, A. J. Bendich, M. P. Gordon, and E. W. Nester. 1974. Agrobacterium tumefaciens DNA and Ps8 bacteriophage DNA not detected in crown gall tumorigenesis. Proc. Natl. Acad. Sci. USA 71:3672-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, S. H., and E. P. Greenberg. 1991. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc. Natl. Acad. Sci. USA 88:11115-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, A. R. J., N. R. Thomson, B. Bycroft, G. S. A. B. Stewart, P. Williams, and G. P. C. Salmond. 1998. A pheromone-independent CarR protein controls carbapenem antibiotic synthesis in the opportunistic human pathogen Serratia marcescens. Microbiology 144:201-209. [DOI] [PubMed] [Google Scholar]
- 8.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez, J. E., and M. M. Marketon. 2003. Quorum sensing in nitrogen-fixing Rhizobia. Microbiol. Mol. Biol. Rev. 67:574-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajdukiewicz, P., Z. Svab, and P. Maliga. 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25:989-994. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, G. 2000. Evidence for Agrobacterium-induced apoptosis in maize cells. Mol. Plant-Microbe Interact. 13:649-657. [DOI] [PubMed] [Google Scholar]
- 13.Herlache, T. C., H. S. Zhang, C. L. Reid, S. Carle, D. Zheng, P. Basaran, M. Thaker, A. T. Burr, and T. J. Burr. 2001. Mutations that affect Agrobacterium vitis-induced grape necrosis also alter its ability to cause a hypersensitive response on tobacco. Phytopathology 91:966-972. [DOI] [PubMed] [Google Scholar]
- 14.Higgins, C. F. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152:205-210. [DOI] [PubMed] [Google Scholar]
- 15.Jin, Q., W. Hu, I. Brown, G. McGhee, P. Hart, A. L. Jones, and S. Y. He. 2001. Visualization of the secreted Hrp and Avr proteins along the Hrp pilus during type III secretion in Erwinia amylovora and Pseudomonas syringae. Mol. Microbiol. 40:1129-1139. [DOI] [PubMed] [Google Scholar]
- 16.Kalogeraki, V. S., and S. C. Winans. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69-75. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan, H. B., J. Lorio, W. S. Kim, G. Jiang, K. Y. Kim, M. DeBoer, and S. G. Pueppke. 2003. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant-Microbe Interact. 16:617-625. [DOI] [PubMed] [Google Scholar]
- 18.Marie, C., W. J. Deakin, V. Viprey, J. Kopcinska, W. Golinowski, H. B. Krishnan, X. Perret, and W. J. Broughton. 2003. Characterization of nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant-Microbe Interact. 16:743-751. [DOI] [PubMed] [Google Scholar]
- 19.Marketon, M. M., S. A. Glenn, A. Eberhard, and J. E. Gonzalez. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 185:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 21.Pearson, J. P., C. van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellock, B. J., M. Teplitski, R. P. Boinay, W. D. Bauer, and G. C. Walker. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 184:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 24.Santos, R., D. Herouart, S. Sigaud, D. Touati, and A. Puppo. 2001. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 14:86-89. [DOI] [PubMed] [Google Scholar]
- 25.Shadel, G. S., R. Young, and T. O. Baldwin. 1990. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J. Bacteriol. 172:3980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slock, J., D. van Riet, D. Kolibachuk, and E. P. Greenberg. 1990. Critical regions of the Vibrio fischeri LuxR protein defined by mutational analysis. J. Bacteriol. 172:3974-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vannini, A., C. Volpari, C. Gargioli, E. Muragli, R. Cortese, R. De Francesco, P. Neddermann, and S. Di Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 30.Wood, D. W., J. C. Setubal, R. Kaul, et al. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, R.-G., T. Pappas, J. L. Brace, P. C. Miller, T. Qulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, D., H. S. Zhang, S. Carle, G. Hao, M. R. Holden, and T. J. Burr. 2003. A luxR homolog, aviR, in Agrobacterium vitis is associated with induction of necrosis on grape and a hypersensitive response on tobacco. Mol. Plant-Microbe Interact. 16:650-658. [DOI] [PubMed] [Google Scholar]
- 33.Zhu, J., Y. Chai, Z. Zhong, S. Li, and S. C. Winans. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium haukuii. Appl. Environ. Microbiol. 69:6949-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance and dimerization. Proc. Natl. Acad. Sci. USA 98:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]