Abstract
In the plant-beneficial soil bacterium Pseudomonas fluorescens CHA0, the production of biocontrol factors (antifungal secondary metabolites and exoenzymes) is controlled at a posttranscriptional level by the GacS/GacA signal transduction pathway involving RNA-binding protein RsmA as a key regulatory element. This protein is assumed to bind to the ribosome-binding site of target mRNAs and to block their translation. RsmA-mediated repression is relieved at the end of exponential growth by two GacS/GacA-controlled regulatory RNAs RsmY and RsmZ, which bind and sequester the RsmA protein. A gene (rsmE) encoding a 64-amino-acid RsmA homolog was identified and characterized in strain CHA0. Overexpression of rsmE strongly reduced the expression of target genes (hcnA, for a hydrogen cyanide synthase subunit; aprA, for the main exoprotease; and phlA, for a component of 2,4-diacetylphloroglucinol biosynthesis). Single null mutations in either rsmA or rsmE resulted in a slight increase in the expression of hcnA, aprA, and phlA. By contrast, an rsmA rsmE double mutation led to strongly increased and advanced expression of these target genes and completely suppressed a gacS mutation. Both the RsmE and RsmA levels increased with increasing cell population densities in strain CHA0; however, the amount of RsmA showed less variability during growth. Expression of rsmE was controlled positively by GacA and negatively by RsmA and RsmE. Mobility shift assays demonstrated specific binding of RsmE to RsmY and RsmZ RNAs. The transcription and stability of both regulatory RNAs were strongly reduced in the rsmA rsmE double mutant. In conclusion, RsmA and RsmE together account for maximal repression in the GacS/GacA cascade of strain CHA0.
Pseudomonas fluorescens CHA0 is a root-colonizing biocontrol strain which suppresses soil-borne plant diseases caused by phytopathogenic fungi (19, 22). Disease suppression is mainly due to the antifungal metabolites 2,4-diacetylphloroglucinol, pyoluteorin, and hydrogen cyanide (HCN) which are produced by the bacterium at the end of exponential growth (23, 24, 32, 43). Biosynthesis of these compounds and of the exoenzymes phospholipase C and the exoprotease AprA strictly depends on the GacS/GacA two-component system, which operates a switch from primary to secondary metabolism in various gram-negative bacteria and which can also be involved in pathogenicity to plants and animals, in ecological fitness, and in stress tolerance (18).
GacS/GacA control of secondary metabolites and exoenzymes was shown to occur in strain CHA0 at the posttranscriptional level involving the RNA-binding protein RsmA as a key regulatory element (7). RsmA is presumed to interact with specific ribosome-binding sites present in target genes and thereby prevent translation. Such translational repression can be alleviated by the action of the small regulatory RNAs RsmY and RsmZ, whose expression is controlled by the GacS/GacA system in response to signal molecules produced by CHA0 at the end of exponential growth (19, 47). RsmY and RsmZ bind multiple copies of the RsmA protein (47, 48) and, by a titration effect, may thus render the ribosome-binding site of target genes accessible for the translation machinery.
RsmA-like proteins are highly conserved in various eubacteria (2, 4, 6, 7, 12, 14, 25, 26, 36, 38, 40, 53, 54). In Escherichia coli, for instance, the RsmA homolog CsrA regulates carbon flux, biofilm formation, and motility, and its effect is antagonized by the UvrY (= GacA)-controlled regulatory RNAs CsrB and CsrC (28, 38, 52). Interestingly, CsrA can act as a repressor or as an activator of translation, depending on the target mRNA. Binding of CsrA to the untranslated leader of the glgCAP mRNA prevents ribosome binding and promotes mRNA decay (5, 27, 28), whereas binding of CsrA to the 5′ segment of flhDC mRNA increases messenger stability and expression (51). In Salmonella enterica serovar Typhimurium, CsrA controls genes involved in cell invasion, and its effect is antagonized by the SirA (= GacA)-regulated CsrB RNA (2, 3). The RsmA protein of the plant pathogen Erwinia carotovora subsp. carotovora controls the production of several virulence factors, including pectolytic enzymes, proteases, and cellulases (8). RsmA-mediated repression is relieved by the small regulatory RNA RsmB expressed under ExpS/ExpA (= GacS/GacA) control (10, 30). In the opportunistic human pathogen Pseudomonas aeruginosa, RsmA posttranscriptionally controls the production of secondary metabolites directly as well as indirectly by modulating the quorum-sensing circuitry (35, 36). A GacA-dependent regulatory RNA closely related to RsmZ of P. fluorescens strain CHA0 (19) and to PrrB of P. fluorescens F113 (1) antagonizes the RsmA effect (21).
Genetic evidence indicates that, in P. fluorescens CHA0, RsmA is not the only negative control element in the GacS/GacA cascade: whereas mutational inactivation of gacS or gacA drastically reduces the expression of genes required for HCN, AprA, and 2,4-diacetylphloroglucinol production, inactivation of the rsmA gene suppresses the gacS defect only partially (7). Furthermore, genomic data indicate that in the closely related strain P. fluorescens Pf-5, two rsmA-like genes exist (http://pseudo.bham.ac.uk). In the present study, we describe RsmE, a homolog of RsmA, which participates in posttranscriptional control of GacS/GacA-dependent genes in P. fluorescens CHA0.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. Bacteria were usually grown on nutrient agar and in nutrient yeast broth (44). When indicated, Triton X-100 was added to liquid cultures at a final concentration of 0.05% to avoid cell aggregation. Antibiotics and mercuric chloride, when required, were added to the growth medium at the following concentrations: tetracycline, 25 μg/ml for E. coli and 100 μg/ml for P. fluorescens; ampicillin, 100 μg/ml; spectinomycin, 50 μg/ml; gentamicin, 10 μg/ml; mercuric chloride, 10 μg/ml for E. coli; and kanamycin, 25 μg/ml for E. coli and 50 μg/ml for P. fluorescens. To counterselect E. coli donor cells in matings with P. fluorescens, chloramphenicol was used at 10 μg/ml. When relevant, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added to plates at a final concentration of 0.02%. Routine incubation temperatures were 37°C for E. coli and 30°C for P. fluorescens. P. fluorescens was grown at 35°C to improve its capacity to accept heterologous DNA, i.e., in electrotransformation or triparental matings with E. coli.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant characteristicsa or sequence (5′ → 3′) | Source or reference |
|---|---|---|
| E. coli | ||
| BL21(DE3) | F−ompT hsdSB(rB,− mB−)gal dcm (λDE3) | Novagen |
| DH5α | recA1 endA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 Δ(lacZYA-argF)U169 (φ80dlacZΔM15) | 39 |
| HB101 | hsdS recA proA2 leu-6 ara-14 galK2 lacY1 xyl-5 mtl-1 rpsL20 thi-1 supE44 | 39 |
| P. fluorescens | ||
| CHA0 | Wild type | 50 |
| CHA19 | ΔgacS | 55 |
| CHA89 | gacA::Kmr | 23 |
| CHA207 | hcnA′-′lacZ | 7 |
| CHA805 | aprA′-′lacZ | 7 |
| CHA806 | ΔgacS, aprA′-′lacZ | 19 |
| CHA1003 | rsmE::Ω-Hg Hgr | This study |
| CHA1005 | rsmE::Ω-Hg aprA′-′lacZ, Hgr | This study |
| CHA1007 | rsmA::Ω-Km rsmE::Ω-Hg ΔgacS aprA′-′lacZ, Kmr Hgr | This study |
| CHA1008 | rsmA::Ω-Km rsmE::Ω-Hg ΔgacS, Kmr Hgr | This study |
| CHA1009 | rsmA::Ω-Km rsmE::Ω-Hg, Kmr Hgr | This study |
| CHA1020 | rsmA::Ω-Km aprA′-′lacZ, Kmr | This study |
| CHA1021 | rsmA::Ω-Km rsmE::Ω-Hg aprA′-′lacZ, Kmr Hgr | This study |
| CHA1022 | ΔgacS hcnA′-′lacZ | This study |
| CHA1023 | rsmA::Ω-Km hcnA′-′lacZ, Kmr | This study |
| CHA1025 | rsmE::Ω-Hg hcnA′-′lacZ, Hgr | This study |
| CHA1027 | rsmA::Ω-Km rsmE::Ω-Hg hcnA′-′lacZ, Kmr Hgr | This study |
| CHA1028 | rsmA::Ω-Km rsmE::Ω-Hg ΔgacS, hcnA′-′lacZ, Kmr Hgr | This study |
| CHA1076 | rsmA::Ω-Km, Kmr | This study |
| CHA1134 | mini-Tn7 GmrrsmE′-′lacZ | This study |
| CHA1136 | gacA::Kmr mini-Tn7 GmrrsmE′-′lacZ | This study |
| CHA1138 | rsmE::Ω-Hg mini-Tn7 GmrrsmE′-′lacZ, Hgr | This study |
| CHA1161 | rsmA::Ω-Km mini-Tn7 GmrrsmE′-′lacZ, Kmr | This study |
| CHA1162 | rsmA::Ω-Km rsmE::Ω-Hg mini-Tn7 GmrrsmE′-′lacZ, Kmr Hgr | This study |
| Plasmids | ||
| pET28a | Expression vector, PT7, Kmr | Novagen |
| pBLS II KS, SK | pBluescript cloning vectors, Apr | Stratagene |
| pHP45ΩSm/Sp | Source of transcription/translation stop cassette; Apr Smr Spr | 37 |
| pHP45ΩHg | Source of transcription/translation stop cassette; Apr Hgr | 37 |
| pME497 | Mobilizing plasmid, Apr | 7 |
| pME3087 | Suicide vector, MCS, Tcr | 50 |
| pME3274 | Suicide plasmid derived from pME3087 for deletion of gacS by gene replacement | 55 |
| pME3280a | Chromosomal integration vector, mini-Tn7 Gmr, MCS, Apr | 55 |
| pME6000 | Cloning vector, pBBR1MCS derivative, Tcr | 32 |
| pME6001 | Cloning vector, pBBR1MCS derivative, Gmr | 7 |
| pME6078 | P. fluorescens CHA0 rsmA (encoding RsmA6H) in pME6032 | 19 |
| pME6081 | Suicide plasmid for gene replacement containing rsmA::Ω-Km, Tcr Kmr | 7 |
| pME6091 | Transcriptional rsmZ-lacZ fusion | 19 |
| pME6359 | Ptac-rsmZ fusion at the +1 site; Tcr | 19 |
| pME6702 | Ptac-phlA′-′lacZ, Tcr | C. Gigot-Bonnefoy and D. Haas, unpublished data |
| pME6834 | pME6000 derivative carrying rsmE in 3-kb insert; Tcr | This study |
| pME6850 | pME6001 derivative carrying rsmE, Gmr | This study |
| pME6851 | pME6001 derivative carrying rsmE under Plac control, Gmr | This study |
| pME6879 | Suicide plasmid for gene replacement containing rsmE::Ω-Hg Tcr Hgr | This study |
| pME6916 | Transcriptional rsmY-lacZ fusion | 47 |
| pME6918 | Ptac-rsmY fusion at the +1 site; Tcr | 47 |
| pME6919 | Template for in vitro transcription with rsmY under PT7 | 47 |
| pME6920 | Template for in vitro transcription with rsmZ under PT7 | 47 |
| pME6926 | Template for in vitro transcription with carA under PT7 | 47 |
| pME7013 | P. fluorescens CHA0 rsmE (encoding RsmE6H) in pET28a | This study |
| pME7545 | pME3280a derivative containing rsmE′-′lacZ, Gmr Apr | This study |
| pNM481 | ′lacZ fusion vector, Apr | 34 |
| pUK21 | Cloning vector, MCS, Kmr | 49 |
| Oligonucleotides | ||
| MB20.1 | GGCTCGCTATCGCGAAAGG | |
| MB21.1 | GATATTTATTGCCTATGT | |
| RSMEHIS6 | TTAAATGGTGATGGTGATGGTGGGGGGTTTCGCGTTTGTCC | |
| RSMEUP | ACGTGGATCCATGATCTTCTCCTTGAT (BamHI site underlined) | |
| RSMEDOWN | ACGTGGATCCTGAACAGCATGAGCAC (BamHI site underlined) | |
| RSME-1 | ACGTGGATCCTGACCATGAACGATGAAATC (BamHI site underlined) | |
| RSME-2 | ACGTAAGCTTCAGCATGATCTTCTCCTTG (HindIII site underlined) |
Ap, ampicillin; Gm, gentamicin; Hg, mercuric chloride; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Tc, tetracycline; MCS, multiple cloning site.
DNA manipulation and cloning procedures.
DNA manipulations were carried out as described by Sambrook and Russell (39). Small-scale preparations of plasmid DNA were carried out by the cetyltrimethylammonium bromide method (11), large-scale preparations were performed with JetStar-Tips (Genomed, Basel, Switzerland). Chromosomal DNA was prepared according to Gamper et al. (15). DNA fragments were purified from agarose gels with the GeneClean DNA extraction kit (Bio 101, La Jolla, Calif.). Transformation of E. coli and P. fluorescens was carried out by electroporation (13). Nucleotide sequences were determined with a Big Dye Terminator Cycle sequencing kit and an ABI-Prism 373 automatic sequencer (Applied Biosystems). Nucleotide sequences were analyzed with the programs of the University of Wisconsin Genetics Computer Group package (version 9.1).
RNA manipulations and Northern blots.
RNA preparations from P. fluorescens strains and Northern blots were done as described previously (48). RNA half-lives were estimated after addition of 200 μg of rifampin per ml to cultures. RNA was extracted at given time points and analyzed by Northern blotting.
Construction of a gene bank from P. fluorescens CHA0 and isolation of the rsmE gene.
Chromosomal DNA from CHA0 was digested partially with Sau3A. Fragments of 2 to 4 kb were gel purified and ligated to pME6000, digested with BamHI, and treated with alkaline phosphatase. To avoid destruction by the host's restriction system, pME6000 DNA was prepared from CHA0. Ligation mixtures were purified as described (46) and stored in aliquots at −20°C until used to electro-transform strain CHA805. Among ca. 5,000 transformants obtained, 6 which showed a white colony phenotype on nutrient agar containing X-Gal were retained. Sequence analysis showed that these clones had a common gene (rsmE) in the pME6000 insert. One clone, pME6834, was kept for further study.
Construction of plasmids and gene replacement mutants.
The rsmE subclones pME6850 and pME6851 were generated by PCR-amplifying rsmE from pME6834 with primers MB20.1 and MB21.1. The 0.53-kb PCR product was blunted with T4 DNA polymerase and cloned into the SmaI site of pME6001 to give pME6850, which carries rsmE in the opposite orientation from that of the vector promoter Plac. To place the gene under Plac control, rsmE was excised from pME6850 on a 0.53-kb BamHI-EcoRV fragment and recloned into pME6001 between the XbaI, made blunt by T4 DNA polymerase treatment, and BamHI sites. This generated pME6851. For overexpression and purification of RsmE, a histidine tag was added at its C terminus by PCR with primers RSMEHIS6 and T7 with pME6850 as the template. The 0.4-kb PCR product was blunted with T4 DNA polymerase, digested with BamHI, cloned into pBLS II KS between the BamHI and HincII sites, excised with XbaI and XhoI, and inserted into pET28a under the control of the T7 promoter. The resulting overexpression construct was named pME7013.
Chromosomal mutations in rsmE, rsmA, and gacS were generated by gene replacement as described previously (24, 42). The suicide plasmid pME6879 used to generate an rsmE::Ω-Hg mutation was constructed as follows. Two PCR products flanking the rsmE gene were obtained from pME6834 with primers RSMEUP plus T3 and RSMEDOWN plus T7, respectively. The resulting 1.1-kb upstream fragment and the 1.7-kb downstream fragment were digested with HindIII and BamHI and with BamHI and EcoRI, respectively, ligated with a 5-kb BamHI-BamHI fragment carrying the Ω-Hg cassette from pHP45Ω-Hg, and cloned into pUK21. The Ω-Hg cassette flanked by the rsmE up- and downstream regions was excised from this pUK21 derivative on an 8-kb SpeI-SpeI fragment and cloned into the XbaI site of pME3087 to produce pME6879. Plasmid pME6879 was introduced into CHA0, CHA805, and CHA207 to generate the rsmE::Ω-Hg mutants CHA1003, CHA1005, and CHA1025, respectively. The rsmA gene was mutated with pME6081 (7) in strains CHA1003, CHA805, CHA1005, CHA207, CHA1025, and CHA0 to give the corresponding rsmA::Ω-Km mutants CHA1009, CHA1020, CHA1021, CHA1023, CHA1027, and CHA1076, respectively. Finally, the suicide plasmid pME3274 (55) served to delete the gacS gene in strains CHA1021, CHA1009, CHA207, and CHA1027, resulting in CHA1007, CHA1008, CHA1022, and CHA1028, respectively (Table 1).
Expression of rsmE was measured with a translational rsmE′-′lacZ fusion, which was constructed as follows. The first two codons of rsmE and 2.3 kb of its upstream region were PCR amplified from CHA0 chromosomal DNA with primers RSME-1 and RSME-2. The resulting fragment was cleaved with BamHI and HindIII and cloned into pNM481. From the resulting construct, a 5.4-kb BamHI-XhoI fragment carrying rsmE′-′lacZ was excised, blunted with T4 DNA polymerase, and ligated to the SmaI-linearized vector pME3280a, which delivers a mini-Tn7 with passenger DNA into the unique Tn7 attachment site of the P. fluorescens chromosome. The resulting plasmid, pME7545, was used as described previously (55) to integrate its mini-Tn7 with rsmE′-′lacZ and a gentamicin resistance gene into the chromosome of strains CHA0, CHA89, CHA1003, CHA1076, and CHA1009 to give strains CHA1134, CHA1136, CHA1138, CHA1161, and CHA1162, respectively.
Detection of RsmE and RsmA by Western blotting.
Erlenmeyer flasks containing 20 ml of NYB amended with 0.05% Triton X-100 were inoculated 1:100 and grown at 30°C with shaking. At given time points after inoculation, cells equivalent to an optical density at 600 nm of 0.4 U per ml were centrifuged, washed with 0.9% (wt/vol) NaCl, resuspended in 20 μl of loading buffer (50 mM Tris-HCl, pH 6.8, 2% [wt/vol] sodium dodecyl sulfate, 0.1% [wt/vol] bromophenol blue, 15% [vol/vol] glycerol, 5% [vol/vol] β-mercaptoethanol) and immediately treated at 100°C for 10 min. Of each sample, 15 μl was loaded on a 16% acrylamide-bisacrylamide gel containing Tricine and sodium dodecyl sulfate (41). After electrophoresis at 70 V during 4 h, proteins were electrotransferred onto polyvinylidene difluoride membranes (Immobilon P, Millipore) at 50 mA and 4°C for 1 h. The RsmA and RsmE proteins were detected with polyclonal antibodies raised against purified Yersinia enterocolitica RsmA (36) and a secondary antibody coupled to peroxidase. Membranes were developed with the ECL Western blotting analysis system (Amersham-Pharmacia) following the manufacturer's instructions.
Purification of histidine-tagged protein fusions of RsmE (RsmE6H) and RsmA (RsmA6H).
The RsmE6H and RsmA6H proteins were overexpressed in E. coli BL21/pME7013 and E. coli DH5α/pME6078, respectively, and purified by Ni-nitrilotriacetic acid affinity chromatography (Qiagen) as described previously (19). The protein eluates were dialyzed against 10 mM Tris-acetate (pH 8.0) at 4°C and stored at −20°C. Protein contents were estimated with the Bradford method with bovine serum albumin as the standard. The purity of the preparations was ≥90% as judged from sodium dodecyl sulfate-Tricine-polyacrylamide gel electrophoresis.
Mobility shift assays.
Radioactively labeled transcripts of RsmY and RsmZ were synthesized from linearized pME6919 and pME6920, respectively, with a T7 transcription kit (Fermentas) in the presence of [α-33P]UTP, following the manufacturer's instructions. Unlabeled competitor RNAs (RsmY, RsmZ, or carA leader) were synthesized following the same protocol but with unlabeled UTP from linearized pME6919, pME6920, or pME6926, respectively. RNA was purified by phenol-chloroform extraction and desalted with Sephadex G-25 minicolumns (Amersham Biosciences). RNA concentrations were estimated by UV absorption at 260 nm. Binding reactions contained [α-33P]UTP-labeled RsmY or RsmZ RNA and purified RsmA6H or RsmE6H at various concentrations (see the legend to Fig. 6 for details). Assays were also carried out in the presence of various unlabeled RNA competitors (see the legend to Fig. 7 for details). In this case, RsmA6H or RsmE6H was added last to the binding reaction containing competing RNAs. The reaction mixtures (10 μl) were incubated at 30°C for 30 min to allow complex formation. Samples were then fractionated on native 10% polyacrylamide gels (4 h at 10 mA), and radioactive bands were visualized by autoradiography after drying the gels (47, 48).
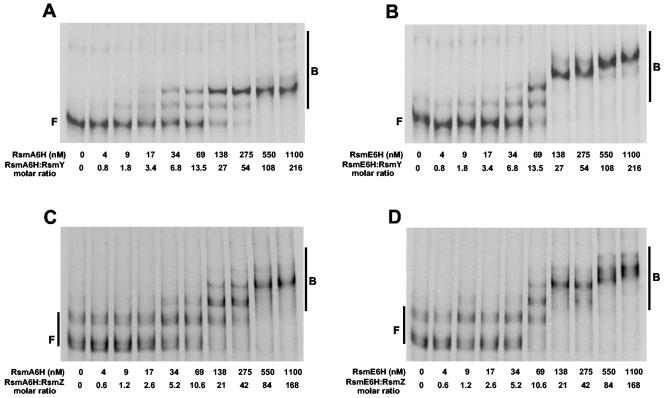
FIG. 6.
Interaction of RsmA6H and RsmE6H with the regulatory RNAs RsmY and RsmZ. [α-33P]UTP-labeled RsmY (5 nM) and RsmZ (6.5 nM) were incubated with different concentrations of purified RsmA6H or RsmE6H before fractionation on nondenaturing gels and autoradiography. The positions of free (F) and bound (B) RNA species are indicated. (A) RsmY versus RsmA6H. (B) RsmY versus RsmE6H. (C) RsmZ versus RsmA6H. (D) RsmZ versus RsmE6H.
FIG. 7.
Competition of RsmY and RsmZ RNAs for binding to RsmE6H. [α-33P]UTP-labeled RsmY (5 nM) and RsmZ (6.5 nM) and different unlabeled RNA competitors (RsmY, RsmZ, and the leader of carA mRNA) were incubated with RsmE6H (275 nM) before fractionation on nondenaturing gels and autoradiography. (A) Competition of unlabeled RNAs with RsmY-RsmE6H complexes. (B) Competition of unlabeled RNAs with RsmZ-RsmE6H complexes. F, free transcripts; B, transcripts bound to RsmE6H.
β-Galactosidase assays.
P. fluorescens strains were grown in 20 ml of NYB (in 50-ml Erlenmeyer flasks) with shaking at 180 rpm and 30°C; 0.05% Triton X-100 was routinely added to avoid cell aggregation. β-Galactosidase activities were quantified by the Miller method (33), with cells permeabilized with 5% (vol/vol) toluene.
Nucleotide sequence accession number.
The nucleotide sequence of rsmE has been assigned GenBank accession number AY547272.
RESULTS
Discovery of rsmE, a homolog of the posttranscriptional regulator gene rsmA.
To identify new regulatory elements involved in the GacS/GacA signal transduction pathway of P. fluorescens CHA0, we searched for genes which, when present on a multicopy plasmid, would downregulate the expression of typical GacS/GacA-controlled genes, such as the translational reporter gene fusions aprA′-′lacZ and hcnA′-′lacZ in strains CHA805 and CHA207, respectively. To this end, a CHA0 gene expression library was constructed in the broad-host-range vector pME6000 and introduced into CHA805. Among several thousand transformants, six were identified which strongly reduced the expression of aprA′-′lacZ as judged by their white color on medium containing X-Gal.
Sequence analysis revealed that these clones had a common gene in their pME6000 insert. The deduced amino acid sequence of this gene, which we named rsmE, had 71% identical amino acids with the posttranscriptional regulator RsmA of CHA0 (Fig. 1). To ensure that rsmE was indeed responsible for repression of aprA′-′lacZ in CHA805, a subclone, pME6851, which carries only the rsmE gene under Plac control was constructed (see Materials and Methods for details). The expression of both aprA′-′lacZ and hcnA′-′lacZ was strongly reduced in CHA805/pME6851 and CHA207/pME6851 compared to CHA805 and CHA207 transformed with the empty vector pME6001, in that both reporter genes were repressed more than 10-fold (data not shown).
FIG. 1.
Alignment of the deduced amino acid sequences of RsmA (19) and RsmE (this study). Asterisks indicate identical amino acids.
To test if strain CHA0 encodes additional RsmA/E homologs, Southern blots were performed under nonstringent conditions with rsmA and rsmE as molecular probes. However, we found no evidence for further homologs of these genes (data not shown). Moreover, in the total genomic sequence of the closely related P. fluorescens strain Pf-5 (http://pseudo.bham.ac.uk), we identified two proteins which are 100% identical to RsmA and RsmE but found no additional homologs. Taken together, these data strongly suggest that the genome of P. fluorescens CHA0 encodes two, but not more, closely related regulators of the RsmA family.
Complete suppression of a gacS mutation by an rsmA rsmE double mutation.
Inactivation of either gacS or gacA strongly reduces the expression of hcnA, aprA, and phlA (7, 55). When the chromosomal rsmA gene is inactivated in a ΔgacS background, the effect of the gacS mutation on aprA′-′lacZ is suppressed only partially, indicating that RsmA is not the only negative regulator operating in the Gac/Rsm cascade (7). To test whether RsmE acts as an additional negative regulator, we evaluated the effect of an rsmE mutation on the expression of GacS/GacA-controlled genes. As expected, expression of hcnA′-′lacZ was strongly reduced in the gacS mutant CHA1022 compared to the expression of this chromosomal fusion in the wild-type CHA207 (Fig. 2A). Inactivation of rsmA or rsmE in strains CHA1023 and CHA1025, respectively, resulted in a slight increase in hcnA expression. However, when both rsmA and rsmE were mutated, hcnA expression was fourfold higher than in the wild type, irrespective of the presence (CHA1027) or the absence (CHA1028) of a functional gacS gene (Fig. 2A).
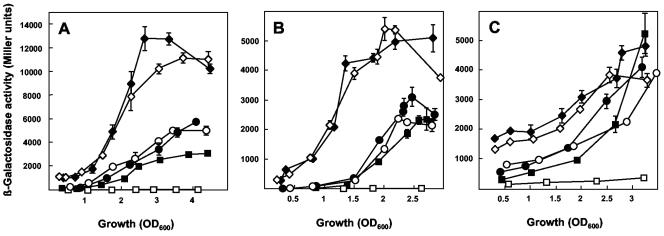
FIG. 2.
Impact of rsmE, rsmA, and gacS mutations on the expression of hcnA, aprA, and phlA. (A) β-Galactosidase expression of a chromosomal hcnA′-′lacZ fusion was determined in CHA207 (solid squares, wild-type context), CHA1022 (open squares, gacS mutant), CHA1023 (open circles, rsmA mutant), CHA1025 (solid circles, rsmE mutant), CHA1027 (solid diamonds, rsmA rsmE double mutant), and CHA1028 (open diamonds, rsmA rsmE gacS triple mutant). (B) Expression of a chromosomal aprA′-′lacZ fusion in CHA805 (solid squares), CHA806 (open squares), CHA1020 (open circles), CHA1005 (solid circles), CHA1021 (solid diamonds), and CHA1007 (open diamonds). (C) Expression of a phlA′-′lacZ fusion on pME6702 in CHA0 (solid squares), CHA19 (open squares), CHA1076 (open circles), CHA1003 (solid circles), CHA1009 (solid diamonds), and CHA1008 (open diamonds). Each value is the average from three different cultures ± standard deviation.
Similar results were found with the aprA′-′lacZ reporter (Fig. 2B). The expression of aprA was only marginally increased in the rsmA (CHA1020), and the rsmE mutant (CHA1005) compared with the wild-type CHA805 and was very low in the ΔgacS mutant CHA806. Again, inactivation of both rsmA and rsmE resulted in a three- to fourfold increase in a gacS-positive (CHA1021) as well as in a gacS-negative (CHA1007) background (Fig. 2B).
Finally, expression of a plasmid-borne phlA′-′lacZ fusion (driven by the tac promoter) in the different genetic backgrounds followed a pattern similar to that described above for the chromosomal hcnA′-′lacZ and aprA′-′lacZ fusions, although the effects of the rsmA and rsmE mutations were less pronounced, possibly due to the particular phlA construct used. Importantly, the rsmA rsmE double mutation fully suppressed a gacS defect with respect to phlA expression (Fig. 2C).
We conclude from these data that in the absence of both the RsmA and RsmE proteins, the presence of a functional GacS/GacA system is no longer required for hcnA, aprA, or phlA expression.
Expression of RsmA and RsmE.
Although the calculated molecular masses of RsmA (6.95 kDa) and RsmE (7.01 kDa) are similar, the two proteins could be separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in Tricine buffer, visualized in Western blots with polyclonal antibodies against RsmA (purified from Yersinia enterocolitica), and identified by the use of rsmA and rsmE mutants (Fig. 3). This allowed us to study the expression of the two proteins during growth. The expression of both RsmA and RsmE increased with increasing cell density; however, the amount of RsmA showed considerably less variation (Fig. 4). The expression of RsmE was also followed by measuring the expression of a chromosomal rsmE′-′lacZ fusion (integrated at the Tn7 attachment site); throughout growth, expression in the gacA mutant CHA1136 was lower than that in the wild-type background CHA1134 (Fig. 5). Inactivation of rsmA or rsmE in mutants CHA1161 and CHA1138, respectively, resulted in a moderate increase in rsmE′-′lacZ expression, whereas expression was highest in the rsmA rsmE double mutant CHA1162 (Fig. 5). The Western blot data (Fig. 3) are in agreement with the regulatory effects of GacA and RsmA on RsmE levels.
FIG. 3.
Production of the RsmE and RsmA proteins in P. fluorescens mutants. (A) Western blot detection of the RsmE and RsmA proteins from 20-ml cultures grown in 125-ml flasks. Samples from strains CHA0 (wild type, wt), CHA89 (gacA), CHA1076 (rsmA), CHA1003 (rsmE), and CHA1009 (rsmA rsmE) were taken in late exponential phase (4 h after inoculation) and in stationary phase (8 h after inoculation) for gel electrophoresis and immunodetection. (B) Control of protein load. A portion of the gel showing polypeptides of 6 to 20 kDa, after protein transfer and staining with Coomassie blue. Note that under the conditions used, the slightly larger but more hydrophobic RsmE protein migrated ahead of RsmA.
FIG. 4.
Cellular levels of RsmE and RsmA proteins in P. fluorescens CHA0 during growth in 50-ml flasks containing 20 ml of NYB and 0.05% Triton X-100. Samples for immunoblot analysis were taken at increasing cell densities. The protein load in each well was similar (not shown).
FIG. 5.
Expression of a chromosomal rsmE′-′lacZ fusion in the wild-type context (CHA1134, solid squares), in a gacA mutant (CHA1136, open squares), in an rsmA mutant (CHA1161, open circles), in an rsmE mutant (CHA1138, solid circles), and in an rsmA rsmE double mutant (CHA1162, solid diamonds). Each value is the average from three different cultures ± standard deviation.
Interaction of RsmE6H and RsmA6H with RsmY and RsmZ.
To investigate whether RsmE can be captured by RsmY and RsmZ, we prepared in vitro transcripts of both RNAs and performed mobility shift assays with RsmE6H and, in parallel, with RsmA6H. Both proteins were overexpressed in E. coli and purified as histidine-tagged fusions. Binding of RsmA6H and RsmE6H to RsmY (5 nM) was detected as two distinct complexes at 34 to 69 nM (Fig. 6A and B) and higher-order complexes became visible at ≥550 nM RsmA6H and ≥138 nM RsmE6H, suggesting that RsmY may have a somewhat higher binding capacity for RsmE6H than for RsmA6H. The interactions of RsmY with RsmA6H and RsmE6H were also compared (Fig. 6C and D). As observed previously (47), in vitro transcription of rsmZ gave two different-sized transcripts. Again, binding of both proteins to RsmZ (6.5 nM) was similar, and the formation of higher-order complexes appears to occur more readily with RsmE6H than with RsmA6H.
Competition experiments were performed to assess the specificity of RsmE6H binding to RsmY and RsmZ. As shown in Fig. 7, unlabeled RsmY and RsmZ transcripts were both able to compete with RsmE6H-RsmY and RsmE6H-RsmZ complexes, whereas the same amount of a similarly sized RNA lacking the putative RsmA/E binding elements (i.e., the untranslated leader of the P. fluorescens carA gene) did not modify the binding of RsmE6H to RsmY or RsmZ. We conclude from these experiments and from data obtained earlier with RsmA6H (47) that both regulatory RNAs bind RsmA and RsmE specifically and with similar affinity.
RsmA and RsmE positively control RsmY and RsmZ levels.
To assess the effects of RsmA and RsmE on the transcription of the two regulatory RNAs, we measured their expression with plasmid-encoded transcriptional lacZ fusions made at the +1 transcription start site (Fig. 8). Expression of rsmY-lacZ and rsmZ-lacZ was similar in the wild type and in the rsmA and rsmE mutants CHA1076 and CHA1003, respectively. In agreement with previous results (19, 47), both fusions gave strongly reduced expression in the gacA mutant CHA89. Interestingly, in the rsmA rsmE double mutant CHA1009 as well as in the rsmA rsmE gacS triple mutant CHA1008, the expression of both fusions was equally low. We conclude from this that RsmA and RsmE are required together with GacA for the transcription of both RNAs.
FIG. 8.
Impact of rsmE, rsmA, and gacA mutations on rsmY and rsmZ transcription. (A) β-Galactosidase expression of an rsmY-lacZ transcriptional fusion on pME6916 in the wild-type CHA0 (solid squares), the gacA mutant CHA89 (open squares), the rsmE mutant CHA1003 (solid circles), the rsmA mutant CHA1076 (open circles), the rsmA rsmE double mutant CHA1009 (solid diamonds), and the rsmA rsmE gacS triple mutant CHA1008 (open diamonds). (B) β-Galactosidase expression of an rsmZ-lacZ transcriptional fusion on pME6091 in CHA0 (solid squares), CHA89 (open squares), CHA1003 (solid circles), CHA1076 (open circles), CHA1009 (solid diamonds), and CHA1008 (open diamonds).
RsmA and RsmE also affected the stability of RsmY and RsmZ (Fig. 9). The estimated half-lives of both RNAs were >20 min in the wild type and <10 min in the rsmA rsmE double mutant, indicating that RsmY and RsmZ are protected from decay by the RsmA and RsmE proteins in vivo.
FIG. 9.
RsmA and RsmE proteins stabilize RsmY and RsmZ RNAs. (A) RsmY and RsmZ transcript decay in the wild-type strain P. fluorescens CHA0 (wt) and in the rsmA rsmE double mutant CHA1009 was determined by Northern blotting after blocking transcription with rifampin. The amount of RNA loaded was 0.5 μg for the wild type and 3 μg for the rsmA rsmE double mutant. As both rsmY and rsmZ are poorly expressed in the rsmA rsmE background, the stability of RsmY and RsmZ was studied in CHA1009 expressing rsmY or rsmZ from the tac promoter of pME6918 (rsmY) or pME6359 (rsmZ). (B) Densitometric analysis of RsmY stability in CHA0 (solid circles) and CHA1009 (open circles). (C) Densitometric analysis of RsmZ stability in CHA0 (solid circles) and CHA1009 (open circles).
DISCUSSION
In the present work, we have searched for novel negative regulators involved in the expression of the hcnA and aprA genes in the biocontrol strain P. fluorescens CHA0, and this has led to the discovery of the RNA-binding protein RsmE. This protein belongs to the RsmA/CsrA family of posttranscriptional regulators, which control a large spectrum of physiological processes ranging from carbon flux to the expression of bacterial traits important for beneficial or deleterious host-microbe interactions (2, 6, 7, 8, 14, 25, 26, 36, 38, 40). In P. fluorescens CHA0, the RsmA protein posttranscriptionally represses the expression of exoproduct genes during exponential growth, probably by obstructing ribosome access to the Shine-Dalgarno sequences of target mRNAs. This repression is relieved by GacS/GacA-dependent regulatory RNAs such as RsmY and RsmZ, which sequester the RsmA regulator (19, 47).
What is the role of RsmE in this regulatory network? Several lines of evidence indicate that the overall function of RsmE is similar to that of RsmA and that both proteins are required together for maximal translational repression of GacS/GacA-controlled target genes. (i) Chromosomal deletion of either rsmA or rsmE resulted in slightly increased expression of three target genes tested, i.e., hcnA, aprA, and phlA (Fig. 2). (ii) Deletion of both rsmA and rsmE strongly increased and advanced expression of the target genes (Fig. 2). (iii) Overexpression of rsmA or rsmE from a vector promoter (plac) strongly reduced target gene expression (7; K. Starke and D. Haas, unpublished data). (iv) RsmA and RsmE specifically bound to the regulatory RNAs RsmY and RsmZ in vitro (Fig. 6 and 7). However, the expression profiles of RsmA and RsmE are different. RsmA was present in considerable amounts throughout growth, whereas little RsmE was made at low cell densities (Fig. 4). Moreover, rsmE expression was regulated positively by GacA and negatively by RsmA and RsmE (Fig. 3 and 5). The rsmA gene appears to be cotranscribed with the upstream lysC (aspartokinase) gene and possibly also with the alaS (alanyl-tRNA synthetase) gene (our unpublished observations), complicating detailed analysis of rsmA expression. The observation that RsmE levels were highest at the end of growth (Fig. 4) suggests that RsmE could play a role in termination of GacA-controlled gene expression.
RsmA and RsmE stabilize both RsmY and RsmZ in vivo (Fig. 9), probably by protecting them from degradation by cellular RNases. In Erwinia carotovora subsp. carotovora, RsmA also increases the half-life of the RsmB riboregulator (9), whereas the stability of the CsrB RNA in E. coli does not appear to be affected by the RNA-binding protein CsrA (16). In Pseudomonas aeruginosa, RsmZ RNA is absent from stationary-phase cells (21). The half-lives of these regulatory RNAs are critical parameters in the GacS/GacA cascade, as they will determine the duration of the “on” phase.
RsmA and RsmE not only affect the stability of RsmY and RsmZ but are also required for good promoter activity of the rsmY and rsmZ genes (Fig. 8). Similarly, the E. coli RsmA homolog CsrA controls the transcription of the csrB and csrC riboregulator genes. CsrA appears to have a positive regulatory effect on the expression of the gacS homolog barA, suggesting that CsrA control of csrB and csrC may, at least in part, be indirect, via GacS and GacA (45). It is currently unknown whether gacA expression is controlled by RsmA and RsmE in P. fluorescens. In P. aeruginosa, however, RsmA positively controls rsmZ transcription without affecting gacA expression (21).
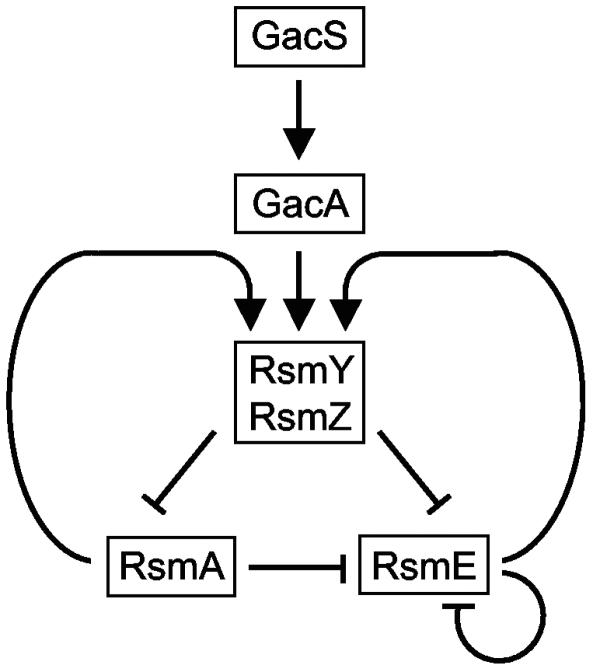
The circuit diagram presented in Fig. 10 summarizes our current understanding of the regulatory interactions operating in the Gac/Rsm cascade of P. fluorescens CHA0. Genetic evidence strongly suggests that GacS activates GacA by a phosphorelay mechanism in response to a bacterial signal (55). Although GacA is crucial for rsmY and rsmZ expression, GacA binding to the promoter regions of rsmY and rsmZ has not been demonstrated yet. RsmY and RsmZ bind multiple copies of RsmA and RsmE in vitro and antagonize the regulatory effects of these RNA binding proteins on secondary metabolite production in vivo (19, 47) (Fig. 2, 6, and 7). RsmE expression is regulated negatively by RsmA and RsmE and positively by GacA (Fig. 5). RsmA and RsmE are required for RsmY and RsmZ transcription and stability (Fig. 8 and 9). It will be of particular interest to analyze how this transcriptional activation by RsmA and RsmE is achieved.
FIG. 10.
Regulatory interactions operating in the Gac/Rsm cascade of P. fluorescens CHA0. See text for details. →, positive effect; ⊣, negative effect.
RsmA/CsrA redundancy in some bacterial species is evident from genomic data, but the present study is the first to examine this phenomenon experimentally. Whereas in enteric bacteria (e.g., E. coli, Salmonella enterica, and Erwinia spp.) and in P. aeruginosa there is only a single rsmA gene (20), in silico analysis reveals two rsmA/E homologs in P. fluorescens Pf-5, three in P. fluorescens SBW25, P. fluorescens Pf0-1, P. putida KT2440, and P. syringae pv. syringae B728a, four in P. syringae pv. tomato DC3000, and at least five in P. syringae pv. phaseolicola 1448A (http://pseudo.bham.ac.uk; http://pseudomonas-syringae.org). The genomic neighborhood of rsmA appears to be conserved in pseudomonads and consists of lysC located upstream and several tRNA genes located downstream. By contrast, the neighborhood of rsmE-like genes is variable, depending on the bacterial species. Thus, rsmE might have been recruited later in evolution and might reinforce the posttranscriptional control machinery steered by GacS/GacA. The roles of multiple rsmA/E homologs in some Pseudomonas spp. remain to be determined.
In conclusion, the fact that an rsmA rsmE double mutation fully suppresses a gacS defect (Fig. 2) indicates that RsmA and RsmE together represent the major negative control elements in the GacS/GacA cascade of P. fluorescens CHA0.
Acknowledgments
We thank Bérénice Humair, Aude Bachelard, and Nicolas González for help with β-galactosidase measurements and protein purification and Paul Williams for supplying anti-RsmA antibodies.
This project was supported through grants from the Swiss National Science Foundation for Scientific Reserach (project 3100A0-100180), the Roche Research Foundation, and the EU project ECOSAFE (QLK2-2000-31759).
REFERENCES
- 1.Aarons, S., A. Abbas, C. Adams, A. Fenton, and F. O'Gara. 2000. A regulatory RNA (PrrB) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113. J. Bacteriol. 182:3913-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635-646. [DOI] [PubMed] [Google Scholar]
- 4.Ang, S., Y. T. Horng, J. C. Shu, P. C. Soo, J. H. Liu, W. C. Yi, H. C. Lai, K. T. Luh, S. W. Ho, and S. Swift. 2001. The role of RsmA in the regulation of swarming motility in Serratia marcescens. J. Biomed. Sci. 8:160-169. [DOI] [PubMed] [Google Scholar]
- 5.Baker, C. S., I. Morozov, K. Suzuki, T. Romeo, and P. Babitzke. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44:1599-1610. [DOI] [PubMed] [Google Scholar]
- 6.Barnard, F. M., M. F. Loughlin, H. P. Fainberg, M. P. Messenger, D. W. Ussery, P. Williams, and P. J. Jenks. 2004. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 51:15-32. [DOI] [PubMed] [Google Scholar]
- 7.Blumer, C., S. Heeb, G. Pessi, and D. Haas. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA 96:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, A., Y. Cui, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl. Environ. Microbiol. 61:1959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee, A., Y. Cui, and A. K. Chatterjee. 2002. RsmA and the quorum-sensing signal, N-[3-oxohexanoyl]-L-homoserine lactone, control the levels of rsmB RNA in Erwinia carotovora subsp. carotovora by affecting its stability. J. Bacteriol. 184:4089-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, Y., A. Chatterjee, and A. K. Chatterjee. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and harpinEcc. Mol. Plant-Microbe Interact. 14:516-526. [DOI] [PubMed] [Google Scholar]
- 11.Del Sal, G., G. Manfioletti, and C. Schneider. 1988. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 16:9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubey, A. K., C. S. Baker, K. Suzuki, A. D. Jones, P. Pandit, T. Romeo, and P. Babitzke. 2003. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 185:4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farinha, M. A., and A. M. Kropinski. 1990. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol. Lett. 58:221-225. [DOI] [PubMed] [Google Scholar]
- 14.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpression of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291:353-360. [DOI] [PubMed] [Google Scholar]
- 15.Gamper, M., B. Ganter, M. Polito, and D. Haas. 1992. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J. Mol. Biol. 226:943-957. [DOI] [PubMed] [Google Scholar]
- 16.Gudapaty, S., K. Suzuki, X. Wang, P. Babitzke, and T. Romeo. 2001. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J. Bacteriol. 183:6017-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas, D., C. Keel, and C. Reimmann. 2002. Signal transduction in plant-beneficial rhizobacteria with biocontrol properties. Antonie van Leeuwenhoek 81:385-395. [DOI] [PubMed] [Google Scholar]
- 18.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 19.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heeb, S., K. Heurlier, C. Valverde, M. Cámara, D. Haas, and P. Williams. 2004. Post-transcriptional regulation in Pseudomonas spp. via the Gac/Rsm regulatory network, p. 239-255. In J.-L. Ramos (ed.), Pseudomonas, vol. 2. Kluwer Academic Publishers, New York, N.Y. [Google Scholar]
- 21.Heurlier, K., F. Williams, S. Heeb, C. Dormond, G. Pessi, D. Singer, M. Cámara, P. Williams, and D. Haas. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the post-transcriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keel, C., and C. Défago. 1997. Interactions between beneficial soil bacteria and root pathogens: mechanisms and ecological impact, p. 27-46. In A. C. Gange and V. K. Brown (ed.), Multitrophic interactions in terrestrial systems. Blackwell Science, London, England.
- 23.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Défago, G., and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laville, J., C. Blumer, C. von Schroetter, V. Gaia, G. Défago, C. Keel, and D. Haas. 1998. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 180:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawhon, S. D., J. G. Frye, M. Suyemoto, S. Porwollik, M. McClelland, and C. Altier. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633-1645. [DOI] [PubMed] [Google Scholar]
- 26.Liaw, S. J., H. C. Lai, S. W. Ho, K. T. Luh, and W. B. Wang. 2003. Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis. J. Med. Microbiol. 52:19-28. [DOI] [PubMed] [Google Scholar]
- 27.Liu, M. Y., H. Yang, and T. Romeo. 1995. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177:2663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, M. Y., and T. Romeo. 1997. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 179:4639-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, M. Y., G. Gui, B. Wei, J. F. Preston 3rd., L. Oakford, U. Yuksel, D. P. Giedroc, and T. Romeo. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272:17502-17510. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., Y. Cui, A. Mukherjee, and A. K. Chatterjee. 1998. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol. Microbiol. 29:219-234. [DOI] [PubMed] [Google Scholar]
- 31.Maurhofer, M., C. Keel., D. Haas, and G. Défago. 1994. Pyoluteorin production by Pseudomonas fluorescens strain CHA0 is involved in the suppression of Pythium damping-off of cress but not of cucumber. Eur. J. Plant Pathol. 100:221-232. [Google Scholar]
- 32.Maurhofer, M., C. Reimmann, P. Schmidli-Sacherer, S. Heeb, D. Haas, and G. Défago. 1998. Salicylic acid biosynthesis genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88:678-684. [DOI] [PubMed] [Google Scholar]
- 33.Miller, V. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Minton, N. P. 1984. Improved plasmid vectors for isolation of translational gene fusions. Gene 31:269-273. [DOI] [PubMed] [Google Scholar]
- 35.Pessi, G., and D. Haas. 2001. Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200:73-78. [DOI] [PubMed] [Google Scholar]
- 36.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. G. Holden, M. Cámara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 38.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the noncoding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Scarpari, L. M., M. R. Lambais, D. S. Silva, D. M. Carraro, and H. Carrer. 2003. Expression of putative pathogenicity-related genes in Xylella fastidiosa grown at low and high cell density conditions in vitro. FEMS Microbiol. Lett. 222:83-92. [DOI] [PubMed] [Google Scholar]
- 41.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 42.Schnider, U., C. Keel, C. Blumer, J. Troxler, G. Défago, and D. Haas. 1995. Amplification of the housekeeping sigma factor in Pseudomonas fluorescens CHA0 enhances antibiotic production and improves biocontrol abilities. J. Bacteriol. 177:5387-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Bonnefoy, C. Reimmann, R. Notz, G. Défago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanisich, V. A., and B. W. Holloway. 1972. A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. (Camb.) 19:91-108. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, K., X. Wang, T. Weilbacher, A.-K. Pernestig, Ö. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuits of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, M. R. 1994. Simple, effective cleanup of DNA ligation reactions prior to electro-transformation of E. coli. BioTechniques 16:988-990. [PubMed] [Google Scholar]
- 47.Valverde, C., S. Heeb, C. Keel, and D. Haas. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50:1361-1379. [DOI] [PubMed] [Google Scholar]
- 48.Valverde, C., M. Lindell, E. Gerhart, H. Wagner, and D. Haas. 2004. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J. Biol. Chem. 279:25066-25074. [DOI] [PubMed] [Google Scholar]
- 49.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 50.Voisard, C., C. Bull, C. Keel, J. Laville, M. Maurhofer, U. Schnider, G. Défago, G., and D. Haas. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Publishers, Weinheim, Germany.
- 51.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245-256. [DOI] [PubMed] [Google Scholar]
- 52.Weilbacher, T., K. Suzuki, A. K. Dubey, X. Wang, S. Gudapaty, I. Morozov, C. S. Baker, D. Georgellis, P. Babitzke, and T. Romeo. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48:657-670. [DOI] [PubMed] [Google Scholar]
- 53.White, D., M. E. Hart, and T. Romeo. 1996. Phylogenetic distribution of the global regulatory gene csrA among eubacteria. Gene 182:221-223. [DOI] [PubMed] [Google Scholar]
- 54.Whittle, G., G. A. Bloomfield, M. E. Katz, and B. F. Cheetham. 1999. The site-specific integration of genetic elements may modulate thermostable protease production, a virulence factor in Dichelobacter nodosus, the causative agent of ovine footrot. Microbiology 145:2845-2855.10537206 [Google Scholar]
- 55.Zuber, S., F. Carruthers, C. Keel, A. Mattard, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]