Abstract
The aim of this study was to identify and characterise Bacillus cereus from a unique national collection of 564 strains associated with 140 strong-evidence food-borne outbreaks (FBOs) occurring in France during 2007 to 2014. Starchy food and vegetables were the most frequent food vehicles identified; 747 of 911 human cases occurred in institutional catering contexts. Incubation period was significantly shorter for emetic strains compared with diarrhoeal strains A sub-panel of 149 strains strictly associated to 74 FBOs and selected on Coliphage M13-PCR pattern, was studied for detection of the genes encoding cereulide, diarrhoeic toxins (Nhe, Hbl, CytK1 and CytK2) and haemolysin (HlyII), as well as panC phylogenetic classification. This clustered the strains into 12 genetic signatures (GSs) highlighting the virulence potential of each strain. GS1 (nhe genes only) and GS2 (nhe, hbl and cytK2), were the most prevalent GS and may have a large impact on human health as they were present in 28% and 31% of FBOs, respectively. Our study provides a convenient molecular scheme for characterisation of B. cereus strains responsible for FBOs in order to improve the monitoring and investigation of B. cereus-induced FBOs, assess emerging clusters and diversity of strains.
Keywords: Bacillus cereus, epidemiology, food-borne infections, outbreak, virulence factors, genotyping
Introduction
The Bacillus cereus sensu lato group includes the following closely related spore-forming species: B. cereus sensu stricto, B. thuringiensis, B. cytotoxicus, B. weihenstephanensis, B. mycoides, B. pseudomycoides and B. anthracis [1]. The first four species are known to be involved in food poisoning [1]. B. thuringiensis is also mainly known as a biopesticide due to production of insecticidal toxins [2]. B. anthracis is highly virulent in mammals and is the causative agent of anthrax [3]. B. cytotoxicus is a newly identified group of strains that induce severe food poisoning. They are characterised by the production of cytotoxin K-1 (CytK-1) and a relatively high genomic diversity compared with other B. cereus strains [1].
B. cereus is currently the second most frequently found causative agent of confirmed and suspected food-borne outbreaks (FBOs) in France after Staphylococcus aureus [4]. Depending on the evidence implicating a food vehicle source during epidemiological and microbiological FBO investigations, the outbreaks are referred as a strong-evidence or weak-evidence FBO. Briefly, an FBO is defined as ‘strong-evidence’ when the following information is known and reported: food vehicle, food source, the link between outbreak cases and the food vehicle, place of exposure, and contributory factors. When several parts of the information are missing, the FBO is considered as ‘weak-evidence’ FBO [5].
Between 2006 and 2014 in France, B. cereus was recorded as the second or third major cause in weak-evidence FBOs. In 2014, B. cereus represented the second cause in weak-evidence FBOs, with 1,902 human cases for 224 FBOs, and the second cause of strong-evidence FBOs, with 23 FBOs accounting for 447 human cases and 18 hospitalisations [4]. The increase in B. cereus-induced FBOs is partly due to the input of national health and food safety authorities in the epidemiological and microbiological investigations of suspected FBOs. Indeed, B. cereus strains isolated from foodstuff suspected of being involved in an FBO are now usually collected by the laboratory for food safety in ANSES. To illustrate this, during 1996 to 2005, only 94 strong-evidence and 196 weak-evidence FBOs were reported, whereas for 2014 alone, 23 and 241 strong- and weak-evidence FBOs were notified, respectively showing the high input of the authorities. Nevertheless, the number of total human B. cereus cases is likely to be underestimated because individuals with gastrointestinal infections rarely seek medical advice and if they do, stools sample are not always asked for by physicians.
B. cereus can induce two types of gastrointestinal disease, leading to emetic or diarrhoeal syndromes. The symptoms associated with B. cereus infection are generally mild and self-limiting, but more serious and even fatal cases have been described in France and around the world [6]. The emetic syndrome is characterised by vomiting and nausea, usually 30 minutes to 6 hours after ingestion, and can be confused with FBOs caused by Staphylococcus aureus. This syndrome is due to the ingestion of a thermostable toxin known as cereulide, pre-formed in food before ingestion of contaminated foods. The emetic B. cereus strains represent a cluster of strains characterised by the presence of the plasmid-located ces gene encoding an enzyme involved in cereulide synthesis [7].
Diarrhoeic symptoms are characterised by abdominal cramps and watery diarrhoea within 8 to 16 hours after ingestion of contaminated foods. These diarrhoeal symptoms and incubation periods can be easily confused with those caused by Clostridium perfringens food poisoning. More precise information about diarrhoeic strains is thus necessary to discriminate between possible causative agents and allow better diagnosis during FBOs. The diarrhoeal syndrome occurs after ingestion of vegetative cells or spores of diarrhoeic strains. This syndrome is generally attributed to at least three enterotoxins: haemolysin BL (Hbl), which has three components B, L1 and L2; non-haemolytic enterotoxin (Nhe) with its three components Nhe-A, Nhe-B and Nhe-C, and cytotoxin K (CytK). Two forms of cytotoxin K have been described, CytK-1 and CytK-2, the former being more cytotoxic than the latter [8]. In addition, B. cereus produces other toxins such as haemolysin II (HlyII), metalloproteases such as InhA1 and InhA2, and the cell wall peptidase FM (CwpFM), which may also be involved in pathogenicity [9-11]. The pathogenic spectrum of B. cereus ranges from strains used as probiotics to strains that are lethal to humans and it remains difficult to predict the pathogenic potential of a strain. Apart from strains encoding ces or cytK-1 genes, which are virulent and well described in the literature [8,12], the pathogenicity of B.cereus diarrhoeal strains is not fully understood and there are currently no specific markers to unambiguously differentiate between pathogenic and harmless strains. Indeed, the genetic studies carried out to date have been inconclusive and, regardless of the diseases they cause, all strains seem to carry genes encoding at least one of the known diarrhoeal toxins [13]. However, highly toxic strains do not necessarily overproduce these toxins [14]. The aim of this study was therefore to identify and characterise B. cereus strains from a unique national collection of 564 strains strongly related to 140 FBOs that occurred in France during 2007 to 2014 in order to improve the monitoring and investigation of B. cereus-induced FBOs, assess the risk of emerging clusters of strains and identify strain variability.
Methods
Epidemiological data
The epidemiological data related to each FBO were mainly collected through interviews or questionnaires by local health authorities. The suspected food in each FBO was traced by the local services of the French Ministry of Agriculture and Food (DDPP, Department for protection of populations). Collected data included a record of the type of suspected food, preparation location and date, type of packaging, number of human cases, symptoms and incubation periods. Then, a database of ANSES (French Agency for Food, Environmental and Occupational Health and Safety) was built, gathering epidemiological data as well as analytical results of B. cereus enumeration in food, strain characterisation and toxin production.
Strain collection
For each FBO, all bacterial strains from suspected food were isolated by plating leftovers on selective media plates allowing the discrimination of B. cereus from other bacterial pathogens (S. aureus, C. perfringens, etc). Identification and numeration of one to five B. cereus strains per FBO were conducted by plating the strains on selective B. cereus agar media (MYP agar media: mannitol-phenol red-egg yolk medium (Biokar) according to the International Organization for Standardization (ISO) 7932 standard method or BACARA (BioMérieux), previously certified commercial alternative method (AES 10/10–07/10). All isolates were tested for haemolytic activity on sheep blood agar [15], lecithinase production on MYP agar media and starch hydrolysis on plate count agar (BioMérieux).
DNA extraction
DNA was extracted after overnight incubation of the strains at 30 °C on trypticase soy agar with 0.6% yeast extract (Sigma-Aldrich) using the DNeasy Blood and Tissue Kit (Qiagen). DNA was quantified by absorbance at 260 nm on a Nanodrop1000 spectrophotometer (Thermo scientific).
Coliphage M13 sequence-based PCR typing
To study strain diversity and discriminate between strains isolated in samples within the same FBO, B. cereus strains were typed using coliphage M13 sequence-based PCR (M13-PCR) derived from an RAPD technique and adapted from [16]. The PCR mix contained 40 ng of DNA template, 0.9 mM dNTP mix (Roche Diagnostics), 4 mM MgCl2, 2 µM primer (GAGGGTGGCGGCTCT), 2.5 U Goldstar DNA polymerase, and Goldstar buffer (Eurogentec). Thermal cycling using the Veriti Thermal Cycler (Applied Biosystems) included a denaturation step at 94 °C for 3 min, followed by 35 cycles of 1 min at 94 °C, 1 min at 40 °C, 8 min at 68 °C and an elongation step at 68 °C for 8 min. The amplified DNA was analysed by SDS-PAGE electrophoresis. The M13-PCR patterns were visualised using ChemiDoc XRS imaging system. Then, DNA profiles were analysed with BioNumerics 7.1 software (Applied Maths).
panC gene sequencing
B. cereus strains were assigned to the seven known phylogenetic groups according to partial sequencing of the panC gene [17]. The sequencing was carried out by a commercial facility (Eurofins MWG Operon). The classification into the phylogenetic groups was performed using the algorithm described in [17]. The two typing methods panC gene sequencing and M13-PCR typing were used for separate objectives. This study did not explore the correlation between the two methods.
Virulence gene detection
The presence of potential virulence genes cytK-1, cytK-2, hblA, hblC, hblD, nheA, nheB, nheC, hlyII and ces [10,13] was evaluated by PCR. As the genetic diversity of B. cytotoxicus strains possessing cytK-1 is substantial, the primers used to detect the other virulence genes were not suitable for those particular strains. The PCR was performed with the Veriti Thermal Cycler. The final reaction mixture (25 µL) contained 200 µM dNTPs, 1X PCR buffer, 1 U FastStart Taq DNA Polymerase (Roche), 200–1,000 nM primers, and 2 µL (ca 10 ng) template DNA. The amplification protocol comprised initial denaturation at 94 °C for 5 min followed by 30 cycles of 94 °C for 30 s, 58 °C for 60 s, and 72 °C for 90 s and final extension at 72 °C for 7 min. PCR products were analysed by SDS-PAGE electrophoresis.
Enterotoxin quantification
The production of the enterotoxins Nhe and Hbl was tested using two immunological tests, the BCET-RPLA Toxin detection kit (Oxoïd) and Tecra kit (BDE VIA, 3M-Tecra), respectively, after culture in brain heart infusion broth (Biomérieux) for 6 hours at 30 °C with stirring [18].
Database and statistical analysis
Strain characterisation results and epidemiological data were entered into a central database using BioNumerics software. The distribution of mean incubation periods, i.e. the time between ingesting contaminated food and symptom onset, was characterised using R 3.1 software and the ‘fitdistrplus’ package [19]. The log-normal was fitted to data according to maximum-likelihood estimation. To study seasonal variation in the occurrence of FBOs, the distribution of FBO dates was analysed throughout the year according to a previously described method [20].
Results
Epidemiological and clinical data
We studied a collection of 564 B. cereus strains associated with 140 FBO that occurred in France during 2007 to 2014. In 66 of the FBOs, B. cereus was isolated concomitantly with other bacterial species (including S. aureus and C. perfringens) during microbiological investigations, making it impossible to affirm that B. cereus was the cause of these FBOs. Our study therefore focused on 339 B. cereus strains isolated from food samples analysed during 74 FBOs where no other pathogenic bacteria were detected in the food during microbiological investigations (Table 1). These 74 FBOs resulted in 911 human cases. Data on sex and age of the cases were not always available and could therefore not be included in the study.
Table 1. Epidemiological and microbiological data of food-borne outbreaks associated solely with Bacillus cereus, France, 2007–2014 (74 outbreaks, 339 strains).
| FBO | Year | Incriminated food | Human cases n |
Incubation period in hours | Symptoms | Strain patterns identified n |
Outbreak settinga | CFU/g | Genetic signature |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2007 | Semolina | 5 | 0–3 | Vomiting | 1 | Commercial catering | 1.20E + 07 | GS3 |
| 2 | 2007 | Shrimp | 12 | 21–24 | Vomiting, diarrhoea | 1 | Commercial catering | 6.80E + 04 | GS1 |
| 3 | 2007 | Tomatoes | 4 | 0–3 | Vomiting, diarrhoea | 1 | Commercial catering | 7.00E + 02 | GS4 |
| 4 | 2008 | Semolina | 40 | 12–15 | Diarrhoea | 1 | Staff canteen | 1.20E + 03 | GS1 |
| 5 | 2008 | Tabbouleh and minced beef | NK | NK | NK | 1 | Commercial catering | 5.00E + 03 | GS2 |
| 6 | 2008 | Mixed salad, goulash mixed beef and mashed potatoes | 19 | NK | Vomiting, diarrhoea | 4 | Medico-social institute | 6.00E + 02 | GS1; GS2; GS7; GS12 |
| 7 | 2008 | Mashed potatoes and boiled potatoes | 28 | NK | Vomiting, diarrhoea | 2 | Medico-social institute | 9.20E + 05 | GS7; GS8 |
| 8 | 2008 | Mixed salad (rice and corn) | 2 | NK | Abdominal pains, vomiting | 1 | Staff canteen | 1.90E + 03 | GS2 |
| 9 | 2008 | Rice salad | 13 | 12–15 | Abdominal pains, vomiting, other | 1 | Medico-social institute | 2.00E + 03 | GS2 |
| 10 | 2008 | Semolina | 61 | 3–6 | Abdominal pains, vomiting | 1 | School canteen | 1.00E + 04 | GS7 |
| 11 | 2008 | Semolina and lamb | 4 | 0–3 | Vomiting | 1 | Commercial catering | 5.50E + 04 | GS3 |
| 12 | 2008 | Mashed potatoes, mashed celery, roast pork, sauce and pasta | 5 | 6–9 | Diarrhoea | 2 | Medico-social institute | 1.50E + 05 | GS4; GS7 |
| 13 | 2008 | Cream caramel and smoked salmon | 11 | 9–12 | Diarrhoea, other | 3 | Commercial catering | 3.00E + 03 | GS2; GS8 |
| 14 | 2008 | Fruit salad | 70 | NK | NK | 1 | Staff canteen | 6.30E + 03 | GS3 |
| 15 | 2008 | Tandoori chicken | 10 | 6–9 | Vomiting, diarrhoea | 2 | Commercial catering | 4.60E + 03 | GS6 |
| 16 | 2008 | Wheat | 3 | 9–12 | Diarrhoea | 3 | Commercial catering | 1.60E + 06 | GS1; GS4 |
| 17 | 2009 | Tiramisu | 15 | 0–3 | Vomiting, diarrhoea | 1 | Company canteen | 8.00E + 02 | GS9 |
| 18 | 2009 | Fish in coconut milk | 2 | 0–3 | Nausea, other | 1 | Commercial catering | 1.10E + 04 | GS1 |
| 19 | 2009 | Mashed potatoes | 24 | NK | Vomiting, diarrhoea | 1 | School canteen | 4.00E + 02 | GS7 |
| 20 | 2009 | Cantonese rice | 2 | 0–3 | Vomiting, other | 1 | Family | 1.60E + 05 | GS3 |
| 21 | 2009 | Mashed potatoes, roast beef and French beans | 7 | 6–9 | Vomiting, diarrhoea | 3 | Medico-social institute | 1.90E + 03 | GS3; GS5 |
| 22 | 2009 | Quenelle of pike | 15 | 0–3 | Vomiting, diarrhoea, other | 1 | Staff canteen | 1.20E + 03 | GS6 |
| 23 | 2009 | Sandwich (tomato, carrots, chicken) | 7 | 0–3 | Abdominal pains, nausea | 4 | Commercial catering | 5.00E + 03 | GS1; GS2; GS6; GS10 |
| 24 | 2009 | Chicken sauce | 15 | NK | Vomiting,- diarrhoea | 1 | Commercial catering | 5.00E + 02 | GS3 |
| 25 | 2009 | Squid sauce | 3 | 9–12 | Diarrhoea | 1 | Staff canteen | 2.10E + 05 | GS12 |
| 26 | 2009 | Sauteed shrimp | 4 | 0–3 | Vomiting, diarrhoea | 7 | Commercial catering | 1.90E + 04 | GS1; GS4; GS6 |
| 27 | 2009 | Semolina and peas | 7 | 3–6 | Nausea, diarrhoea, other | 5 | Staff canteen | 2.00E + 07 | GS2; GS5 |
| 28 | 2010 | Salad | 44 | NK | Vomiting, diarrhoea, other | 3 | School canteen | 1.00E + 03 | GS2 |
| 29 | 2010 | Pasta gratin | 2 | 0–3 | vomiting - diarrhoea | 1 | Family | 1.50E + 07 | GS3 |
| 30 | 2010 | Sausage and rice salad | 8 | 0–3 | Vomiting, diarrhoea | 1 | Family | 3.00E + 03 | GS3 |
| 31 | 2010 | Paella | 27 | 6–9 | Diarrhoea | 1 | Medico-social institute | 2.80E + 04 | GS2 |
| 32 | 2010 | Samosa and marinated shrimp tail | 3 | 0–3 | Diarrhoea | 13 | Commercial catering | 2.90E + 05 | GS1; GS2; GS4; GS5; GS6; GS10 |
| 33 | 2010 | Chicken | 8 | 3–6 | Vomiting, diarrhoea | 1 | Family | 6,50E + 04 | GS3 |
| 34 | 2010 | Tabbouleh | 11 | NK | Abdominal pains, other |
1 | Medico-social institute | NK | GS2 |
| 35 | 2010 | Mashed potatoes and mashed vegetables | 19 | NK | Vomiting, diarrhoea, other | 1 | Medico-social institute | 1.20E + 04 | GS1 |
| 36 | 2010 | Pasta salad and rice salad | 20 | 0–3 | Vomiting, diarrhoea | 7 | Family | 9.60E + 07 | GS1; GS3; GS4; GS5; GS6 |
| 37 | 2011 | Mixed dish, soup, mixed ham, mixed apple and lasagne bolognese | 19 | 6–9 | Vomiting, diarrhoea | 2 | Medico-social institute | 3.10E + 03 | GS3 |
| 38 | 2011 | Shrimp | 3 | 0–3 | Abdominal pains, vomiting, other | 2 | Commercial catering | 1.90E + 03 | GS1 |
| 39 | 2011 | Moussaka | 1 | 3–6 | Abdominal pains | 3 | Commercial catering | 8.20E + 04 | GS1; GS4; GS5 |
| 40 | 2011 | Spaghetti | 18 | 12–15 | Vomiting, diarrhoea | 2 | School canteen | 1.00E + 03 | GS8 |
| 41 | 2011 | Couscous, semolina, lamb, vegetable dish | 19 | 9–12 | Nausea, diarrhoea | 2 | Medico-social institute | 2.30E + 03 | GS4; GS11 |
| 42 | 2011 | Carrots | 3 | 3–6 | Vomiting, diarrhoea, other | 1 | Commercial catering | 5.80E + 03 | GS2 |
| 43 | 2011 | Mashed potatoes | 10 | NK | Vomiting, diarrhoea | 1 | School canteen | 7.80E + 04 | GS4 |
| 44 | 2011 | Mashed celery | 15 | 12–15 | Vomiting, diarrhoea | 1 | Staff canteen | 1.00E + 05 | GS7 |
| 45 | 2011 | Tomatoes and fish | 3 | 12–15 | Vomiting, diarrhoea | 1 | Medico-social institute | 5.50E + 03 | GS2 |
| 46 | 2011 | Miso soup | 1 | NK | NK | 1 | Family | 1.50E + 03 | GS9 |
| 47 | 2011 | Mixed salad | 3 | 0–3 | Vomiting, diarrhoea | 1 | Medico-social institute | 2.00E + 03 | GS2 |
| 48 | 2011 | Tomato, corn, courgette dish | 9 | 6–9 | Abdominal pains, vomiting | 1 | School canteen | 4.00E + 03 | GS2 |
| 49 | 2011 | Samosa | 9 | 0–3 | Nausea, other | 1 | Commercial catering | 1.,00E + 09 | GS6 |
| 50 | 2011 | Rice and shellfish dish and fish | 6 | 3–6 | Abdominal pains, nausea | 2 | Staff canteen | 2.70E + 03 | GS5; GS6 |
| 51 | 2012 | Apricot compote, mashed carrots and mashed broccoli | 8 | 9–12 | Vomiting | 1 | School canteen | 7.00E + 02 | GS1 |
| 52 | 2012 | Paella | 2 | 0–3 | Vomiting, diarrhoea, other | 3 | Commercial catering | 2.10E + 04 | GS1; GS3; GS10 |
| 53 | 2012 | Pasta | 60 | 0–3 | Vomiting, diarrhoea | 3 | School canteen | 5.80E + 04 | GS5 |
| 54 | 2012 | Mixed salad | 8 | 18–21 | Abdominal pains, vomiting, other | 1 | Family | 4.00E + 02 | GS2 |
| 55 | 2012 | Chicken | NK | NK | Other | 3 | Commercial catering | 4.00E + 03 | GS2; GS5 |
| 56 | 2012 | Lamb meat | 5 | 6–9 | Vomiting, diarrhoea | 1 | Staff canteen | 2.30E + 03 | GS2 |
| 57 | 2012 | Mashed fish | 18 | 9–12 | Vomiting, diarrhoea | 1 | Medico-social institute | 4.00E + 02 | GS7 |
| 58 | 2012 | Diced mixed vegetables | 14 | 9–12 | Vomiting, diarrhoea | 1 | Medico-social institute | 4.00E + 02 | GS2 |
| 59 | 2012 | Millefeuille pastry | 2 | 3–6 | Nausea | 1 | Commercial catering | 2.00E + 03 | GS2 |
| 60 | 2012 | Onion soup | 5 | 9–12 | Vomiting | 1 | School canteen | 4.00E + 02 | GS2 |
| 61 | 2013 | Semolina | 3 | 3–6 | Vomiting, diarrhoea | 2 | Family | 1.00E + 04 | GS5; GS10 |
| 62 | 2013 | Grilled pork | 2 | 6–9 | Vomiting, diarrhoea | 2 | Family | 1.80E + 04 | GS1; GS9 |
| 63 | 2013 | Cheese-topped dish of seafood, pasta | 15 | 6–9 | Diarrhoea, other | 4 | Staff canteen | 6.50E + 03 | GS1; GS3; GS4 |
| 64 | 2013 | Mashed potatoes | 12 | 3–6 | Vomiting, diarrhoea, other | 2 | Medico-social institute | 2.90E + 03 | GS1; GS3 |
| 65 | 2013 | Pineapple | 5 | NK | Other | 2 | School canteen | 4.50E + 02 | GS1; GS9 |
| 66 | 2013 | Mashed spinach | 13 | 6–9 | Vomiting, diarrhoea | 3 | Medico-social institute | 1.00E + 04 | GS1; GS4 |
| 67 | 2013 | Vegetable soup | 10 | 15–18 | Vomiting, diarrhoea | 1 | Medico-social institute | 9,10E + 02 | GS2 |
| 68 | 2013 | Mixed salad | NK | 6–9 | Abdominal pains | 1 | School canteen | 5.50E + 02 | GS2 |
| 69 | 2013 | Spinach | 8 | 0–3 | Vomiting, diarrhoea, other | 2 | Staff canteen | 3.60E + 02 | GS5; GS10 |
| 70 | 2013 | Mixed pie | 19 | 12–15 | Vomiting, diarrhoea | 1 | Medico-social institute | 4.00E + 02 | GS1 |
| 71 | 2014 | Mashed parsnips | 11 | 0–3 | Vomiting | 2 | School canteen | 4,00E + 02 | GS3 |
| 72 | 2014 | Shrimp | 6 | 0–3 | Abdominal pains, vomiting | 2 | School canteen | 7.70E + 03 | GS1 |
| 73 | 2014 | Polenta | 25 | 18–21 | Abdominal pains, diarrhoea | 1 | Medico-social institute | 9.00E + 03 | GS5 |
| 74 | 2014 | Semolina and ginger (spice) | 11 | 0–3 | Vomiting, diarrhoea | 2 | Family | 1.50E + 06 | GS3; GS6 |
FBO: food-borne outbreak; NK: not known.
a Medico-social institutes included centres for disabled people, leisure centres, retirement homes and other community facilities.
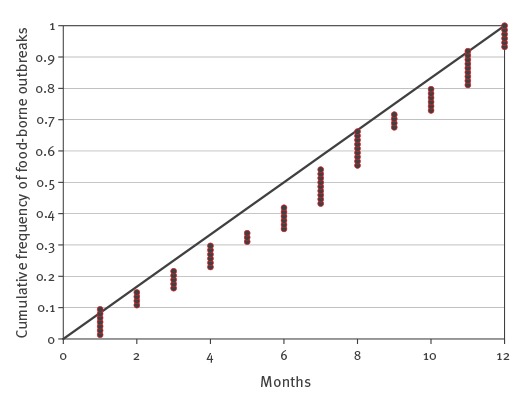
Over the eight years of the survey, the occurrence of FBOs was not subject to any seasonal effect (Figure 1). Emetic and diarrhoeal symptoms of human cases were often present at the same time and were reported for 57% of FBOs (42/74), whereas abdominal pains, diarrhoeic or emetic syndromes alone occurred in 4% (36/911), 12% (109/911) and 13% (118/911) total human cases, respectively.
Figure 1.
Distribution of food-borne outbreaks associated to Bacillus cereus by month of outbreak compared to a theoretical uniform distribution, France, 2007–2014
For the theoretical uniform distribution, each dot represents a food-borne outbreak.
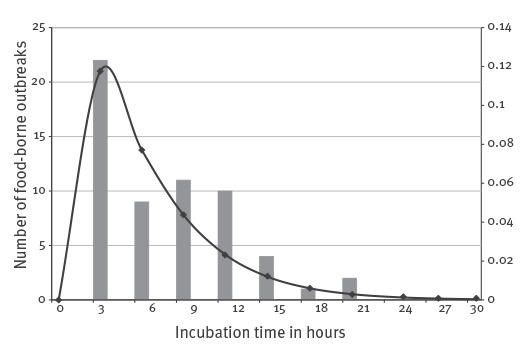
Between 400 and 108B. cereus CFU/g were found in the incriminated foods. Levels lower than 105 CFU/g were observed in 48/57 FBOs due to diarrhoeal strains and in 11/17 FBOs due to emetic strains (Table 1). The incubation period (time between ingestion of contaminated food and symptom onset) varied from less than 3 hours to 21 hours (Figure 2). The mean incubation period was 5.7 hours (standard deviation (SD) 1.3) and could vary within the same FBO (Table 1). However, the incubation period was significantly shorter for emetic strains (carrying the ces gene) – mean: 2.6 hours (SD: 2.1) – compared with diarrhoeal strains (mean: 6.6 hours (SD: 1.4).
Figure 2.
Distribution of food-borne outbreaks by incubation periods for the entire Bacillus cereus collection, France, 2007–2014
Bars represent the number of food-borne outbreaks for each incubation time.
A single food source was incriminated for 57% of FBOs (42/74), of which 14/42 were associated with starchy food, 8/42 and 7/42 FBOs with vegetables and with mixed dishes composed of starchy food or vegetables, respectively (Table 1). Only 14% (10/74) of FBOs were associated with foodstuffs of animal origin.
Furthermore, 60% of FBOs (44/74) occurred in institutional catering, involving 82% (747/911) of the human cases. FBOs were poorly reported in a family context, which represented 13% of the FBOs (10/74) and 7% (64/911) of the human cases (Table 1). The remaining 27% (20/74) of FBOs occurred in a commercial catering context, involving 11% (100/911) of cases.
Strain characterisation
Phenotypic analysis of the strains showed that 92% (312/339) of the strains produced lecithinase. Haemolytic activity on sheep blood agar was detected for 87% (295/339). Some 48% (163/339) of strains were able to hydrolyse starch (data not shown). The panC gene sequences were used to assign B. cereus strains to one of the seven previously described phylogenetic groups I to VII (Table 2). Group I was not represented in the strains analysed. Group III was the most represented (46%; 156/339). Groups IV and II represented 24% (81/339) and 19% (64/339), respectively. The distribution of strains in groups VII, VI and V were 5% (17/339), 4% (14/339) and 2% (7/339), respectively.
Table 2. Genetic signatures of Bacillus cereus strains according to gene detection and panC phylogenetic groups, France, 2007–2014 (n = 159).
| Genetic signature | Number of strains | Genes detected | panC phylogenetic groups | |||||
|---|---|---|---|---|---|---|---|---|
| cytk-1 | cytk-2 | ces | hlyII | nheABC | hblCDA | |||
| GS1 | 34 | Neg | Neg | Neg | Neg | Pos | Neg | II -III - IV |
| GS2 | 28 | Neg | Pos | Neg | Neg | Pos | Pos | IV |
| GS3 | 25 | Neg | Neg | Pos | Neg | Pos | Neg | III |
| GS4 | 18 | Neg | Pos | Neg | Neg | Pos | Neg | II - III |
| GS5 | 18 | Neg | Neg | Neg | Pos | Pos | Pos | II - III |
| GS6 | 10 | Neg | Pos | Neg | Pos | Pos | Pos | II - IV |
| GS7 | 8 | Pos | ND | ND | ND | ND | ND | VII |
| GS8 | 6 | Neg | Neg | Neg | Neg | BC | AD | VI |
| GS9 | 4 | Neg | Pos | Neg | Pos | Pos | Neg | II - III |
| GS10 | 5 | Neg | Neg | Neg | Neg | Pos | Pos | IV - V |
| GS11 | 1 | Neg | Pos | Pos | Neg | Pos | Neg | III |
| GS12 | 2 | Neg | Neg | Neg | Pos | Pos | Neg | II |
AD: only hblA and hblD detected; BC: only nheB and nheC detected; ND: primers used are unable to detect these genes in GS7 group strains; Neg: negative; Pos: positive.
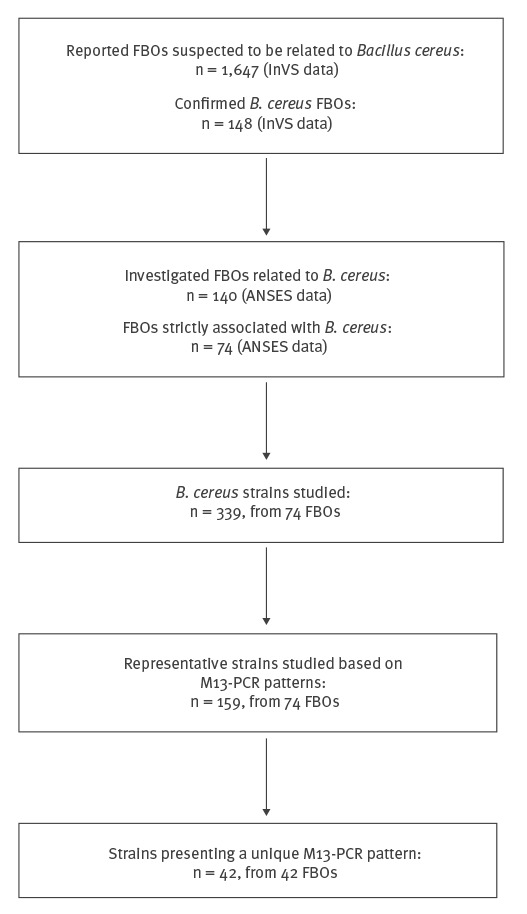
M13-PCR typing and genetic characterisation were conducted on all 339 B. cereus isolates from the 74 FBOs in order to discriminate different patterns and genetic profiles. Up to five isolates from each FBO were subjected to M13-PCR typing. For 42 FBOs, a unique M13 pattern was identified among all isolates recovered from samples within the same FBO (such as FBO number 5, Figure 3A). In the remaining 32 FBOs, several M13 patterns were observed in samples within the same FBO (such as FBO number 6 with four different M13 patterns, Figure 3B). Thus, a total of 159 representative strains gathering 42 strains from the 42 FBOs of unique M 13 pattern and 117 strains representative of the M 13 pattern diversity from the remaining 32 FBOs, were selected for further characterisation (Figure 4).
Figure 3.
Coliphage M13 sequence-based PCR typing of selected Bacillus cereus strains isolated from various samples in two food-borne outbreaks, France, 2007–2014 (n = 11)
B: Bacillus; FBO: food-borne outbreak.
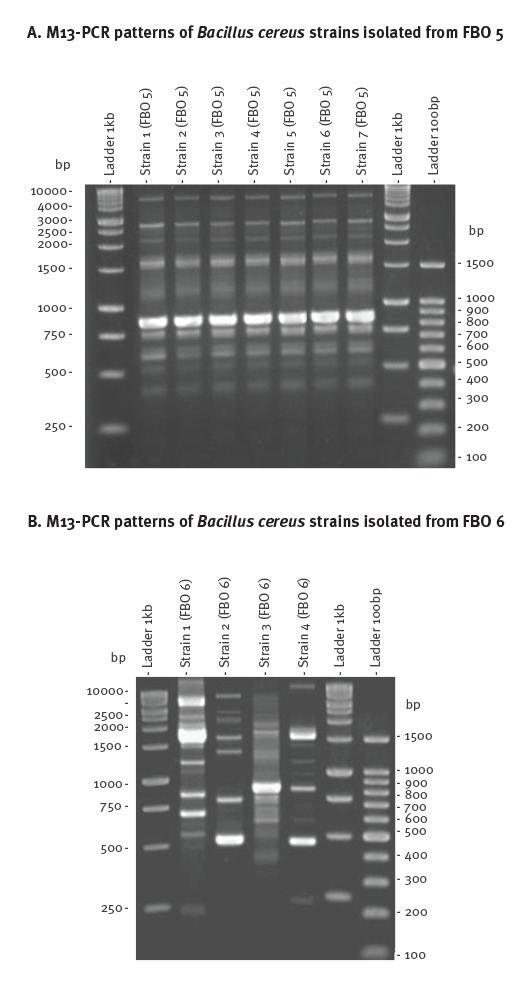
Figure 4.
Selection of food-borne outbreaks and panel of Bacillus cereus strains studied, France, 2007–2014 (n = 159)
ANSES: French Agency for Food, Environmental and Occupational Health and Safety; FBO: food-borne outbreak; InVS: French Institute for Public Health surveillance, Santé publique France
The presence of major virulence genes was investigated (Table 2). The ces gene was detected in 16% (25/159) of the B. cereus strains, meaning they were emetic strains. All the emetic strains belonged to phylogenic group III. The cytK-1 gene was detected in 5% (8/159) of strains, strictly associated with group VII and classified as B. cytotoxicus strains.
The most frequently distributed genes were those encoding enterotoxin Nhe, namely nheC, nheB and nheA genes detected in respectively 100% (159/159), 99% (157/159) and 96% (153/159) of the tested strains. The hblA, hblD and hblC genes encoding enterotoxin Hbl were detected in 44% (70/159), 44% (70/159) and 40% (64/159) of the strains, respectively. The cytK-2 gene was detected in 42% (67/159) of strains and 23% (37/159) of strains carried hlyII.
These genetic features allowed to cluster the strains into 12 pathogenicity or ‘genetic signatures’ (GSs), GS1 to GS12 (Table 2). Some 84% (133/159) of the strains belonged to GS1 to GS6. The most frequent GS encountered in the collection was GS1, which accounted for 21% (34/159) of strains. In GS1, only Nhe-encoding genes were detected. The ces-positive strains were all placed in GS3 (except a single one in GS11) and possessed nhe genes in addition to the ces gene. GS11 also displayed the cytK-2 gene. GS7 contained all the B. cytotoxicus strains carrying the cytK-1 gene. GS8 was characterised by strains carrying nheB and nheC genes, and hblA and hblD genes. All the strains in this group belonged to phylogenetic group VI (Table 2). Several GSs defined in this study were associated with a single panC phylogenetic group, i.e. GS2 (IV), GS203 (III) GS7 (VII), GS8 (VI), GS11 (III) and GS12 (II).
Discussion
Food-borne infections are a common yet distressing and sometimes life-threatening problem for millions of people throughout the world [21]. B. cereus is reported to be the fourth major cause of notified FBOs in the European Union and the second in France [4,5]. However, B. cereus-associated outbreaks are likely to be underestimated, as they usually remain undiagnosed and therefore under-reported. If B. cereus is suspected, several identification tests can be performed: morphology tests on selective media, resistance to polymyxin B, lecithinase synthesis, haemolytic capacity, mannitol fermentation and starch hydrolysis [22]. These tests do not, however, reveal whether the isolated strains are pathogenic nor their genetic features.
The main strengths of our study are the unique national B. cereus strain collection linked to strong-evidence FBOs, the long period covered and an accurate epidemiological and strain characterisation. The study of symptoms does not readily allow the identification of the pathogen causing the FBO because gastroenteritis symptoms are also characteristic of other food-borne pathogens, especially S. aureus or C. perfringens [22]. However, phenotypic analysis and species discrimination allowed us to collect isolates and epidemiological data from 140 FBOs, of which 74 were strictly associated with B. cereus and affected 911 human cases. Considering food safety issues, this provides confirmation that B. cereus must be considered an important food-borne pathogen, and underlines the need to improve monitoring.
For 32 of these 74 FBOs, several strain patterns were distinguished from samples of a single FBO and it was not possible to discriminate which strain or which combination of strains was responsible for the outbreak, highlighting the need for accurate data on the diversity of the isolated strains during FBO investigation. In contrast, for 42 of the 74 FBOs, a unique strain pattern was identified for each FBO, providing a valuable strain collection for further analysis of the correlation between B. cereus genotypic features and associated diseases. Thus, the design of this study strengthens the interpretation of results and avoids bias regarding the bacterial agent causing the FBO.
Our study described 74 FBOs in which only B. cereus was recovered. Nevertheless, a limitation of our study is the exhaustivity of the studied FBOs during the period, as the French institute for public health surveillance (InVS, since 2016 Santé publique France) notified 148 FBOs between 2007 and 2014, in which B. cereus was the confirmed causative agent (Figure 4). The number FBOs notified to InVS was slightly higher than that of FBOs for which strains were received in ANSES and could be explained by the absence of microbiological investigation of such FBOs or the absence of isolation or sending B. cereus strains for further analysis.
Starchy food and vegetables were the most common food vehicles identified in our study. A previous study in commercial cooked chilled foods containing vegetables had shown high B. cereus contamination levels in raw vegetables [23]. Thus, particular attention should be taken during sampling and epidemiological investigation into potential B. cereus contamination of vegetables and starchy food. In our study, 60% (44/74) of FBOs occurred in an institutional catering context. In the family context, 40% (26/64) of the cases were caused by emetic strains. Incorrect cooling of food during preparation or the conservation of cooked dishes at room temperature is thought to be the cause of cereulide production [24]. Moreover, the severity of symptoms associated with emetic strains might explain an increased reporting of these strains in the family context, compared with diarrhoeic strains which may remain undiagnosed and therefore under-reported.
Epidemiological and clinical data show that the type of symptom could not be specifically associated with the presence of emetic or diarrhoeic strains. Indeed, 57% (n=42) of the 74 FBOs shared both diarrhoeic and emetic syndromes although they were caused by only one type of strain. This may be partially explained by the fact that the emetic GS3 strains strongly produce Nhe enterotoxin (data not shown). We suspect that emetic strains may be ingested concomitantly with cereulide preformed in food, increasing pathogenicity and causing a mix of symptoms.
A significant difference was observed for the incubation period according to the type of strain. This is in accordance with previous findings showing that rapid onset of an emetic syndrome indicates intoxication by cereulide [25]. In contrast, ingestion of diarrhoeic bacteria can induce pathology via the production of enterotoxins in the small intestine, leading to a longer incubation period [26]. In some FBOs, the strains had short incubation periods (0–3 hours) without involvement of emetic strains. We hypothesise that those strains might be responsible for rapid vomiting despite absence of the ces gene as previously described [27], or alternatively that the emetic toxin was concomitantly ingested with the contaminated food in addition to a ces-negative strain, or that unknown factors were responsible for vomiting symptoms.
Diarrhoeal diseases are often associated with B. cereus counts of 105 to 108 cells or spores [28]. In our study, concentrations below 103 CFU/g were found in 12 of 57 foods related to diarrhoeal FBOs. This challenges the concept of a minimum infectious dose for B. cereus in diarrhoeal FBOs. A mathematical model based on systematic data collection of B. cereus concentrations in food implicated in outbreaks could be developed for dose–response assessment, in order to quantify infectivity associated with single cells [29]. Levels of at least 105 CFU/g have generally been reported in the incriminated food linked to an emetic syndrome [30]. In our study, levels of as few as 400 CFU/g were implicated. This could be explained by cereulide’s strong resistance to various treatments, underlining the importance of quantifying cereulide in foods. We cannot exclude the possibility that the CFUs recovered from leftover food accurately corresponded to the initial ingested CFUs. Indeed, food processing and storage before tests may have injured vegetative bacteria. However, we suspect that the spores, which are resistant to storage, are likely to be responsible for food-borne infections.
The genetic diversity of B. cereus strains involved in FBOs was revealed in our study by characterisation of strains based on the detection of the genes encoding cereulide, diarrhoeic toxins (Nhe, Hbl, CytK-1 and CytK-2) and Haemolysin (HlyII) and by phylogenetic classification. A total of 12 pathogenicity signatures based on genetic features of the strains were identified. Emetic strains were clustered in GS3, and possessed both the ces gene and the nhe genes. This corroborates with the M13 patterns, showing a high clonality of the GS3 group. Surprisingly, all the GS3 strains were unable to hydrolyse starch, although they were mostly found in starchy foods, as published elsewhere [31]. An atypical ces-positive strain was classified in GS11, characterised by the presence of the cytk-2 gene and the absence of Nhe production, despite detection of nhe genes (data not shown). This strain was detected once in the analysis of FBO 41, together with a strain belonging to GS4. Such atypical emetic strains have been described [25].
The diarrhoeic strains were more polymorphic than the emetic strains, displaying nine different genetic signatures, although six accounted for 84% (105/125) of the strains. Genes encoding Nhe were present in all GSs, but had variable Nhe production (data not shown), suggesting that other factors may be involved in pathogenicity. GS1 (nhe genes only) and GS2 (nhe, hbl and cytK2) were the most prevalent GSs and may have a large impact on human health: they were present in 28% (20/74) and 31% (23/74) of FBOs, respectively. This is consistent with previous findings showing 28% and 24% of B. cereus strains belonging to GS1 and GS2, respectively [13]. Unlike GS1 strains, which were divided into three different phylogenetic groups, all GS2 strains belonged to phylogenetic group IV. These strains produce high concentrations of Hbl, are strongly cytotoxic to Caco2 cells and are more prevalent among strains responsible for food poisoning [12]. These characteristics might partially explain the pathogenic potential of strains of GS2, although a synergistic effect of Hbl and Nhe on pathogenicity was not observed [32].
GS7 contained all the B. cytotoxicus strains carrying the cytK-1 gene, which were related to phylogenetic group VII. Strains carrying cytk-1 were mainly found in vegetable purees, corroborating results of a study showing that 35% of B. cereus strains found in commercial potato products taken on retail level or from catering establishments, possess cytk-1 [33].
Several studies suggest that the pathogenic potential of group VI strains is very low [12]. In our study, these GS8 strains were involved in two FBOs in association with other strains belonging to GS2 and GS7, (FBO 7 and 13, respectively). Thus, it was not proven that GS8 strains were responsible for the symptoms. However, FBO 40, with 18 human cases, was caused by a unique GS8 strain, suggesting a virulence potential of this group [12].
Taken together, assignation of the strains according to genetic signature showed a high genetic diversity of B. cereus strains involved in FBOs and their pathogenic potential. Our results underline that B. cereus is a food-borne pathogen with a substantial impact on human health that should be investigated when a FBO is suspected. We propose an approach based on reported symptoms and incubation period. Particular attention should be given to vegetables and starchy food during the sampling as part of the investigation. We recommend collecting at least five colonies from each food sample potentially contaminated with B. cereus, with different morphologies, as several B. cereus with different genetic characteristics may be present in the same food product.
Acknowledgements
The authors would like to warmly thank all the district veterinary and food analysis laboratories for carrying out B. cereus detection and transmitting isolates together with epidemiological data to the Laboratory for Food Safety. We also address special thanks to Dr Marie Laure De Buyser, who initiated the strain collection and the Central Veterinary Services Laboratory Unit of the Laboratory for Food safety.
Conflict of interest: None declared.
Authors’ contributions: BG participated in the design of the study and the draft of all the manuscript, he conducted the microbial analysis of confirmation and characterisation of the strains, as well collected the epidemiological data as part of a PhD work. SH participated in the design and coordination of the study, and the draft of all the manuscript. LG carried out the statistical analysis on the clinical and epidemiological data and took part in the draft of the manuscript. SCS took part in building of the strain collection and molecular characterisation. MLV and JG took part in the confirmation of isolates and PCR gene detection. SP was in charge of the enterotoxins production. VM and JAH participated in the design of the study and took part in the draft of the manuscript. NR participated in the design of the study, data interpretation and the draft of all the manuscript. AB participated in the design and coordination of the study, and the draft of all the manuscript.
References
- 1. Guinebretière M-H, Auger S, Galleron N, Contzen M, De Sarrau B, De Buyser M-L, et al. Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus Group occasionally associated with food poisoning. Int J Syst Evol Microbiol. 2013;63(Pt 1):31-40. 10.1099/ijs.0.030627-0 [DOI] [PubMed] [Google Scholar]
- 2. Drobniewski FA. Bacillus cereus and related species. Clin Microbiol Rev. 1993;6(4):324-38. 10.1128/CMR.6.4.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mock M, Fouet A. Anthrax. Annu Rev Microbiol. 2001;55(1):647-71. 10.1146/annurev.micro.55.1.647 [DOI] [PubMed] [Google Scholar]
- 4.Santé publique France. Données relatives aux toxi-infections alimentaires collectives déclarées en France en 2013. [Data on collective food-borne outbreaks reported in France in 2013]. Saint-Maurice: Santé publique France. [Accessed Dec 2015]. French. Available from: http://invs.santepubliquefrance.fr/Dossiers-thematiques/Maladies-infectieuses/Risques-infectieux-d-origine-alimentaire/Toxi-infections-alimentaires-collectives/Donnees-epidemiologiques
- 5.European Food Safety Authority, European Centre for Disease Prevention and Control (EFSA, ECDC). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2013. EFSA Journal 2015;13(1):3991. doi: 10.2903/j.efsa.2015.3991,165 pp. [DOI]
- 6. Dierick K, Van Coillie E, Swiecicka I, Meyfroidt G, Devlieger H, Meulemans A, et al. Fatal family outbreak of Bacillus cereus-associated food poisoning. J Clin Microbiol. 2005;43(8):4277-9. 10.1128/JCM.43.8.4277-4279.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ehling-Schulz M, Vukov N, Schulz A, Shaheen R, Andersson M, Märtlbauer E, et al. Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl Environ Microbiol. 2005;71(1):105-13. 10.1128/AEM.71.1.105-113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fagerlund A, Ween O, Lund T, Hardy SP, Granum PE. Genetic and functional analysis of the cytK family of genes in Bacillus cereus. Microbiology. 2004;150(Pt 8):2689-97. 10.1099/mic.0.26975-0 [DOI] [PubMed] [Google Scholar]
- 9. Ramarao N, Sanchis V. The pore-forming haemolysins of bacillus cereus: a review. Toxins (Basel). 2013;5(6):1119-39. 10.3390/toxins5061119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cadot C, Tran SL, Vignaud ML, De Buyser ML, Kolstø AB, Brisabois A, et al. InhA1, NprA, and HlyII as candidates for markers to differentiate pathogenic from nonpathogenic Bacillus cereus strains. J Clin Microbiol. 2010;48(4):1358-65. 10.1128/JCM.02123-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tran SL, Guillemet E, Gohar M, Lereclus D, Ramarao N. CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J Bacteriol. 2010;192(10):2638-42. 10.1128/JB.01315-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guinebretière M-H, Velge P, Couvert O, Carlin F, Debuyser M-L, Nguyen-The C. Ability of Bacillus cereus group strains to cause food poisoning varies according to phylogenetic affiliation (groups I to VII) rather than species affiliation. J Clin Microbiol. 2010;48(9):3388-91. 10.1128/JCM.00921-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang IC, Shih DY-C, Huang T-P, Huang Y-P, Wang J-Y, Pan T-M. Establishment of a novel multiplex PCR assay and detection of toxigenic strains of the species in the Bacillus cereus group. J Food Prot. 2005;68(10):2123-30. [DOI] [PubMed] [Google Scholar]
- 14. Jeßberger N, Krey VM, Rademacher C, Böhm M-E, Mohr A-K, Ehling-Schulz M, et al. From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front Microbiol. 2015;6:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beecher DJ, Wong AC. Identification of hemolysin BL-producing Bacillus cereus isolates by a discontinuous hemolytic pattern in blood agar. Appl Environ Microbiol. 1994;60(5):1646-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guinebretiere M-H, Nguyen-The C. Sources of Bacillus cereus contamination in a pasteurized zucchini purée processing line, differentiated by two PCR-based methods. FEMS Microbiol Ecol. 2003;43(2):207-15. [DOI] [PubMed] [Google Scholar]
- 17. Guinebretière M-H, Thompson FL, Sorokin A, Normand P, Dawyndt P, Ehling-Schulz M, et al. Ecological diversification in the Bacillus cereus Group. Environ Microbiol. 2008;10(4):851-65. 10.1111/j.1462-2920.2007.01495.x [DOI] [PubMed] [Google Scholar]
- 18. Guinebretière M-H, Broussolle V, Nguyen-The C. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J Clin Microbiol. 2002;40(8):3053-6. 10.1128/JCM.40.8.3053-3056.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pouillot R, Delignette-Muller ML. Evaluating variability and uncertainty separately in microbial quantitative risk assessment using two R packages. Int J Food Microbiol. 2010;142(3):330-40. 10.1016/j.ijfoodmicro.2010.07.011 [DOI] [PubMed] [Google Scholar]
- 20. Guillier L, Thébault A, Gauchard F, Pommepuy M, Guignard A, Malle P. A risk-based sampling plan for monitoring of histamine in fish products. J Food Prot. 2011;74(2):302-10. 10.4315/0362-028X.JFP-10-234 [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization (WHO). WHO estimates of the global burden of foodborne diseases; foodborne disease burden epidemiology reference group 2007-2015. Geneva: WHO; 2015. Available from: http://apps.who.int/iris/bitstream/10665/199350/1/9789241565165_eng.pdf
- 22.Cadel Six S, De Buyser M-L, Vignaud ML, Dao TT, Messio S, Pairaud S, et al. Toxi-infections alimentaires collectives à Bacillus cereus : bilan de la caractérisation des souches de 2006 à 2010. [Bacillus cereus food poisoning outbreaks: strain characterization results, 2006-2010]. Bull Epidemiol Hebd. 2012;(Hors-série):45-9. French. Available from: https://pro.anses.fr/bulletin-epidemiologique/Documents/BEP-mg-BE50-art14.pdf
- 23. Choma C, Guinebretière MH, Carlin F, Schmitt P, Velge P, Granum PE, et al. Prevalence, characterization and growth of Bacillus cereus in commercial cooked chilled foods containing vegetables. J Appl Microbiol. 2000;88(4):617-25. 10.1046/j.1365-2672.2000.00998.x [DOI] [PubMed] [Google Scholar]
- 24. Agata N, Ohta M, Yokoyama K. Production of Bacillus cereus emetic toxin (cereulide) in various foods. Int J Food Microbiol. 2002;73(1):23-7. 10.1016/S0168-1605(01)00692-4 [DOI] [PubMed] [Google Scholar]
- 25. Ehling-Schulz M, Guinebretiere M-H, Monthán A, Berge O, Fricker M, Svensson B. Toxin gene profiling of enterotoxic and emetic Bacillus cereus. FEMS Microbiol Lett. 2006;260(2):232-40. 10.1111/j.1574-6968.2006.00320.x [DOI] [PubMed] [Google Scholar]
- 26. Ceuppens S, Rajkovic A, Hamelink S, Van de Wiele T, Boon N, Uyttendaele M. Enterotoxin production by Bacillus cereus under gastrointestinal conditions and their immunological detection by commercially available kits. Foodborne Pathog Dis. 2012;9(12):1130-6. 10.1089/fpd.2012.1230 [DOI] [PubMed] [Google Scholar]
- 27. Schmid D, Rademacher C, Kanitz EE, Frenzel E, Simons E, Allerberger F, et al. Elucidation of enterotoxigenic Bacillus cereus outbreaks in Austria by complementary epidemiological and microbiological investigations, 2013. Int J Food Microbiol. 2016;232:80-6. 10.1016/j.ijfoodmicro.2016.05.011 [DOI] [PubMed] [Google Scholar]
- 28. Granum PE, Lund T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol Lett. 1997;157(2):223-8. 10.1111/j.1574-6968.1997.tb12776.x [DOI] [PubMed] [Google Scholar]
- 29. Teunis PFM, Kasuga F, Fazil A, Ogden ID, Rotariu O, Strachan NJC. Dose-response modeling of Salmonella using outbreak data. Int J Food Microbiol. 2010;144(2):243-9. 10.1016/j.ijfoodmicro.2010.09.026 [DOI] [PubMed] [Google Scholar]
- 30.Gilber RJ, Kramer JM. Bacillus cereus food poisoning.). In: Cliver DC, Cochrane BA Editors. Progress in Food Safety.(proceeding of symposium) Madison (WI): Food Research Institute, University of Wisconsin-Madison, Madison; 1986. p. 85-93. [Google Scholar]
- 31. Chon JW, Kim JH, Lee SJ, Hyeon JY, Song KY, Park C, et al. Prevalence, phenotypic traits and molecular characterization of emetic toxin-producing Bacillus cereus strains isolated from human stools in Korea. J Appl Microbiol. 2012;112(5):1042-9. 10.1111/j.1365-2672.2012.05277.x [DOI] [PubMed] [Google Scholar]
- 32. Sastalla I, Fattah R, Coppage N, Nandy P, Crown D, Pomerantsev AP, et al. The Bacillus cereus Hbl and Nhe tripartite enterotoxin components assemble sequentially on the surface of target cells and are not interchangeable. PLoS One. 2013;8(10):e76955. 10.1371/journal.pone.0076955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Contzen M, Hailer M, Rau J. Isolation of Bacillus cytotoxicus from various commercial potato products. Int J Food Microbiol. 2014;174:19-22. 10.1016/j.ijfoodmicro.2013.12.024 [DOI] [PubMed] [Google Scholar]


