Abstract
Cutaneous T-cell lymphoma (CTCL) is the most common type of primary cutaneous lymphoma. Here we report that CTCL patients show increased interleukin-15 (IL-15) in a clinical stage-dependent manner. Mechanistically, we show that Zeb1 is a transcriptional repressor of IL-15 in T-cells and that hypermethylation of the Zeb1 binding region within the IL-15 promoter, as seen in CTCL patients, prevents Zeb1 binding and causes increased transcription of IL-15. Using a transgenic mouse model of IL-15, we provide evidence that overexpression of IL-15 induces a spontaneous CTCL that mimics the human neoplasm. Excessive autocrine production of IL-15 in T-cells inhibits an HDAC1-mediated negative autoregulatory loop, resulting in the upregulation of HDAC1 and HDAC6, and transcriptional induction of the onco-miR-21. Interruption of IL-15 downstream signaling with isotype-specific HDAC inhibitors halts (HDAC1) or significantly delays (HDAC6) the progression of CTCL in vivo and provides pre-clinical evidence supporting a hierarchical model of oncogenic signaling in CTCL.
Introduction
Among primary cutaneous lymphomas, cutaneous T-cell lymphoma (CTCL) is by far the most common type.(1) The majority of CTCL consists of two related CD4+ mature T-cell neoplasms; Mycosis Fungoides (MF) and Sezary Syndrome (SS). MF originates from a clonal expansion of skin-homing CD4+ memory T cells, usually presents with limited skin involvement, and is initially characterized by an indolent clinical course with stepwise progression toward greater tumor burden in the skin, followed in a subset of patients by extracutaneous dissemination. SS is a more aggressive type of CTCL that can present de novo or as an advanced stage progression of MF and is characterized by erythroderma, lymphadenopathy, and circulating clonal atypical CD4+ T-cells. Survival in MF/SS varies according to clinical stage, which is defined according to a composite tumor, node, metastasis, and blood (TNMB) classification, as revised by the EORTC/ISCL International Society of Cutaneous Lymphomas.(2) Patients with “early stage” disease have skin-limited involvement with superficial patches or plaques and an expected survival of >10 years. Patients with “advanced stage” disease, which includes all SS patients, have skin tumors, erythroderma, and/or extracutaneous involvement, and median survival < 5 years. While discrete but highly overlapping molecular signatures associated with MF and SS have been described, the cancer-initiating events and the oncogenic drivers in CTCL remain largely unknown, and there are no curative therapies. Several pan-inhibitors of histone deacetylase (HDAC) have been approved for treatment of CTCL, yet none are curative and each is associated with significant toxicity.(3, 4) In the experimental setting, the term CTCL has been used synonymously with MF/SS.
Cytokines can affect T-cell proliferation and survival during various stages of T-cell development and homeostasis. In CTCL, cytokines such as IL-4, IL-7, IL-13, IL-15, IL-16, IL-17 and IL-31 are present and/or dysregulated during various stages of disease progression.(5–10) Since IL-15 was first proposed to be essential for lymphoma cell growth in vitro, numerous studies have confirmed the pathogenic role of IL-15 in T-cell lymphoma. (5, 6, 11–14) IL-15 is a pleiotropic cytokine that utilizes a non-transducing, private α-chain and a common IL-2/IL-15 βγ-signal transducing complex to function as a growth and survival factor for T-cells and NK cells.(15, 16) There is evidence suggesting that IL-15 has a pathogenic role in CTCL. IL-15 is highly stimulatory for CD4+ CTCL cells in vitro(10, 17–19) and strong IL-15 expression in lesional skin is a characteristic feature of CTCL since normal skin does not express significant amounts of this cytokine, and the reported prevalence and intensity of IL-15 expression in inflammatory skin disorders such as atopic dermatitis and psoriasis is variable.(14, 20) In CTCL, IL-15 has been implicated in the recruitment of CD4+ memory T-cells to the skin, induction of T-cell proliferation, and inhibition of apoptotic cell death.(17, 21) Human CD4+ CTCL cells produce IL-15 in culture thereby sustaining cell growth in an autocrine fashion.(10) An increase in autocrine IL-15 production during disease progression has been proposed to promote CTCL cells’ independence from the skin microenvironment and the development of extracutaneous disease.(14) Furthermore, IL-15 can induce skin-homing CD4+ memory cells to acquire a regulatory T-cell phenotype in vitro, which may protect the malignant cells from elimination through immune evasion.(22) Additional data have established that hair follicle derived IL-15 and IL-7 regulate epidermotropism of memory T cells in inflammatory skin diseases and malignant lymphoma.(23)
While the malignant CD4+ T-cells from blood of all SS patients we have tested appear to produce and be activated by IL-15, the role of this cytokine in the pathogenesis of CTCL remains unresolved. Here we show that overexpression of IL-15 in CTCL patients is coincident with epigenetic disruption of Zeb1 binding at the IL-15 promoter. In vitro, disruption of Zeb1 binding results in overexpression of IL-15, which in turn drives HDAC1 and HDAC6 upregulation, and activation of onco-miR-21. In vivo, overexpression of IL-15 in a mouse model recapitulates human CTCL, while pharmacologic inhibition of HDAC1 and HDAC6 prevents the progression of CTCL.
Results
CD4+ T cells from CTCL patients display IL-15 promoter methylation in the Zeb1 repressor binding region
To study the expression of IL-15 in CTCL, lesional skin biopsies and peripheral blood lymphocytes from CTCL patients were analyzed for IL-15 expression. IL-15 protein is strongly expressed in CTCL, with intense and specific positive staining of atypical lymphoid cells in Pautrier’s microabscesses that are pathognomonic for CTCL (Figure 1A), whereas normal human skin and T-cells do not express IL-15 (Supplementary Figure 1A).(17) We then measured expression of IL-15 mRNA in highly enriched peripheral blood CD4+ T-cells (>90% pure as determined by flow cytometry) from CTCL patients with various stages of disease. Relative to normal donor CD4+ cells; peripheral blood CD4+ cells from CTCL patients listed in Supplementary Table 1A expressed significantly more IL-15 transcript, which was proportional to disease severity in patients (mean ± SEM of fold increase in Stage IB vs. Stage IIB vs. Stage III/IV = 2.58 ± 0.70 vs. 5.38 ± 2.09 vs. 7.47 ± 1.78; n = 3 each; P = 0.032, 0.005 and 0.011 respectively; Figure 1B).
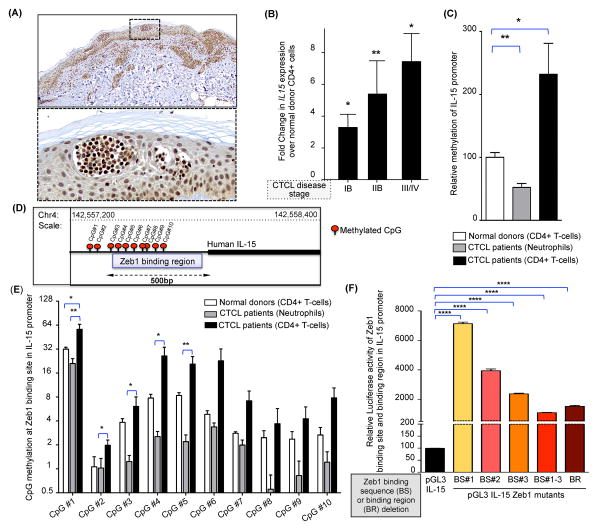
Figure 1. Overexpression of IL-15 in CTCL patient samples.
(A) Representative microscopic images of IL-15 immunohistochemical staining of a skin lesion from a CTCL patient. Scale bar=100μm. Dotted box in the upper panel indicates higher magnification of skin lesion presented in the lower panel. (B) Fold change in IL-15 transcript (mean ± SEM) in CD4+ T cells obtained from blood of patients with progressive stages of CTCL (N=3 each), relative to CD4+ T-cells in normal donor blood (N=3). IL-15 transcript was normalized to 18S and the values for normal donors were arbitrarily set at 1. (C) Graphical representation of IL-15 promoter methylation as determined by pyrosequencing in DNA extracted from sorted CD4+ T-cells and neutrophils of CTCL patients listed in Supplementary Table I. The relative quantity of promoter methylation was compared to purified CD4+ T-cells from normal donors, which is arbitrarily set at 100%. Data shown are mean ± SEM, N=9 for CTCL patients and N=6 for normal donors. (D) Diagram of regulatory region within the human IL-15 promoter, illustrating both the location of the putative Zeb1 binding sites within the CpG rich region and the extent of CpG methylation within this region (CpG1-CpG10) for a typical CTCL patient. (E) Differential methylation of CpG dinucleotides 1 through 10 with in a CpG rich Zeb1 binding region of the IL-15 promoter in CTCL patients (CD4+ T-cells and neutrophils) vs. normal donor CD4+ T-cells; data presented as mean ± SEM, N=9 for patients and N=6 for normal donors. (F) To characterize the transcriptional competence of Zeb1 binding site in IL-15 promoter, the pGL3-IL-15 plasmid construct (presented here as native promoter) was subjected to site directed mutagenesis to create IL-15 promoters lacking Zeb1 binding sites (BS#1, BS#2, BS#3, BS#1–3) or the entire binding region (BR). Relative luciferase activity was measured and normalized to a promoterless PGL3 basic vector (mean ± SEM, N=3 each). For each graphical representation in Figure 1, data are presented as mean ± SEM, *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001 unpaired or paired two-tailed student’s t-test.
Since epigenetic modifications such as DNA methylation can alter gene expression, we selected samples highly enriched in neoplastic T-cells from the blood of SS patients listed in Supplementary Table 1B and asked if promoter methylation of the IL-15 gene might influence its transcriptional regulation. We observed significantly higher overall CpG methylation within the IL-15 promoter in CD4+ T-cells from SS patients vs. normal donors (mean ± SEM of relative % methylation= 232.0 ± 49.18, n=9 vs 100.1 ± 7.619, n=6, P=0.03; Figure 1C). Furthermore, methylation at the IL-15 promoter was higher in malignant CD4+ T-cells compared to non-malignant neutrophils from the same patient (mean ± SEM of relative % methylation = 232.0 ± 49.18 vs 52.31 ± 6.28%, N=9 each, paired t-test, P=0.0025; Figure 1C). Using the same approach, we analyzed methylation of each ‘CpG dinucleotide’ in the IL-15 promoter (Figure 1D). Once again, methylation was higher in CD4+ T-cells from CTCL patients than CD4+ T-cells from normal donors and, within each patient, in CD4+ T-cells versus neutrophils (Figure 1E and Supplementary Figure 1B). This CpG rich region of the human IL-15 promoter contains three putative binding sites for the known transcriptional repressor Zeb1 (Figure 1D and Supplementary Figure 1C).(24, 25)
In order to examine the effect of CpG methylation (at the Zeb1 binding region) on IL-15 transcription, the CpG rich 5′ regulatory region of the IL-15 promoter (0.5kb upstream of transcription start site) was amplified and cloned into PGL3 luciferase vector. As measured by relative luciferase activity, this region of the IL-15 promoter is transcriptionally active in its native form (data not shown). The deletion of the putative Zeb1 binding sites (BS#1, BS#2, BS#3 and BS#1–3) or the entire binding region (BR) in the IL-15 promoter led to a significant increase in IL-15 transcription as determined by relative luciferase activity (mean ± SEM of relative luciferase activity of pGL3 IL-15 vs. BS#1 vs. BS#2 vs. BS#3 vs. BS#1–3 vs. BR vectors = 99.56 ± 0.33 vs. 7135 ± 108.5 vs. 3940 ± 62.36 vs. 2372 ± 24.65 vs. 1099 ± 9.72 vs. 1525 ± 32.00, n=4 each; P<0.0001 each; Figure 1F). We suspect that the deletion of the entire BR is less effective than deleting BS#1–3 because the former likely includes binding sites for transcriptional activators in the regulatory region of the IL-15 gene.
Since CpG methylation of DNA can physically prevent transcription factor binding to a regulatory region of DNA, we hypothesized that CD4+ cells from CTCL patients might display reduced binding of Zeb1 to the IL-15 promoter thereby increasing IL-15 transcription. Indeed, using chromatin immunoprecipitation (ChIP)-PCR, we observed substantial loss of Zeb1 binding at the IL-15 promoter in CD4+ CTCL patient CD4+ cells compared to normal donor CD4+ cells (mean ± SEM of relative Zeb1 binding to IL-15 promoter in normal donor vs. CTCL patient CD4+ T-cells = 101.3 ± 1.48 vs. 25.31 ± 3.78, n=3 each; P<0.0001; Supplementary Figure 1D and Supplementary Table 1C). Of note, the expression levels of Zeb1 binding protein were similar in SS patients, normal donors and patient derived CTCL cell lines (Supplementary Figure 1E–1F). Additionally, silencing Zeb1 in normal donor CD4+ T-cells caused increase in IL-15 transcript (Supplementary Figure 1G). These experiments show that CpG methylation within the Zeb1 binding sites of the IL-15 promoter blocks Zeb1 binding and increases IL-15 expression. This suggests that overexpression of IL-15 in CTCL patient samples is due, at least in part, to this epigenetic event.
IL-15 overexpression in the mouse mimics human CTCL
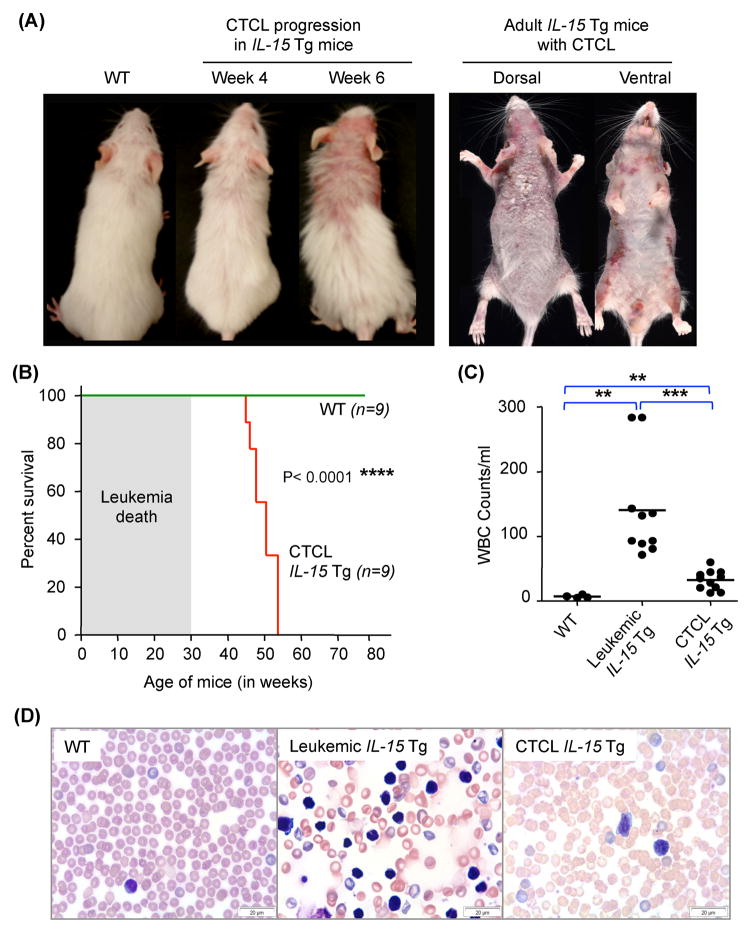
We confirmed that IL-15 is expressed by the neoplastic T-cells in lesional skin and that IL-15 expression is increased in a stage-dependent manner in purified peripheral blood CD4+ T-cells from CTCL patients, compared to normal donors. Next we proceeded to elucidate the role of IL-15 in the development of CTCL using IL-15 transgenic (tg) mice. We had earlier reported that ~30% of the IL-15 tg colony rapidly succumbed to an aggressive leukemia of large granular lymphocytes (LGL).(26, 27) We subsequently observed that the remaining 70% of IL-15 tg mice developed progressive alopecia and extensive cutaneous lesions within 4–6 weeks of birth. The dermatologic features of the IL-15 tg mice display many of those observed in human CTCL, including full body, scaly erythematous plaques/patches, exfoliative dermatitis, ulcerations (Figure 2A), and severe pruritus. The mice that develop the CTCL phenotype display a significantly longer survival compared to those with the aggressive LGL leukemia, but still have a significantly shorter life span compared to wild type (WT) littermate controls (median age of IL-15 tg vs. WT = 50.4 weeks vs. 80 weeks, P<0.0003; Figure 2B). IL-15 tg CTCL mice have only moderate increases in peripheral blood white blood cell (WBC) counts compared to age-matched WT controls or leukemic mice (mean ± SEM of WBC count in WT vs. leukemic IL-15 Tg vs. CTCL IL-15 Tg = 7225 ± 1211 vs. 140750 ± 25103 vs. 32609 ± 4483, n=4, 10 and 11 respectively; Figure 2C). While extreme leukocytosis is not a feature of the IL-15 tg CTCL mice, peripheral blood smears show the presence of atypical lymphocytes with hyperchromatic nuclei, with folded and indented nuclear membrane, which is one of the hematologic manifestations of late stage MF patients and SS patients (Figure 2D).
Figure 2. Overexpression of IL-15 in mice favors development of spontaneous CTCL.
(A) Compared to the WT littermates at week 4 (left most), IL-15 tg mice show progressive development of CTCL with advancing age as shown by a representative mouse pictured at week 4 and week 6. Skin from all IL-15 tg adult mice have severe dermatological signs and symptoms consistent with CTCL (33 weeks old IL-15 tg, right panel). (B) Kaplan-Meier survival curve analysis of IL-15 tg mice compared to WT littermate controls up to 80 weeks of age. Shaded area represents large granular lymphocytic leukemia related deaths, as previously reported.27 Statistical significance was determined by Log-rank test, ****P ≤ 0.0001. (C) White blood cell (WBC) counts from 8–10 week-old, age matched WT control mice and non-leukemic CTCL IL-15 tg mice were compared to leukemic IL-15 tg mice. WBC counts were highest in the leukemic IL-15 tg mice; IL-15 tg mice with CTCL had moderate but significant increases in WBC compared to WT mice. Data is presented as mean ± SEM, **P ≤ 0.01 and ***P ≤ 0.001, unpaired two-tailed student’s t-test. (D) Representative Wright-Giemsa staining of peripheral blood smear from WT, leukemic IL-15 tg and CTCL IL-15 tg mice. Scale bar = 20μM.
To compare the pathology and immunophenotype of the spontaneous CTCL in IL-15 tg mice and human CTCL, we performed a histological analysis of the skin. Human CTCL can be distinguished from many inflammatory skin disorders by the presence of Pautrier’s microabscesses in the epidermis, which reflect the epidermotropic nature of the lymphoid infiltrate, and when found, are a hallmark of the disease.(28) Histological analysis of lesional skin from IL-15 tg mice revealed marked infiltration of the dermal-epidermal junction with atypical lymphocytes, with cells tracking to the upper skin layers and formation of classic Pautrier’s microabscesses (Figure 3A, compare with 1A). Like human CTCL, the cutaneous lymphoid infiltrate of IL-15 tg mice predominantly consisted of both CD3+CD4+ and CD3+CD8+ T-cells, with an approximately 25-fold increase in CD3+ T-cells compared to WT littermate controls (mean ± SEM of absolute cell number in WT vs IL-15 tg skin= 0.15 ± 0.09 vs. 3.80 ± 1.86, n=8 and 14 respectively; P=0.0001; (Figure 3B, Supplementary Figure 2A–B). The cutaneous lymphoid infiltrate in the IL-15 tg mice showed robust expression of the skin homing molecules cutaneous lymphocyte antigen (CLA) and chemokine receptor 4 (CCR4) (Supplementary Figure 2C). In addition, skin CD4+ T-cells from the IL-15 tg mice showed significantly higher expression of the adhesion molecule CD44 (mean ± SEM of CD4+CD44+ expression in WT vs IL-15 tg skin= 12.28 ± 4.35 vs. 34.99 ± 2.44, n=8 and 14 respectively; P<0.0001), as observed in human CTCL cells (Supplementary Figure 2D).(29, 30) Likewise, CD62L expression was either lost or reduced in the CD4+ T-cells of IL-15 tg mice, compared to WT mice (mean ± SEM of CD4+CD62L- expression in WT vs IL-15 tg skin= 70.67 ± 4.74 vs. 88.56 ± 2.23, n=8 and 14 respectively; P=0.0009; Supplementary Figure 2E).(31) T-cell receptor (TCR)-Vβ staining of T-cells isolated from the skin of IL-15 tg mice revealed the simultaneous expansion of select clonal populations of T-cells, none sufficiently large to emerge as a dominant clone, resulting in an oligoclonal pattern (Supplementary Figure 2F). Additionally, CD26 expression was reduced in skin of IL-15 tg mice compared to WT mice (mean ± SEM of CD26 expression in WT vs IL-15 tg skin=33.77 ± 1.235 vs. 6 ± 0.611, n=3 each; P<0.0001; Supplementary Figure 2G). Furthermore, skin resident mononuclear cells overexpressed PLS3, GATA3 and CD164 in IL-15 tg mice compared to WT mice (Supplementary Figure 2H).
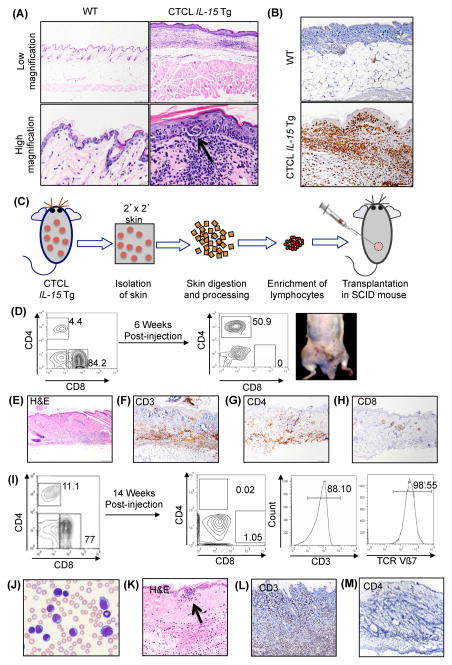
Figure 3. Skin of IL-15 tg mice has pathological features similar to skin of human CTCL.
(A) Representative microphotographs of hematoxylin and eosin (H&E) staining of skin sections from a WT mouse (upper and lower left) and a IL-15 tg mouse (upper and lower right); the latter shows presence of atypical lymphocytic infiltrate called Pautrier’s microabscesses (arrow). (B) Immunohistochemical analysis of skin from an IL-15 tg mouse showed increased presence of CD3+ T-cells (lower panel) when compared to the skin of a matched WT control mouse (upper panel). (C) Experimental scheme for isolation of mononuclear cells from skin of IL-15 tg mice for transplantation into SCID mice recipients. (D) Representative transplant experiment, showing engraftment of IL-15 tg skin mononuclear cells resulting in development of indolent CTCL in the SCID mouse recipient. Approximately 2x106 skin mononuclear cells were transplanted subcutaneously in the right flank of each SCID mouse. The contour plot (left panel) showed FACS analysis of donor mononuclear cells prior to the transplant into the SCID mouse recipient. Six weeks following transplantation, lesions were observed in the recipient’s skin (right), along with the presence of donor T-cells in peripheral blood of the recipient (right contour blot). (E–H) Immunohistochemical staining with H&E, and CD3, CD4 and CD8 antibodies confirmed the presence of donor CD3+CD4+CD8- tumor cell infiltrates in skin sections of the recipient (lower panel). Scale bar = 100μM. (I) Representative transplant experiment, showing engraftment of IL-15 tg skin mononuclear cells resulting in development of aggressive clonal lymphoma in the SCID mouse recipient. Flow cytometric analysis of IL-15 tg donor mononuclear cells immediately prior to the transplant (left, contour plot). Fourteen weeks post-transplant, engrafted cells revealed expansion of CD3+CD4-CD8− cells, clonal for expression of TCR Vβ7 (right, contour plots). (J) Wright-Giemsa staining of peripheral blood smear from the recipient SCID mouse revealed presence of neoplastic cells. Scale bar = 100μM (K) Skin histology shows tumor infiltrates forming pathognomonic ‘Pautrier’s microabscess’ that were (L) CD3+ (M) CD4− and (Supplementary Figure 2I) CD8−.
The validity and impact of a spontaneous murine cancer model are enhanced by the demonstration that the malignant cells are able to engraft, expand, and mimic the primary disease in secondary recipient mice.(32) To test this, we transplanted mononuclear cells isolated from the skin of IL-15 tg (approximately 35 weeks old) into recipient SCID mice, as shown in the experimental schema (Figure 3C). Because the heavy CD3+ cutaneous T-cell infiltrate in the CTCL IL-15 tg mice is a mixture of CD4+ and CD8+ lymphocytes, often with a predominance of CD8+ cells, unsorted cells were transplanted subcutaneously to determine which subset displayed the neoplastic phenotype (Figure 3D, left). We observed two patterns of transplantable CTCL growth in secondary mice. In a subset of secondary recipient mice, six-weeks after transplantation, discrete tumor-like lesions were observed throughout the dorsal skin (Figure 3D, right), caused by cutaneous infiltration with atypical CD3+CD4+CD8− T-cells spanning the dermis and epidermis (Figure 3E–H). In the same mice, a ~12-fold expansion of CD4+ T-cells and a near complete absence of CD8+ T-cells were observed in the peripheral blood. These observations identify the CD4+ T-cell subset as the malignant population in the primary IL-15 tg mouse. In another subset of transplanted mice, the development of secondary skin lesions took longer to evolve (~14 weeks) but resulted in a more aggressive form of secondary CTCL (Figure 3I) with a florid leukemic component due to the expansion of a double negative CD3+CD4-CD8− T-cell population that was clonal for TCR Vβ7 and consisted of morphologically atypical lymphocytes with cerebriform hyperchromatic nuclei similar to classic SS cells (Figure 3J). H&E staining of the skin lesion in this subset of mice also revealed a deep patchy infiltrate of large lymphoid cells, with a residual epidermotropic component, as shown by the formation of Pautrier-like structures (Figure 3K). Similar to the peripheral blood, the cutaneous lymphoid infiltrate was predominantly CD3+CD4-CD8−. (Figure 3L–3M, Supplementary Figure 2I). Thus, in spite of the variable immunophenotype, the transplantation of skin mononuclear cells from primary CTCL mice resulted in a range of secondary T-cell neoplasms that again reflected the clinical and histological spectrum of human CTCL.(33) Notably, secondary recipient mice transplanted with splenic mononuclear cells from IL-15 tg mice did not develop skin lesions or secondary T-cell lymphoma, suggesting that the malignant cells from the primary CTCL mice are highly enriched in, or limited to the skin in some cases (data not shown). Further, these findings support the notion that the neoplastic T-cell population within the skin of the IL-15 tg mice consist of the CD3+CD4+CD8− and CD3+CD4-CD8− T-cells. Additionally, malignant cells from secondary mice overexpressed IL-15Rα and displayed significantly enhanced cell death in the presence of the JAK1/3 inhibitor, Tofacitinib, suggesting that IL-15 signaling is a requisite survival pathway in these cells (Supplementary Figures 2J–K).
IL-15 regulates histone deacetylase expression
CTCL patients show clinical responsiveness to pan-HDAC inhibitors (HDACi), however insight as to the mechanisms of HDAC expression in CTCL is limited.(34) Along with the elevated expression of IL-15 in CTCL patients’ samples we observed that patients’ peripheral blood CD4+ T-cells have increased expression of HDAC1 and HDAC6 transcripts through different stages of the disease when compared to normal donors (Figure 4A). Although not statistically significant, there was a trend for higher expression of HDAC1 and HDAC6 in more advanced stages of CTCL. Similar increases in protein levels of HDAC1 were observed in blood CD4+ cells from SS patients as well as in the SS patient derived cell line, Hut78, compared to normal donors (Supplementary Figure 3A–3B). In order to test the hypothesis that IL-15 modulates HDAC expression, we exposed normal donor CD4+ T-cells to IL-15 in vitro and measured HDAC protein levels. We observed upregulated levels of HDAC1 and HDAC6 in normal CD4+ T-cells after 48 hours of exposure to IL-15 in vitro (Figure 4B–4C). We then examined the intracellular distribution of the HDAC1 and HDAC6 before and after exposing normal CD4+ T-cells to IL-15. We observed that while HDAC1 was nuclear regardless of IL-15 exposure, HDAC6 (that was both cytoplasmic and nuclear before IL-15 incubation) translocated exclusively to the cytoplasm upon incubation with IL-15 (Supplementary Figure 3C–3D). The IL-15-mediated induction of HDACs was not associated with increased CD4+ T-cell growth (Supplementary Figure 3E).
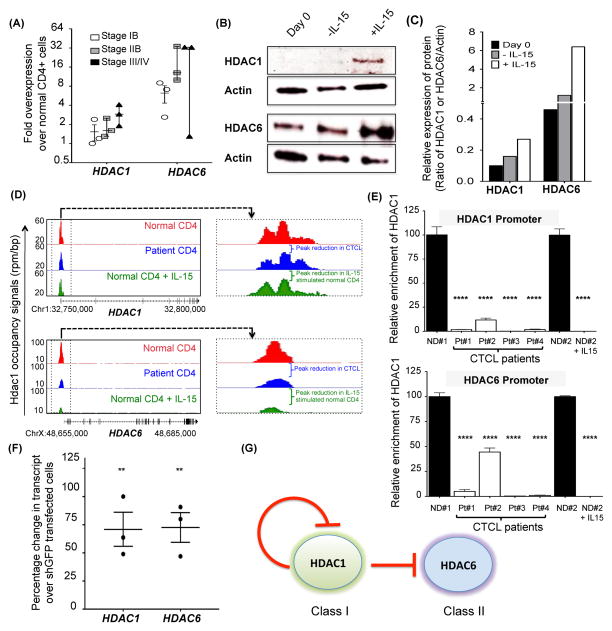
Figure 4. Regulation of HDAC1 and HDAC6 by IL-15 in CTCL and normal CD4+ cells.
(A) Fold change in endogenous HDAC1 and HDAC6 transcript (mean ± SEM, n=3 each) in resting CD4+ T-cells from CTCL patients, relative to resting CD4+ T-cells from normal donor peripheral blood (N=3). For each patient, HDAC1 and HDAC6 transcript were normalized to 18S and the values for normal donors were arbitrarily set at 1. (B) Immunoblot analysis of HDAC1 and HDAC6 in normal donor CD4+ T-cells at day 0 (before IL-15 exposure) and on day 2 (−/+ IL-15). Protein lysates were probed for expression of HDAC1 and HDAC6; β-Actin was used as housekeeping control. (C) Relative expression of HDAC1 and HDAC6 protein of immunoblots in figure 4B by densitometry analysis. (D) Gene tracks showing occupancy of HDAC1 at the HDAC1 and the HDAC6 gene promoters. Normalized tag counts for HDAC1 in normal donor CD4+ T-cells (red) were compared to CTCL patient CD4+ T-cells (blue) and normal donor CD4+ T-cells stimulated with 100ng/ml IL-15 for 24 hours (green). The HDAC1 binding region is zoomed-in on the right panel. (E) Graphical representation of ChIP PCR analysis for binding of HDAC1 to its promoter (top) and to the HDAC6 promoter (bottom) in normal donor (ND) CD4+ T-cells versus CTCL patient CD4+ T-cells. Also included in the same graph is ChIP PCR analysis for binding of HDAC1 to the HDAC1 and the HDAC6 promoter in normal donor (ND) CD4+ T-cells either left unstimulated (−) or exposed to 100 ng/ml of IL-15 for 24 hours (+). Input DNA from each normal donor and patient was used as control. (F) Relative increase in HDAC1 and HDAC6 transcript in normal donor CD4+ T-cells transfected with shHDAC1 plasmids (relative to normal donor CD4+ T-cells transfected with control shGFP plasmid. HDAC1 and HDAC6 transcripts were normalized to 18S and the values for normal donor CD4+ T-cells transfected with control shGFP plasmid (mean ± SEM, N=3 each). (G) Role of HDAC1-mediated regulation of HDAC1 (Class I HDAC) and HDAC6 (Class II HDAC). Under resting conditions, HDAC1 negatively regulates its own transcription as well as transcription of HDAC6. Upon stimulation with IL-15, negative regulation of both of these genes is overcome by reduced occupancy of HDAC1 at the TSS. Unless otherwise stated, data in Figure 4 is presented as mean ± SEM, **P ≤ 0.01, unpaired two-tailed student’s t-test.
In order to determine the mechanism(s) of HDAC1 and HDAC6 induction by IL-15 we first investigated a previously described negative auto-regulatory feedback loop where HDAC1 binds directly to its own promoter and represses transcription.(35) Using antibodies specific for HDAC1, we probed the interaction of HDAC1 with its own promoter in CTCL CD4+ T-cells, resting normal donor CD4+ T-cells, and IL-15-stimulated normal donor CD4+ T-cells by “ChIP-seq”, which combines chromatin immunoprecipitation (ChIP) with sequencing of DNA associated with the protein of interest, i.e. HDAC1. ChIP-seq data showed that CTCL patients’ CD4+ T-cells displayed decreased occupancy of HDAC1 at its own promoter when compared to resting normal donor CD4+ T-cells (Supplementary Table 1D and Figure 4D–4E). We also observed reduced binding of HDAC1 to its promoter in IL-15 stimulated normal CD4+ T-cells when compared to resting normal donor CD4+ T-cells (Figure 4D–4E), suggesting that the overexpression of HDAC1 seen in CTCL patients’ CD4+ T-cells is due, at least in part, to IL-15-induced decrease in HDAC1 occupancy at the HDAC1 promoter. In order to investigate HDAC1’s role as a transcriptional repressor for HDAC1 and HDAC6, normal donor T-cells were transfected with shHDAC1 and evaluated for changes in HDAC1 and HDAC6 transcripts by RT-PCR. Silencing of HDAC1 in resting normal donor CD4+ cells leads to increased transcription of both HDAC1 and HDAC6 mRNA, suggesting HDAC1 is a transcriptional repressor of both HDAC1 and HDAC6 in CTCL patients’ CD4+ T-cells and in normal donor CD4+ T-cells upon IL-15 stimulation (Figure 4F). Thus, decreased binding of HDAC1 to its own promoter and to the HDAC6 promoter in IL-15-expressing CTCL CD4+ T-cells in vivo, and in normal CD4+ T-cells following stimulation with IL-15 in vitro, results in their increased expression, while knock down of HDAC1 results in upregulation of both HDAC1 and HDAC6 in normal resting CD4+ T-cells. These data confirm that HDAC1 is a physiologic transcriptional repressor of HDAC1(35) and show, for the first time, that it is also a physiologic transcriptional repressor of HDAC6. Further, we show that chronic stimulation of CTCL CD4+ T-cells by IL-15 disrupts the negative auto-regulatory loop of HDAC1 thereby contributing to the upregulation of HDAC1 and HDAC6 in vivo (Figure 4G).
Given the broad effects of HDACs on the signaling pathways involved in cancer cell growth and survival(36), we measured the downstream effects of IL-15-induced increase in HDAC expression in normal CD4+ T-cells. For example, we confirmed overexpression of HDAC1 in normal donor CD4+ T-cells stimulated with IL-15, and the consequent silencing of its downstream target p21 (Supplementary Figure 4A).(37) Since lymphocyte migration is regulated by HDAC6(38) and important in CTCL, we showed that knockdown of HDAC6 by shRNA in normal donor CD4+ T-cells reduced cell migration towards IL-15, when compared to controls (mean ± SEM of T-cell migration in shGFP control vs. shHDAC6 Clone#3 vs. shHDAC6 Clone#5= 49.67 ± 3.28 vs. 20.00 ± 2.88 vs. 26.67 ± 1.66, n=3 each; P= 0.0025 and 0.0033, respectively; Supplementary Figure 4B). Knockdown of HDAC1 did not affect T-cell migration suggesting that HDAC1 does not influence IL-15-induced migration of T-cells (Supplementary Figure 4B). These observations are consistent with the notion that the IL-15-HDAC1/6 axis contributes to the pathogenesis of CTCL through multiple mechanisms.
Genome-wide mapping of HDAC1 in CTCL
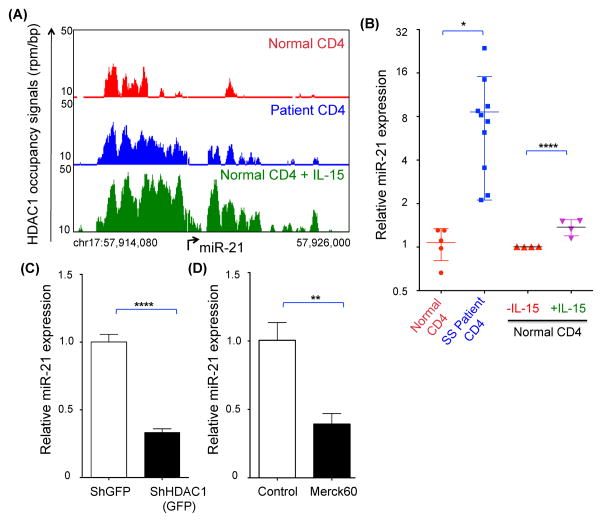
Although HDAC1 is overexpressed in CTCL, the relationship of its overexpression to gene regulation has not been clearly defined. Here we interrogated the epigenetic landscape of HDAC1 in highly enriched malignant CD4+ T-cells from a SS patient and from normal donor CD4+ T-cells (the latter with or without IL-15 stimulation). We found that there are parallels in HDAC1 occupancy between CD4+ T-cells from a SS patient and from normal donor CD4+ T-cells stimulated with IL-15. Overall HDAC1 occupancy at intron and 5′-UTR regions were similar among these two cell populations when compared to resting normal donor CD4+ T-cells, while no changes in HDAC1 occupancy were noted among these three CD4+ T-cell populations in seven other genomic locations that were analyzed (i.e., distal promoter, proximal promoter, exon, 3′-UTR, proximal downstream, distal downstream and distal intergenic regions). Specifically, CD4+ T-cells from the SS patient and from the normal donor CD4+ T-cells exposed to IL15 showed decreased HDAC1 occupancy at the intron location and increased overall genomic HDAC1 occupancy at the 5′-UTR locations compared to resting normal donor CD4+ T-cells (Supplementary Figure 5A). To determine whether HDAC1 binding patterns were indicative of a broader trend in transcriptional regulation, we examined the relationship between HDAC1 enriched sites with enhancer mark H3K27Ac to measure levels of gene expression on a global scale.(39) As shown, the "Promoter Plots” show the tag distribution across all human RefSeq promoters (25,143 regions), the "Active Regions Plots” show the tag distribution at all H3K27Ac/HDAC1 peak locations (35,725 regions) and the “Super Enhancers (SE) Plots” show the tag distribution at all H3K27Ac and HDAC1 SE locations (1,525 regions). Consistent with HDAC1’s known role as a repressor of gene transcription, our data suggest that occupancy patterns of HDAC1 (resting normal donor CD4+ T-cells > normal donor CD4+ T-cells stimulated with IL-15 > SS patient CD4+ T-cells) are inversely proportional to histone H3K27Ac (CTCL patient CD4+ T-cells > normal donor CD4+ T-cells stimulated with IL-15 > normal donor CD4+ T-cells), which is a mark for active enhancers (Supplementary Figure 5B).(39) While there was significant overlap in occupancy between the three types of samples, unique patterns were observed (Supplementary Figure 5C). Using Ingenuity Pathway Analysis (IPA)(40) to functionally annotate HDAC1 target genes, we identified the top biological processes regulated by HDAC1 in normal (with or without IL-15) CD4+ T-cells and in CD4+ T-cells from the SS patient (Supplementary Figure 5D). IPA identified super clusters of high-scoring functions in each of the three data sets revealing dominance of a gene expression network in IL-15-stimulated normal donor CD4+ T-cells and in CD4+ T-cells from the SS patient (Supplementary Figure 5D, orange and green panels, respectively). Interestingly, cell cycle is the most enriched pathway in normal resting CD4+ T-cells suggesting role for HDAC1 in maintaining the physiologic cell cycle gene network (Supplementary Figure 5D, blue panel). Importantly, while the comparison of genome wide HDAC1 landscape and consequent pathway analysis in CD4+ cells from the SS patient, normal donor and IL-15 stimulated normal donor is revealing, our very small sample size (n=1 each) limits our interpretation of these data.
HDAC1 is positive regulator of oncomiR miR-21
We have previously shown the involvement of HDAC1 in the regulation of microRNA (miR) expression.(27) While a considerably large proportion of HDAC1 occupancy was observed in the resting normal donor CD4+ T-cell genome, we identified a few gene loci where a higher proportion of HDAC1 occupancy was observed in the SS patient CD4+ T-cells and in IL-15-stimulated normal donor CD4+ T-cells (data not shown). One such region of interest was the promoter of the microRNA-21 (miR-21), a bona fide ‘oncomiR’(41) that is overexpressed in CTCL patients (Figure 5A, Supplementary Figure 5E and Supplementary Table 1D).(42–44) Consistent with this observation, we found significantly increased levels of miR-21 in the SS patients’ CD4+ T-cells and in IL-15-stimulated normal donor CD4+ T-cells compared to resting normal donor CD4+ T-cells (Figure 5B and Supplementary Table 1C). Circulating CD4+ T-cells in CTCL patients with MF also exhibited increased levels of miR-21 compared to normal donor cells (Supplementary figure 5F and Supplementary Table 1C). Interestingly, similar observations were made when comparing circulating normal CD4+ T cells in WT to IL-15 tg mice with early stage disease. Circulating CD4+ cells (from peripheral blood) as well as skin resident cells in IL-15 tg mice overexpressed miR-21 in comparison to WT counterparts (Supplementary figure 5G). To test if the occupancy of HDAC1 on miR-21 promoter affects its transcription, we silenced HDAC1 levels in the CTCL cell line Hut78 using Sh-RNA directed against HDAC1 and showed downregulation of miR-21 (Figure 5C). Similarly, treatment of Hut78 cells with an HDAC1/2 inhibitor (Merck60) caused decreased expression of miR-21 (Figure 5D). Therefore, along with the known role of HDAC1 as transcriptional repressor, we determined that HDAC1 is a positive regulator of mir-21 both in CTCL patient CD4+ T-cells and in IL-15-stimulated normal donor CD4+ T-cells.
Figure 5. HDAC1 as enhancer of oncomiR, miR-21.
(A) Gene track of HDAC1 occupancy at miR-21 promoter in normal donor, CTCL patient and IL-15-stimulated normal donor CD4+ T-cells (↱ = TSS). (B) Relative expression of miR-21 in normal donor and CTCL patient CD4+ T-cells. Mature miR-21 transcript was normalized to RNU43 and the values are presented as mean ± SEM, N=5 for normal donor CD4+ T-cells and N=10 for CTCL patient CD4+ T-cells. On the right side is relative expression of miR-21 in normal donor CD4+ T-cells that were either exposed to PBS (control) or 100ng/ml IL-15 in vitro for 24 hours. CD4+ T cells from four different normal donors were exposed to IL-15 as shown by symbols of different colors. Mature miR-21 transcript was normalized to RNU43 and the values are presented as mean ± SEM, N=3 for each normal donor. (C) Relative expression of miR-21 in normal donor CD4+ T-cells transfected with either ShGFP (control) or ShHDAC1 GFP plasmids. Cells were harvested at 24 hours for expression of mature miR-21 transcript and normalized to RNU43. The values are presented as mean ± SEM, N=3 for each condition. (D) Relative expression of miR-21 in the human CTCL cell line Hut78 cells treated with either DMSO (control) or 150nM Merck60 (an HDAC1/2 inhibitor). Cells were harvested at 24 hours for expression of mature miR-21 transcript and normalized to RNU43. The values are presented as mean ± SEM, N=3 for each condition.
Inhibition of HDAC activity prevents progression of CTCL in vivo
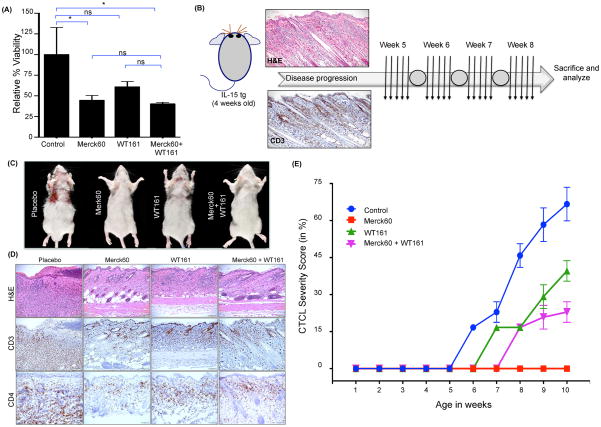
Pan-HDAC inhibitors (HDACis) show strong anti-tumor activity in a subset of CTCL patients.(45) Considering that overexpression of IL-15 leads to increased HDAC1 and HDAC6 in our preclinical model, we used specific HDACis to target HDAC1/2 (Merck60)(46) and HDAC6 (WT161)(47) either individually or in combination. Hut78 cells were more sensitive to cell death with the HDAC1/2 specific inhibitor than with the HDAC6 inhibitor or with the combination (mean ± SEM of relative viability in Hut78 control vs. Merck60 vs. WT161 vs. combination treatment = 100.0 ± 16.36 vs. 44.65 ± 2.97 vs. 60.94 ± 3.24 vs. 40.32 ± 0.90, n=4 each, P < 0.0001; Figure 6A). To test the relative in vivo efficacy of these inhibitors, 4-week old IL-15 tg mice with early evidence of CTCL (Supplementary figure 6A–D) were treated with 50 mg/kg of either the HDAC1/2 specific inhibitor or the HDAC6 specific inhibitor or in combination, 5 days/week for 4 weeks (Figure 6B). As indicated by our CTCL severity score (Supplementary figure 7), while placebo-treated IL-15 tg control mice progressively developed the typical cutaneous lesions, the HDAC1/2 specific inhibitor Merck60 alone halted and even regressed clinical progression of CTCL in the IL-15 tg mice (mean ± SEM of relative CTCL severity (Supplementary Figure 7A–E) in Placebo vs. Merck60 vs. WT161 vs. combination treatment groups, P=0.038; Figure 6C–E). IL-15 tg mice treated with the HDAC6 specific inhibitor WT161 showed delay in progression of CTCL compared to placebo treated mice, but WT161 was less effective than Merck60 (Figure 6C–E). These observations were further corroborated by histopathological analysis of skin sections from treated mice, as IL-15 tg mice treated with Merck60 had the least CD3+CD4+ infiltrates in the skin compared to the other treatment groups (Figure 6D). Taken together, our preclinical data show that targeting HDAC1/2 with Merck60 effectively inhibits progression of early CTCL in the IL-15 tg mice and support a similar approach in the treatment of human CTCL with the hope of reducing toxicities associated with the use of pan-HDACis (Figure 6E).
Figure 6. Targeting HDACs for treatment of CTCL in vitro and in vivo.
(A) The human CTCL cell line Hut78 was treated with 150nM HDAC inhibitors to test the efficacy of specific HDAC inhibition on cell viability. Relative percent viability of treated cells was measured by the MTS assay 72 hours after treatment and the values for control treated cells were arbitrarily set at 100% (mean ± SEM, N=4 for each condition). (B) Treatment schema for approximately 4 week old CTCL IL-15 tg mice with either Merck60 and/or WT161 (N=4/group). Representative H&E and CD3 immunohistochemical staining in skin tissue of CTCL IL-15 tg mice on day 0 (week 4) of treatment. Each arrow indicates a daily dose of drug(s) or placebo, and each circle indicates a 2-day weekend drug holiday (see Methods for details), followed by sacrifice and analysis 2 days after the discontinuation of therapy. (C) Representative autopsy photographs taken upon sacrifice at the end of treatment showing progression of CTCL or lack thereof in each treatment group. (D) Representative histological findings across the four treatment groups showing the extent of CTCL infiltrates (H&E staining, top row). Skin sections from each treatment groups were also stained for CD3+ and CD4+ T-cell infiltrates (middle and bottom row, respectively). (E) CTCL severity of IL-15 tg mice treated with either Merck60 and/or WT161 (N=4/group). Unless otherwise stated, data is presented as mean ± SEM, nsP > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, unpaired two-tailed student’s t-test.
Discussion
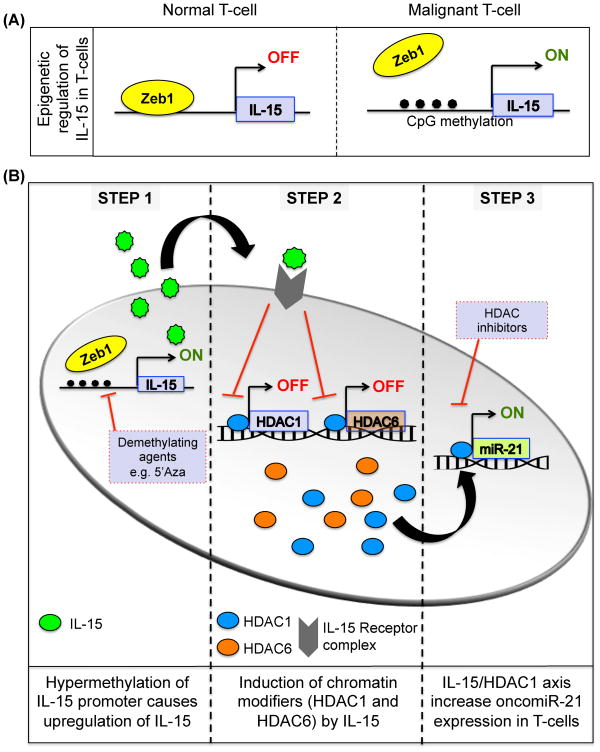
IL-15 has been previously implicated in T-cell lymphoproliferative disorders(48, 49), and overexpression of IL-15 in clinical samples obtained from CTCL patients has been documented by several groups.(10, 14, 17) However, why IL-15 expression is increased in CTCL and what the potential role of this cytokine is in the initiation and progression of CTCL remain poorly understood. With the intent of gaining insight into mechanisms of IL-15 overexpression and its role in CTCL pathogenesis, we first confirmed the relevance of IL-15 by showing stage-dependent overexpression in lesional skin and peripheral blood CD4+ T-cells from CTCL patients. We elucidated at least one mechanism by which IL-15 can be overexpressed in vivo, showed how this cytokine induces specific HDACs in CTCL CD4+ T-cells, and then investigated the biological and clinical aspects of a spontaneous epidermotropic CTCL that develops in mice that constitutively overexpress IL-15. We demonstrated that the CTCL in IL-15 tg mice closely resembles human CTCL, based on clinical, histopathological, and immunophenotypical criteria. We also found shared molecular aberrations between the CTCL in IL-15 tg mice and human CTCL, which could be replicated in normal human CD4+ T-cells exposed to IL-15 in vitro. Finally, we interrogated the animal model with the purpose of identifying mechanisms and pathways amenable to selective pharmacological targeting for preclinical validation, demonstrating some success at halting the progression and inducing the regression of CTCL in vivo using an HDAC1-specific inhibitor (Figure 7).
Figure 7. Schematic illustration of the proposed deregulated IL-15 signaling pathway in T-cells.
(A) Epigenetic regulation of IL-15 transcription by Zeb1 in normal and malignant T-cells. Excessive methylation at the Zeb1 binding site promotes the aberrant production of IL-15 from CD4+ T cells. (B) Autocrine IL-15 production and consequent signaling in CD4+ T cells induces expression of chromatin modifiers HDAC1 and HDAC6. Upregulation of HDAC1 positively regulates expression of oncomir miR-21. Collectively, the deregulation of these enzymes by aberrant IL-15 signaling contributes to malignant transformation of a normal T-cell.
Mechanism of IL-15 overexpression in CTCL
While the IL-15 tg mouse provides a novel and informative experimental platform to study the oncogenic effects of IL-15 in CTCL and explore new treatments, it cannot by itself offer insight into the mechanisms of IL-15 overexpression in patients. Therefore, we examined the structure of the IL-15 gene in CTCL patients and studied the epigenetic regulation of its transcription. Mutations, insertions and deletion of the transcriptional checkpoints in the IL-15 gene have previously been implicated in the constitutive overexpression of this cytokine in T-cell lymphoma cell lines, suggesting that structural alterations of the gene can modulate IL-15 production.(50–52) Although we did not find any genetic alterations in the IL-15 promoter isolated from CTCL patients’ CD4+ T-cells (data not shown), we did observe an epigenetic alteration (i.e., hypermethylation) at the IL-15 promoter in these cells when compared to normal donor CD4+ T-cells. Hypermethylation of CpG islands in the 5′ regulatory region of genes can alter gene expression through mechanisms that do not involve changes in nucleotide sequence.(53, 54) Genes such as BCL7a, PTPRG, THBS4, MGMT, p73, p16, p15, CHFR, SHP-1 and TMS1 display widespread promoter hypermethylation in CTCL patients’ CD4+ T-cells.(55, 56) In addition to gene silencing, hypermethylation can also result in gene activation by inhibiting access of transcriptional repressors to a promoter’s regulatory regions.(57, 58) Hypermethylation of repressive sequences in the hTERT gene has been shown to upregulate hTERT expression in high-risk HPV induced cervical cancer.(59) Our data show that hypermethylation of CpG islands in the IL-15 promoter within CTCL patients’ CD4+ T-cells prevents the transcriptional repressor Zeb1 (also known as TCF8) from binding and negatively regulating IL-15 expression, thus providing an explanation for the overexpression of IL-15 in CTCL patients’ CD4+ T-cells and offering the first evidence for the epigenetic regulation of its expression. This role is also consistent with other well-characterized functions for Zeb1 in T-cells, such as rapid shut-off of IL-2 transcription after antigen recognition and negative regulation of CRTAM (Class-I MHC-restricted T-cells associated molecule).(60) The fact that loss of genetic material at 10p11.2, where the Zeb1 gene is located, as well as loss of functional somatic mutations, have been observed in a subset of CTCL patients, further supports a tumor suppressor role for Zeb1 in CTCL, highlighting multiple alternative mechanisms for the loss of negative IL-15 regulation.(61–63) The patients tested in our study were positive for Zeb1 protein, indicating that genetic deletions of Zeb1 could be ruled out as a causative mechanism for IL-15 overexpression. Finally, loss of Zeb1 has also been observed in primary cells from another type of mature T-cell neoplasm, adult T-cell leukemia (ATL) and genetic disruption of Zeb1 in mice results in the development of CD4+ T-cell lymphomas with a 100% penetrance.(64) Interestingly, overexpression of another gene HMGA1 in mice induces upregulation of IL-15 leading to the development of T-cell lymphomas.(65) These results, together with our finding that knockdown of Zeb1 causes upregulation of IL-15 expression in resting normal human CD4+ T-cells, suggested that the binding of Zeb1 within the IL-15 promoter is critical for inhibition of IL-15 transcription (Figure 7A). The methylation analysis conducted using neutrophils from each CTCL patient as a control also shows that IL-15 promoter methylation is relatively specific for the CTCL patients’ CD4+ T-cells, suggesting a model whereby aberrant epigenetic regulation of IL-15 expression in CD4+ T-cells generates an autocrine loop allowing IL-15 to sustain malignant CD4+ cell growth as well as methylation of its IL-15 promoter.(27) The mechanisms leading to the hypermethylation of the IL-15 promoter in human CTCL cells are currently unknown, but could be related to excessive methyltransferase activity.(27)
The IL-15 tg mouse as a model of CTCL
Although the clinical combination of dermatitis, alopecia, pruritus, and skin ulcers is not specific nor sufficient to define a CTCL phenotype in an animal model, our analysis of the histopathological and immunophenotypical features of the T-cell lymphoproliferative disorder of IL-15 tg mice revealed very compelling parallels with human CTCL. The cutaneous T-cell infiltrate in the IL-15 tg mice displayed evident epidermotropism, revealed by the dense infiltrate in the dermal-epidermal junction and by the presence of Pautrier’s microabscesses that are pathognomonic for the disease. Furthermore, the T-cells isolated from the skin of the mice expressed a set of homing receptors and adhesion molecules that are very typical of the neoplastic, skin homing T-cells of human CTCL. Transplant experiments further support the conclusion that the T-cell neoplasm of the IL-15 tg mice predominantly affects the skin, as in human CTCL, because secondary CTCL in SCID mice developed only when T-cells, isolated from the skin, but not from the spleen, of affected primary IL-15 tg mice were adoptively transplanted. The presence of a mixed CD4+/CD8+ cutaneous T-cell infiltrate, and even dominance of CD8+ T-cells in the primary IL-15 tg mouse, with oligoclonal rather than monoclonal expansions, also parallels the pattern seen in skin biopsies of patients with early stage CTCL, where the neoplastic CD4+ T-cells are often the minority, CD8+ T-cells are overrepresented, and clonality can be difficult to detect.(66–68) Interestingly, while the presence of a heavy CD8+ T-cell infiltrate in early, human CTCL has been traditionally interpreted as the sign of a host anti-tumor response, it is also possible that it may instead reflect poly- or oligoclonal, multi-lineage T-cell expansions in the very early phases of the disease, in response to autocrine production or cross-presentation of IL-15 by cognate cells. In advanced stage CTCL, it is well known that neoplastic CD4+ T-cells acquire complex genetic aberrations, expand, outgrow and possibly suppress CD8+ T-cells, developing autocrine growth and metastatic potential. Likewise, secondary CTCL produced by the transplantation of skin lymphocytes from primary IL-15 tg mice in our model are more homogeneously CD4+, grow as tumors in the SCID mice, and disseminate, as noted in the late stages of human CTCL. The early development of MF-like disease compared to longer latency for leukemic CTCL in transplanted mice could be attributed to the extent to which accumulation of mutation over time causes emergence of more aggressive, clonal leukemic variant.
Regulation of HDACs under chronic inflammatory conditions such as CTCL
The oncogenic effect of IL-15 during the transformation process is mediated by a variety of signaling pathways.(15) Chronic exposure to pro-inflammatory cytokines such as IL-15 can have profound impact on the epigenetic signature of the cellular genome(15, 27). Data from CTCL patients provide clues into the complex epigenome of neoplastic T-cells, including altered histone code(69) and aberrant methylation.(55) Of histone modifiers, HDACs, especially isoform HDAC1, have been found to be overexpressed in several malignancies and to participate in pathogenesis by repressing tumor suppressor genes.(69, 70) The isoform HDAC1 belongs to the class I family of HDACs and is located in the nucleus. Interaction partners for HDAC1 include HDAC2, p53, RB, MYOD, NF-κB, DNMT1, DNMT3a, MBD2, Sp1, BRCA1, MeCP2, ATM, Smad7, and STAT1 and STAT2.(70, 71) Recently, HDAC1 has become of interest as a potential cancer target because it is recruited by the MDM2 protein and subsequently promotes the degradation of the tumor suppressor gene, p53.(72, 73) Indeed, in patients with CTCL, HDAC inhibition via pharmacological inhibitors has yielded very robust therapeutic efficacy.(34, 74) While increased expression of HDAC1 and HDAC6 may have prognostic significance in CTCL patient(75), factors inducing their upregulation are primarily unknown. In this study, we observed a correlation between overexpression of IL-15 and expression of HDAC1 and HDAC6 in CTCL patients’ CD4+ T-cells, and identified IL-15 as a factor capable of inducing HDAC expression in normal donor CD4+ T-cells in vitro. HDAC overexpression in normal CD4+ T cells likely contributes to their malignant transformation, since HDACs have been shown to act as bona fide oncogenes in many types of human cancer, by regulating the expression of genes involved in cancer initiation and progression through deacetylation of histones(76) as well as non-histone proteins such as p53.(77) Furthermore, induction of HDACs by IL-15 can further amplify tissue inflammation and cell transformation since HDACs positively regulate expression of other pro-inflammatory cytokines such as IL-6, IL-8, and TNF.(78, 79)
Most noteworthy, while studying regulation of HDACs by IL-15, detailed analysis of HDAC1 binding revealed a previously unknown function of HDAC1 as a negative regulator of HDAC6 in normal CD4+ cells. Research from other groups has shown that HDAC1 is associated with a variety of complexes involved in gene silencing, including ones that contain the switch independent protein 1 (Sin3), NuRD (nucleosome remodeling and histone deacetylase), Co-REST (co-repressor for REI silencing transcription factor), Rb, Sp1 and PRC2 proteins.(80) HDAC1 is incorporated into the NuRD complex that mediates DNA methylation by recruiting DNMT1 and chromatin remodeling with recruitment of helicase/ATPase family members.(80) Although overexpression and improper recruitment of HDACs have been found in cancers(81), the identity of the genomic landscape of HDAC1 in malignancies remains unknown.
Exploring the mechanistic aspects of HDAC regulation by IL-15, we observed upregulation of HDAC1 and HDAC6 in normal T-cells following exposure to IL-15. The involvement of HDAC1 in CTCL is further underscored by our observation that 1) IL-15 overexpressing human CTCL CD4+ T-cells display decreased binding of HDAC1 at its own promoter, implying loss of the negative auto-regulatory loop, which normally controls HDAC1 expression; and 2) that similar changes in HDAC1 binding can be induced by stimulating normal CD4+ T-cells with IL-15 in vitro. Aberrant IL-15 signaling may therefore lead to upregulation of HDAC1 in CTCL cells via loss of the negative auto-regulatory loop(35). IL-15 also caused similar loss of HDAC1 occupancy at the HDAC6 promoter, resulting in increased HDAC6 in normal CD4+ T-cells stimulated with IL-15. HDAC6 has been shown to increase angiogenesis and facilitate cell migration, thereby, enhancing the metastatic potential.(82) In lymphocytes particularly, HDAC6 has been shown to increase cell migration independent of its deacetylase activity.(38)
While HDAC1 is primarily found in repressor complexes, we have uncovered a previously unknown function of HDAC1 as an activator of oncomir-21, thus playing dual roles in oncogenesis. Detailed analysis of HDAC1 binding revealed increased presence of HDAC1 in miR-21 loci in CTCL patients’ CD4+ T-cells, and in normal donor CD4+ T-cells when stimulated with IL-15 and compared to resting normal donor CD4+ T-cells. With the emerging role of miR-21 in variety of cancers, it is possible that HDAC1 forms an activating complex to induce miR-21. Interfering with HDAC1 signaling using an HDAC1-specific inhibitor or knockdown of HDAC1 using shRNA supported the importance of HDAC1 in the positive regulation of miR-21. Considering that STAT3 activates miR-21 in CTCL patients and that IL-15 activates both miR-21 and also STAT3 signaling, it will be interesting to further explore the complex interplay between HDAC1 in IL-15 mediated oncogenesis.(15, 44) Thus, overexpression of IL-15 in normal T-cells can cause malignant transformation by: 1) increasing cell proliferation/survival via repression of tumor suppressor genes through the induction of core histone deacetylase HDAC1 at their promoter regions, 2) induction of miR-21, a bona fide oncomiR and, 3) possible extravasation of T-cells by increasing their migration through HDAC6-mediated deacetylation of tubulin (Figure 7B). The clinical relevance of these findings is supported by the observation that both pan and isotype-specific HDACis can prevent progression of CTCL in the IL-15 tg mice and in man.(34)
Selective targeting of HDACs in IL-15 tg mice impedes progression of CTCL
Although some of the highest response rates to single agent pan-HDACis have been observed in CTCL patients, the overall efficacy of such inhibitors remains limited, in part due to toxicity.(34) Both vorinostat and romidepsin are FDA approved HDACis with favorable clinical activity in the treatment of CTCL; however, response rates are only 30 to 35%.(34, 83, 84) In addition to the moderate response rates observed with these drugs, the fact that vorinostat and romidepsin are not HDAC isotype specific but rather pan-HDAC and a class 1 inhibitor, respectively, prevents the identification of the key signaling pathways that must be targeted to improve the efficacy and possibly limit the toxicity of this class of drugs. IL-15 tg mice treated with the HDAC6 specific inhibitor WT161 showed delay in progression of CTCL compared to placebo treated mice, but WT161 was less effective than the HDAC1/2 inhibitor, Merck60. Interestingly, the combination of two compounds was less effective than either agent alone in preventing CTCL. As our experimental work largely demonstrates a halt in the progression of early stage CTCL, we believe this targeted therapeutic might be best applied in a comparable clinical setting.
Conclusion
Human CTCL begins as an indolent neoplasm characterized by the progressive and predominant accumulation of malignant CD4+ T-cells in the upper layers of the epidermis, and gradually proceeds to affect larger body surface areas and ultimately to leukemic and visceral spread. Once patients progress to tumor stage, survival is significantly compromised. The key mechanisms initiating and promoting the stepwise progression in CTCL have not been identified, but a set of prior observations and the data presented here support an important role for the dysregulation of IL-15. The parallel investigation of CTCL in IL-15 tg mice and in CTCL patient samples performed in this study showed a similar pattern where initially small populations of CD4+ T-cells are attracted to and retained in the skin microenvironment, and their docking to epidermal Langerhans cells produces the histologic hallmark of CTCL: Pautrier’s microabscesses. In CTCL patients, the oncogenetic aberrations, and even the malignant nature of this “founder” population are incompletely understood because of the difficulty to isolate these cells in the midst of a dominant reactive cellular infiltrate. In CTCL patients, over a variable interval, the continuous presence of microenvironmental factors sustaining lymphocyte trafficking to the skin, combined with the selective drive exerted by repeated exposure to less-than-completely effective therapies, leads to the emergence of clonal populations of T-cells with chromosomal instability, complex genetic aberrations, autocrine growth, constitutive activation of NF-κB signaling, and metastatic potential. Finally we show that a mechanism of IL-15 overexpression in CTCL patients could be related to promoter hypermethylation and the failure of the transcriptional repressor Zeb1 to have access to the IL-15 regulatory region. This is a novel finding that not only raises interesting questions about the epigenetic regulation of IL-15 in normal individuals and CTCL patients, but also supports the consideration of utilizing a therapeutic combination of HDAC inhibitors and demethylating agents in CTCL.(85–87)
In summary, we provide in vivo evidence that IL-15 likely can have a causal role in the pathogenesis of at least some cases of CTCL, in part via the epigenetic inhibition of the transcriptional repressor, Zeb1, which in turn leads to overexpression of IL-15 and activation of specific HDACs. IL-15 tg CTCL mice provide a novel model for studying the development of CTCL and evaluating potential therapies. Selective inhibition of HDAC1/2 produces a comparable halt in the progression of experimental CTCL as that seen with the pan-HDAC inhibitors, and thus may provide an equally potent yet less toxic alternative in the clinic.
Methods
Human T-cell isolation
Peripheral blood mononuclear cells were obtained from patients after informed consent in accordance with the Declaration of Helsinki. The Institutional Review Board of Henry Ford Hospital and the Ohio State University Comprehensive Cancer Center approved this study. Normal donor cells were obtained from Red Cross Blood Bank. The CD4+ T-cells from peripheral blood were purified (approximately 95% or more) by negative selection using RosetteSep™ human CD4 depletion (STEMCELL, Vancouver, Canada) per manufacturer’s instructions.
Mouse skin T-cell isolation
All animal studies were performed under approved protocols following the Ohio State University Institutional Animal Care and Use Committee. Mice were housed in a barrier facility. Age and sex matched WT, IL-15 tg and SCID mice (The Jackson laboratory, Bar Harbor, ME) were used. All procedures were done using sterile techniques and tools. Before harvesting skin, all mice were sprayed with 70% isopropanol (Ricca Chemical Company, Arlington, TX). Wild-type mice were shaved prior to collection of skin sample. Skin was peeled off the mouse skin both at the dorsal and ventral side of the skin (approximately 8–12cm2). The skin were removed and digested in 10mg/ml of collagenase (Sigma Aldrich, St. Louis, MO) diluted 1:1000 in RPMI 1640 medium (GIBCO, Grand Island, NY). The tissues were minced using sterile scissors and tissues were digested thoroughly for 30 minutes at 37°C (vigorously shaking at 300 rpm for a total of 60 minutes). The supernatant suspension was filtered using a 70-μm nylon mesh cell strainer (BD Bioscience, Bedford, MA) and then washed with RPMI 1640 medium with 2% FBS at 1500rpm for 10 minutes. Lymphocytes were isolated using Ficoll-Paque PLUS (GE Healthcare, Uppsala, Sweden) gradient centrifugation and then washed with PBS (GIBCO, Grand Island, NY) with 2% FBS. The single cell suspension was then stained for use in flow cytometry and cell sorting.
Skin mononuclear cell transplantation
After isolation, skin cells were washed twice in 10mL of PBS and resuspended in 200μL PBS for transplantation. Injections were administered subcutaneously in the dorsal inferior half of the body using a 1 mL BD™ Tuberculin syringe with 27 G x ½ BD PrecisionGlide™ detachable needle after shaving with an electric razor. Mice were observed daily for development of lesions or tumors, and re-shaved once every 30 days. Photographs were taken when observable lesions were noteworthy.
Antibody staining and flow cytometry
Peripheral blood, spleen, and skin samples were harvested from moribund mice. Single cell suspensions were prepared for all samples. The following flurochrome-conjugated monoclonal anitbodies (mAb) were purchased from BD Pharmingen, San Jose, CA and were used for flow cytometry: CD8a (clone 53–6.7), CD3e (clone 145-2C11), CD4 (clone RM4–5), CLA/CD162 (clone 2PH1), CD62L (clone MEL-14), CD69 (clone H1.2F3), yδ TCR (clone GL3), Vβ2 (clone B20.6), Vβ3 (clone KJ25), Vβ4 (clone KT4), Vβ5.1 (clone MR9-4), Vβ6 (clone RR4-7), Vβ7 (clone TR310), Vβ8.1 (clone MR5-2), Vβ8.3 (clone 1B3.3), Vβ9 (clone MR10-2), Vβ10b (clone B21.5), Vβ11 (clone RR3-15), Vβ12 (clone MR11-1), Vβ13 (clone MR12-3), Vβ14 (clone 14-2), and Vβ17a (clone KJ23).
Immunofluorescence staining
Skin sections were frozen in OCT Compound (Sakura Finetek, Torrance, CA), cyrosectioned into 8 μm sections and placed on glass slides. Skin sections were fixed for 5 minutes in cold acetone, washed twice with PBS and incubated with protein block (Dako, Carpinteria, CA) for 30 minutes at room temperature. Sections were washed twice with PBS and stained for CCR4 or CLA (CD162). Goat polyclonal anti-CCR4 antibody (Abcam 1664) was used at 1/100 dilution in antibody diluent (Dako, Carpinteria, CA) for an hour. Sections were washed 7-times with PBS supplemented with 2% FBS, followed by incubation with 1/500 dilution of donkey anti-Goat IgG H&L (PE) secondary antibody (ab7004) for an hour at room temperature. CLA/CD162 staining was performed similarly by incubating skin sections with 1/00 dilution of mAb CD162 (clone 2PH1) for an hour at room temperature. Skin sections were washed 7-times with PBS and mounted with a drop of VECTASHIELD mounting medium (Vector Laboratories, Burlingame, CA).
Isolation of RNA, cDNA preparation and PCR
Purified cells were pelleted and resuspended in TRIzol Reagent (Invitrogren, Carlsbad, CA). Total RNA was prepared as previously described.(27) RNA was quantified by NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and used for cDNA synthesis. cDNA synthesis was achieved by SuperScript First-Strand Synthesis System for RT-PCR kit (Invitrogen Carlsbad, CA) using random hexamers per manufacturer’s protocol. Real-time PCR was performed in a 20 μL volume using 1 μl cDNA, 20X Taqman Assay for each gene and 2X Taqman Universal Fast PCR Master Mix (Applied Biosystems). An ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) was used for standard fast-protocol for taqman PCR. Taqman probe ID for the genes can be provided upon request.
Cloning human IL-15 promoter
Human IL-15 was PCR amplified from CD4+ normal donor genomic DNA for construction of promoter-containing luciferase reporter plasmids. Regions of IL-15 gene that contained 500 bp of the promoter region upstream of TSS was amplified and cloned in TOPO® TA Cloning® vector. The vector was introduced into TOP 10® One Shot competent bacteria; positive clones were selected and verified by restriction digest with KpnI and XhoI. Released band was purified using QIAquick Gel Extraction Kit (Qiagen, Valencia, CA) per manufacturer’s instructions. The gel purified DNA fragments containing IL-15 promoter were re-inserted into pGL3-basic luciferase vector (Promega, Madison, WI) and verified by restriction enzyme digestion.
Transfection and luciferase assay
Approximately 5 x106 Jurkat cells (obtained from the ATCC) were transfected with 2.5 μg of methylated or non-methylated plasmid constructs and 0.5 μg of renilla luciferase plasmid using amaxa human T-cell transfection kit (Lonza, Allendale, NJ). Transfection was performed using manufacturers protocol using V-24 program. Cells were harvested and lysed 16–24 hours post-transfection for luciferase assay using Dual-Luciferase® Reporter Assay System (Promega).
In vitro culture of CD4+ T-cells with IL-15
Purified CD4+ T cells from normal donors were seeded in a 24 well plate at a density of 1 x 106 cells per well. Cells were incubated at 37°C and 5% CO2 in RPMI 1640 and chronically stimulated with IL-15 at a concentration of 100 ng/mL. Cells were harvested at indicated time point of incubation for either immunoblot analysis or ChIP assay. For growth analysis of T-cells in response to IL-15, long-term culture was maintained by replacing 0.5 ml of old medium with fresh medium containing IL-15.
Transfection and migration assay
Approximately 10x106 freshly purified CD4+ cells were transfected with 10 μg of shRNA plasmid as described above. Forty-eight hours after transfection, cells were FACS sorted for GFP expression. GFP+ cells were seeded in upper half of a cell permeable membrane cassette and cells were allowed to migrate towards lower chamber containing serum free medium supplemented with 50ng/ml of IL-15 for 12 hours. Cells in lower chamber were counted by trypan blue exclusion and relative percentage migration was calculated by subtracting spontaneous migration.
In vitro drug treatment and cell viability assay
Hut78 cells (obtained from the ATCC) were seeded at the density of 1x105 cells/well in 96-well plate in triplicates. Drugs were added at the indicated time point to cells on day 0. Cell viability was determined by MTS assay using CellTiter 96 Aqueous One Solution Reagent (Promega) at 48–72 hours per manufacturer’s instructions.
HDACi therapy in CTCL mice
In order to evaluate the effects of inhibiting HDAC1/2 and HDAC6 in progression of CTCL, 4-week old IL-15 tg mice were treated with 50mg/kg of JQ12 and/or WT161 respectively. Control mice were treated with equal volume of 10% DMSO in saline. Mice were given intra-peritoneal injection 5 days/week for 4 weeks. Mice were sacrificed and analyzed 2 days after cessation of treatment. The mice were observed daily for changes in severity of lesions and photographs were taken at the beginning and end of the therapy. Mice were euthanized and analyzed for malignant infiltration in skin by histology analysis.
Statistical analysis
Two-sample t test was used to compare two independent groups and paired t test was used to compare two paired groups. Data transformation was performed if the original distribution is non-normal. ANOVA models or generalized linear models were used to compare three or more groups. Survival curves were estimated by Kaplan-Meier method and treatment groups were compared by the log-rank test. P values were adjusted for multiple comparisons by Holm’s procedure. A p value of < 0.05 was considered significant.
Supplementary Material
Significance.
To date, CTCL pathogenesis remains unknown and there are no curative therapies. Our findings not only demonstrate a critical role for IL-15-mediated inflammation in cutaneous T-cell lymphomagenesis, but also uncover a new oncogenic regulatory loop in CTCL involving IL-15, HDAC1, HDAC6 and miR-21 that show differential sensitivity to isotype-specific HDAC inhibitors.
Footnotes
Authors have no conflict of interest to declare.
Uncategorized References
- 1.Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005;105(10):3768–85. doi: 10.1182/blood-2004-09-3502. [DOI] [PubMed] [Google Scholar]
- 2.Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC) Blood. 2007;110(6):1713–22. doi: 10.1182/blood-2007-03-055749. [DOI] [PubMed] [Google Scholar]
- 3.Duvic M, Talpur R, Ni X, Zhang C, Hazarika P, Kelly C, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109(1):31–9. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whittaker SJ, Demierre MF, Kim EJ, Rook AH, Lerner A, Duvic M, et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(29):4485–91. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- 5.Yamanaka K, Clark R, Rich B, Dowgiert R, Hirahara K, Hurwitz D, et al. Skin-derived interleukin-7 contributes to the proliferation of lymphocytes in cutaneous T-cell lymphoma. Blood. 2006;107(6):2440–5. doi: 10.1182/blood-2005-03-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer EM, Shin DB, Nattkemper LA, Benoit BM, Klein RS, Didigu CA, et al. IL-31 is produced by the malignant T-cell population in cutaneous T-Cell lymphoma and correlates with CTCL pruritus. J Invest Dermatol. 2013;133(12):2783–5. doi: 10.1038/jid.2013.227. [DOI] [PubMed] [Google Scholar]
- 7.Krejsgaard T, Litvinov IV, Wang Y, Xia L, Willerslev-Olsen A, Koralov SB, et al. Elucidating the role of interleukin-17F in cutaneous T-cell lymphoma. Blood. 2013;122(6):943–50. doi: 10.1182/blood-2013-01-480889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guenova E, Watanabe R, Teague JE, Desimone JA, Jiang Y, Dowlatshahi M, et al. TH2 cytokines from malignant cells suppress TH1 responses and enforce a global TH2 bias in leukemic cutaneous T-cell lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(14):3755–63. doi: 10.1158/1078-0432.CCR-12-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geskin LJ, Viragova S, Stolz DB, Fuschiotti P. Interleukin-13 is overexpressed in cutaneous T-cell lymphoma cells and regulates their proliferation. Blood. 2015;125(18):2798–805. doi: 10.1182/blood-2014-07-590398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asadullah K, Haeussler-Quade A, Gellrich S, Hanneken S, Hansen-Hagge TE, Docke WD, et al. IL-15 and IL-16 overexpression in cutaneous T-cell lymphomas: stage-dependent increase in mycosis fungoides progression. Exp Dermatol. 2000;9(4):248–51. doi: 10.1034/j.1600-0625.2000.009004248.x. [DOI] [PubMed] [Google Scholar]
- 11.Wasik MA. IL-13 as a novel growth factor in CTCL. Blood. 2015;125(18):2737–8. doi: 10.1182/blood-2015-02-626432. [DOI] [PubMed] [Google Scholar]
- 12.Vowels BR, Lessin SR, Cassin M, Jaworsky C, Benoit B, Wolfe JT, et al. Th2 cytokine mRNA expression in skin in cutaneous T-cell lymphoma. J Invest Dermatol. 1994;103(5):669–73. doi: 10.1111/1523-1747.ep12398454. [DOI] [PubMed] [Google Scholar]
- 13.Krejsgaard T, Ralfkiaer U, Clasen-Linde E, Eriksen KW, Kopp KL, Bonefeld CM, et al. Malignant cutaneous T-cell lymphoma cells express IL-17 utilizing the Jak3/Stat3 signaling pathway. J Invest Dermatol. 2011;131(6):1331–8. doi: 10.1038/jid.2011.27. [DOI] [PubMed] [Google Scholar]
- 14.Dobbeling U, Dummer R, Laine E, Potoczna N, Qin JZ, Burg G. Interleukin-15 is an autocrine/paracrine viability factor for cutaneous T-cell lymphoma cells. Blood. 1998;92(1):252–8. [PubMed] [Google Scholar]
- 15.Mishra A, Sullivan L, Caligiuri MA. Molecular pathways: interleukin-15 signaling in health and in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(8):2044–50. doi: 10.1158/1078-0432.CCR-12-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood. 2001;97(1):14–32. doi: 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- 17.Leroy S, Dubois S, Tenaud I, Chebassier N, Godard A, Jacques Y, et al. Interleukin-15 expression in cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome) The British journal of dermatology. 2001;144(5):1016–23. doi: 10.1046/j.1365-2133.2001.04192.x. [DOI] [PubMed] [Google Scholar]
- 18.Marzec M, Halasa K, Kasprzycka M, Wysocka M, Liu X, Tobias JW, et al. Differential effects of interleukin-2 and interleukin-15 versus interleukin-21 on CD4+ cutaneous T-cell lymphoma cells. Cancer research. 2008;68(4):1083–91. doi: 10.1158/0008-5472.CAN-07-2403. [DOI] [PubMed] [Google Scholar]
- 19.Qin JZ, Zhang CL, Kamarashev J, Dummer R, Burg G, Dobbeling U. Interleukin-7 and interleukin-15 regulate the expression of the bcl-2 and c-myb genes in cutaneous T-cell lymphoma cells. Blood. 2001;98(9):2778–83. doi: 10.1182/blood.v98.9.2778. [DOI] [PubMed] [Google Scholar]
- 20.Han GW, Iwatsuki K, Inoue M, Matsui T, Nishibu A, Akiba H, et al. Interleukin-15 is not a constitutive cytokine in the epidermis, but is inducible in culture or inflammatory conditions. Acta Derm Venereol. 1999;79(1):37–40. doi: 10.1080/000155599750011679. [DOI] [PubMed] [Google Scholar]
- 21.Marzec M, Liu X, Kasprzycka M, Witkiewicz A, Raghunath PN, El-Salem M, et al. IL-2- and IL-15-induced activation of the rapamycin-sensitive mTORC1 pathway in malignant CD4+ T lymphocytes. Blood. 2008;111(4):2181–9. doi: 10.1182/blood-2007-06-095182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasprzycka M, Zhang Q, Witkiewicz A, Marzec M, Potoczek M, Liu X, et al. Gamma c-signaling cytokines induce a regulatory T cell phenotype in malignant CD4+ T lymphocytes. J Immunol. 2008;181(4):2506–12. doi: 10.4049/jimmunol.181.4.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, et al. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat Med. 2015;21(11):1272–9. doi: 10.1038/nm.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Lee S, Teh CE, Bunting K, Ma L, Shannon MF. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. International immunology. 2009;21(3):227–35. doi: 10.1093/intimm/dxn143. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Tillo E, de Barrios O, Siles L, Amendola PG, Darling DS, Cuatrecasas M, et al. ZEB1 Promotes invasiveness of colorectal carcinoma cells through the opposing regulation of uPA and PAI-1. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(5):1071–82. doi: 10.1158/1078-0432.CCR-12-2675. [DOI] [PubMed] [Google Scholar]
- 26.Fehniger TA, Suzuki K, Ponnappan A, VanDeusen JB, Cooper MA, Florea SM, et al. Fatal leukemia in interleukin 15 transgenic mice follows early expansions in natural killer and memory phenotype CD8+ T cells. J Exp Med. 2001;193(2):219–31. doi: 10.1084/jem.193.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra A, Liu S, Sams GH, Curphey DP, Santhanam R, Rush LJ, et al. Aberrant overexpression of IL-15 initiates large granular lymphocyte leukemia through chromosomal instability and DNA hypermethylation. Cancer Cell. 2012;22(5):645–55. doi: 10.1016/j.ccr.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edelson RL. Cutaneous T cell lymphoma: the helping hand of dendritic cells. Ann N Y Acad Sci. 2001;941:1–11. [PubMed] [Google Scholar]
- 29.Cornejo MG, Kharas MG, Werneck MB, Le Bras S, Moore SA, Ball B, et al. Constitutive JAK3 activation induces lymphoproliferative syndromes in murine bone marrow transplantation models. Blood. 2009;113(12):2746–54. doi: 10.1182/blood-2008-06-164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whittaker S. Biological insights into the pathogenesis of cutaneous T-cell lymphomas (CTCL) Seminars in oncology. 2006;33(1 Suppl 3):S3–6. doi: 10.1053/j.seminoncol.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 31.Borowitz MJ, Weidner A, Olsen EA, Picker LJ. Abnormalities of circulating T-cell subpopulations in patients with cutaneous T-cell lymphoma: cutaneous lymphocyte-associated antigen expression on T cells correlates with extent of disease. Leukemia. 1993;7(6):859–63. [PubMed] [Google Scholar]
- 32.O’Connor OA, Toner LE, Vrhovac R, Budak-Alpdogan T, Smith EA, Bergman P. Comparative animal models for the study of lymphohematopoietic tumors: strengths and limitations of present approaches. Leuk Lymphoma. 2005;46(7):973–92. doi: 10.1080/10428190500083193. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg JM, Jaworsky C, Benoit BM, Telegan B, Rook AH, Lessin SR. The clonal nature of circulating Sezary cells. Blood. 1995;86(11):4257–62. [PubMed] [Google Scholar]
- 34.Rangwala S, Zhang C, Duvic M. HDAC inhibitors for the treatment of cutaneous T-cell lymphomas. Future medicinal chemistry. 2012;4(4):471–86. doi: 10.4155/fmc.12.6. [DOI] [PubMed] [Google Scholar]
- 35.Schuettengruber B, Simboeck E, Khier H, Seiser C. Autoregulation of mouse histone deacetylase 1 expression. Mol Cell Biol. 2003;23(19):6993–7004. doi: 10.1128/MCB.23.19.6993-7004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnstone RW, Licht JD. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell. 2003;4(1):13–8. doi: 10.1016/s1535-6108(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 37.Wilting RH, Yanover E, Heideman MR, Jacobs H, Horner J, van der Torre J, et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29(15):2586–97. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cabrero JR, Serrador JM, Barreiro O, Mittelbrunn M, Naranjo-Suarez S, Martin-Cofreces N, et al. Lymphocyte chemotaxis is regulated by histone deacetylase 6, independently of its deacetylase activity. Mol Biol Cell. 2006;17(8):3435–45. doi: 10.1091/mbc.E06-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu PY, Hsu HK, Lan X, Juan L, Yan PS, Labanowska J, et al. Amplification of distant estrogen response elements deregulates target genes associated with tamoxifen resistance in breast cancer. Cancer Cell. 2013;24(2):197–212. doi: 10.1016/j.ccr.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 42.Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sezary syndrome. Cell Death Dis. 2011;2:e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandoval J, Diaz-Lagares A, Salgado R, Servitje O, Climent F, Ortiz-Romero PL, et al. MicroRNA expression profiling and DNA methylation signature for deregulated microRNA in cutaneous T-cell lymphoma. J Invest Dermatol. 2015;135(4):1128–37. doi: 10.1038/jid.2014.487. [DOI] [PubMed] [Google Scholar]
- 44.van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, et al. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. J Invest Dermatol. 2011;131(3):762–8. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- 45.Batty N, Malouf GG, Issa JP. Histone deacetylase inhibitors as anti-neoplastic agents. Cancer Lett. 2009;280(2):192–200. doi: 10.1016/j.canlet.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 46.Methot JL, Chakravarty PK, Chenard M, Close J, Cruz JC, Dahlberg WK, et al. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2) Bioorganic & medicinal chemistry letters. 2008;18(3):973–8. doi: 10.1016/j.bmcl.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 47.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malamut G, El Machhour R, Montcuquet N, Martin-Lanneree S, Dusanter-Fourt I, Verkarre V, et al. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease-associated inflammation and lymphomagenesis. The Journal of clinical investigation. 2010;120(6):2131–43. doi: 10.1172/JCI41344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu S, Fischer L, Gokbuget N, Schwartz S, Burmeister T, Notter M, et al. Expression of interleukin 15 in primary adult acute lymphoblastic leukemia. Cancer. 2010;116(2):387–92. doi: 10.1002/cncr.24729. [DOI] [PubMed] [Google Scholar]
- 50.Gazdar AF, Carney DN, Bunn PA, Russell EK, Jaffe ES, Schechter GP, et al. Mitogen requirements for the in vitro propagation of cutaneous T-cell lymphomas. Blood. 1980;55(3):409–17. [PubMed] [Google Scholar]
- 51.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(12):7415–9. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bamford RN, Battiata AP, Burton JD, Sharma H, Waldmann TA. Interleukin (IL) 15/IL-T production by the adult T-cell leukemia cell line HuT-102 is associated with a human T-cell lymphotrophic virus type I region /IL-15 fusion message that lacks many upstream AUGs that normally attenuates IL-15 mRNA translation. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(7):2897–902. doi: 10.1073/pnas.93.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Das PM, Singal R. DNA methylation and cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22(22):4632–42. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 54.Lewis J, Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991;285(2):155–9. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- 55.van Doorn R, Zoutman WH, Dijkman R, de Menezes RX, Commandeur S, Mulder AA, et al. Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(17):3886–96. doi: 10.1200/JCO.2005.11.353. [DOI] [PubMed] [Google Scholar]
- 56.Contassot E, French LE. Epigenetic causes of apoptosis resistance in cutaneous T-cell lymphomas. J Invest Dermatol. 2010;130(4):922–4. doi: 10.1038/jid.2009.427. [DOI] [PubMed] [Google Scholar]
- 57.Bahar Halpern K, Vana T, Walker MD. Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development. J Biol Chem. 2014;289(34):23882–92. doi: 10.1074/jbc.M114.573469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pipaon C, Real PJ, Fernandez-Luna JL. Defective binding of transcriptional repressor ZEB via DNA methylation contributes to increased constitutive levels of p73 in Fanconi anemia cells. FEBS Lett. 2005;579(21):4610–4. doi: 10.1016/j.febslet.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 59.de Wilde J, Kooter JM, Overmeer RM, Claassen-Kramer D, Meijer CJ, Snijders PJ, et al. hTERT promoter activity and CpG methylation in HPV-induced carcinogenesis. BMC Cancer. 2010;10:271. doi: 10.1186/1471-2407-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rojas-Marquez C, Valle-Rios R, Lopez-Bayghen E, Ortiz-Navarrete V. CRTAM is negatively regulated by ZEB1 in T cells. Mol Immunol. 2015;66(2):290–8. doi: 10.1016/j.molimm.2015.03.253. [DOI] [PubMed] [Google Scholar]
- 61.Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015 doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vermeer MH, van Doorn R, Dijkman R, Mao X, Whittaker S, van Voorst Vader PC, et al. Novel and highly recurrent chromosomal alterations in Sezary syndrome. Cancer research. 2008;68(8):2689–98. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- 63.Williams TM, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, et al. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254(5039):1791–4. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 64.Hidaka T, Nakahata S, Hatakeyama K, Hamasaki M, Yamashita K, Kohno T, et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood. 2008;112(2):383–93. doi: 10.1182/blood-2008-01-131185. [DOI] [PubMed] [Google Scholar]
- 65.Fedele M, Pentimalli F, Baldassarre G, Battista S, Klein-Szanto AJ, Kenyon L, et al. Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene. 2005;24(21):3427–35. doi: 10.1038/sj.onc.1208501. [DOI] [PubMed] [Google Scholar]
- 66.Ponti R, Quaglino P, Novelli M, Fierro MT, Comessatti A, Peroni A, et al. T-cell receptor gamma gene rearrangement by multiplex polymerase chain reaction/heteroduplex analysis in patients with cutaneous T-cell lymphoma (mycosis fungoides/Sezary syndrome) and benign inflammatory disease: correlation with clinical, histological and immunophenotypical findings. The British journal of dermatology. 2005;153(3):565–73. doi: 10.1111/j.1365-2133.2005.06649.x. [DOI] [PubMed] [Google Scholar]
- 67.Hoppe RT, Medeiros LJ, Warnke RA, Wood GS. CD8-positive tumor-infiltrating lymphocytes influence the long-term survival of patients with mycosis fungoides. Journal of the American Academy of Dermatology. 1995;32(3):448–53. doi: 10.1016/0190-9622(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 68.Vermeer MH, van Doorn R, Dukers D, Bekkenk MW, Meijer CJ, Willemze R. CD8+ T cells in cutaneous T-cell lymphoma: expression of cytotoxic proteins, Fas Ligand, and killing inhibitory receptors and their relationship with clinical behavior. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2001;19(23):4322–9. doi: 10.1200/JCO.2001.19.23.4322. [DOI] [PubMed] [Google Scholar]
- 69.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26(37):5420–32. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 70.Kelly Richard DW, Cowley Shaun M. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts 2013. 2013 Jun 01;:741–9. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- 71.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. The Journal of clinical investigation. 2014;124(1):30–9. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arora A, Gera S, Maheshwari T, Raghav D, Alam MJ, Singh RK, et al. The dynamics of stress p53-Mdm2 network regulated by p300 and HDAC1. PLoS One. 2013;8(2):e52736. doi: 10.1371/journal.pone.0052736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21(22):6236–45. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang C, Richon V, Ni X, Talpur R, Duvic M. Selective induction of apoptosis by histone deacetylase inhibitor SAHA in cutaneous T-cell lymphoma cells: relevance to mechanism of therapeutic action. J Invest Dermatol. 2005;125(5):1045–52. doi: 10.1111/j.0022-202X.2005.23925.x. [DOI] [PubMed] [Google Scholar]
- 75.Marquard L, Gjerdrum LM, Christensen IJ, Jensen PB, Sehested M, Ralfkiaer E. Prognostic significance of the therapeutic targets histone deacetylase 1, 2, 6 and acetylated histone H4 in cutaneous T-cell lymphoma. Histopathology. 2008;53(3):267–77. doi: 10.1111/j.1365-2559.2008.03109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 77.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 78.Leoni F, Zaliani A, Bertolini G, Porro G, Pagani P, Pozzi P, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Villagra A, Sotomayor EM, Seto E. Histone deacetylases and the immunological network: implications in cancer and inflammation. Oncogene. 2010;29(2):157–73. doi: 10.1038/onc.2009.334. [DOI] [PubMed] [Google Scholar]
- 80.Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14(6):735–51. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 81.Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans. 2013;41(3):741–9. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- 82.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277(1):8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 83.Olsen EA, Kim YH, Kuzel TM, Pacheco TR, Foss FM, Parker S, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25(21):3109–15. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 84.Piekarz RL, Frye R, Turner M, Wright JJ, Allen SL, Kirschbaum MH, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(32):5410–7. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh BN, Zhou H, Li J, Tipton T, Wang B, Shao G, et al. Preclinical studies on histone deacetylase inhibitors as therapeutic reagents for endometrial and ovarian cancers. Future Oncol. 2011;7(12):1415–28. doi: 10.2217/fon.11.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31(2):141–9. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 87.Thurn KT, Thomas S, Moore A, Munster PN. Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer. Future Oncol. 2011;7(2):263–83. doi: 10.2217/fon.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.