Abstract
HAMP domains are signal transduction domains typically located between the membrane anchor and cytoplasmic signaling domain of the proteins in which they occur. The prototypical structure consists of two helical amphipathic sequences (AS-1 and AS-2) connected by a region of undetermined structure. The Escherichia coli aerotaxis receptor, Aer, has a HAMP domain and a PAS domain with a flavin adenine dinucleotide (FAD) cofactor that senses the intracellular energy level. Previous studies reported mutations in the HAMP domain that abolished FAD binding to the PAS domain. In this study, using random and site-directed mutagenesis, we identified the distal helix, AS-2, as the component of the HAMP domain that stabilizes FAD binding. AS-2 in Aer is not amphipathic and is predicted to be buried. Mutations in the sequence coding for the contiguous proximal signaling domain altered signaling by Aer but did not affect FAD binding. The V264M residue replacement in this region resulted in an inverted response in which E. coli cells expressing the mutant Aer protein were repelled by oxygen. Bioinformatics analysis of aligned HAMP domains indicated that the proximal signaling domain is conserved in other HAMP domains that are not involved in chemotaxis or aerotaxis. Only one null mutation was found in the coding sequence for the HAMP AS-1 and connector regions, suggesting that these are not active signal transduction sites. We consider a model in which the signal from FAD is transmitted across a PAS-HAMP interface to AS-2 or the proximal signaling domain.
In Escherichia coli, the periplasmic sensor domain of chemoreceptors is connected by a transmembrane helix and a linker to the cytoplasmic output domain (17). The latter is a 190- to 260-Å-long hairpin structure in the serine chemoreceptor, Tsr, with two antiparallel helices (25, 56). This signal output domain is highly conserved in chemotaxis receptors and in the aerotaxis receptor Aer (9, 40, 42). Signaling is effected through regulation of the CheA histidine kinase by the output domain (11, 26). Binding of aspartate to the sensing domain of the aspartate chemoreceptor Tar results in a 1- to 2-Å inward movement of the transmembrane helix (13, 17, 36). Signal transduction has been proposed to be a piston-like movement of the transmembrane and linker segments, conveying a conformational change to the output domain (13, 36).
The linker regions were recently proposed to be conserved signal transduction domains in histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins (MCPs), and phosphatases and were renamed HAMP domains (5, 12, 29, 58). There is limited sequence identity, but all HAMP domains are predicted to consist of two helical amphipathic sequences (AS-1 and AS-2) connected by a sequence of undefined secondary structure (3, 4, 12, 30). The proposed structure of the HAMP domain is supported by biochemical analysis of the Tar HAMP domain from Salmonella enterica serovar Typhimurium (12).
Genetic analysis supports a signal transduction role for the HAMP domain. Mutations in HAMP domains of the serine chemoreceptor Tsr, the nitrate sensor NarX, and the osmosensor EnvZ bias the signal transmitted by the sensor protein (2, 3, 14, 24, 38). Cysteine substitutions in the Tar AS-1 and AS-2 helices that perturb receptor function are strongly segregated to the buried helical faces, indicating that the packing face of each α-helix is critical for normal receptor activity (12). Furthermore, seven residues in the connector are also essential for the proper folding, stability, and function of Tar (12).
Hybrid sensors have been constructed in which the sensor module and HAMP domain of a receptor transmit a signal to the output module of another protein (3, 7, 8, 18, 27, 41, 51, 53, 55). The signal transduced by the hybrid protein is determined by the sensor module to which the HAMP domain is fused (3, 7, 51, 55). Sensor proteins with hybrid HAMP domains that combine HAMP elements from different proteins are not functional, probably due to the sequence diversity in different HAMP domains (3).
The E. coli aerotaxis transducer, Aer, represents a unique type of chemoreceptor in which the N-terminal PAS sensory domain and an F1 segment that links the PAS domain to a membrane anchor sequence are located in the cytoplasm (8, 9, 40, 41). A postmembrane linker sequence connects the membrane anchor to the C-terminal signal output domain. The PAS domain binds a flavin adenine dinucleotide (FAD) cofactor that is believed to reflect the redox status of the electron transport system (47, 48). It is proposed that the Aer PAS sensory domain generates signals to regulate the activity of the output domain upon changes in the redox potential of FAD. In the present study, we confirmed that the linker in the Aer protein is a member of the HAMP domain family, and, using genetic analysis, we investigated structure-function relationships of the Aer HAMP domain in aerotaxis signaling by identification of the amino acid residues that are essential for aerotactic signaling.
MATERIALS AND METHODS
Strains and construction of plasmids.
E. coli cells were grown at 30°C in Luria-Bertani (LB) medium (16) supplemented with thiamine (0.5 μg ml−1) and ampicillin (100 μg ml−1). Plasmids containing aer mutations were expressed in E. coli strains BT3312 (Δaer Δtsr) (41) and UU1117 (Δaer) (9), derivatives of RP437 (wild type for chemotaxis) (39). BT3339 [Δ(tsr)7021 Δ(tar-tap)5201 trg::Tn10] was constructed by inactivating the aer gene in HCB339 (59), using the pKO3aer-2::kan vector as described previously (40). BT3340 [Δ(tsr)7021 Δ(tar-tap)5201 trg::Tn10 aer-2::kan recA::cat] was constructed by inactivating the recA gene in BT3339, using P1 transduction of recA::cat from BW10724 (34, 35, 52), and confirmed by UV sensitivity assays.
The pGH1 plasmid (40) expresses a wild-type aer gene inserted into the pTrc99A expression vector (Amersham-Pharmacia), whereas pAVR2 (41) expresses the aer gene with a translational fusion of an N-terminal six-histidine (His6) tag in the pProEX HTa expression vector (Life Technologies). Protein expression is controlled by the Ptrc promoter and the lacIq repressor gene in both plasmids.
The pQH16 plasmid containing an aer gene with a deletion of the HAMP domain coding region (codons 206 to 279) was derived from pGH1 by inverse PCR using Pfu Turbo DNA polymerase (Stratagene). The sense primer (5′-GCGGAGCTCAACGCCCATACCCAGCAGACAGTT-3′) and the antisense primer (5′-GTCTTCGAAACAGGCGCTTGC-3′) included restriction sites SacI and BstBI (underlined), respectively, which introduced silent mutations to facilitate cloning.
Construction of single-residue substitutions in the Aer HAMP domain.
Single-residue substitutions of the selected codons in the Aer HAMP domain were constructed by oligonucleotide-directed mutagenesis of pAVR2, using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The mutated plasmids were isolated from E. coli XL1Blue cells, and the single-amino-acid replacements were verified by automated DNA sequencing. PCR random mutagenesis (23) was also used to generate single missense mutations in the coding sequence for the Aer HAMP and proximal signaling domains (residues 206 to 279). A 230-bp DNA fragment encoding the Aer fragment comprising amino acids 204 to 281 (Aer204-281) was amplified with Taq DNA polymerase (Roche Molecular Biochemicals) in the presence of 0.05 mM MnCl2, using sense primer 5′-GGTTTCGAATGGCAGATTGTG-3′ and antisense primer 5′-GCTGAGCTCATCGGTGCCTTTCGCCAGCGT-3′. The PCR product was digested, cloned into the BstBI-SacI sites of pQH16 (pTrc99A Δaer206-279), and transformed into BT3312 cells, resulting in a library of E. coli cells expressing mutant Aer proteins with single-amino-acid replacements.
Phenotype screening of aer mutants.
Single colonies of BT3312 transformants were transferred from the LB agar plates to 0.28% semisolid agar plates containing succinate (30 mM) and ampicillin (100 μg ml−1) for phenotype screening (9). Cells with aberrant phenotypes were grown in LB medium supplemented with ampicillin to an optical density at 600 nm of 0.40, and 5 μl of each culture was inoculated onto a fresh succinate semisolid agar plate to verify the phenotype. Succinate plates supplemented with various concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG) were used to distinguish the abnormal phenotypes caused by altered expression levels. Western blot analysis with the anti-His6-Aer2-166 antisera (41) was used to screen for mutants with a null phenotype that expressed a full-length Aer mutant protein. The mutation in each plasmid was identified by sequencing the entire aer coding region.
Behavioral studies.
An aerotaxis capillary assay was performed as described by Zhulin et al. (60). Briefly, cells grown to an optical density at 600 nm of 0.40 to 0.45 were loaded into an open-ended, flat capillary tube. The formation of the aerotactic bands was observed under a dark-field microscope and videotaped. The distance from the center of the aerotactic band to the meniscus was measured on the monitor 30 min after the cells were loaded into the capillary. Values in millimeters include a 71-fold magnification. The aerotactic temporal assay was used to quantify responses to increased (0 to 21%) and decreased (21 to 0%) oxygen concentrations (40). Each mutant strain was analyzed on two different days, each with three replicate assays. Succinate swarm plates (9) were used to screen phenotypes as mentioned above.
Determination of FAD binding.
The increase in membrane-bound FAD when Aer was overexpressed was used as a reporter for Aer FAD binding. Our modification of the original method of Bibikov et al. (8) was reported previously (41).
Analysis of secondary structure.
Putative secondary structures of the HAMP domains were analyzed with the Jpred2 (15) and PSIPRED (22) programs and the protein sequence analysis (PSA) server (43, 44, 57). Sequence amphipathicity was determined with the Kyte-Doolittle hydropathicity plot (28).
RESULTS
Prediction of the secondary structure of the Aer HAMP domain.
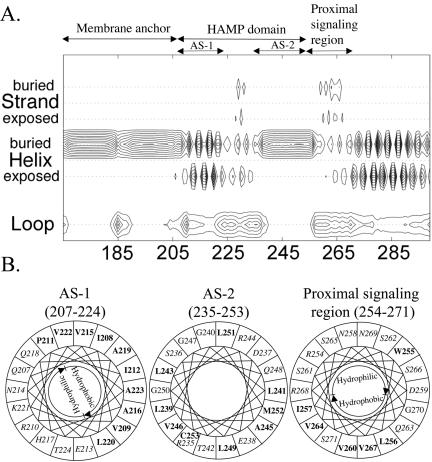
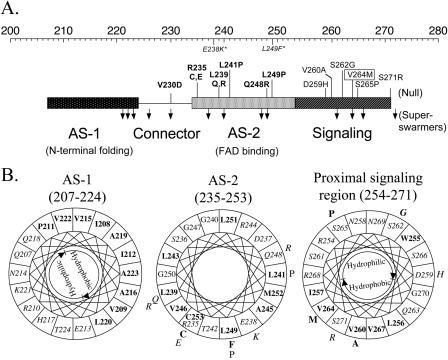
The Aer linker region is a member of the HAMP domain family (Pfam accession number PF00672; see http://pfam.wustl.edu/cgi-bin/getdesc?name=HAMP) (6). This classification was confirmed by a PSI-BLAST search (1) done with Aer residues 205 to 265, which include the sequence between the membrane anchor and the proximal signaling domain (30), used as the query sequence. Since there is no three-dimensional structure available for any of the HAMP domains, we analyzed their putative secondary structures (Fig. 1 and 2A). Our analysis predicted that the HAMP domain of Aer contains two α-helical segments found in other HAMP domains (Fig. 1). Figure 2A represents the individual probabilities that each residue is incorporated into a β-strand, an α-helix, or a loop. The buried helix that terminates at residue 206 is the membrane anchor. The probability of a short loop (approximately residues 183 to 187) in the center of the anchor is compatible with an anchor that consists of two antiparallel transmembrane helices separated by a short periplasmic loop. However, there is no experimental evidence that addresses this prediction. Distal to the anchor is the HAMP domain with (i) an amphipathic helix (AS-1) that has a high probability that one face of the helix is buried, (ii) a connector of undetermined structure having lower probabilities of a β-strand or an α-helix, and (iii) a distal helix that aligns with the amphipathic sequence (AS-2) in the Tar HAMP domain but that is predicted to be buried in Aer (Fig. 1) (see also reference 12). When the HAMP domain of Aer was mapped manually on helical wheels (Fig. 2B) and the amphipathicity of the sequence was analyzed, the Aer distal helix had hydrophobic residues on the face corresponding to the hydrophilic face of AS-2 in Tar. This phenomenon is consistent with a buried helix. Three putative helix-breaking residues (glycine) were also found in this region (Fig. 2B), suggesting structural flexibility. The proximal segment of the signaling domain (residues 254 to 271) was predicted to be an amphipathic helix by use of Jpred2 and PSIPRED (Fig. 1) but had an undefined structure in the PSA analysis (Fig. 2A).
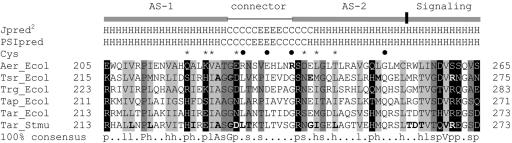
FIG. 1.
Representative sequence alignment of HAMP domain sequences found in MCPs. The consensus sequence, determined with the program Consensus (http://www.bork.embl-heidelberg.de/Alignment/consensus.html), is indicated beneath the alignment and represented as follows: uppercase, conserved residues; lowercase, similar residues; s, small (ACDGNPSTV); h, hydrophobic (ACFGILMPTVWY); l, aliphatic (ILV); p, polar (CDEHKNQRST). Conserved residues are indicated as follows: dark gray shading, small residues; light gray shading, hydrophobic residues; white font on a black background, polar residues. Residues known to produce a null phenotype when mutated are shown in bold. Secondary structures of the Aer HAMP domain predicted by Jpred2 and PSIPRED analyses are shown above the aligned sequences. H, α-helix; C, coil; E, β-strand. Locations of AS-1, the connector, AS-2, and proximal signaling domains of Tar in S. enterica serovar Typhimurium are indicated. The residues in Aer that were selected for cysteine substitution are indicated with an asterisk or a filled circle, depending on whether the residue was conserved or nonconserved, respectively. The essential Arg235 residue of Aer is in bold. Ecol, E. coli; Stmu, S. enterica serovar Typhimurium.
FIG. 2.
Secondary-structure analysis of the Aer HAMP domain. (A) Contour plot detailing secondary-structure probabilities (PSA server) (43, 44, 57). Rows designate the secondary-structure state; columns indicate each residue position. The probability of each structural state is depicted with contour lines with probability increments of 0.1. (B) Putative helical segments of the Aer HAMP domain revealed by mapping the residues on helical wheels. The hydrophobic residues are shown in bold, and the hydrophilic residues are in italics within the helical wheels.
Algorithms may differ slightly in the predicted starts and finishes of the secondary structure elements (32, 33). For this study, the boundaries for the Aer HAMP domain proximal and distal helices are defined to align with the boundaries published for the secondary structure of the HAMP domain in the Tar chemoreceptor (12), with residues 207 to 220 comprising AS-1, residues 235 to 253 comprising AS-2, and residues 221 to 234 comprising the connector.
Selection of HAMP domain residues for cysteine replacement mutagenesis.
The HAMP domains of the chemoreceptors from E. coli and S. enterica serovar Typhimurium have many similar amino acid residues in the HAMP sequence (Fig. 1), implying a common function in chemotactic signal transduction. However, there are notable differences in the sequence of the HAMP domain of Aer (Fig. 1) that suggest a different and perhaps unique role for the Aer HAMP domain. Since Aer has a distinctive topology and a HAMP domain that is essential for FAD binding and signaling (8, 41), we substituted cysteine for residues at four positions in the Aer HAMP domain that are not conserved in other HAMP domains (Fig. 1 and Table 1). Additionally, Aer residues that aligned with the functionally important residues from 21 HAMP domains (2, 3, 12, 14, 20, 21, 23, 24, 37, 50; see reference 30 for this sequence alignment) were also chosen for site-directed mutagenesis (Fig. 1 and Table 1).
TABLE 1.
Phenotypes of BT3312 cells expressing the site-specific cysteine substitutions in the Aer HAMP domain
| Plasmida | Protein | Succinate plate assay results | Capillary assay results | Response time (s)b
|
Ratio of FAD (induced/uninduced)c | Residue subtituted is Aer specificd | |
|---|---|---|---|---|---|---|---|
| O2 increase | O2 decrease | ||||||
| pProEX HTa | Vector | − | − | 0 | 0 | 0.96 ± 0.18 | |
| pAVR2 | His6-Aer2-506 | + | + | 173.8 ± 10.4 | 26.6 ± 3.0 | 4.98 ± 2.62 | |
| pQH30 | His6-AerQ218C | + | + | >180e | 26.3 ± 10.2 | + | No |
| pQH31 | His6-AerK221C | + | + | >180e | 33.0 ± 5.6 | + | No |
| pQH32 | His6-AerV222C | + | + | 159.3 ± 24.9 | 31.8 ± 5.5 | + | No |
| pQH33 | His6-AerE226C | + | + | 97.2 ± 13.8 | 23.3 ± 5.4 | + | No |
| pQH34 | His6-AerR227C | + | + | >180e | 35.3 ± 7.3 | + | Yes |
| pQH36 | His6-AerE231C | + | + | >180e | 27.8 ± 2.4 | + | Yes |
| pQH38 | His6-AerR235C | − | − | 0 | 0 | 1.02 ± 0.01 | Yes |
| pQH39 | His6-AerR235E | − | − | 0 | 0 | 0.69 ± 0.03 | Yes |
| pQH40 | His6-AerR235K | + | + | 70.0 ± 7.9 | 12.2 ± 1.5 | + | Yes |
| pQH43 | His6-AerD237C | + | + | 103.7 ± 41.0 | 14.5 ± 2.2 | + | No |
| pQH44 | His6-AerL239C | + | + | 174.2 ± 9.2 | 25.3 ± 7.4 | + | No |
| pQH46 | His6-AerT242C | + | + | >180e | 23.2 ± 1.5 | + | No |
| pQH47 | His6-AerG250C | + | + | 46.9 ± 17.6 | 25.1 ± 5.7 | 2.94 ± 0.62 | Yes |
All plasmids were derived from the pProEX HTa expression vector.
The data represent means ± standard deviations of results for two independent experiments, each with three trials.
All Aer mutants were induced with IPTG to at least 2,500 copies per cell, which is eightfold higher than Aer expressed chromosomally (300 copies cell−1). Membranes from cells with the pProEx vector alone (aer) contained ≈500 copies of FAD cell−1. A 20% increase (ratio, 1.2) in total membrane FAD after IPTG induction was used to indicate FAD binding to Aer. +, aerotaxis activity was evidence that Aer was expressed and that it bound FAD in these strains.
Yes, the residue is not conserved in HAMP domains other than Aer.
Response times were variable within a range of 190 to ≈250 s.
Phenotypes of the cysteine replacement mutants.
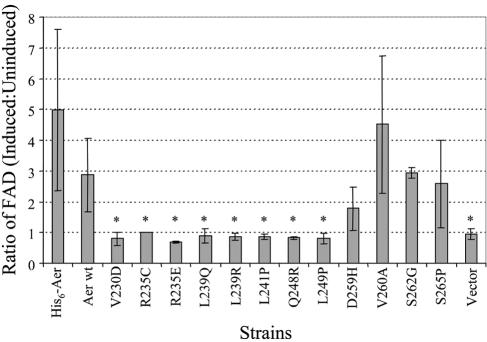
The R235C substitution in the HAMP domain inhibited FAD binding to the Aer protein (Fig. 3), and cells expressing the His6-tagged Aer protein with the R235C mutation (His6-AerR235C) had a null aerotaxis phenotype (Table 1). Replacement of arginine by the positively charged lysine in His6-AerR235K retained half of the aerotaxis activity of His6-Aer (wild type for aerotaxis). However, negatively charged glutamic acid in His6-AerR235E abolished aerotaxis activity (Table 1) and FAD binding (Fig. 3) even when overexpressed at high levels by induction with 1 mM IPTG. Under such conditions, the intracellular concentrations of His6-AerR235C and His6-AerR235E were similar to that of His6-Aer (carried by pAVR2) induced by 1 mM IPTG (Fig. 3).
FIG. 3.
Ratios of the membrane FAD content in the Aer HAMP domain null mutants under induced versus uninduced conditions. Ratios over 1.4 indicate binding of FAD. An asterisk denotes a significant difference (P < 0.01) in results compared with the parental strains. pAVR2 (His6-Aer) was the parent for the His6-AerR235C and His6- AerR235E mutants, and pGH1 (Aer wt) was the parent for the remaining Aer HAMP mutants.
The E226C, D237C, and G250C mutations in Aer decreased the duration of aerotaxis responses to changes in oxygen concentration in a temporal aerotaxis assay (Table 1). Other cysteine substitutions in the HAMP domain did not affect the Aer phenotype on succinate semisoft agar or in temporal aerotaxis assays.
Phenotypes of the randomly generated HAMP domain mutants.
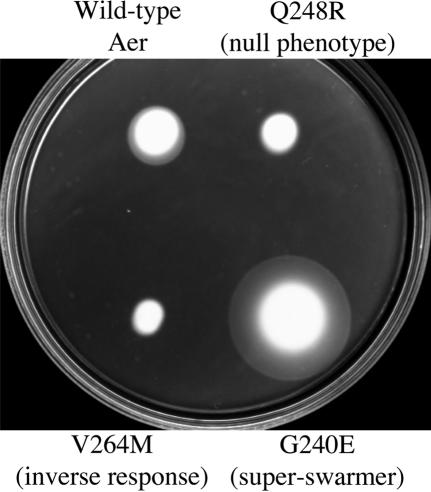
A library of DNA fragments containing single-codon substitutions in the sequence corresponding to the Aer206-279 region was generated and fused to the remainder of the aer gene. Over 1,000 E. coli BT3312 transformants were screened for phenotype on succinate semisolid agar plates (9, 40). Nearly 15% of the tested colonies exhibited a null aerotaxis phenotype by forming a cylinder-shaped swarm without an aerotactic ring (Fig. 4), resembling the phenotype of BT3312 cells (Δaer Δtsr). The null phenotypes of the V230D, L239Q, L239R, L241P, Q248R, L249P, D259H, V260A, S262G, V264M, S265P, and S271R mutants were confirmed by aerotaxis temporal assays. The mutant strains did not restore aerotactic rings on succinate semisolid agar plates supplemented with IPTG at various concentrations (data not shown). Two mutant proteins (AerG225E and AerE238G) with a nonaerotactic phenotype restored an aerotactic ring when the proteins were overexpressed by induction with 50 μM to 1 mM IPTG (data not shown), suggesting that the null phenotype exhibited by these two mutants may be due to low levels of protein expression.
FIG. 4.
Different phenotypes of BT3312 cells expressing the mutant Aer proteins on succinate semisolid agar plates without addition of IPTG.
Of particular interest, the swarm size of the AerV264M mutant was slightly smaller than those of the other nonaerotactic mutants (Fig. 4). An aerotaxis temporal assay revealed that cells from the BT3312 strain with AerV264M (BT3312/AerV264M) had a strong inverse response to an increase or decrease in oxygen concentration; i.e., cells swam smoothly when they became anoxic but tumbled when 21% oxygen was reintroduced (Table 2).
TABLE 2.
Behavioral assays of Aer mutants exhibiting super-swarming or an inverse response
| Plasmida | Protein | Swarm sizeb,c (mm) | Band distancec (mm) | Response time (s)c,d
|
Aer expressione | |
|---|---|---|---|---|---|---|
| O2 increase | O2 decrease | |||||
| pGH1 | Wild-type Aer | 20.0 ± 0.0 | 18.5 ± 2.1 | 143.0 ± 23.4 | 25.7 ± 3.6 | +++ |
| pQH68 | AerK221E | 39.6 ± 8.1 | 11.5 ± 0.7 | 131.7 ± 15.1 | 25.2 ± 4.2 | +++ |
| pQH69 | AerV222A | 31.5 ± 1.5 | 16.3 ± 1.8 | 93.7 ± 31.5 | 30.8 ± 4.4 | +++ |
| pQH70 | AerA223V | 39.6 ± 7.4 | 13.0 ± 4.2 | 102.7 ± 8.7 | 30.3 ± 0.6 | +++ |
| pQH58 | AerE226V | 45.5 ± 2.5 | NDf | 59.0 ± 7.4 | 24.8 ± 2.3 | +++ |
| pQH74 | AerV230A | 31.3 ± 6.7 | 21.5 ± 2.1 | 70.7 ± 13.0 | 11.2 ± 2.0 | + to ++ |
| pQH76 | AerD237E | 40.0 ± 8.5 | 43.0 ± 11.3 | 26.2 ± 8.3 | 18.4 ± 3.8 | ++ |
| pQH81 | AerG240E | 39.8 ± 6.0 | 13.5 ± 2.1 | 103.0 ± 10.4 | 22.7 ± 3.1 | +++ |
| pQH84 | AerG247R | 34.5 ± 10.6 | 16.5 ± 2.1 | 114.0 ± 14.9 | 16.0 ± 3.5 | ++ to +++ |
| pQH85 | AerQ248L | 40.8 ± 7.4 | 12.5 ± 3.5 | 84.5 ± 0.7 | 18.0 ± 4.0 | +++ |
| pQH93 | AerS261L | 31.5 ± 9.2 | 13.8 ± 1.8 | 150.0 ± 0.0 | 30.0 ± 2.0 | +++ |
| pQH95 | AerV264A | 30.4 ± 1.2 | 15.5 ± 3.5 | 58.2 ± 14.8 | 21.0 ± 3.4 | +++ |
| pQH96 | AerV264M | 12.2 ± 1.6 | No band | −(135.3 ± 9.0) | −(17.0 ± 1.8) | +++ |
| pQH98 | AerS266C | 43.1 ± 4.4 | 10.0 ± 0.0 | 92.5 ± 18.2 | 26.3 ± 4.4 | +++ |
| pQH100 | AerE272G | 41.2 ± 1.6 | 17.0 ± 0.0 | 89.3 ± 10.2 | 29.8 ± 6.3 | +++ |
All plasmids were derived from the pTrc99A expression vector. Cells expressing pQH96 had an inverse aerotaxis response. Cells expressing all other plasmids had a superswarming phenotype.
Swarm sizes were normalized to 20 mm for pGH1.
The data represent means ± standard deviations of results from more than two independent experiments.
Negative values represent inverse responses.
+, ++, +++, graduated levels of proteins detected in the whole-cell extracts of strains without induction.
Not determined.
A novel aerotaxis phenotype was also observed. Eighteen Aer mutants formed larger-than-normal swarm sizes with sharp aerotactic rings on succinate semisolid agar plates (Fig. 4 and Table 2). The lower surfaces of the mutant colonies spread much faster than those of the wild-type control colonies, forming a truncated cone shape with a more gradual slope. We designated these mutants “superswarmers.”
Plasmids containing Aer HAMP mutations expressing a null phenotype were transformed into the recA strain BT3340, and transformants were screened for spontaneous aerotactic pseudorevertants. Only one pseudorevertant was isolated; the T285I mutation suppressed the V260A mutation. We concluded that selective mutagenesis would be required for pseudoreversion analysis of Aer HAMP mutations that could address domain interactions in Aer. The results of such a study, published recently, supported the idea of interactions between the PAS and HAMP domains (54). One of these mutations, N34D, is specific for the HAMP C253R mutant, supporting the notion that these domains are in close proximity.
FAD binding and expression of the HAMP domain null-phenotype mutants.
The FAD levels in each Aer mutant with a null phenotype were analyzed under induced and uninduced conditions as described previously (8, 41). Six mutant proteins did not bind FAD (Fig. 3). Conversely, the D259H, V260A, S262G, and S265P mutant Aer proteins bound FAD, suggesting that they perturbed aerotaxis by means other than interference with FAD binding.
Aer HAMP mutants that did not bind FAD showed low expression in the absence of IPTG. This finding raised the question of whether their null phenotype might be a result of defects in protein expression. However, this possibility was ruled out, as they could not restore aerotactic signaling or FAD binding when they were induced to express Aer over a range of concentrations that exceeded eightfold that of Aer expressed from the chromosome. We also confirmed that the decreased Aer protein level was caused by single-residue substitutions in Aer HAMP rather than any accidental mutations introduced elsewhere in the plasmids by recloning the mutated HAMP DNA fragments and by reverting the substitutions to the wild-type codon sequences by using site-directed mutagenesis. RT-PCR revealed that transcription of all the mutant aer genes was normal (data not shown). These data collectively suggested that the V230D, R235C, R235E, L241P, Q248R, and L249P substitutions in Aer changed the protein conformation to a state that was unfavorable for FAD binding and thus interfered with aerotaxis signaling, whereas the L239Q and L239R substitutions caused a null phenotype, possibly by altering the stability of the protein structure.
Phenotypes and protein expression of the superswarmers.
Growth curves for the superswarmer strains were similar to those expressing wild-type Aer, suggesting that the large swarm size was not caused by increased growth rate. All the superswarming strains showed normal tumbling frequencies, but a majority had responses to an increase or decrease in oxygen concentration in temporal assays that were significantly reduced in duration (Table 2). In a capillary assay, most superswarmers formed bands closer to the meniscus than did the wild-type control (Table 2), implying that they migrated to a higher oxygen concentration than wild-type cells. Cells expressing the AerA223V and AerQ248L proteins formed sharp bands within 3 min, in contrast to an average of 10 min required by the BT3312/pGH1 cells (wild-type Aer). The AerV230A and D237E mutant strains formed aerotactic bands far away from the meniscus and showed slightly decreased levels of the proteins expressed (Table 2). When the expression was induced with IPTG, these mutant cells formed bands closer to the meniscus and exhibited a wild-type swarming pattern on succinate semisolid agar plates (data not shown). This behavior mirrored that of cells expressing wild-type protein, which migrated closer to the meniscus as the level of Aer expression increased (data not shown), a finding that suggests that the superswarming phenotype of these mutant strains could result from lowered levels of protein expression.
DISCUSSION
Genetic analysis of the Aer HAMP and proximal signaling domains.
Previous studies identified mutations in the F1 and HAMP domains that abolished FAD binding to Aer (8, 41, 54). The present, more extensive mutational analysis of the HAMP domain reveals that all mutations that abolished FAD binding are localized in the sequence coding for the AS-2 helix, except for V230D in the connector (Fig. 5A). In addition, this study identified mutations clustered in the coding sequence for the proximal signaling domain that impaired signaling and aerotaxis but that did not alter FAD binding (Fig. 3 and 5A). The phenotype resulting from these mutations clearly indicates a signal transduction role for this domain. Of particular interest is the AerV264M mutant, which has an inverse aerotaxis response (Table 2) in which E. coli migrates away from oxygen as though it were a repellent. The V264 residue is located on the putative hydrophobic face of the signaling domain (Fig. 5B). Possible mechanisms for inverse responses have been discussed previously (3, 23, 46).
FIG. 5.
Mutations altering Aer activity mapped onto regions of the HAMP and proximal signaling domains. (A) Segment of Aer representing helical (rectangle) and undefined connector (line) regions of the HAMP and contiguous signaling domains. The top scale represents the amino acid residue numbers. Mutations which abolished FAD binding are in bold, those that bound FAD but had null activity are in plain text, and those that caused superswarming are represented by downward arrows. V264M inverted the response and is shown boxed. Mutations in italics and followed with an asterisk denote two null mutants reported previously by Bibikov et al. (8). (B) Null mutations mapped onto the helical wheels from Fig. 2. Individual mutations are placed outside of the wheel. See the legend to Fig. 2 for the key.
Mutations causing superswarming (Fig. 4) were distributed throughout the HAMP and proximal signaling domains (Fig. 5A). Aer proteins with these mutations exhibited shorter temporal responses to a rise or fall in oxygen concentration and aberrant “aerophilic” behavior in an oxygen spatial gradient (Table 2). Growth rates, swimming speeds, and tumbling biases were unaffected, as were chemotactic responses mediated by Tsr and Tar (data not shown). Some superswarmer strains, such as BT3312/AerV230A and BT3312/AerD237E, had low levels of protein expression and resumed normal aerotaxis responses after the protein level was increased by IPTG induction. One possible explanation for this finding is that wild-type Aer is inhibitory to aerotactic swarming at the expression levels shown in Table 2. In this case, a lower Aer expression would increase the size of the aerotactic colonies on succinate semisolid agar. However, most superswarmer strains expressed the mutant Aer proteins at levels similar to that expressed by pGH1, which contained wild-type Aer. The mechanism by which these unusual aerotactic responses translated into superswarming behavior on swarm plates is unclear and requires further analysis.
Structure-function relationships.
The Aer HAMP domain has a predicted secondary structure (Fig. 1 and 2) similar to the structure predicted for other HAMP domains (3, 12), with the exception that the AS-2 helix is predicted to be buried rather than amphipathic in Aer (Fig. 2). Seven of the nine AS-2 residue replacements that abolished FAD binding fall within a 120° sector on the helix surface (Fig. 5B), and this may be a face that interacts with the PAS domain. The C253 residue was shown by second-site suppressor analysis to be in proximity to PAS residue N34 (54) and also cross-links at the dimer interface (31). This finding suggests that the AS-2-PAS face is close to the AS-2-AS-2′ dimer interface. The structure of the proximal signaling domain is uncertain, with Jpred2 (15) and PSIPRED (22) analyses predicting a continuous helix (Fig. 1) and PSA predicting a nonhelical region (43, 44, 57) (Fig. 2A). The inability to resolve the tertiary structure of the Tsr HAMP domain by X-ray crystallography suggests that MCP HAMP domains may be flexible (25). Additionally, the total length of the Tsr receptor measured by electron microscopy is 310 Å (56), 70 Å less than that predicted by Kim et al., assuming an extended HAMP domain (25). The 70-Å shortfall could be due to folding of the HAMP domain.
In silico analysis.
Sequence comparison of the Aer HAMP with a family of 21 HAMP domains (Fig. 1) (see reference 30 for full alignment) suggests that the HAMP sequence may extend beyond the presently assigned endpoint of AS-2 at residue 253 to include the proximal signaling domain (residues 254 to 265). Assigning definitive boundaries to HAMP domains will require determination of the HAMP structure. Unique residues in the Aer HAMP domain may be associated with unique functions of the Aer domain, such as the PAS-HAMP interaction that stabilizes the native PAS fold and FAD binding and possibly propagation of the aerotaxis signal from PAS to HAMP domains (19, 54). Residues that are conserved in other HAMP domains but not in Aer include R227, E231, G247, L249 (41, 54), and C253 (Fig. 1). Of these, L249 (Fig. 5A) and C253 (54) are determinants for FAD binding. The unique residues may also account for exclusion of Aer in early alignments of HAMP sequences (5, 58).
Signal transduction pathway.
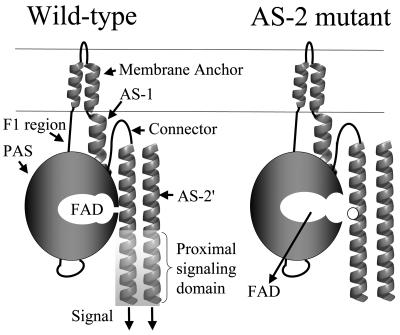
The schematic of Aer (Fig. 6) incorporates recent findings that relate structure and signaling function in Aer. FAD most likely binds to the PAS domain of Aer (8, 41, 54) but binds only when the HAMP domain AS-2 helix is present (this study). Native folding of the PAS domain requires the membrane anchor and the HAMP domain (19). The HAMP and PAS domains may interact directly, as supported by data showing suppression of a C253R substitution in the HAMP domain by an N34D replacement in the PAS domain (54). The proposed model shows the AS-2 helix stabilizing the PAS conformation that binds FAD, consistent with the present work and with Aer folding studies (19; see also reference 8). The AS-2 helix is physically close to AS-2′ from the cognate monomer, since native Cys253 residues cross-link in vivo in response to an oxidant (31), and the interaction of AS-2 with both the PAS domain and AS-2′ is consistent with the prediction that AS-2 is buried in Aer (Fig. 2). Few nonaerotactic mutations were isolated in the coding sequence for the AS-1 and connector components of the HAMP domain, suggesting that contacts between these components and the PAS domain are not as important in signaling by Aer.
FIG. 6.
Schematic summarizing the data described in the text. With the exception of V230D in the connector, all HAMP mutations that abolished FAD binding affected residues (white circle) in the AS-2 region. The signaling domain of Aer controls the direction of rotation of the flagellar motor by modulating autophosphorylation of CheA and phosphorylation of CheY (see Fig. 1 in reference 31).
The accumulated evidence for direct interaction between the PAS and HAMP domains suggests a possible signal transduction pathway. The FAD cofactor is oxidized and reduced in response to redox changes in the electron transport system (45, 49). Oxidation and reduction of FAD may result in a conformational change in the PAS domain that is transmitted across the PAS-HAMP interface to the AS-2 helix or the proximal signaling domain, where it is propagated through the signaling domain to modulate CheA autophosphorylation (10). This model can accommodate a PAS domain interface with the HAMP domain from the same monomer or the cognate monomer. However, at present, there is no definitive evidence for a transverse signaling pathway such as this rather than a linear pathway through the membrane anchor (Fig. 6). In its simplest form, the model does not address other possible interactions in a trimer of dimers or higher-order signaling complexes that contain Aer, although the model can be adapted to include such interactions, which may also be important in signaling.
Acknowledgments
We thank Igor Zhulin for assistance with bioinformatics analyses, Kylie Watts for critical analysis and helpful discussions. We are grateful to Debbie Thomas, Jack Scarbrough, and Joanne Ragoonanan for their technical assistance and Alexander Repik for his technical suggestions.
This work was supported by grants from the National Institute of General Medical Sciences (GM29481) and Loma Linda University to B.L.T.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., and J. S. Parkinson. 1988. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell 55:817-826. [DOI] [PubMed] [Google Scholar]
- 3.Appleman, J. A., L.-L. Chen, and V. Stewart. 2003. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 185:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 6.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32(Database issue):D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumgartner, J. W., C. Kim, R. E. Brissette, M. Inouye, C. Park, and G. L. Hazelbauer. 1994. Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J. Bacteriol. 176:1157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borkovich, K. A., N. Kaplan, J. F. Hess, and M. I. Simon. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. USA 86:1208-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourret, R. B., K. A. Borkovich, and M. I. Simon. 1991. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu. Rev. Biochem. 60:401-441. [DOI] [PubMed] [Google Scholar]
- 12.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chervitz, S. A., and J. J. Falke. 1996. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. USA 93:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins, L. A., S. M. Egan, and V. Stewart. 1992. Mutational analysis reveals functional similarity between NARX, a nitrate sensor in Escherichia coli K-12, and the methyl-accepting chemotaxis proteins. J. Bacteriol. 174:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. Jpred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 16.Davis, R. W., Botstein, D., and Roth, J. R. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, X., J. W. Baumgartner, and G. L. Hazelbauer. 1997. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J. Bacteriol. 179:6714-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J. Bacteriol. 186:6782-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, T., and M. Inouye. 1993. Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. J. Mol. Biol. 232:484-492. [DOI] [PubMed] [Google Scholar]
- 21.Jin, T., and M. Inouye. 1994. Transmembrane signaling. Mutational analysis of the cytoplasmic linker region of Taz1-1, a Tar-EnvZ chimeric receptor in Escherichia coli. J. Mol. Biol. 244:477-481. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 23.Jung, K.-H., and J. L. Spudich. 1998. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: insights into the color-sensing mechanism. J. Bacteriol. 180:2033-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalman, L. V., and R. P. Gunsalus. 1990. Nitrate- and molybdenum-independent signal transduction mutations in narX that alter regulation of anaerobic respiratory genes in Escherichia coli. J. Bacteriol. 172:7049-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 26.Kofoid, E. C., and J. S. Parkinson. 1988. Transmitter and receiver modules in bacterial signaling proteins. Proc. Natl. Acad. Sci. USA 85:4981-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krikos, A., M. P. Conley, A. Boyd, H. C. Berg, and M. I. Simon. 1985. Chimeric chemosensory transducers of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:1326-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 29.Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568-585. [DOI] [PubMed] [Google Scholar]
- 30.Ma, Q. 2001. HAMP domain and signaling mechanism of the Aer protein. Ph.D. dissertation. Loma Linda University, Loma Linda, Calif.
- 31.Ma, Q., F. Roy, S. Herrmann, B. L. Taylor, and M. S. Johnson. 2004. The Aer protein of Escherichia coli forms a homodimer independent of the signaling domain and flavin adenine dinucleotide binding. J. Bacteriol. 186:7456-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 33.McGuffin, L. J., and D. T. Jones. 2003. Benchmarking secondary structure prediction for fold recognition. Proteins 52:166-175. [DOI] [PubMed] [Google Scholar]
- 34.Metcalf, W. W., and B. L. Wanner. 1993. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 129:27-32. [DOI] [PubMed] [Google Scholar]
- 35.Metcalf, W. W., and B. L. Wanner. 1993. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA′ elements. J. Bacteriol. 175:3430-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ottemann, K. M., W. Xiao, Y. K. Shin, and D. E. Koshland, Jr. 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285:1751-1754. [DOI] [PubMed] [Google Scholar]
- 37.Park, H., and M. Inouye. 1997. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J. Bacteriol. 179:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, H., S. K. Saha, and M. Inouye. 1998. Two-domain reconstitution of a functional protein histidine kinase. Proc. Natl. Acad. Sci. USA 95:6728-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 43.Stultz, C. M., R. Nambudripad, R. H. Lathrop, and J. V. White. 1997. Predicting protein structure with probabilistic models, p. 447-506. In N. Allewell and C. Woodward (ed.), Protein structural biology in bio-medical research, vol. 22B. JAI Press, Greenwich, Conn. [Google Scholar]
- 44.Stultz, C. M., J. V. White, and T. F. Smith. 1993. Structural analysis based on state-space modeling. Protein Sci. 2:305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, B. L. 1983. Role of proton motive force in sensory transduction in bacteria. Annu. Rev. Microbiol. 37:551-573. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, B. L., and M. S. Johnson. 1998. Rewiring a receptor: negative output from positive input. FEBS Lett. 425:377-381. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, B. L., M. S. Johnson, and K. J. Watts. 2003. Signal transduction in prokaryotic PAS domains, p. 15-50. In S. T. Crews (ed.), PAS proteins: regulators and sensors of development and physiology. Kluwer Academic Publishers, Norwell, Mass.
- 48.Taylor, B. L., A. Rebbapragada, and M. S. Johnson. 2001. The FAD-PAS domain as a sensor for behavioral responses in Escherichia coli. Antioxid. Redox Signal. 3:867-879. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, B. L., I. B. Zhulin, and M. S. Johnson. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 50.Tokishita, S., A. Kojima, and T. Mizuno. 1992. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of membrane-located protein kinase, EnvZ. J. Biochem. (Tokyo) 111:707-713. [DOI] [PubMed] [Google Scholar]
- 51.Utsumi, R., R. E. Brissette, A. Rampersaud, S. A. Forst, K. Oosawa, and M. Inouye. 1989. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science 245:1246-1249. [DOI] [PubMed] [Google Scholar]
- 52.Wanner, B. L., and W. W. Metcalf. 1992. Molecular genetic studies of a 10.9-kb operon in Escherichia coli for phosphonate uptake and biodegradation. FEMS Microbiol. Lett. 79:133-139. [DOI] [PubMed] [Google Scholar]
- 53.Ward, S. M., A. Delgado, R. P. Gunsalus, and M. D. Manson. 2002. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol. Microbiol. 44:709-719. [DOI] [PubMed] [Google Scholar]
- 54.Watts, K. J., Q. Ma, M. S. Johnson, and B. L. Taylor. 2004. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J. Bacteriol. 186:7440-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weis, R. M., T. Hirai, A. Chalah, M. Kessel, P. J. Peters, and S. Subramaniam. 2003. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 185:3636-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White, J. V., C. M. Stultz, and T. F. Smith. 1994. Protein classification by stochastic modeling and optimal filtering of amino-acid sequences. Math. Biosci. 119:35-75. [DOI] [PubMed] [Google Scholar]
- 58.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 59.Wolfe, A. J., M. P. Conley, T. J. Kramer, and H. C. Berg. 1987. Reconstitution of signaling in bacterial chemotaxis. J. Bacteriol. 169:1878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhulin, I. B., V. A. Bespalov, M. S. Johnson, and B. L. Taylor. 1996. Oxygen taxis and proton motive force in Azospirillum brasilense. J. Bacteriol. 178:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]