Abstract
Rhizobia form a symbiotic relationship with plants of the legume family to produce nitrogen-fixing root nodules under nitrogen-limiting conditions. We have examined the importance of glutathione (GSH) during free-living growth and symbiosis of Sinorhizobium meliloti. An S. meliloti mutant strain (SmgshA) which is unable to synthesize GSH due to a gene disruption in gshA, encoding the enzyme for the first step in the biosynthesis of GSH, was unable to grow under nonstress conditions, precluding any nodulation. In contrast, an S. meliloti strain (SmgshB) with gshB, encoding the enzyme involved in the second step in GSH synthesis, deleted was able to grow, indicating that γ-glutamylcysteine, the dipeptide intermediate, can partially substitute for GSH. However, the SmgshB strain showed a delayed-nodulation phenotype coupled to a 75% reduction in the nitrogen fixation capacity. This phenotype was linked to abnormal nodule development. Both the SmgshA and SmgshB mutant strains exhibited higher catalase activity than the wild-type S. meliloti strain, suggesting that both mutant strains are under oxidative stress. Taken together, these results show that GSH plays a critical role in the growth of S. meliloti and during its interaction with the plant partner.
All aerobic organisms are exposed to reactive oxygen species (ROS), such as the superoxide anion, hydrogen peroxide (H2O2), and the hydroxyl radical, during normal aerobic metabolism or after exposure to stress conditions. ROS can cause irreversible damage to cellular components if they are not rapidly detoxified by antioxidant defense systems (15). To protect themselves against oxidant damage, cells contain effective defense mechanisms, including scavenging enzymes, such as catalases, superoxide dismutases, and glutathione peroxidases, and antioxidants, such as the tripeptide glutathione γ-glutamyl-l-cysteinylglycine (GSH).
GSH is synthesized by a two-step process. In the first step, glutamate and cysteine are conjugated by γ-glutamyl cysteine synthetase (γECS) to form γ-glutamyl cysteine (γEC). In a second step, glycine is added to γEC to form GSH in a reaction catalyzed by glutathione synthetase (GSHS). The redox-active sulfhydryl group of GSH protects cells from ROS by directly scavenging free radicals and acting as a cofactor for antioxidant enzymes such as glutathione peroxidases. GSH oxidized in this manner forms GSH disulfide, which is recycled back to its reduced form by glutathione reductase.
Initial studies of the requirement for GSH and its role in protection against oxidative stress were performed on bacteria. E. coli mutants devoid of GSH were isolated by means of a transposon insertion in gshA, which encodes γECS (13). From these studies, it was found that not only was GSH nonessential under normal growth conditions, but also, rather surprisingly, these mutants were unaffected in their resistance to compounds which cause oxidative damage. These results contrast with the situation found in mammalian cells, where GSH deficiency results in cellular damage, suggesting an important difference in GSH function between prokaryotes and eukaryotes. Yeast mutants lacking GSH through transposon insertion into the gsh1 gene, which encodes γECS, showed a severely retarded growth phenotype and enhanced sensitivity to oxidative stress (10, 11).
Sinorhizobium meliloti is an aerobic gram-negative bacterium from the rhizosphere able to infect the roots of Medicago species and develop symbiosis. During this process, an exchange of recognition signals occurs between the two partners (20), leading to the development of infection threads and finally to the formation of the root nodule (23). Inside the root nodule, the bacteria differentiate into their symbiotic forms, which are capable of reducing atmospheric nitrogen to ammonium. Soil rhizobia are constantly challenged by a variety of stresses in their natural environments, including elevated temperature, acidity, and osmotic and oxidative shock. Moreover, it has been suggested that during the symbiotic interaction, the bacteria are potentially exposed to oxidative stress during the oxidative burst that follows the infection process (28). In addition, the functional nodule is prone to high levels of ROS due to the high rate of respiration necessary to supply energy required by the nitrogen reduction process. Thus, it appears to be important to understand the roles played by the various antioxidants in both free-living and symbiotic rhizobia.
In this study, we investigated the requirement for GSH in free-living Sinorhizobium meliloti and the effects of depletion of GSH on the ability of the bacteria to produce nitrogen-fixing nodules with their symbiotic partner, Medicago sativa.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C, and S. meliloti strains were grown in LB medium supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LBMC) at 30°C. Antibiotics were added as required at the following concentrations (micrograms per milliliter): ampicillin, 100; chloramphenicol, 10; kanamycin, 100; neomycin, 100; spectinomycin, 100; tetracycline, 10; and gentamicin, 10. For growth curve analysis, high-performance liquid chromatography (HPLC) analysis, and catalase activity, cultures of S. meliloti grown on LBMC were inoculated at an initial optical density at 600 nm (OD600) of 0.1 onto M9 minimal medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| RCR2011 | SU47; wild type; Nod+ Fix+ | 25 |
| Rm1021 | RCR2011 derivative; Smr | 21 |
| SmgshA | Same as Rm1021 but with gshA insertion mutation; Smr Nmr | This study |
| SmgshB | Same as Rm1021 but with gshB insertion mutation; Smr Tcr | This study |
| E. coli | ||
| DH5α | F−supE44 ΔlacU169(Φ80dlacZDM15) hsdR17(rK−mK+) recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| MT607 | pro-82 thi-1 hsdR17 supE44 recA56 | 5 |
| MT616 | MT607(pRK600) | 5 |
| Plasmids | ||
| pLAFR1 | IncP1 cosmid cloning vector; Tcr | 7 |
| pRK600 | ColE1 replicon with RK2 transfer region; Cmr | 5 |
| pBluescript KS(+) | Derivative of pUC19 with f1(+)oriR; Apr | Stratagene |
| pGEM-T | T vector with f1 oriR; Apr | Promega Corp. |
| pGEM-T easy | T vector with f1 oriR; Apr | Promega Corp. |
| pSUP202 | ColE1; Mob+; Tcr Apr Cmr | 30 |
| pKOK4 | Apr Kmr; pSUP202 derivative; source of Kmr | 18 |
| pBBR1MCS-5 | Derivate of pBBR1-MCS; Gmr | 19 |
| pGEMgshA | pGEM-T, with 800-bp PCR-amplified fragment of gshA | This study |
| pSUPgshA | pSUP202 with 800-bp PCR-amplified fragment of gshA | This study |
| pJHgshA | pSUP202 with 800-bp PCR-amplified fragment of gshA and Kmr from pKOK4 | This study |
| pGEMcgshA | pGEM-T with 1,500-bp PCR-amplified fragment of gshA | This study |
| pgshAc | pBBR1MCS-5 with 1,500-bp PCR-amplified fragment of gshA | This study |
| pGMgshB | pGEM-T with 784-bp PCR-amplified fragment of gshB | This study |
| pCMgshB | pSUP202 with 800-bp PCR-amplified fragment of gshB | This study |
Tc, tetracycline; Sm, streptomycin; Ap, ampicillin; Km, kanamycine; Cm, chloramphenicol; Sp, spectinomycin; Gm, gentamycine, Nm, neomycin.
General molecular biology techniques.
Molecular cloning techniques, restriction analysis, ligation, and transformation were performed essentially according to the standard methods (26). DNA probes were labeled using the Prime-a-gene random-priming system (Promega, Charbonnières, France) and [α-32P]dCTP (Amersham Biosciences, Orsay, France). Plasmids and DNA fragments were purified using Nucleospin (Macherey Nagel EURL, Hoert, France) and QIAGEN (Courtaboeuf, France) kits.
Construction of the SmgshA and SmgshB strains and of the pgshAc plasmid.
S. meliloti Rm1021 gshA and gshB DNA fragments were amplified by PCR using oligonucleotides designed against the predicted gshA sequence (5′-GTTCCTGATCCGCACCGAAC-3′ and 5′-GACGGACGTGAAAGAGG-3′) and the gshB sequence (5′-ACCATGTTTCGGGCATCACCATT-3′ and 5′-ATGCATGTTGGAGCGAGAATCG-3′). The 800- (gshA) and 784-bp (gshB) fragments amplified were cloned into pGEM-T and pGEM-T Easy (Promega), respectively. The gshA DNA fragment was subsequently subcloned into the vector pSUP202 using BamHI-HindIII sites. A kanamycin resistance cassette from pKOK4 was added in the PstI site of the gshA DNA fragment. The resulting recombinant plasmid was named pJHgshA. The gshB DNA fragment was subcloned into the vector pSUP202 using an EcoRI site. The resulting recombinant plasmid was named pCMgshB. The recombinant suicide plasmids were transferred to the Rm1021 recipient strain by triparental mating using the MT616 E. coli strain as a helper, as described previously (9). Simple recombinant clones were selected on LBMC media containing spectinomycin and neomycin to select the gshA mutant strain and spectinomycin and tetracycline to select the gshB mutant strain. Recombination at the correct location was checked by Southern hybridization. One S. meliloti mutant clone in which the gshA gene was interrupted and one S. meliloti mutant clone in which the gshB gene was interrupted were selected for further analysis. These two stains were named SmgshA and SmgshB, respectively. The S. meliloti Rm1021 gshA gene containing the promoter region was amplified by PCR using oligonucleotides designed against the predicted gshA sequence (5′-AAGGATCCCCGCGGCGCTCCGTCCG-3′ and 5′-AAAAGCTTGGCGATGAGCCGTCCATCC-3′). The 1,500-bp amplified DNA fragment was cloned into pGEM-T and sequenced. The gshA gene was subsequently subcloned into the vector pBBR1MCS-5 using BamHI and HindIII sites (underlined in the oligonucleotide sequences). The resulting recombinant plasmid was named pgshAc. The recombinant plasmid was transferred to the SmgshA recipient strain by triparental mating using the MT616 E. coli strain as a helper. SmgshA(pgshAc) exconjugants were selected on LBMC medium containing neomycin, spectinomycin, and gentamicin. The strains and plasmids used during the study are shown in Table 1.
HPLC analysis of low-molecular-weight thiols.
Bacterial cells (109) were centrifuged at 5,000 × g for 10 min. The resulting pellet was resuspended in 150 μl of 0.1 N HCl, derivatized using monochlorobimane, and quantified following separation on reverse-phase HPLC according to the protocol described previously (6). Commercial GSH (Sigma, Saint Quentin Fallavier, France) and γEC (Promochem, Molsheim, France) were used as standards.
Catalase activity and in-gel assays.
Crude bacterial extracts were prepared from free-living bacteria by sonication in a phosphate buffer as described previously (14). Catalase was visualized on nondenaturing polyacrylamide gels by inhibition of diaminobenzidine oxidation (3). Total catalase activity was measured spectrophotometrically by following the decomposition of H2O2 at 240 nm (17). The protein concentration was determined with the Bio-Rad (Marnes-la-Coquette, France) protein assay kit using bovine serum albumin as a standard.
Plant nodulation assays.
M. sativa L. var. Europe (alfalfa) was used as a host plant to test nodulation of the S. meliloti strains. Surface-sterilized germinating seedlings were grown in test tubes on modified Fahraeus medium (2) containing 1.5% agarose. Five days after germination, the roots were inoculated with 200 μl of the appropriate strain of bacterial suspension culture/plant. The bacterial suspension culture was prepared from overnight LBMC cultures of the bacteria (containing the appropriate antibiotics), washed three times, and diluted to the desired concentration (5 × 107/ml) with sterile distilled water. The nitrogen fixation capacity of the nodules was determined by the acetylene reduction assay as described previously (14).
Microscopy studies.
For microscopy, nodules were harvested 4 weeks after inoculation. The nodules were fixed, postfixed, and embedded as previously described (27). Sections were performed with a diamond knife in a Reichert Ultracut Ultramicrotome (Leica, Rueil-Malmaison, France). After being stained with uranyl acetate and Reynold's lead citrate, ultrathin sections were examined in a transmission electron microscope (CM12; Philipps, Cambridge, United Kingdom).
Statistical analysis.
All the data presented for thiol concentrations, catalase activity, acetylene reduction assays, and nodule numbers are given as means with standard errors. The significance of the results was assessed using Student's t test.
RESULTS
The GSH synthesis pathway is essential for S. meliloti growth. In a first step in studying the role of GSH in the growth of S. meliloti, in silico analysis of the bacterial genome (8) was performed to find the genes involved in GSH synthesis. The deduced amino acid sequence of the open reading frame SMc00825 showed ∼75% identity with putative Rhizobiales γECS (Agrobacterium tumefaciens, Brucella melitensis biovar Suis1330, and Mesorhizobium sp. strain BNC1) and ∼55% identity with plant γECS (Arabidopsis thaliana and Medicago truncatula). Similarly, the translational product of the open reading frame SMc00419 was found to display 85% protein identity with the GSHS from Rhizobium tropici (AAL91575). In order to investigate the importance of endogenous GSH production by S. meliloti in free-living growth and during interaction with the host plant, alfalfa, the γECS- and GSHS-defective mutant strains (SmgshA and SmgshB, respectively) derived from wild-type (wt) S. meliloti Rm1021 were constructed (see Materials and Methods).
The GSH and γEC contents of the wild type and the glutathione-deficient mutants were determined during growth on M9 minimal medium (Table 2). γEC was not detectable in wild-type Rm1021 under our assay conditions. The GSH concentration was found to be higher during the exponential growth phase and early stationary phase, reaching a maximum of 7 nmol/109 cells after 24 h of growth, which was followed by a decrease in GSH content. As expected, both γEC and GSH were found to be undetectable in the SmgshA strain. Analysis of the thiol content of the mutant strain SmgshA expressing the gshA gene constitutively, SmgshA(pgshAc), revealed the accumulation of both γEC and GSH, which showed growth phase-dependent accumulation similar to that found with the wt Rm1021. The presence of a multicopy plasmid, however, resulted in concentrations of GSH that were twice those detected in the wt Rm1021. GSH was undetectable in the SmgshB mutant, which showed, in contrast, a marked accumulation of γEC (up to 19 nmol/109 cells).
TABLE 2.
Growth-dependent accumulation of γEC and GSH in S. meliloti
| Strains | Accumulationa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 12 h
|
24 h
|
48 h
|
72 h
|
|||||
| γ-EC | GSH | γ-EC | GSH | γ-EC | GSH | γ-EC | GSH | |
| 1021 | —b | 6 ± 1 | — | 7 ± 1 | — | 7 ± 1 | — | 3 ± 0.3 |
| SmgshA | — | — | — | — | — | — | — | — |
| SmgshB | 17 ± 3 | — | 19 ± 2 | — | 19 ± 2 | — | 14 ± 3 | — |
| SmgshA (pgshAc) | 0.4 ± 0.2 | 14 ± 2 | 0.5 ± 0.1 | 14 ± 3 | 0.5 ± 0.1 | 14 ± 3 | — | 8 ± 2 |
γEC and GSH were measured by HPLC as described in Materials and Methods. The data represent the means of three experiments ± standard errors.
—, not detected.
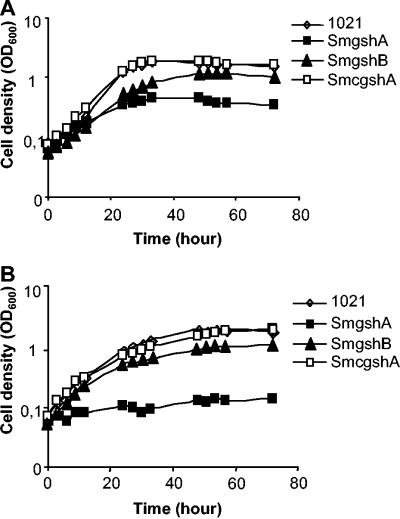
The growth curves of free-living wt S. meliloti Rm1021 and mutant strains SmgshA and SmgshB on M9 following overnight culture on LBMC are shown in Fig. 1A. In comparison to the wt Rm1021, both mutant strains showed a reduced growth rate, with that of SmgshA being the most severely affected. The growth of the wt Rm1021 and the mutant strains on M9 following overnight adaptation to M9 is shown in Fig. 1B. Although the exponential growth phases of all cultures were slightly slower than those shown in Fig. 1A, the most striking result was the complete growth arrest of the SmgshA mutant. Under both culture conditions, growth of the SmgshA strain was fully restored by complementation with a plasmid containing the S. meliloti γECS gene (pgshAc) or by the addition of 1 mM GSH or γEC to the growth medium (data not shown). In parallel, growth of the SmgshB strain was restored by the addition of 100 μM GSH to the growth medium (data not shown). Taken together, these results show that the GSH synthesis pathway is essential for the free-living growth of S. meliloti. However, γEC, which is present at a high concentration in the SmgshB strain, is able to complement the lack of GSH.
FIG. 1.
Growth curves of Sinorhizobium meliloti strains. At time zero, rhizobial cells were diluted with minimal medium (M9) to an OD600 of 0.1. The growth of wild-type Rm1021, mutant strain SmgshA, mutant strain SmgshB, and mutant strain SmgshA complemented with pgshAc was monitored by measuring the OD600. (A) Growth curves of free-living S. meliloti strains on M9 medium following overnight culture on LBMC. (B) Growth curves of free-living S. meliloti strains on M9 medium following overnight adaptation to M9. The data are the means from three experiments.
Since it has been proposed that the oxidized/reduced ratio of GSH in the cells is essential for proper scavenging of ROS, we examined whether low levels of thiol resulted in a change in the GSH redox state. Due to the severely retarded growth phenotype following overnight adaptation to M9, all experiments were performed on cultures grown overnight on LBMC and subsequently subcultured into M9. Both reduced and oxidized GSH and γEC pools were determined during the exponential growth phase. The percentage of the oxidized γEC pool was found to be 34% ± 4% in the SmgshB mutant strain, whereas the oxidized GSH and γEC pools represented just 10% ± 6% in the wt Rm1021 and the complemented SmgshA(pgshAc) strains. This suggests that the SmgshB strain is under oxidative stress.
Catalase activity is modulated by GSH deficiency in S. meliloti.
It has been demonstrated that catalases are induced by oxidative stress in S. meliloti (14, 29), and therefore, we concluded that catalase activity represents a good physiological marker to assess whether the bacteria are under oxidative stress. It had been reported that three distinct hydroperoxidases of S. meliloti are involved in the dismutation of H2O2, two monofunctional catalases (KatA and KatC) and one bifunctional catalase-peroxidase (KatB). Each of these catalases has a distinctive expression profile during growth of the bacteria under free-living conditions and in response to particular stresses (14, 16, 29). katA is expressed during the exponential growth phase and is induced by H2O2. katB encodes a housekeeping catalase-peroxidase that is expressed constitutively. katC is expressed during the stationary phase and is induced by environmental stresses.
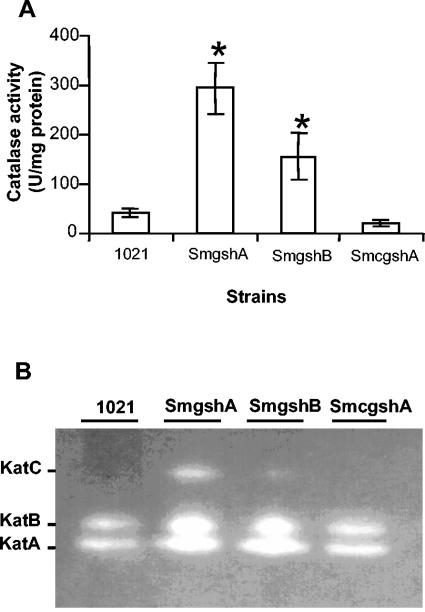
In order to examine whether catalase activity was higher in mutant strains than in the wt Rm1021 strain, the level of total catalase activity was determined in cells during the late exponential growth phase (Fig. 2A). The mutant SmgshA was found to have a sevenfold-higher total catalase activity than the wt Rm1021. The total catalase activity of the SmgshB mutant also exhibited a three- to fourfold increase compared to that of the wt Rm1021. These results showed that catalase activity is enhanced in GSH-deficient mutant strains. In order to extend this analysis, we evaluated the effect of GSH deficiency on the catalase activity profile by using catalase activity gel (Fig. 2B). As previously reported (29), only KatA and KatB are expressed in S. meliloti during the late exponential phase. An increase in KatA and KatB, and in addition the detection of KatC, could be observed in the SmgshA strain compared to wt Rm1021. The SmgshB strain showed an intermediate phenotype in which the activities of the three catalases were found to be high, albeit at lower levels than in the SmgshA strain. Complementation of the SmgshA strain by the plasmid pgshAc resulted in the recovery of a wild-type phenotype in terms of the total catalase activity and catalase profile. The higher level of oxidation of the γEC pool in the SmgshB strain, which correlated with the higher level of catalase activity observed in strains SmgshA and SmgshB, demonstrates that GSH deficiency causes oxidative stress in S. meliloti.
FIG. 2.
Catalase activities of S. meliloti strains. At time zero, rhizobial cells were diluted with M9 medium to an OD600 of 0.1. Cell extracts were prepared from the Rm1021 (1021), SmgshA, and SmgshB strains and the SmgshA strain complemented with pgshAc (SmcgshA) after 24 h. (A) Total catalase activities for the different strains. The data are the means (± standard errors) of triplicates from three experiments.* indicates significant difference (P < 0.05). (B) Catalase activity patterns. Proteins (20 μg) were submitted to electrophoresis on a native polyacrylamide gel (7%) that had been stained for catalase activity. The positions of KatA, KatB, and KatC are indicated according to Sigaud et al. (29).
Glutathione deficiency strongly affects the nodulation process.
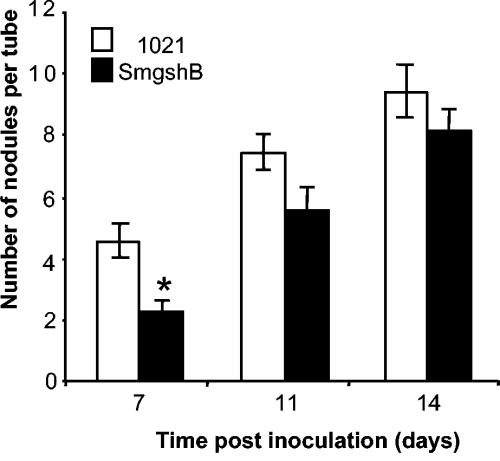
In order to test the implication of GSH in the symbiotic process, the capacities of the SmgshA and SmgshB mutant strains to nodulate and produce nitrogen-fixing nodules were examined by alfalfa inoculation experiments. No nodules were produced by plants inoculated with the SmgshA mutant strain. The nodulation efficiencies following inoculation with Rm1021 and SmgshB are shown in Fig. 3. Seven days postinoculation, significantly fewer nodules were observed on plants inoculated with S. meliloti strain SmgshB than on those inoculated with strain Rm1021. However, this difference was no longer significant 11 and 14 days postinoculation, suggesting that although the nodulation capacity itself was unaffected, the timing of nodule formation was delayed in the SmgshB mutant.
FIG. 3.
Plant nodulation efficiency. Plants were inoculated with the S. meliloti Rm1021 (1021) and SmgshB strains. The total numbers of nodules per tube were assessed 7, 11, and 14 days postinoculation. Each tube contained three plants. The data are the means (± standard errors) of 48 tubes from two experiments; * indicates a significant difference (P < 0.05).
To test the functionality of nodules, their nitrogen-fixing capacities were also determined 4 weeks after inoculation with the Rm1021 and SmgshB strains. Using the acetylene reduction assay, it was found that the nitrogen fixation capacity was severely affected in the SmgshB mutant, with a reduction from 11.0 ± 2 nmol of C2H4/mg of nodules/h in wt Rm1021 to 2.7 ± 0.7 nmol of C2H4/mg of nodules/h in the mutant.
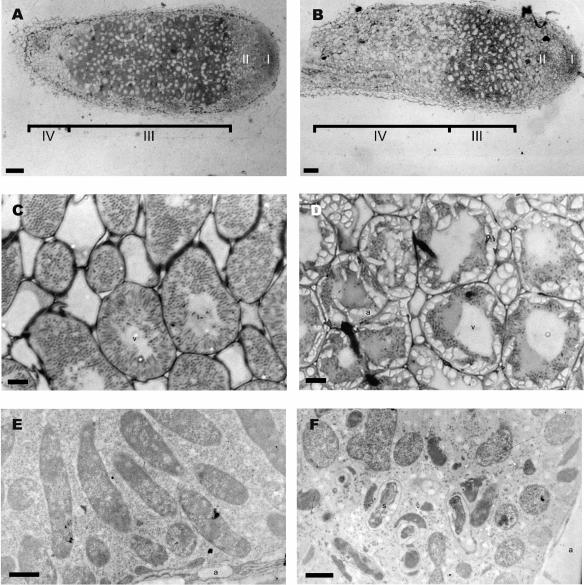
Microscopic analysis of the nodules was performed to visualize the nodule structure (Fig. 4). All the steps in cell differentiation were observed in nodules after infection with Rm1021 (31). The different zones observed in a wt Rm1021 nodule are presented in Fig. 4A. The meristematic zone I was bacterium free. Bacteria released from infection threads in plant cells were surrounded by a peribacteroid membrane in the distal infection zone II (type 1 bacteroids). Bacteroids elongated without dividing in the proximal infection zone II (type 2 bacteroids). In interzone II-III (IZ), bacteroids stopped their elongation and displayed cytoplasmic heterogeneity (type 3 bacteroids). In zone III, the bacteroids showed higher cytoplasmic heterogeneity and corresponded to nitrogen-fixing rhizobia (type 4 bacteroids) (Fig. 4C and E). Finally, symbionts degenerated in the senescent zone IV. In contrast, SmgshB nodules exhibited a large senescent zone IV (Fig. 4B). The fixing zone III was not clearly defined, and amyloplast-rich cells, which are characteristic of the IZ, appeared to be adjacent to senescent zone IV (Fig. 4D). Ultrastructural studies (Fig. 4F) showed that SmgshB bacteria were released but rapidly underwent senescence, often before differentiating into fixing bacteroids. Both nonfixing bacteroids (types 1 to 3) and senescent bacteroids could be observed in the same plant cell (Fig. 4F). Taken together, these results demonstrated that S. meliloti requires GSH in order to achieve an efficient symbiotic interaction with alfalfa, particularly in preventing early senescence.
FIG. 4.
Structure and ultrastructure of 4-week-old nodules. Nodules were incited by Rm1021 (A, C, and E) and SmgshB (B, D, and F) strains. The meristematic (I), infection (II), fixing (III), and senescent (IV) zones and II-III IZ are represented in A and B. a, amyloplast, and s, senescing bacteroid. Scale bars = 100 (A and B), 10 (C and D) and 1 μm (E and F).
DISCUSSION
During this study, the synthesis of glutathione and the importance of this low-molecular-weight thiol were examined in free-living and symbiotic S. meliloti. The functions of gshA and gshB were confirmed by HPLC analysis of the low-molecular-weight thiol contents of the strains with mutations in the gshA and gshB genes, respectively. A mutant strain defective in synthesis of γECS (strain SmgshA) does not accumulate γEC and GSH, and a strain lacking glutathione synthetase (SmgshB strain) accumulates γEC. Therefore, there are no other genes coding for functions able to replace those sequences altered in our mutants. In order to test the importance of the GSH synthesis pathway in S. meliloti, the growth, antioxidant status, and nodulation capacities of the mutant strains SmgshA and SmgshB were compared to those of the wild-type Rm1021. Our study demonstrates that the growth of an S. meliloti strain with the gshA gene mutated was severely impaired, whereas only a slightly reduced growth phenotype was observed in the SmgshB strain. These results indicate that γEC is able to compensate for the absence of glutathione during growth under free-living conditions, and they are remarkably different from the results observed in E. coli. Mutation of gshA and the subsequent deficiency in glutathione or γEC did not modify the growth rate of E. coli, indicating a nonessential role for the GSH synthesis pathway under nonstress conditions (13). In contrast, it was demonstrated that a yeast mutant with γECS deleted was unable to grow, while mutation in GSHS resulted in an impaired growth phenotype (10, 11). Thus, our study demonstrated that S. meliloti behaves in a manner similar to yeast, while it diverges from the behavior of E. coli.
Hydroperoxidases are implicated in the detoxification of H2O2. Our studies showed that a lack of glutathione is associated with an increase in total catalase activity. Gel assays revealed that this increase is due to augmentation in KatA and KatB and the appearance of KatC. The fact that a decrease in GSH content induces both KatA and KatC activities suggests that the mutant strains SmgshA and SmgshB are facing stress conditions. Moreover, the highly oxidized γEC pool present in the SmgshB strain suggests that the threefold-higher γEC concentration does not suffice for the maintenance of the redox state found in the Rm1021 strain. Previous studies have shown that although mutation in the gshA gene of E. coli did not result in a growth phenotype, the bacteria were found to be more sensitive to H2O2 and had a higher level of catalase activity (22). The authors suggested that glutathione may play a role in the regulation of catalases, possibly through its regulation of intracellular H2O2 or through its interaction with redox-sensitive sites in the OxyR regulatory protein. GSH-deficient yeast mutants, as well as R. tropici gshB mutants, showed dramatic sensitivity to H2O2 (12, 24). Altogether, these data suggest that GSH and catalases provide overlapping defenses for protection against H2O2 in both prokaryotic and eukaryotic cells.
The nodulation efficiencies of S. meliloti glutathione pathway-deficient mutants were investigated by inoculating M. sativa plants with the SmgshA and SmgshB strains. Under our experimental procedures, we found no evidence that the mutant SmgshA was capable of nodulating M. sativa. Since the growth of the mutant rhizobium was severely affected, it seemed logical to expect that its nodulation ability would also be negatively affected. However, the stage of early interaction between the mutant strain and alfalfa, such as attachment, curling, and infection thread formation, that might be affected remains to be determined. The emergence of nodules on M. sativa following inoculation with the mutant strain SmgshB was delayed in comparison to wt Rm1021, although the final nodule number was unaffected. Similarly, the number of nodules on Phaseolus vulgaris and Leucaena leucocephala 4 weeks postinoculation with the gshB-deficient strain was found to be similar to that on plants inoculated with the wild-type strain (24). A highly significant feature of our study was the large reduction in the nitrogen fixation capacity of nodules. This phenotype is consistent with the microscopy studies, in which early senescence was observed in the mutant strain SmgshB. The studies do suggest that redox balance is an important factor in nodulation. Superoxide anions and H2O2 are produced during the initial stages of the plant-microbial interaction (28). To protect itself from this oxidative burst, S. meliloti possesses three catalases and two superoxide dismutases (SodA and SodC). Transcriptome analysis of S. meliloti has shown that sodC is induced during the infection process (1), and Sod activity appears to play an important role during the early steps of symbiosis (27). In addition, catalases present a differential pattern of expression during nodule development. katB is constitutively expressed in the nodules, katA is expressed in zone III, and katC is expressed in zones II and IV (16). While single null mutants of katA, katB, and katC have a wild-type nodulation phenotype, katB katC and katA katC double mutants have reduced infection efficiencies and decreased nitrogen fixation capacities, showing that degradation of H2O2 in planta is essential to allow an efficient nodulation process (14, 16, 29). The reduction of nitrogen fixation capacity observed in the gshB mutant strain could be associated with an alteration of the bacterial antioxidant defense, which promotes early senescence. It could also be suggested that the γEC produced in the gshB strain may not be able to fully substitute for the specific GSH function during cell differentiation occurring in the symbiotic process. This deficiency may cause early senescence of nodules, which is reflected by lower nitrogenase activity.
The requirement for rhizobia in the GSH synthesis pathway during growth in free-living conditions seems to be analogous to that in yeast and higher eukaryotes and unlike the situation in E. coli. This dissimilarity in physiology may be paralleled by phylogenetic differences between S. meliloti and E. coli γECS. Indeed, the sequences of gshA from γ-proteobacteria (E. coli) and α-proteobacteria (S. meliloti) fall into two distinct groups (4). The latter group consists primarily of α-proteobacteria, such as Mesorhizobium loti, Agrobacterium tumefaciens, and S. meliloti, and flowering plants, such as A. thaliana, M. truncatula, and Glycine max. The different influences of GSH deficiency on cell growth observed in S. meliloti and E. coli may arise from divergent evolutionary processes of these two bacterial genera, which is demonstrated by γECS gene evolution.
In S. meliloti, it appears that GSH may be replaced by γEC under free-living conditions but not during the symbiotic association with M. sativa. This differential behavior may also be linked to the evolution of the GSH synthesis pathway. Copely and Dhillon (4) suggested that gshB was recruited later during evolution than gshA. In this scenario, bacterial γEC may have served multiple functions, including those carried on by GSH before the recruitment of the gshB gene. However, our study also shows that glutathione itself is important for optimal nitrogen fixation, showing that endogenous γEC could not fully restore some essential functions of GSH which may have been acquired after the recruitment of the gshB gene.
Taken together, our results indicate that the GSH synthesis pathway plays a crucial role in the growth and symbiotic capacity of S. meliloti. The transcriptome analysis of the SmgshA and SmgshB strains should bring significant insights into the different roles of γEC and GSH.
REFERENCES
- 1.Ampe, F., E. Kiss, F. Sabourdy, and J. Batut. 2003. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 4:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boisson-Dernier, A., M. Chabaud, F. Garcia, G. Bécard, C. Rosenberg, and D. G. Barker. 2001. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant-Microbe Interact. 14:695-700. [DOI] [PubMed] [Google Scholar]
- 3.Clare, D. A., M. N. Duong, D. Darr, F. Archibald, and I. Fridovich. 1984. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal. Biochem. 140:532-537. [DOI] [PubMed] [Google Scholar]
- 4.Copely, S. D., and J. K. Dhillon. 2002. Lateral gene transfer and parallel evolution in the history of the glutathione biosynthesis genes. Genome Biol. 3:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frendo, P., D. Gallesi, T. Turnbull, G. Van de Sype, D. Hérouart, and A. Puppo. 1999. Localization of glutathione and homoglutathione in Medicago truncatula is correlated to a differential expression of genes involved in their synthesis. Plant J. 17:215-219. [Google Scholar]
- 7.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 8.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 9.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 10.Grant, C. M., F. H. MacIver, and I. W. Dawes. 1996. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29:511-515. [DOI] [PubMed] [Google Scholar]
- 11.Grant, C. M., F. H. MacIver, and I. W. Dawes. 1997. Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to the accumulation of the dipeptide gamma-glutamylcysteine. Mol. Biol. Cell 8:1699-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant, C. M., G. Perrone, and I. W. Dawes. 1998. Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 253:893-898. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg, J. T., and B. Demple. 1986. Glutathione in Escherichia coli is dispensable for resistance to H2O2 and gamma radiation. J. Bacteriol. 168:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hérouart, D., S. Sigaud, S. Moreau, P. Frendo, D. Touati, and A. Puppo. 1996. Cloning and characterization of the katA gene of Rhizobium meliloti encoding a hydrogen peroxide-inducible catalase. J. Bacteriol. 178:6802-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imlay, J. A. 2003. Pathway of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 16.Jamet, A., S. Sigaud, G. Van de Sype, A. Puppo, and D. Hérouart. 2003. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant-Microbe Interact. 16:217-225. [DOI] [PubMed] [Google Scholar]
- 17.Jones, D. P. 1982. Intercellular catalase function: analysis of the catalytic activity by product formation in isolated liver cells. Arch. Biochem. Biophys. 214:806-814. [DOI] [PubMed] [Google Scholar]
- 18.Kokotek, W., and W. Lotz. 1989. Construction of a lacZ-kanamycin-resistance cassette, useful for site-mutagenesis and as promoter probe. Gene 84:467-471. [DOI] [PubMed] [Google Scholar]
- 19.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBRMCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 20.Long, S. R. 1996. Rhizobium symbiosis: nod factors in perspective. Plant Cell 8:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niel, C., J. B. Guillaume, and M. Bechet. 1977. Mise en evidence de deux enzymes présentant une activité β-galactosidase chez Rhizobium meliloti. Can. J. Microbiol. 23:1178-1181. [PubMed] [Google Scholar]
- 22.Oktyabrsky, O. N., G. V. Smirnova, and N. G. Muzyka. 2001. Role of glutathione in regulation of hydroperoxidase 1 in growing Escherichia coli. Free Radic. Biol. Med. 31:250-255. [DOI] [PubMed] [Google Scholar]
- 23.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riccillo, P. M., C. I. Muglia, F. J. de Bruijn, A. J. Roe, I. R. Booth, and O. M. Aguilar. 2000. Glutathione is involved in environmental stress responses in Rhizobium tropici, including acid tolerance. J. Bacteriol. 182:1748-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg, C., P. Boistard, J. Denarie, and F. Casse-Delbart. 1981. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol. Gen. Genet. 184:326-333. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press Cold Spring Harbor, N.Y.
- 27.Santos, R., D. Hérouart, A. Puppo, and D. Touati. 2000. Critical protective role of bacterial superoxide dismutase in Rhizobium-legume symbiosis. Mol. Microbiol. 38:750-759. [DOI] [PubMed] [Google Scholar]
- 28.Santos, R., D. Hérouart, S. Sigaud, D. Touati, and A. Puppo. 2001. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 14:86-89. [DOI] [PubMed] [Google Scholar]
- 29.Sigaud, S., V. Becquet, P. Frendo, A. Puppo, and D. Hérouart. 1999. Differential regulation of two divergent Sinorhizobium meliloti genes for HPII-like catalases during free-living growth and protective role of both catalases during symbiosis. J. Bacteriol. 181:2634-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Pülher. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 31.Vasse, J., F. de Billy, S. Camut, and G. Truchet. 1990. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]