Abstract
Carbohydrate polymers drive microbial diversity in the human gut microbiota. It is unclear, however, whether bacterial consortia or single organisms are required to depolymerize highly complex glycans. Here we show that the gut bacterium Bacteroides thetaiotaomicron utilizes the most structurally complex glycan known; the plant pectic polysaccharide rhamnogalacturonan-II, cleaving all but one of its 21 distinct glycosidic linkages. We show that rhamnogalacturonan-II side-chain and backbone deconstruction are coordinated, to overcome steric constraints, and that degradation reveals previously undiscovered enzyme families and novel catalytic activities. The degradome informs revision of the current structural model of RG-II and highlights how individual gut bacteria orchestrate manifold enzymes to metabolize the most challenging glycans in the human diet.
Plants containing rhamnogalacturonan-II (RG-II) have been consumed since the archaic humans1. This branched pectic-polysaccharide is the most complex glycan known containing 13 different sugars and 21 distinct glycosidic linkages (Fig. 1)2. The abundance of RG-II in red wine and other processed pectins3 reflects its recalcitrance to degradation. The polysaccharide, however, is degradation as it does not accumulate in the environment, but the organisms that utilize RG-II and the mechanism of depolymerisation remain opaque. Dietary glycans are the predominant nutrients available to the human gut microbiota (HGM)4–6 and are major drivers in defining its community structure7. As RG-II has been a component of the human diet over hundreds of thousands of years we hypothesise that the glycan exerts a selection pressure on the HGM leading to the evolution of organisms that degrade the pectic carbohydrate, consistent with initial studies suggesting that Bacteroides thetaiotaomicron (HGM bacterium) can utilize at least components of the glycan8. To test this hypothesis the mechanism by which B. thetaiotaomicron degrades RG-II was elucidated.
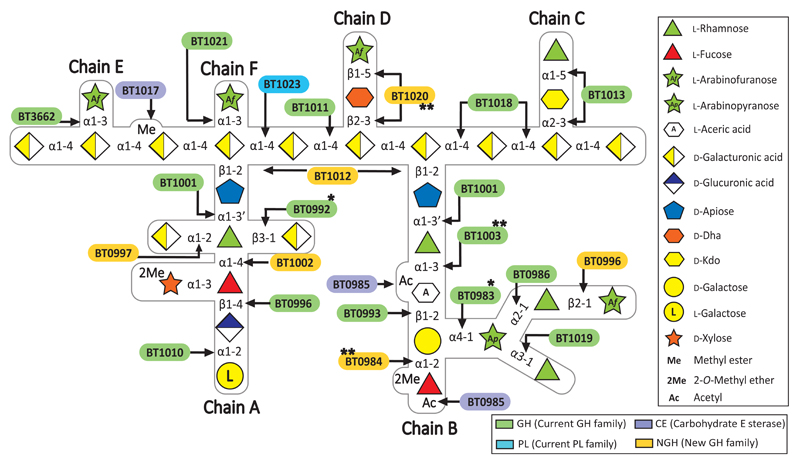
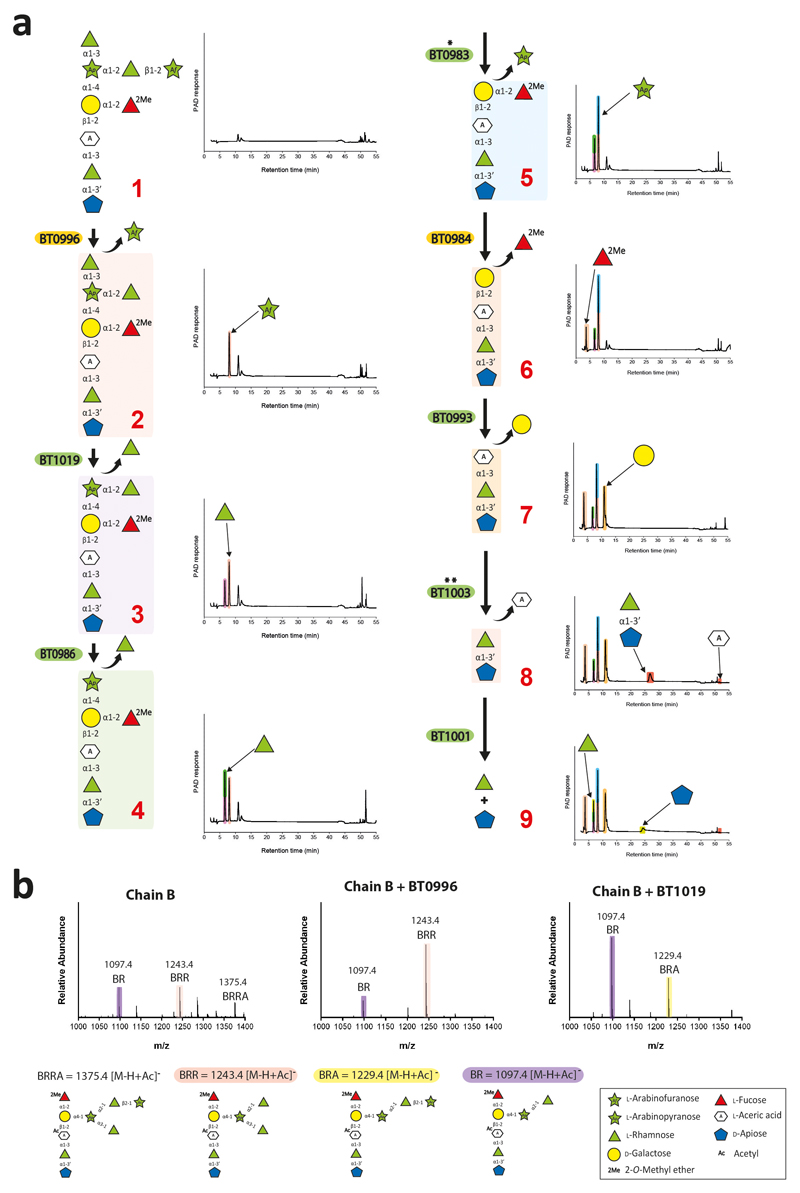
Fig. 1. Schematic of enzymes and PULs involved in RG-II degradation.
Sugars shown using the Consortium for Functional Glycomics notation25. Enzymes are appropriately colour-coded. * signifies a new activity for a GH family. ** are enzymes with novel activities.
Growth of HGM Bacteroides on RG-II
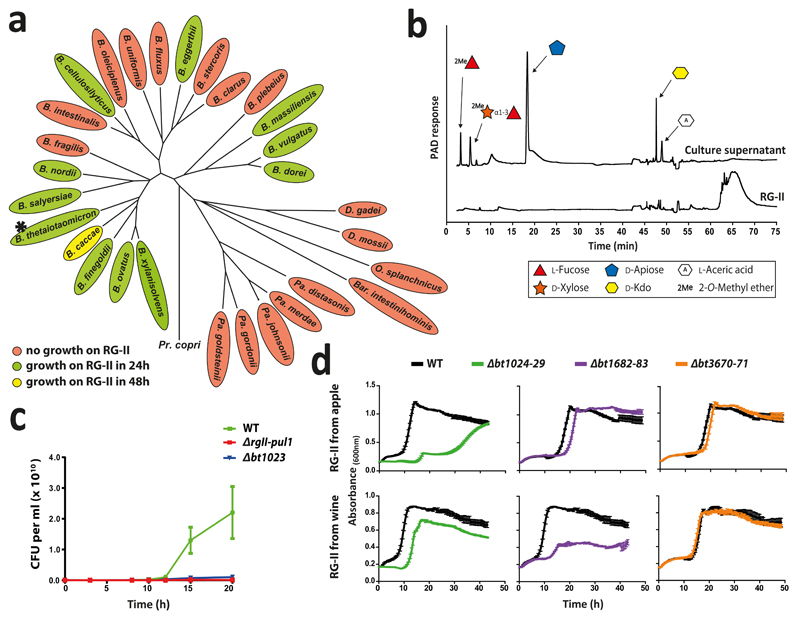
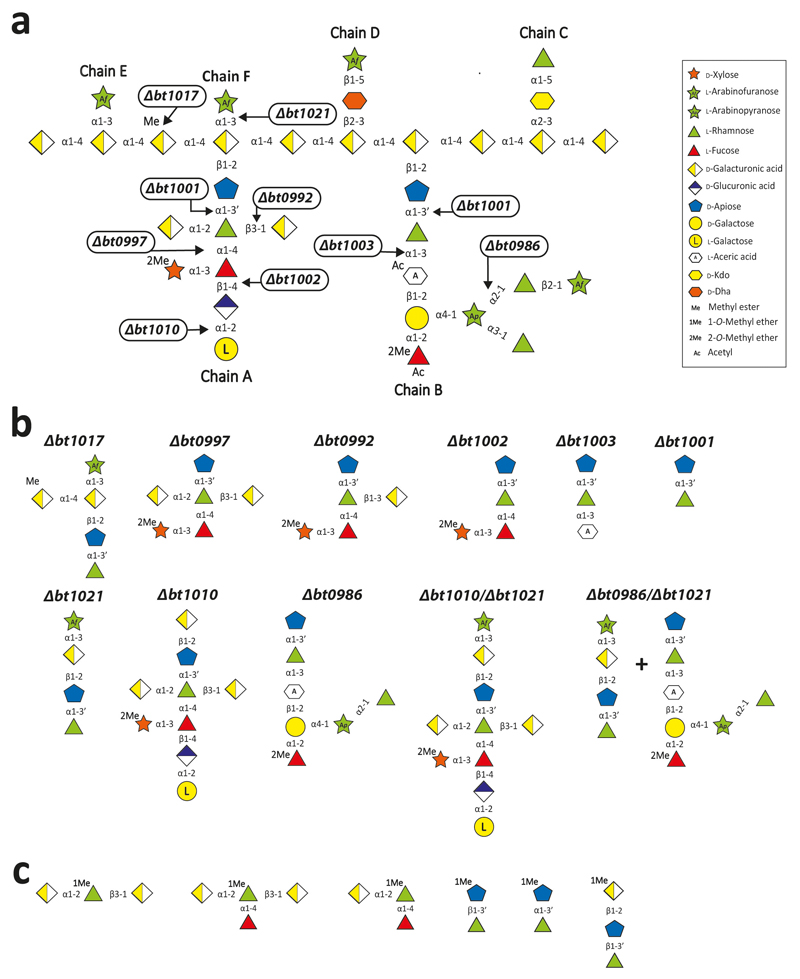
To explore whether single organisms grow on RG-II, the glycan was incubated with Bacteroides species of the HGM. Approximately 30% of these organisms grew on the glycan (Fig. 2a). Only monosaccharides and 2-O-Me-d-Xyl-α1,3-l-Fuc remained in stationary phase cultures (Fig. 2b and Supplementary Table 1) indicating the bacteria cleaved 20 of the 21 distinct glycosidic linkages in the polysaccharide. Transcriptomic analysis of one of these species, B. thetaiotaomicron, showed that RG-II upregulates three polysaccharide utilization loci (PULs), RG-II PUL1-PUL3, (Extended data Fig. 1a)8. Activities of recombinant enzymes encoded by these loci were determined using RG-II or RGII-derived oligosaccharides (Extended Data Fig. 2 and Methods 1.1.2). The data revealed the degradative pathway for each oligosaccharide, leading to a model for RG-II depolymerisation (Fig. 1).
Fig. 2. Growth of Bacteroidetes species on RG-II.
a, Growth of Type strains of HGM Bacteroidetes on RG-II and phylogeny of the organisms (biological replicates, n = 6). b, HPAEC-PAD analysis of stationary-phase culture of B. thetaiotaomicron (* in a). c, wild type B. thetaiotaomicron (WT) and mutants lacking RG-II-PUL1 (Δrg11-pul1) or the polysaccharide lyase BT1023 (Δbt1023) were inoculated into RG-II-media and CFUs determined. d, WT and mutants in which susch-susdh pairs had been deleted were cultured on red wine or apple juice RG-II and growth monitored every 20 min (biological replicates n =6, error bars s.e.m.).
Specificity of enzymes that cleave RG-II
Each glycosidic linkage in RG-II, except for 2-O-Me-d-Xyl-α1,3-l-Fuc, is hydrolysed by a bespoke enzyme (Fig. 1 and 2b). Thus, although the loci encode multiple α-l-rhamnosidases and l-arabinosidases, there is no redundancy in the linkage hydrolysed by these enzymes (Supplementary Tables 2 and 3). This stringent enzymatic specificity likely reflects an apparatus that is tuned to maximise the rate of the degradative process, offsetting the energy required to synthesise such an elaborate catabolic system.
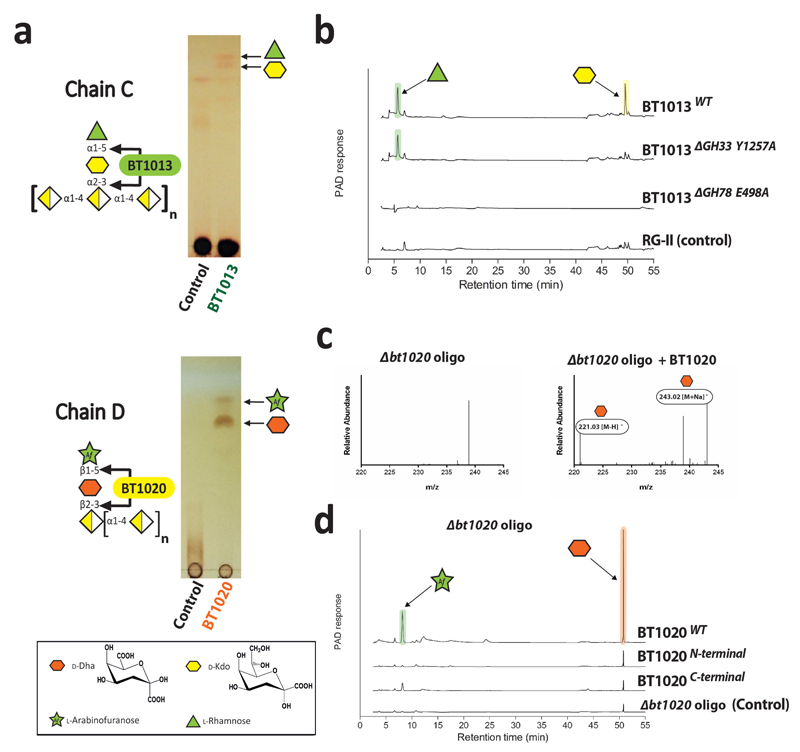
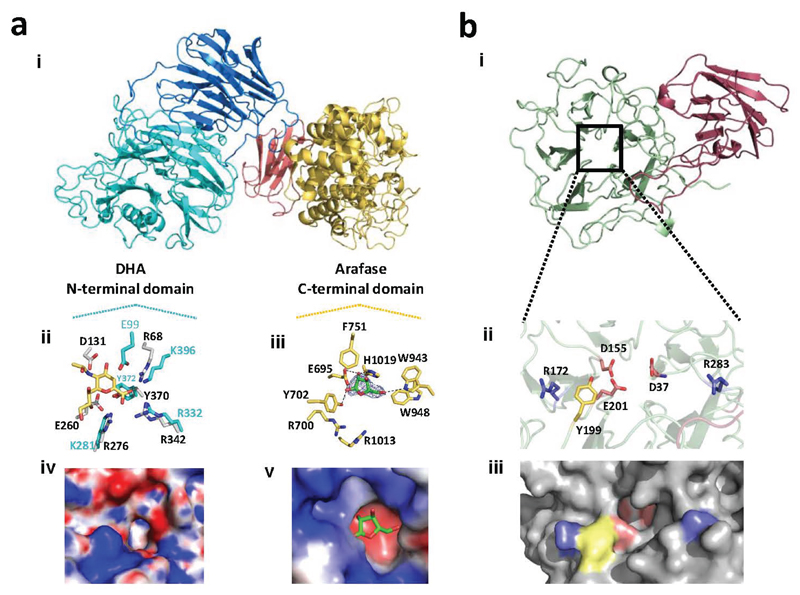
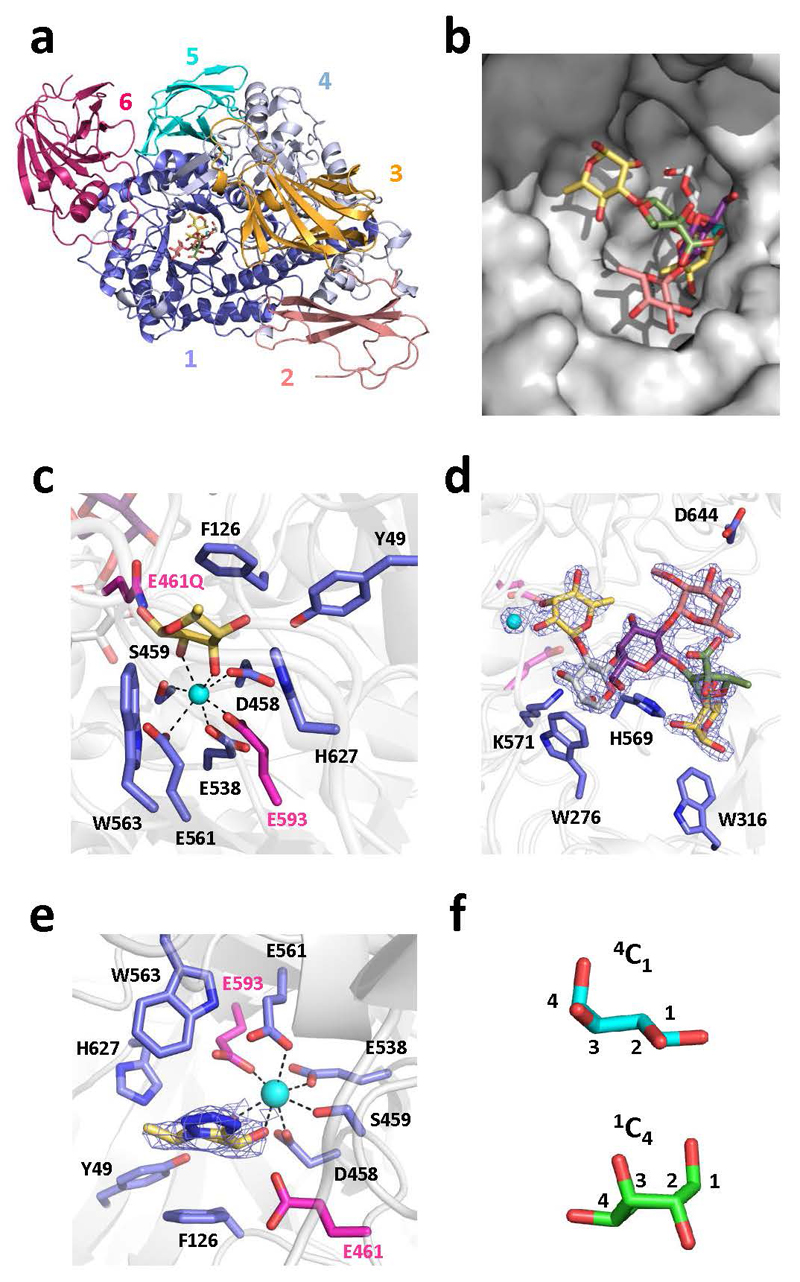
The RG-II degrading system (RG-II-degradome) contains three enzymes each comprising two distinct catalytic modules. BT0996 contains a β-d-glucuronidase and a β-l-arabinofuranosidase that target Chain A and B, respectively (Supplementary Table 3) implying that degradation of the oligosaccharide decorations is coordinated. BT1013 and BT1020 hydrolyse the two linkages in Chain C and D, respectively (Extended Data Fig. 3 and Supplementary Discussion 2). The crystal structure of BT1020 (Extended Data Fig. 4a) reveals an N-terminal 5-bladed β-propeller 2-keto-3-deoxy-d-lyxo-heptulosaric acid (Dha)-hydrolase and C-terminal (α/α)6 barrel β-l-arabinofuranosidase. The β-l-arabinofuranosidase domain (in complex with arabinose) contains a canonical glycoside hydrolase (GH) catalytic apparatus comprising two carboxylate residues. In the Dha-hydrolase active site a tyrosine and glutamate function as the catalytic nucleophile and acid-base residues; resembling the catalytic centre of sialidases that also act on ulosonic acids9. The highly basic nature of the active sites of BT1020 are consistent with the negatively charged species that occupy the two catalytic centres (Supplementary Discussion 3.1).
The CAZy database groups GHs into sequence-based families (designated GHXX)10. Predicting specificity based on family classification, however, is challenging since GH families are often poly-specific with only ~2% of the enzymes characterized10. Analyses of enzymes that deconstruct RG-II advance the functional annotation of existing GH families. The GH2 β-d-galacturonidase, BT0992; GH2 α-arabinopyranosidase, BT0983; and GH127 aceric acid hydrolase, BT1003 (Fig. 3 and Extended Data Fig. 5), represent new activities for their respective families (Supplementary Table 3), while the crystal structure of the α-l-rhamnosidase BT0986 provides unique insights into the mechanism of substrate recognition and catalysis for a GH106 enzyme11 (Extended Data Fig. 6). This 1100 residue protein contains an N-terminal (α/β)8-barrel catalytic domain. The heptaoligosaccharide generated by Δbt0986 bound in the centre of the (α/β)8-barrel in a funnel-like structure extending from the active site pocket. Mutagenesis studies revealed the catalytic residues BT0986 (Supplementary Table 4). Glu593, which is 5 Å from the anomeric carbon likely activates a water molecule that mounts a nucleophilic attack at C1, thus the glutamate is predicted to be the catalytic base. Glu461 is within hydrogen bonding distance of the glycosidic oxygen and is the candidate catalytic acid. In the active site glutamates coordinate calcium, which makes polar interactions with O2 and O3 of the l-rhamnose. The importance of calcium is illustrated by the loss of activity by EDTA or when the glutamates that coordinate the metal were mutated (Supplementary Table 4 and Supplementary Discussion 3.2). The essentiality of calcium has resonance with other inverting enzymes that target α-manno-configured linkages, which often exploit divalent metal ions in substrate binding12.
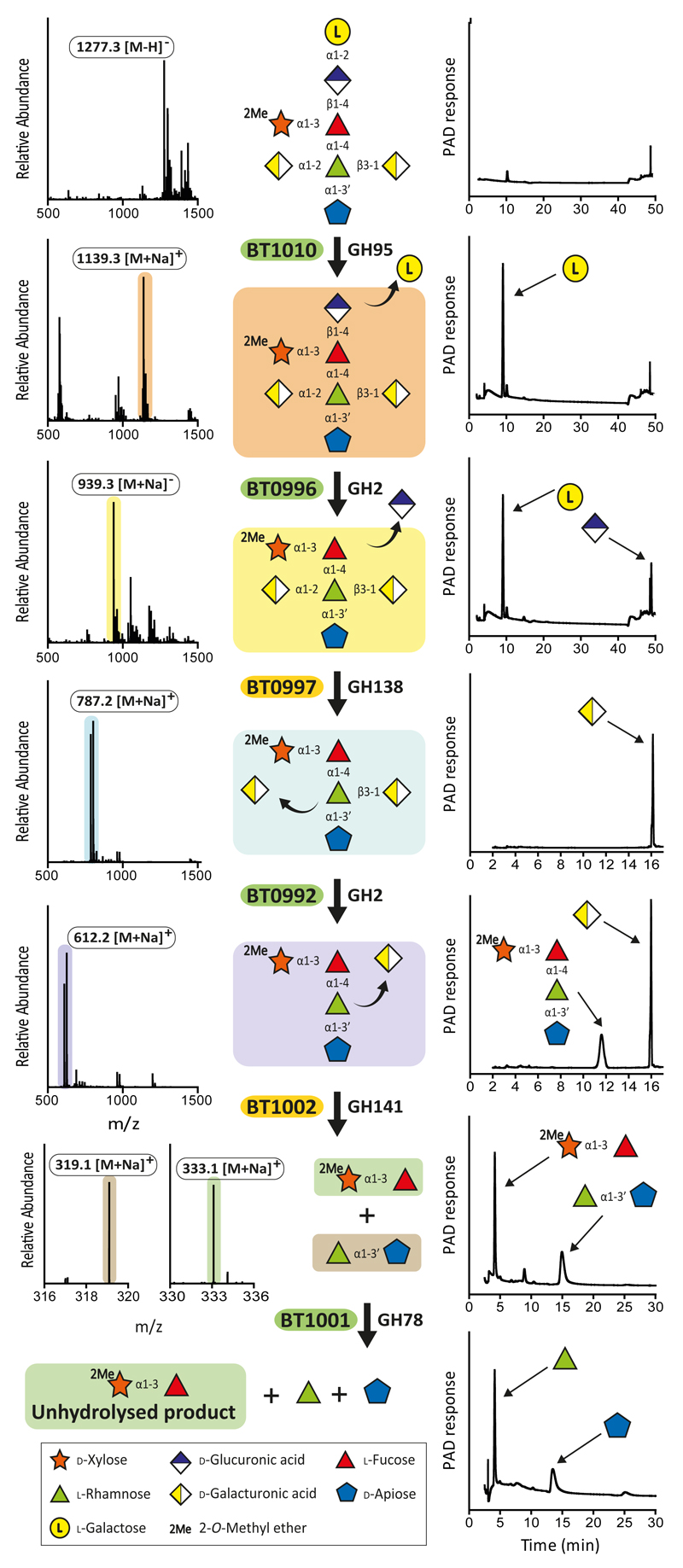
Fig. 3. B. thetaiotaomicron-mediated depolymerisation of RG-II Chain A.
The substrates comprised Chain A released from RG-II by trifluoroacetic acid and the oligosaccharides generated by mutants of B. thetaiotaomicron in which bt0997, bt0992 or bt1002 had been deleted. Individual proteins (1 μM) were incubated with the glycans (5 mM) for 16 h at 37 °C in 20 mM sodium phosphate buffer, pH 7.0. Monosaccharides and oligosaccharides generated were identified by HPAEC-PAD and ESI-MS, respectively. Verification of the model was achieved by reconstituting the pathway using the six enzymes in concert, which showed that the GHs only functioned in the order shown in the figure. The example is from technical replicates n = 3.
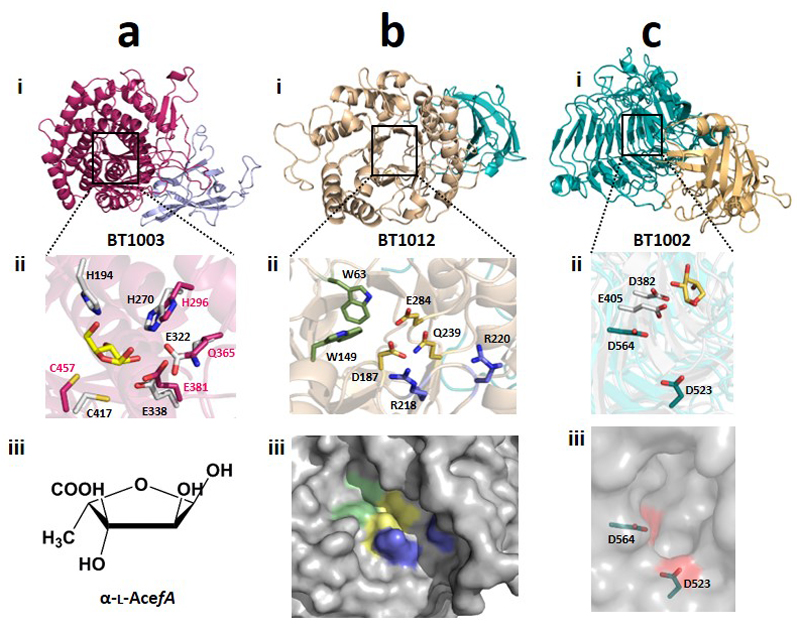
The aceric acid hydrolase activity of BT1003 is intriguing as GH127 was, prior to this work, populated only by β-l-arabinofuranosidases13; l-AceA is 3-C-carboxy-5-deoxy-l-xylose14. The crystal structure and mutagenesis of BT1003 (Extended Data Fig. 7a and Supplementary Table 4) showed that the proposed catalytic acid-base of GH127 arabinofuranosidases is not conserved in BT1003, suggesting substrate participation in glycosidic bond cleavage (Supplementary Discussion 3.5 and Supplementary Fig. 1).
Founding members of new GH families
The RG-II-degradome contains seven enzymes that reveal new GH families (Fig. 1 and Supplementary Table 3), comprising an endo-apiosidase (BT1012, GH140), Dha-hydrolase (N-terminus of BT1020, GH143), two β-l-arabinofuranosidases (C-terminus of BT1020, GH142; N-terminus of BT0996, GH137), α-2-O-Me-l-fucosidase (BT0984, GH139), α-l-fucosidase (BT1002, GH141) and α-galacturonidase (BT0997, GH138). BT1017 represents a new family of pectin methyl-esterases (Extended Data Fig. 8b). Additional novel features of RG-II depolymerisation are the α-2-O-Me-fucosidase (2MeFuc), Dha-hydrolase and AceAase activities, which have not previously been described even though 2MeFuc and Dha exist in other glycans15,16. The widespread presence of these newly discovered enzyme families in Bacteroidetes and other bacterial phyla, and the occurrence of GH139, GH140, GH141 and GH142 in fungi, suggest that these enzymes contribute to glycan degradation in diverse environments.
The generic relevance of RG-II PUL1 in RG-II utilization was supported by bioinformatic analysis. Xenologs of RG-II PUL1 proteins were identified by reciprocal best-BLAST hits in the HGM Bacteroides species (Extended data Fig. 1b). Only species that grew on RG-II contain proteins that display 65-95% identity with the B. thetaiotaomicron enzymes. Of 372 non-HGM Bacteroidetes species searched for candidate RG-II-degradomes, 40 contained PULs that encoded xenologs of all the necessary enzyme activities for RG-II breakdown (Supplementary Table 5). Example organisms include Prevotella bryantii (bovine rumen), Flavobacterium johnsoniae and Niabella soli (soil). This suggests that these species may also depolymerize the glycan (Extended data Fig. 1b and Supplementary Fig. 3).
A model for RG-II degradation
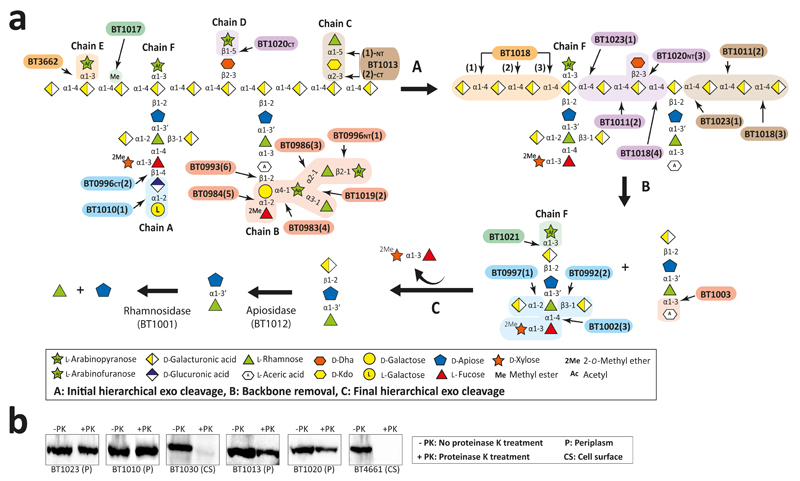
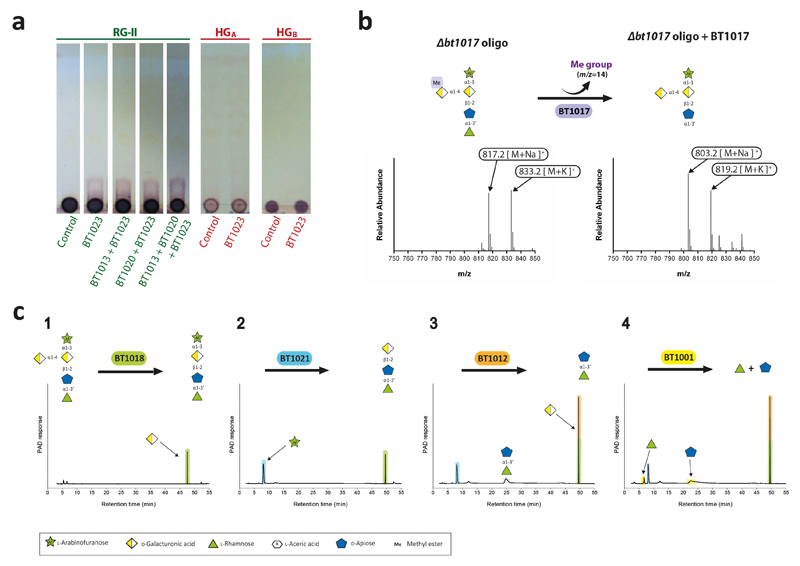
A hierarchical model for the RG-II degradome is proposed (Fig. 4). Chain C and E, the l-Araf of Chain D and the terminal region of Chain A and B are cleaved without hydrolysis of the glycan backbone (Supplementary Table 6). The steric constraints imposed by RG-II are consistent with this incomplete degradation. We propose that cleavage of the backbone increases enzyme access, enabling further degradation of the side-chains. The complete deconstruction of these side-chains is tightly linked to disassembly of the backbone. BT1023, a polysaccharide lyase, initiates backbone cleavage, which is critical for RG-II utilization as the Δbt1023 mutant grew poorly on the glycan (Fig. 2c). The enzyme cleaved only the homogalacturonan within RG-II (Extended Data Fig. 8a) indicating that side-chains are specificity determinants for BT1023. The completion of backbone degradation was mediated by removal of methyl esters (BT1017) and Dha (BT1020) (Extended Data Fig. 8b and 3cd), in harness with exo-acting glycosidases (Supplementary Table 3). Enzyme cell localization studies indicate that depolymerisation is exclusively periplasmic (Fig. 4b and Supplementary Discussion 4).
Fig. 4. Model of RG-II disassembly by B. thetaiotaomicron.
a, displays the degradative model. Enzyme cohorts that degrade a specific region of RG-II are coloured the same and the values in parenthesis indicate the predicted order in which they act, based on data (technical replicates n = 3) in Supplementary Table 6, b, cellular localization of key RG-II degrading enzymes based on their resistance to proteinase K when expressed by B. thetaiotaomicron. BT4661 and BT1030 are known surface glycan binding proteins. The example is from biological replicates n = 3.
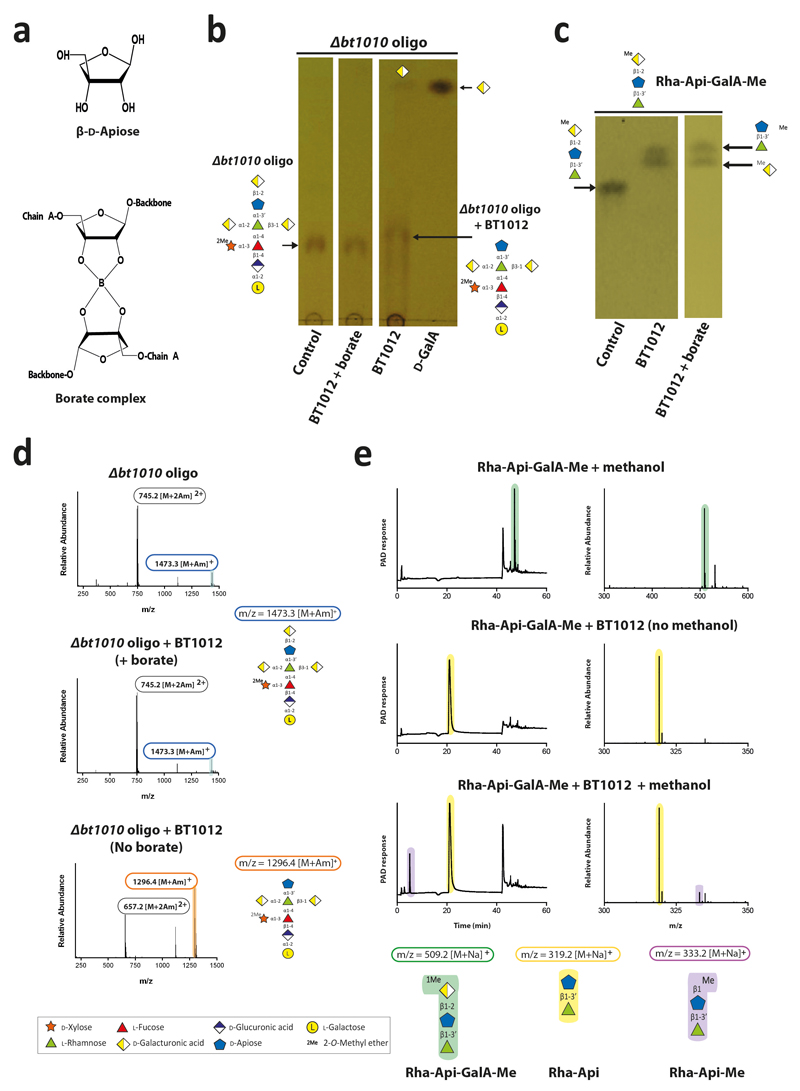
After backbone depolymerization, d-Apif of Chains A and B, linked to the remaining d-GalAs, are released by BT1012 prior to cleavage of l-Rhap-d-Apif by BT1001 (Supplementary Table 6). Thus, features of the α-l-rhamnosidase distal to the +1 subsite prevent binding of substrates with a degree of polymerization >2. The crystal structure of the apiosidase (BT1012; Extended Data Fig. 7bi) reveals a (α/β)8-barrel catalytic domain and a C-terminal β-sandwich domain. The predicted catalytic residues, Asp187 and Glu284, which are essential for activity (Supplementary Table 4), are separated by ~5.5 Å, consistent with a double displacement mechanism and retention of anomeric configuration (confirmed experimentally, Extended Data Fig. 7bii). Two arginines in the +1 subsite likely contribute to d-GalA binding through formation of ion pairs, consistent with mutagenesis data (Supplementary Table 4). The -1 (active site) and +1 subsites are narrow preventing binding of d-Apif linked to borate and d-GalA appended to the backbone at O-1 or O-4 (Extended Data Fig. 7biii), explaining why the apiosidase functions late in the degradative process (Fig. 4a).
The enzymes that depolymerize Chains A and B cleave their target sugars only when they are at the non-reducing termini (Supplementary Table 6). Thus, both chains are depolymerized through sequential-acting exo-glycosidases. Of particular note is BT1002, a α-l-fucosidase that targets Fuc substituted at O-3 with 2-O-Me-α-d-Xyl (Fig. 3). The active site pocket of the hydrolase is likely to be significantly more open than in GH95 and GH29 fucosidases that are unable to tolerate O-3 substitutions17,18. This is consistent with the topology of the catalytic centre of BT1002, which comprises an extended pocket (Extended Data Fig. 7c), which may bind the disaccharide 2-O-Me-d-Xyl-α1,3-l-Fuc (Supplementary Discussion 3.3).
Removal of the borate steric barrier
RG-II in planta is a dimer mediated by a borate diester linkage between d-Apif in Chain A of each monomer15. How the RG-II-degradome overcomes the steric constraints imposed by this dimer is a critical question. Previous studies19 proposed that removal of l-Gal-d-GlcA, destabilises borate-mediated dimerization. We show that Chain A-derived oligosaccharides, lacking the terminal l-Gal and d-GlcA, contained d-Apif at their reducing end (Extended Data Fig. 2). In contrast Δbt1010 generated intact Chain A with d-GalA attached to d-Apif. These data indicate that the apiosidase was only active against Chain A after l-Gal and d-GlcA had been removed, and thus the loss of this disaccharide destabilized the interaction of d-Apif with borate. Thus the apiosidase was not active against Chain A in this mutant, indicating that borate remained bound to d-Apif preventing access to the apiosidase. In vitro the activity of the apiosidase against intact Chain A was sensitive to borate (Extended Data Fig. 9bd) but the oxyanion did not inhibit the enzyme against l-Rha-d-Api-d-GalA (Extended Data Fig. 9c). These data indicate that borate only bound to d-Apif in full length Chain A, explaining how initial degradation of this side chain relieved steric constraints imposed by the oxyanion.
The species-dependent variation of the terminal region of Chain B in RG-II19,20, (Supplementary Discussion 1.0) may explain why RG-II PUL3 encodes several putative GHs that may contribute to degradation of all RG-II structures. Thus, BT3662 removed Chain E (Fig. 1 and Supplementary Table 2), present in grape RG-II but absent in the glycan from other plants. The crystal structure of BT3662, which hydrolyses α-Araf linkages, (Extended Data Fig. 4b), comprises an N-terminal five-bladed β-propeller catalytic domain. The active site pocket, which contains all the features of a typical GH43 α-l-arabinofuranosidase21, abuts onto a channel containing Tyr199 in the +1 subsite and two arginines in the distal regions. Mutagenesis data (Supplementary Table 4) suggest the basic residues sequester the acidic backbone of RGII into the substrate binding cleft, facilitating optimal interactions of the leaving group GalA with Tyr199. RG-II PUL2 only encodes a SusCh/SusDh pair (outer membrane glycan transporter)22, BT1683 and BT1682. The Δ1683/Δbt1682 mutant grew on apple but not wine RG-II, while the Δbt1024/Δbt1029 variant (deletion of susch/susdh in RG-II PUL1) metabolized the glycan from wine but slowly from apple (Fig. 2d). These data suggest that the multiple SusCh/SusDh pairs encoded by the RG-II PULs are required to import plant-specific variants of RG-II.
New features of RG-II structure
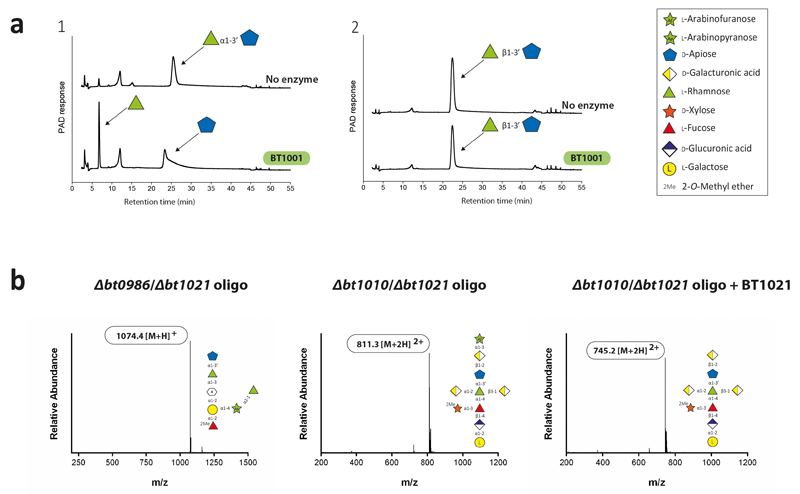
The specificity of the RG-II-degradome shows that the current model of the glycan requires revision. BT1001, which cleaves the Rha-Api linkage in Chains A and B, was shown to be a α-l-rhamnosidase (Extended data Fig. 10a; Supplementary Tables 3 and 7). Thus, the Rha-Api linkage in Chain A and B is α and not β as previously reported19. The change in the stereochemistry of this l-Rha linkage is likely to have a substantial impact on our knowledge of the conformations adopted by RG-II.
Analysis of the RG-II-degradome revealed a new side chain (Chain F) consisting of a α-Araf linked O-3 to the backbone GalA substituted at O-2 with Chain A. This arabinose was evident in the oligosaccharides generated by B. thetaiotaomicron mutants Δbt1017 Extended data Fig. 8bc), Δbt1021, Δbt1010/Δbt1021 and Δbt0986/Δbt1021 (Extended Data Fig. 10b; Supplementary Discussion 5). Chain E, also comprising a single arabinose linked to the backbone, is distinct from Chain F as they are removed by BT3662 and BT1021, respectively (Supplementary Tables 3 and 6). Although methyl-esterification of backbone GalA(s) is known, the location and extent of these decorations were unclear23. The Δbt1017 mutant, (lacks the pectin methyl-esterase), generated an oligosaccharide with a methyl-esterified GalA linked to O-4 of the GalA decorated with Chain A and F (Extended Data Fig. 8bc). This suggests that methyl-esterification occurs on a specific backbone GalA proximal to Chain A and F.
The crystal structure of the β-l-arabinofuranosidase of BT0996 revealed electron density for Chain B (Fig. 5 and Supplementary Discussion 3.4). The conformation of the oligosaccharide was stabilized through polar interactions within the nonasaccharide and confirmed the α-linkage between l-Rhap and d-Apif (Fig. 5d). Significantly, the l-Arap, substituted at O-2 and O-3 by l-Rhap, was in the unusual 1C4 conformation, consistent with NMR data24. This unfavourable conformation is stabilised by polar interactions between the 3-linked l-Rhap and the rest of the chain. Removing this α-l-1,3-Rhap by BT1019 likely allows the Arap to adopt the normal 4C1 conformer and is thus a substrate for the GH2 α-arabinopyranosidase BT0983, which hydrolyses equatorial glycosidic bonds. The relaxed 4C1 conformation of Arap was captured in the structure of the Chain B-derived unbranched heptasaccharide (lacks the 3- linked l-Rhap) bound to BT0986 (Extended data Fig. 6df).
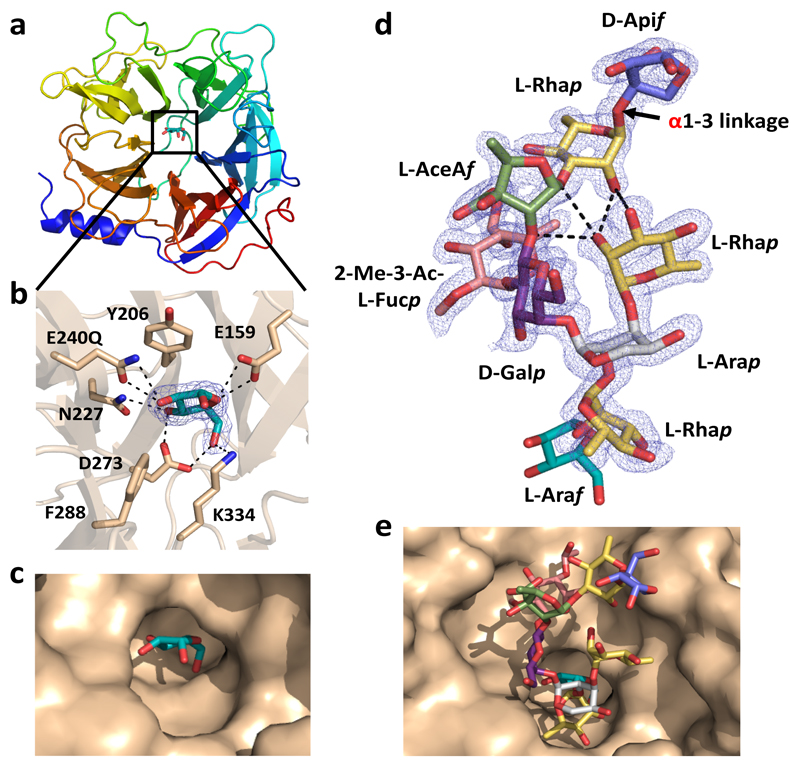
Fig. 5. Crystal structure of N-terminal catalytic domain of BT0996 (BT0996-N) in complex with RG-II-Chain B.
a, schematic of BT0996-N rainbow colour ramped from the N- (blue) to C- (red) termini. Black box; l-Araf-containing active site. b, residues interacting with l-Araf, which include the putative catalytic amino acids [Glu240 (shown as Gln240) and Glu159]. c, surface representation of the active site-pocket containing l-Araf. d, crystal structure of Chain B in complex with BT0996-N. The α linkage between l-Rhap and d-Apif is shown in red. e, shows Chain B (sugars coloured as in d) in the funnel-like substrate binding site of BT0996-N. In c,d polar interactions are broken black lines and the blue mesh is the electron density map (2Fo – Fc) of the ligands at 1.5 σ.
This study reveals the elaborate and highly specific enzyme system by which B. thetaiotaomicron utilizes RG-II, a highly complex glycan. Our data show that RG-II PUL1 encoded several proteins annotated as hypothetical are enzymes that are the founding members of new GH and esterase families. The discovery of seven new enzyme families, catalytic functions not previously reported, and new specificities for existing GH families illustrates the novelty of the RG-II degradome, and how the unique structural features of this glycan have driven the evolution of new catalysts. This contrasts with recent studies of the glycan-degrading apparatus of other HGM organisms, which exclusively utilize enzymes from existing CAZy families21–25. The model of RG-II degradation proposed, coupled with the unveiling of novel enzymes and activities, now provides a framework to discover and dissect pectic polysaccharide degradation in environments extending beyond the human gut microbiota. The true extent of RGII degradation in nature can now be accessed.
Methods
1.1. Preparing substrates
RG-II from wine and apple were selected for this work as they contain all the features of this glycan that are absent in the pectic polysacchahride from some other plant sources, Supplementary Discussion 1.0. Thus, the enzyme system that depolymerizes wine and apple RG-II is capable of degrading the glycan from all other plant sources.
1.1.1. Purifying RG-II
RG-II from wine was purified as described previously26. To prepare RG-II from apple a concentrate of the juice (25 kg box -equivalent of ~300 L of apple juice) was concentrated 1000 x using a Millipore peristaltic pump (3 L capacity) and Millipore Pellicon 2 (10000 molecular weight cut off, MWCO) filter cassette module. Concentrated material was mixed with 5 volumes of pure ethanol to precipitate carbohydrates, which was then centrifuged at 20000 x g at room temperature. The pellet was dissolved in 1 L of ultrapure water and re-precipitated with 5 volumes of ethanol. The precipitate was collected by centrifugation and this process was repeated another four times. The final pellet was redissolved in 0.5 L of ultrapure water and freeze-dried to obtain crude RG-II apple material.
HPAEC analysis showed that the crude apple RG-II was contaminated with arabinan and galactan. Thus a mixture of enzymes targeting specifically arabinan and galactan polymers was added to the RG-II material. These recombinant enzymes were expressed and purified by IMAC as described in Section 1.2.1. The enzyme mixture comprised GH43 endo-arabinanases BT0360 and BT0367 that target branched and linear arabinan, respectively; GH51 l-arabinofuranosidases BT0368 and BT0348 that hydrolyse α1,5-backbone and α1,3-side chains, respectively; the GH2 β1,4 galactosidase BT4667; and GH53 endo-β1,4-galactanase BT4668. Around 300 nM of each enzyme was incubated for 24 h at 37 °C with crude RG-II (4% w/v) in 20 mM sodium phosphate buffer, pH 7.0. The enzyme-treated RG-II was fractionated on a XK50column containing Fast flow DEAE Sepharose (column capacity 300 ml, GE healthcare) at a flow rate of ~10 ml/min. The column was washed with 300 ml of 50 mM sodium acetate buffer, pH 4.5, and then eluted batch-wise with 3 x column volumes of 50 mM sodium acetate containing 0.17 M, 0.3 M and 0.6 M NaCl. The RG-II eluted in the 0.3 M NaCl fraction based on glycosyl residue composition analyses, the types of sugars released by wild type B. thetaiotaomicron cultured on this material, and the inability of the ΔrgII-pul1 mutant to grow on this fraction. The fraction containing RG-II was dialysed against ultrapure water using 3.5 kDa cut off dialysis tubing. Progress of dialysis was monitored by use of the conductivity meter HI 99300 (Hanna instruments). The desalted RG-II was freeze-dried and kept at room temperature.
1.1.2. RGII derived oligosaccharides
B. thetaiotaomicron strains containing specific gene deletions were inoculated into minimal medium5 containing 1% RGII and grown in glass tubes for 48 h at 37 °C, in an anaerobic cabinet (Whitley A35 Workstation; Don Whitley, UK) to an OD600nm of 2.0. Cells were harvested by centrifugation initially at 2400 x g for 10 min and later at 17000 x g for another 10 min. The resulting supernatant was filtered through a 1.2 μm syringe filter (VWR) and separated on a Bio-Gel P2 (Biorad) size-exclusion column (SIZE OF COLUMN?) eluted with 50 mM acetic acid at 0.2ml/min. Fractions (200 μl) were collected and analysed by TLC using orcinol/sulfuric acid to reveal the resolved sugars. Fractions containing oligosaccharides of interest were pooled and concentrated by freeze-drying using a CHRIST Gefriertrocknung ALPHA 1-2 freeze-dryer (Helmholtz-Zentrum Berlin) at -50°C.
The chemical synthesis of selected oligosaccharides used protocols described previously: l-Rhap-α1,3’-d-Apif-O-Me and l-Rhap-β1,3’-d-Apif-O-Me27; d-GalA-α1,2-[d-GalA-β1,3]-[l-Fuc-α1,4]-l-Rha-O-Me and d-GalA-α1,2-[d-GalA-β1,3]-l-Rha-O-Me28; l-Rhap-β1,3’-d-Apif-β1,2-d-GalA-O-Me29
1.2. Biochemical studies
1.2.1. Producing recombinant proteins for biochemical assays
DNAs encoding enzymes lacking their signal peptides were amplified by PCR using appropriate primers. The amplified DNAs were cloned into NcoI/XhoI, Nco/BamHI, NdeI/XhoI or NdeI/BamHI restricted pET21a or pET28a, as appropriate. The encoded recombinant proteins generally contained a C-terminal His6-tag although, where appropriate, the His-tag was located at the N-terminus of the protein. To express the recombinant genes, Escherichia coli strains BL21(DE3) or TUNER, harbouring appropriate recombinant plasmids, were cultured to mid-exponential phase in Luria Bertani broth at 37 °C. Recombinant gene expression was induced by the addition of 1 mM (strain BL21(DE3)) or 0.2 mM (TUNER) isopropyl β-d-galactopyranoside (IPTG) , and the culture was grown for a further 5 h at 37 °C or 16 h at 16 °C, respectively. The recombinant proteins were purified to >90% electrophoretic purity by immobilized metal ion affinity chromatography (IMAC) using Talon™, a cobalt-based matrix, and eluted with 100 mM imidazole, as described previously4. To generate seleno-methionine (Se-Met) proteins for structure resolution, E. coli cells were cultured as described previously30, and the proteins were purified using IMAC as described above. For crystallization, the Se-Met proteins were further purified by size exclusion chromatography. After IMAC, fractions containing the purified proteins were buffer-exchanged, using PD-10 Sephadex G-25M gel-filtration columns (GE Healthcare), into 10 mM Na-Hepes buffer, pH 7.5, containing 150 mM NaCl and were then subjected to gel filtration using a HiLoad 16/60 Superdex 75 column (GE Healthcare) at a flow rate of 1 ml/min. For crystallization trials, purified proteins were concentrated using an Amicon 10-kDa molecular mass centrifugal concentrator and washed three times with 5 mM DTT (for the Se-Met proteins) or water (for native proteins).
1.2.2. Site-Directed Mutagenesis
Site-directed mutagenesis was carried out employing a PCR-based NZY-Mutagenesis kit (NZYTech Ltd) using the plasmids encoding the appropriate enzymes as the template. The mutated DNA clones were sequenced to ensure that only the appropriate DNA change was introduced after the PCR.
1.2.3. Glycoside hydrolase assays
Spectrophotometric quantitative assays for l-rhamnosidases (BT0986; BT1001 and BT1019), l-arabinofuranosidases (BT0983; BT0996; BT1021 and BT3662), d-galacturonidases (BT0992; BT0997 and BT1018), the d-glucuronidase (BT0996) and d-galactosidase (BT0993) were monitored by the formation of NADH, at A340nm using an extinction coefficient of 6230 M−1 cm−1, with an appropriately linked enzyme assay system. The assays were adapted from purchased Megazyme International assay kits. These kits were as follows: the l-Rhamnose assay kit (K-RHAMNOSE); l-arabinose/D-Galactose assay kit (K-ARGA); d-Glucuronic acid/D-Galacturonic acid assay kit (K-URONIC)). The activity of BT0984 was monitored by using an excess of BT0993 and quantifying d-galactose release as mentioned above. BT1003 and BT1012 activity was measured by using an excess of BT1001 and linking it to rhamnose release as described above. Substrate depletion assays were used to determine the activity of enzymes BT1002 and BT1012, whilst the formation of l-galactose from RG-II was used to determine the activity of BT1010. Briefly, aliquots of the enzyme reaction were removed at regular intervals and, after boiling for 10 min to inactivate the enzyme and centrifugation at 13000 x g, the amount of the substrate remaining or product produced was quantified by HPAEC using standard methodology. The reaction substrates and products were bound to a Dionex CarboPac PA1 column and were eluted with an initial isocrartic flow of 100 mM NaOH then a 0-200 mM sodium acetate gradient in 100 mM NaOH at a flow rate of 1.0 ml min-1, using pulsed amperometric detection. Linked assays were checked to make sure that the relevant enzyme being analysed was rate limiting by increasing its concentration and ensuring a corresponding increase in rate was observed. A single substrate concentration was used to calculate catalytic efficiency (kcat/KM) and was checked to be <<KM by halving and doubling the substrate concentration and observing an appropriate increase or decrease in rate. The equation V0 = (kcat/KM)[S][E] was used to calculate kcat/KM unless substrate depletion was used then the calculation was as follows ln(kcat/KM) = (S0/St)/[E] where [E] is enzyme concentration and S substrate31. All reactions were carried out in 20 mM sodium phosphate buffer, pH 7.0, with 150 mM NaCl and performed in at least technical triplicates. The activity of selected enzymes against complex oligosaccharides was also evaluated using mass spectrometry as described below. The substrates used were RGII from wine or apple, RGII-derived oligosaccharides generated using B. thetaiotaomicron mutants, by partial acid treatment of RG-II, or by chemical synthesis (prepared as described above). TLC was used to provide a qualitative profile of the activity of selected enzymes. Around 4 μl of the reaction was spotted on silica gel TLC plates and the plates were developed in ascending butanol:acetic acid:water 2:1:1. Carbohydrate products were detected by spraying with 0.5% orcinol in 10% sulphuric acid and heating to 100 °C for 10 min.
1.3. Mass spectrometry (MS) of oligosaccharides
1.3.1. Infusion electrospray MS Analysis
The structures of the desalted oligosaccharides (in 10 mM ammonium acetate, pH 7.0) were analysed via negative ion mode infusion/offline electrospray ionization mass spectrometry (ESI-MS) following dilution (typically 1:1 [v:v]) with 5% trimethylamine in acetonitrile.
Electrospray data was acquired using an LTQ-FT mass spectrometer (Thermo) with a FT-MS resolution setting of 100,000 at m/z = 400 and an injection target value of 1,000,000. Infusion spray analyses were performed on 5-10 µl of samples using medium ‘nanoES‘ spray capillaries (Thermo) for offline nanospray mass spectrometry in negative ion mode at 1 kV.
1.3.2. Nano electrospray LC- MS Analysis
Oligosaccharide samples were diluted (typically 1:50 [v:v]) with 0.1% formic acid (aq) and injected (0.5 µl) onto a capillary 3 μ particle, 200 Å pore ProntoSIL C18AQ trapping column (nanoLCMS Solutions LLC, USA) in-line with a 75 µm X 100 mm BEH130 C18 capillary column (Waters, UK) running on a NanoAcquity UPLC system (Waters, UK). The gradient conditions were 0.1-15% Buffer B in Buffer A in 15 min, 15 - 50% Buffer B in Buffer A over 24 min and 50 - 90% Buffer B in Buffer A over 5 min, with 15 min re-equilibration (0.1% Buffer B in Buffer A) at a flow rate of 0.4 µl/min. Buffer A: 0.1% formic acid in water; Buffer B: 0.1% formic acid in acetonitrile.
Nanoelectrospray data was acquired using an LTQ-FT mass spectrometer (Thermo). Survey MS scans were performed over the mass range m/z = 150 – 2000 in data-dependent mode. Data was acquired with a FT-MS resolution setting of 100,000 at m/z = 400 and a Penning trap injection target value of 1,000,000. The top five ions in the survey scan were automatically subject to collision-induced dissociation MS/MS in the linear ion trap region of the instrument at an injection target value of 100,000, using a normalized collision energy of 30% and an activation time of 30 ms (activation Q = 0.25).
1.4. Growth of B. thetaiotaomicron and generation of mutants
The selection of B. thetaiotaomicron to analyse the mechanism by which RG-II is degraded in the HGM was based on the following criteria: 1) B. thetaiotaomicron is a component of the HGM in a wide range of individuals; 2) the bacterium was previously shown to be capable of growing on RG-II and the genetic loci activated by the pectic polysaccharide identified; 3) a genetic system for the bacterium has been developed enabling routine targeted gene deletion, which was critical in identifying the enzymes that depolymerised RG-II.
1.4.1. Growth of B. thetaiotaomicron
B. thetaiotaomicron was routinely cultured under anaerobic conditions at 37 °C using an anaerobic cabinet (Whitley A35 Workstation; Don Whitley, UK) in culture volumes of 0.2, 2 or 5 ml) of TYG (tryptone-yeast extract-glucose medium) or minimal medium containing 1% of an appropriate carbon source plus 1.2 mg/ml porcine hematin (Sigma-Aldrich) as previously described5. The growth of the cultures were routinely monitored at OD600nm using a Biochrom WPA cell density meter (Cambridge, UK) for the 5 ml cultures or a Gen5 v2.0 Microplate Reader (Biotek) for the 0.2 and 2 ml cultures.
1.4.2. Constructing mutants of RGII-PUL1 in B. thetaiotaomicron
The mutants of the complete RGII-PUL1 knock out and single gene clean deletions were introduced by counter selectable allelic exchange using the pExchange vector as described32.
1.5. Crystallization, Data Collection, Structure Solution and Refinement
Crystallization: All protein concentrations were around 10 mg/ml except BT1012 which was at 4 mg/ml. BT3662 was crystallised in 20% polyethylene glycol (PEG) 6000 and 0.1 M NaHepes pH 6.5. BT1012 was crystallised in 40 % (v/v) MPD, 0.2 M ammonium phosphate and Tris pH 8.5. Selenomethionine (Se-Met)-containing BT0996 was crystallised in 20 % (w/v) PEG 8000, 0.1 M Tris pH 8.5. Additionally Se-Met-containing BT0996 were crystallised in 20% (w/v) PEG 3350 and 0.2 M Lithium Acetate for ligand soaking in mother liquor containing 25 mg/ml of Chain B for up to 16 h. The same process was repeated for inactive mutant BT0996-E240Q. BT1002 protein was crystallised in 15% (v/v) ethylene glycol, 15% (w/v) PEG 8000, 30 mM MgCl2, 30 mM CaCl2, 50mM NaHepes and 50 mm MOPS pH 7.5. Se-Met-containing BT1002 was crystallised in 10% (w/v) PEG 3350, 0.1 M Hepes pH 7.5 and 0.2 M l-Proline. BT1003 was crystallised in 20% (w/v) PEG 3350 and 0.2 M potassium acetate. BT0986 was crystalized 15% (w/v) PEG 550 MME, 15% (w/v) PEG 20000, 0.25 M rhamnose, 50mM hepes and 50 mm MOPS pH 7.5 in absence or presence of 6 mM d-rhamnopyranose tetrazole. The inactive mutant BT0986-D461Q was crystalized in 15% (w/v) PEG 550 MME, 15% (w/v) 20000, 50 mM Imidazol, 50 mM MES pH 6.5, 30 mM NaF, 30 mM NaBr, 30 mM NaI in presence of 5 mM of RGII Chain B. Se-Met-containing BT0986 was crystallised in 20% (w/v) PEG 3350, 0.2 M sodium sulphate, 0.25 M rhamnose and 0.1 M bis-tris propane pH 6.5. Se-Met containing BT1020 was crystallised in 14% (v/v) ethylene glycol, 14% (w/v) PEG 8000, 30 mM of each di-, tri, tetra- and penta-ethylene glycol, 50 mM hepes and 50 mM MOPS pH 7.5. BT1020 was crystallised in 15% (v/v) ethylene glycol, 15% (w/v) PEG 8000, 20 mM d-glucose, 20 mM d-mannose, 20 mM d-galactose, l-fucose, d-xylose, N-acetyl-d-glucosamine, 300 mM l-arabinose, 44.5 mM imidazole, 55.5 mM MES pH 6.5. All proteins were initially screened by the sitting drop method with a protein volume of 0.1 or 0.2 µl and a reservoir ratio of 1:1 or 2:1 using a robotic nanodrop dispensing system (mosquito™ LCP; TTPLabTech).
Cryo-protection: BT1002, BT1003, Se-Met-BT0986, BT0986 in presence of rhamnotetrazole, Se-Met-containing BT1020 were cryo-protected by supplementing the mother liquor with 20% (v/v) PEG 400. 20% ethylene glycol (v/v) were used for BT3362 and 20% (v/v) glycerol for BT0996. All other samples did not require supplemental cryo-protection.
Data collection and processing: All data were integrated using XDS33 apart from BT1003 which was integrated with DIALS34 and BT0986 with the rhamnotertazole which was integrated with xia2 3dii35. The data were scaled with either XDS33 or Aimless36.
Structure solution and refinement: The phase problem for BT1002, BT1020, BT0986 and BT0996 was solved by Se-Met-SAD using hkl2map37 and the shelx pipeline38. Buccaneer39 and/or arp-warp40 were used for automated model building. BT0986 in the presence of rhamnotetrazole and BT0986-D461Q in the presence of a Chain B-deribed heptasaccharide were solved with molrep41 using BT0986 native as search model. All the other data were solved by molecular replacement using phaser42 and the PDB model 3QZ4 for BT3662, 3KZS for BT1012 and an ensemble of PDB models 3WKX and 4QJY for BT1003. Buccaneer39 and/or arp-warp40 were as well used when needed to improve the initial molecular replacement solution. Recursive cycles of model building in coot43 and refinement in refmac544 were performed to produce the final model. For all models solvent molecules were added using coot43 and checked manually; five percent of the observations were randomly selected for the Rfree set. All models were validated using coot43 and molprobity45. The data statistics and refinement details are reported in Supplementary Table 8.
1.7. Comparative genomics analysis
Polysaccharide utilization loci (PULs) similar to the RG-II PULs were searched for in 372 Bacteroidetes genomes (complete list provided in Supplementary Table 5). The identification of similar PULs was based on PUL alignments. Gene composition and order of Bacteroidetes PULs were computed using the PUL predictor described in PULDB46. Then, in a manner similar to amino-acid sequence alignments, the predicted PULs were aligned to the RG-II PULs according to their modularity as proposed in the RADS/RAMPAGE method47. Modules taken into account include CAZy families, sensor-regulators and susCD-like genes. Finally, PUL boundaries and limit cases were refined by BLASTP-based analysis. The novel glycoside hydrolase families discovered in this study are listed in the main paper.
1.8. Protein cellular localization
Cellular localisation of proteins was carried out as described previosuly4. Briefly, B. thetaiotaomicron cultures where grown overnight (OD600nm 2.0) in 5 ml minimal media containing 1% apple RGII. The next day, cells were harvested by centrifugation at 5000 g for 10 min and resuspended in 2 ml phosphate buffered saline (PBS). Proteinase K (0.5 mg/ml final concentration) was added to 1 ml of the suspension and the other half left untreated (control). Both samples were incubated at 37 °C overnight followed by centrifugation (5000 x g for 10 min) to collect cells. To eliminate residual proteinase K activity, cell pellets were resuspended in 1 ml of 1.5 M trichloroacetic acid in PBS and incubated on ice for 30 min. Precipitated mixtures were then centrifuged (5000 g, 10 min) and washed twice in 1 ml ice cold acetone (99.8%). The resulting pellets were allowed to dry in a 40 °C heat block for 5 min and dissolved in 250 μ l Laemmli buffer. Samples were heated for 5 min at 98 °C and mixed by pipetting several times before resolving by SDS/PAGE using 7.5% gels. Electrophoresed proteins were transferred to nitrocellulose membranes by Western blotting followed by immunochemical detection using primary rabbit polyclonal antibodies (Eurogentec) generated against various proteins and secondary goat anti-rabbit antibodies (Santa Cruz Biotechnology). For BT1010 and BT1013 whose anti-sera failed to produce the desired reactivity, a C-terminal FLAG peptide (DYKDDDDK) was incorporated at the C-terminals of both native proteins expressed by B. thetaiotaomicron through counter-selectable allelic exchange32. This allowed for their detection using rabbit anti-flag antibodies (Sigma) as primary antibodies.
Extended Data
Extended Data Fig. 1. Sequence conservation and genomic organization of components of RG-II PUL1 across Bacteroidetes species.
a, PULs activated by RG-II. Genes encoding proteins of known or predicted functionalities are colour coded. The arrows indicate the orientation of each gene. b for Bacteroidetes species (in rows), xenologs of B. thetaiotaomicron (Bt) RG-II PUL1 proteins (in columns) were identified by reciprocal best-blastp hits. These organisms were coloured to reflect growth on RG-II; blue, growth in 24 h; orange, growth in 48 h; red, no growth; black, growth on RG-II was not evaluated. Bt proteins that contribute to RG-II degradation are grey and the family or predicted function are defined as GHXX, GH family; PL1, polysaccharide lyase family 1; CE, carbohydrate esterase; PME, pectin methylesterase. bi, xenologs are in the same column as the corresponding Bt protein and % identity is in a red colour-scale to 24% identity. bii, genomic organization of xenologs into gene clusters/loci. The top row shows 55 Bt proteins numbered according to relative position of the gene on the genome. The xenologs of the Bt genes are numbered as they appear in their respective cluster/locus. For each species Bt RG-II PUL1 was split into separate clusters when the contiguous xenologs were separated by ≤30 unrelated genes. Each cluster has a distinct background color (green, blue, pink and yellow). Proteins in B. xylanisolvens XB1A, not annotated previously as ORFs identified here by tblastn are marked with a dagger. Split proteins in B. cellulosilyticus DSM 14838 due to incomplete genome assembly were numbered based on the B. cellulosilyticus WH2 genome (marked with an asterisk). Bt enzymes that were split into two distinct single module enzymes in other species are denoted with a ‘plus’ sign in the PUL. Supplementary Fig. 3 shows a complete depiction of the PUL organization in the selected species.
Extended Data Fig. 2. The generation of RG-II derived oligosaccharides by mutants of B. thetaiotaomicron.
a, shows the site of action of the enzyme that was eliminated or not functional in the corresponding mutant. b, displays the structures of the oligosaccharides generated by each mutant. These molecules were isolated by size-exclusion chromatography and used as substrates to elucidate the mechanism of RG-II degradation. c, shows the structure of bespoke chemically synthesised oligosaccharides that were used as substrates to dissect the mechanism of RG-II depolymerization.
Extended Data Fig. 3. Analysis of the disassembly of Chain C and Chain D.
All the reactions were carried out standard conditions described, and samples labelled “Control” signifies that the glycan was not enzyme treated. a, wild type (WT) BT1013 and BT1020 were incubated with RG-II and the oligosaccharides generated by the B. thetaiotaomicron Δbt1020 mutant (Δbt1020 oligo) grown for 50 h, respectively, and the reactions were analysed by TLC. b, WT and mutants of BT1013 were incubated with RG-II and the products were subjected to HPAEC-PAD analysis. In the mutants ΔGH78 E496A and ΔGH33 Y1257A are mutants of the predicted catalytic residues of the GH78 rhamnosidase and GH33 sialidase catalytic modules, respectively. c, Δbt1020 oligo was incubated with WT BT1020 and the products analysed by HPAEC-PAD and mass spectrometry. d, the activity of the N- (residues Asp26 to Asp613) and C-terminal (residues Lys614 to Leu1107) regions of BT1020 were compared with the WT enzyme using HPAEC-PAD analysis. Arabinose and rhamnose were identified through HPAEC-PAD by co-migration with the appropriate standard, while the identity of Dha was consistent with the mass spectrometry data. The data displayed are examples from biological replicates n = 3.
Extended Data Fig. 4. Crystal structures of BT1020 and BT3662.
a, and b, show the structure of BT1020 and BT3662, respectively. ai, schematic of BT1020 in which the domains from N- to C-termini are coloured cyan (Dhase catalytic domain), blue (structural β-sandwich domain 1), yellow (β-l-Arafase catalytic domain), salmon (structural β-sandwich domain 2). aii and aiii depict the key active site residues in stick format of the Dhase and β-l-Arafase catalytic domains, respectively. In aii the BT1020 residues (carbon and lettering coloured cyan) are overlayed with the GH33 Clostridium perfringens sialidase NanI (PDB code 2VK7) in which the carbons and lettering are light grey and black, respectively. The ligand, N-acetyl-neuramic acid, is derived from the NanI structure and is shown in yellow. In aiii the active site amino acids (carbons coloured yellow) that interact with the bound l-Araf (carbons in green) are shown with polar contacts depicted by broken black lines. aiv and av show a charged surface representation of the active site pockets of the Dhase and β-l-Arafase catalytic domains (with l-Araf bound to the β-l-Arafase), respectively. Note the highly basic feature at the active sites consistent with the negatively charged Dha located in the active site or +1 subsite of the two catalytic domains. bi, schematic of BT3662 in which the five-bladed β-propeller catalytic domain and the β-sandwich domain are shown in green and red, respectively. In bii, key residues are shown in stick format. The carbons of the amino acids coloured salmon pink are catalytic residues; the yellow tyrosine is positioned in the +1 subsite and the blue arginines are in the distal regions that interact with the d-GalA-containing backbone. Biii, solvent exposed surface representation of Bii using the same colour format for the amino acids highlighted. The active site housing the catalytic residues is located in a pocket that abuts onto a shallow channel containing the arginines and tyrosine.
Extended Data Fig. 5. Mechanism by which B. thetaiotaomicron depolymerizes Chain B of RGII.
The oligosaccharides generated by Δbt1003 and Δbt0986, RG-II and chemically synthesised molecules were used to determine which enzymes acted on Chain B. a, enzymes identified were added sequentially to Chain B (isolated by mild acid treatment of RGII). The reactions were carried out under standard conditions. The sugars released were identified and quantified by HPAEC-PAD. b, shows examples of the use of mass spectrometry to monitor the enzymatic disassembly of Chain B. The example shown are from biological replicates n = 3.
Extended Data Fig. 6. Crystal structure of BT0986.
a, a schematic of the enzyme is displayed, revealing the (α/β)8-barrel catalytic domain (yellow) that is interrupted with three β-sandwich domain (blue), while the C-terminal domain (salmon) also folds into a β-sandwich. b, active site of BT0986 bound to rhamnose. Catalytic amino acids are coloured magenta while the other amino acids are blue and the rhamnose yellow. c, transition state mimic rhamnopyranose tetrazole bound in the active site of BT0986. The same colouring was used as in c. The blue mesh surrounding the ligand represents the 2Fo-Fc electron density map (1.3 Å resolution) at 1.5 σ. In b and c the calcium ion in the active site is shown as a cyan sphere and its polar contacts with amino acids and ligands are indicated by black dashed lines. d, Chain B-derived heptasaccharide generated by the mutant bacterium Δbt0986 bound in the substrate binding site of BT0986 shown as a surface representation. e, shows the interactions of the Δbt0986 heptasaccharide with BT0986. Amino acids that interact with the oligosaccharide are coloured blue and the sugars in both d and e are as depicted in Fig. 5. The blue mesh is the electron density of the oligosaccharide. f, conformation of the arabinopyranose in Chain B (green) and in the Δbt0986 heptasaccharide (cyan). The carbons are numbered.
Extended Data Fig. 7. Crystal structure of BT1003, BT1012 and BT1002.
All amino acids are in stick format and the position of the active site in the schematics of the respect enzymes is indicated by a black box. a, structure of the GH127 AceAase. ai, schematic of the enzyme revealing the catalytic domain (α/α)6 barrel (red) and β-sandwich domain (blue). aii, overlay of the key active site residues of the AceAase, coloured red, and the GH127 β-l-arabinofuranosidase HypBA1 from Bifidobacterium longum (PDB 3WKX), coloured white grey. The proposed catalytic nucleophile, Cys457 in BT1003, is conserved in the two enzymes; the catalytic acid/base in HpyBA1 (Glu322), however, is a glutamine in the AceAase. The ligand, coloured yellow is β-l-Araf derived from the crystal structure of the β-l-arabinofuranosidase. ciii, structure of aceric acid. b, structure of the apiosidase BT1012. bi schematic of the enzyme in which the (α/β)8-barrel catalytic domain is coloured beige and the C-terminal β-sandwich domain blue. bii, the yellow amino acids are the key residues in the active site (-1 subsite) that are proposed to play a direct catalytic role (D187 and E284) or substrate binding function (Q239). The pair of arginine residues coloured blue in the +1 subsite are likely to contribute to d-GalA binding through interactions with the carboxylate of the uronic acid. The aromatic residues coloured green are in the -2 subsite. biii, solved exposed surface representation of the substrate binding region of BT1012. The residues highlighted in cii are displayed in the appropriate colour. c, structure of BT1002. ci, schematic of the enzyme in which the C-terminal β-parallel helical catalytic domain and the N-terminal β-sandwich domain are coloured green and yellow, respectively. cii overlay of BT1002 and its closest structural homolog, a GH120 β-xylosidase (PDB code 3VSU), highlighting the position of the catalytic amino acids coloured green (BT1002) or light grey (β-xylosidase). The xylose in the active site of the β-xylosidase is shown to orientate the close but not identical position of the catalytic centre of the two enzymes. ciii, solvent exposed surface of the active site pocket of BT1002 in which the catalytic residues are shown in stick format and their location on the surface depicted in red. The pocket is elongated hinting that the 2-O-Me-xylose appended to the l-fucose may be housed in the catalytic centre of the α-l-fucosidase.
Extended Data Fig. 8. Identification of the enzymes that disassemble the backbone of RGII.
a, BT1023 was incubated with RG-II or homogalacturonan (1% w/v) in 50 mM CAPSO buffer, pH 9.0, containing 2 mM CaCl2. RG-II substrate was either untreated or had been previously incubated with BT1013 and/or BT1020. The reactions were analysed by TLC. Lanes labelled “control” were the appropriate glycan incubated with the appropriate buffer but without the inclusion of enzyme. b, the oligosaccharide generated by the Δbt1017 mutant (Δbt1017 oligo) was incubated with BT1017 and the reaction product was analysed by mass spectrometry. c,sequential degradation of the product generated in b by the enzymes indicated under standard conditions. The sugars released by these other enzymes (reactions 1 to 4) were identified and quantified by HPAEC-PAD. The structure of the oligosaccharide sugars followed the notation described in Fig. 1 and Extended data Fig. 2. The example shown are from technical replicates n = 3.
Extended Data Fig. 9. The influence of borate on the retaining apiosidase BT1012.
a, oligosaccharide generated by the B. thetaiotaomicron mutant Δbt1010 (Δbt1010 oligo) was incubated with BT1012 under standard conditions in the presence and absence of 50 mM borate and the products analysed by TLC. b, the same experiment was carried out except that the substrate was Rha-β1,3’-Api-α1,2-GalA-Me. c, mass spectrometric analyse of the products generated in a. d, BT1012 was incubated under standard conditions with Rha-β1,3’-Api-α1,2-GalA-Me in the presence and absence of 2.5 M methanol. The reactions were analysed by HPAEC-PAD and mass spectrometry. The example shown are from technical replicates n = 3.
Extended Data Fig. 10. Modification of the structure of RG-II.
a, recombinant BT1001 and BT1012 were incubated with bespoke chemically synthesised oligosaccharides using standard conditions. b, The enzymatic disassembly of RG-II was used to investigate the structure of RG-II. Mass spectrometry combined with HPAEC-PAD were used to analyse the structure of the oligosaccharides (defined as oligo) generated by the mutants Δbt1010/Δbt1021 and Δbt1010/Δbt0986 before and after treatment with recombinant BT1021 using standard conditions. The enzyme reactions were analysed by HPAEC-PAD.
Supplementary Material
Acknowledgment
This work was supported in part by a grant to H.J.G. and B.H. from the European Research Council (Grant No. 322820). B.H. was also funded by Agence Nationale de la Recherche under grant number ANR 12-BIME-0006-01. H.J.G was also supported by Biotechnology and Biological Research Council (grant numbers BB/K020358/1 and BB/K001949/1), the Wellcome Trust (grant No. WT097907MA) and, with X.Z., M-C.R. and F.B. were funded by the European Union Seventh Framework Programme under the WallTraC project (Grant Agreement number 263916). M.A.O and B.U. were supported in part by grant DE-FG02-12ER16324 from The Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy. I.M. was in receipt of a Marie Skłodowska-Curie Fellowship (grant No: 707922). GJD is a Royal Society Ken Murray Research Professor. We thank Diamond Light Source for access to beamline I02, I04-1 and I24 (mx1960, mx7854 and mx9948) that contributed to the results presented here, and to Drs T. Doco and S. J. Charnock who supplied the partially purified apple RG-II.
Footnotes
Conflict of Interest: The authors declare that they have no conflicts of interest with the contents of this article
Author Contributions
Enzyme characterisation was carried out by D.N., A.R., A.C., A.S.L., I.V., A.L., D.W.A., Y.Z. and X.Z. Crystallographic studies by A.C., A.B., A.S.L., D.N. and I.V. Purification of RGII and Oligosaccharide products by M.A.O., A.R., D.N., A.C, A.L., A.S.L., F.B. and MC.R. HPLC analysis by A.R., D.N., A.C. and A.S.L., whilst Mass Spectrometry analysis was by A.R. and J.G. Chemical synthesis was by S.N. and R.A.F. Growth analysis on purified RGII performed by D.N. and A.R. Gene deletion strains were created and characterised by D.N. Co-culturing experiments were carried out by J.B. Phylogenetic reconstruction and metagenomic analysis: N.T. and B.H. Bacterial growth and transcriptomic experiments: Y.X. and E.C.M. Experiments were designed by D.N., A.R., A.C., A.S.L., E.C.M and H.J.G. The manuscript was written by H.J.G. with contributions from G.J.D., M.A.O, B.U., E.C.M. and W.S.Y. Figures were prepared by A.R., A.L., D.N. and A.S.L.
Data Availability. The crystal structure datasets generated (coordinate files and structure factors) have been deposited in the Protein Data Bank (and are listed in Supplementary Table 8). The authors declare that the data supporting the findings of this study are available within the paper and the Supplementary Information
References
- 1.Wißing C, et al. Isotopic evidence for dietary ecology of late Neandertals in North-Western Europe. Quaternary International. 2016;411:327–345. [Google Scholar]
- 2.Pellerin P, et al. Structural characterization of red wine rhamnogalacturonan II. Carbohydr Res. 1996;290:183–197. doi: 10.1016/0008-6215(96)00139-5. [DOI] [PubMed] [Google Scholar]
- 3.Apolinar-Valiente R, et al. Polysaccharide Composition of Monastrell Red Wines from Four Different Spanish Terroirs: Effect of Wine-Making Techniques. Journal of Agricultural and Food Chemistry. 2013;61:2538–2547. doi: 10.1021/jf304987m. [DOI] [PubMed] [Google Scholar]
- 4.Cuskin F, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsbrink J, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogowski A, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 2015;6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martens EC, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amaya MF, et al. Structural insights into the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Structure. 2004;12:775–784. doi: 10.1016/j.str.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis BG, et al. Tetrazoles of manno- and rhamno-pyranoses: Contrasting inhibition of mannosidases by 4.3.0 but of rhamnosidase by 3.3.0 bicyclic tetrazoles. Tetrahedron. 1999;55:4489–4500. [Google Scholar]
- 12.Speciale G, Thompson AJ, Davies GJ, Williams SJ. Dissecting conformational contributions to glycosidase catalysis and inhibition. Curr Opin Struct Biol. 2014;28:1–13. doi: 10.1016/j.sbi.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita K, et al. Molecular cloning and characterization of a beta-l-Arabinobiosidase in Bifidobacterium longum that belongs to a novel glycoside hydrolase family. J Biol Chem. 2011;286:5143–5150. doi: 10.1074/jbc.M110.190512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spellman MW, McNeil M, Darvill AG, Albersheim P, Henrick K. Isolation and characterization of 3-C-carboxy-5-deoxy-l-xylose, a naturally occurring, branched-chain, acidic monosaccharide. Carbohydrate Research. 1983;122:115–129. [Google Scholar]
- 15.Guerardel Y, et al. The nematode Caenorhabditis elegans synthesizes unusual O-linked glycans: identification of glucose-substituted mucin-type O-glycans and short chondroitin-like oligosaccharides. Biochem J. 2001;357:167–182. doi: 10.1042/0264-6021:3570167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russa R, Urbanik-Sypniewska T, Choma A, Mayer H. Identification of 3-deoxy-lyxo-2-heptulosaric acid in the core region of lipopolysaccharides from Rhizobiaceae. FEMS Microbiol Lett. 1991;68:337–343. doi: 10.1016/0378-1097(91)90379-o. [DOI] [PubMed] [Google Scholar]
- 17.Nagae M, et al. Structural basis of the catalytic reaction mechanism of novel 1,2-alpha-l-fucosidase from Bifidobacterium bifidum. J Biol Chem. 2007;282:18497–18509. doi: 10.1074/jbc.M702246200. [DOI] [PubMed] [Google Scholar]
- 18.Sulzenbacher G, et al. Crystal structure of Thermotoga maritima alpha-l-fucosidase. Insights into the catalytic mechanism and the molecular basis for fucosidosis. J Biol Chem. 2004;279:13119–13128. doi: 10.1074/jbc.M313783200. [DOI] [PubMed] [Google Scholar]
- 19.O'Neill MA, Ishii T, Albersheim P, Darvill AG. Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu Rev Plant Biol. 2004;55:109–139. doi: 10.1146/annurev.arplant.55.031903.141750. [DOI] [PubMed] [Google Scholar]
- 20.Pabst M, et al. Rhamnogalacturonan II structure shows variation in the side chains monosaccharide composition and methylation status within and across different plant species. Plant J. 2013;76:61–72. doi: 10.1111/tpj.12271. [DOI] [PubMed] [Google Scholar]
- 21.Cartmell A, et al. The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J Biol Chem. 2011;286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glenwright AJ, et al. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature. 2017;541:407–411. doi: 10.1038/nature20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourlard T, Pellerin P, Morvan C. Rhamnogalacturonans I and II are pectic substrates for flax-cell methyltransferases. Plant Physiology and Biochemistry. 1997;35:623–629. [Google Scholar]
- 24.Glushka JN, et al. Primary structure of the 2-O-methyl-alpha-l-fucose-containing side chain of the pectic polysaccharide, rhamnogalacturonan II. Carbohydr Res. 2003;338:341–352. doi: 10.1016/s0008-6215(02)00461-5. [DOI] [PubMed] [Google Scholar]
- 25.Raman R, et al. Advancing glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology. 2006;16:82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- 26.Buffetto F, et al. Recovery and fine structure variability of RGII sub-domains in wine (Vitis vinifera Merlot) Ann Bot. 2014;114:1327–1337. doi: 10.1093/aob/mcu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chauvin AL, Nepogodiev SA, Field RA. Synthesis of an apiose-containing disaccharide fragment of rhamnogalacturonan-II and some analogues. Carbohydr Res. 2004;339:21–27. doi: 10.1016/j.carres.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 28.Chauvin AL, Nepogodiev SA, Field RA. Synthesis of a 2,3,4-triglycosylated rhamnoside fragment of rhamnogalacturonan-II side chain A using a late stage oxidation approach. J Org Chem. 2005;70:960–966. doi: 10.1021/jo0482864. [DOI] [PubMed] [Google Scholar]
- 29.Nepogodiev SA, Fais M, Hughes DL, Field RA. Synthesis of apiose-containing oligosaccharide fragments of the plant cell wall: fragments of rhamnogalacturonan-II side chains A and B, and apiogalacturonan. Org Biomol Chem. 2011;9:6670–6684. doi: 10.1039/c1ob05587a. [DOI] [PubMed] [Google Scholar]
- 30.Charnock SJ, et al. The X6 “thermostabilizing” domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry. 2000;39:5013–5021. doi: 10.1021/bi992821q. [DOI] [PubMed] [Google Scholar]
- 31.Matsui I, et al. Subsite structure of Saccharomycopsis alpha-amylase secreted from Saccharomyces cerevisiae. J Biochem. 1991;109:566–569. doi: 10.1093/oxfordjournals.jbchem.a123420. [DOI] [PubMed] [Google Scholar]
- 32.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterman DG, et al. Diffraction-geometry refinement in the DIALS framework. Acta Crystallogr D Struct Biol. 2016;72:558–575. doi: 10.1107/S2059798316002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter G. xia2: an expert system for macromolecular crystallography data reduction. Journal of Applied Crystallography. 2010;43:186–190. [Google Scholar]
- 36.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. Journal of Applied Crystallography. 2004;37:843–844. [Google Scholar]
- 38.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowtan K. Fitting molecular fragments into electron density. Acta Crystallogr D Biol Crystallogr. 2008;64:83–89. doi: 10.1107/S0907444907033938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Vagin AA, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 45.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terrapon N, Lombard V, Gilbert HJ, Henrissat B. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics. 2015;31:647–655. doi: 10.1093/bioinformatics/btu716. [DOI] [PubMed] [Google Scholar]
- 47.Terrapon N, Weiner J, Grath S, Moore AD, Bornberg-Bauer E. Rapid similarity search of proteins using alignments of domain arrangements. Bioinformatics. 2014;30:274–281. doi: 10.1093/bioinformatics/btt379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.