Abstract
AI-2 is a quorum-sensing signaling molecule proposed to be involved in interspecies communication. In Escherichia coli and Salmonella enterica serovar Typhimurium, extracellular AI-2 accumulates in exponential phase, but the amount decreases drastically upon entry into stationary phase. In S. enterica serovar Typhimurium, the reduction in activity is due to import and processing of AI-2 by the Lsr transporter. We show that the Lsr transporter is functional in E. coli, and screening for mutants defective in AI-2 internalization revealed lsrK and glpD. Unlike the wild type, lsrK and glpD mutants do not activate transcription of the lsr operon in response to AI-2. lsrK encodes the AI-2 kinase, and the lsrK mutant fails to activate lsr expression because it cannot produce phospho-AI-2, which is the lsr operon inducer. glpD encodes the glycerol-3-phosphate (G3P) dehydrogenase, which is involved in glycerol and G3P metabolism. G3P accumulates in the glpD mutant and represses lsr transcription by preventing cyclic AMP (cAMP)-catabolite activator protein (CAP)-dependent activation. Dihydroxyacetone phosphate (DHAP) also accumulates in the glpD mutant, and DHAP represses lsr transcription by a cAMP-CAP-independent mechanism involving LsrR, the lsr operon repressor. The requirement for cAMP-CAP in lsr activation explains why AI-2 persists in culture fluids of bacteria grown in media containing sugars that cause catabolite repression. These findings show that, depending on the prevailing growth conditions, the amount of time that the AI-2 signal is present and, in turn, the time that a given community of bacteria remains exposed to this signal can vary greatly.
Quorum sensing is a cell-to-cell signaling process that enables bacteria to collectively control gene expression, thereby synchronizing activities that are productive only at a high population density. This process is accomplished through the production, secretion, and detection of small chemical signals called autoinducers. Production and detection of most autoinducers are restricted to organisms in a species. In contrast, one autoinducer, designated AI-2, and its synthase, LuxS, are widely distributed in the bacterial kingdom, and AI-2 controls a variety of traits in different bacteria (18, 48, 55). These unique characteristics of AI-2 have led to the hypothesis that AI-2 is used for interspecies communication.
AI-2 was initially identified for its control of the expression of bioluminescence in the marine bacterium Vibrio harveyi (1). Genetic and biochemical analyses of mutants defective in AI-2 production showed that AI-2 is made from S-adenosylmethionine (39), which is used as a methyl donor in a variety of cellular processes which yield S-adenosylhomocysteine. S-Adenosylhomocysteine is subsequently metabolized to adenine and S-ribosylhomocysteine. S-Ribosylhomocysteine is the substrate for LuxS, which cleaves it to produce homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD). DPD cyclizes spontaneously and undergoes further rearrangements to form AI-2. The structure of AI-2 bound to the V. harveyi AI-2 binding protein LuxP was determined, and the results showed that V. harveyi AI-2 is a furanosyl borate diester, indicating that borate adds to the hydrated cyclized DPD molecule (8).
The biosynthetic pathways leading to production of DPD have been shown to be identical in Escherichia coli, Salmonella enterica serovar Typhimurium, V. harveyi, Vibrio cholerae, Enterococcus faecalis, Neisseria meningitidis, Porphyromonas gingivalis, and Staphylococcus aureus (39, 54). While these findings indicate that all LuxS-containing bacteria make DPD by the same metabolic pathway, it is not clear whether any bacteria besides V. harveyi use the furanosyl borate diester form of AI-2 or if different bacteria use a variety of rearranged species of DPD as the active AI-2 signal. Recent work with the AI-2 binding protein from S. enterica serovar Typhimurium suggests that the latter is true. Specifically, analysis of the S. enterica serovar Typhimurium AI-2 bound to the S. enterica serovar Typhimurium AI-2 binding protein LsrB showed that it does not contain boron. Rather, cyclized, hydrated DPD, with a stereochemistry different from that of V. harveyi AI-2, is the active ligand for S. enterica serovar Typhimurium (35).
Since the discovery of AI-2 in V. harveyi, other organisms have been shown to use AI-2 to regulate genes specifying diverse functions, such as genes encoding virulence factors in Actinobacillus actinomycetemcomitans (19, 20), enterohemorrhagic E. coli (EHEC) O157:H7 (43), P. gingivalis (7, 9), Streptococcus pyogenes (32), V. cholerae (29, 34, 57), and Vibrio vulnificus (27); motility in Campylobacter jejuni (15), EHEC O157:H7, and enteropathogenic E. coli O127:H6 (21, 45); cell division in E. coli W3110 and EHEC O157:H7 (13, 44); antibiotic production in Photorhabdus luminescens (14); biofilm formation and carbohydrate metabolism in Streptococcus gordonii (33); and an AI-2 ATP binding cassette (ABC)-type transporter in S. enterica serovar Typhimurium (50). These reports indicate that different bacteria use AI-2 to control an assortment of niche-specific genes. However, the mechanism of AI-2 detection and the signal transduction pathway linking AI-2 detection to target gene expression have been established only in V. harveyi, V. cholerae, and S. enterica serovar Typhimurium.
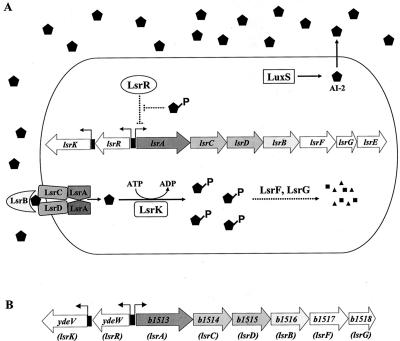
Surprisingly, unlike canonical autoinducers which accumulate in the stationary phase, in most bacteria examined, extracellular AI-2 activity peaks in mid- to late exponential phase and declines precipitously in stationary phase. In S. enterica serovar Typhimurium the rapid disappearance of AI-2 is a consequence of its import by an ABC transporter designated the Lsr transporter (luxS regulated). The lsr operon contains seven genes, lsrACDBFGE, and its transcription is activated by AI-2 (Fig. 1A) (50). The first four genes, lsrACDB, encode components of the AI-2 transporter apparatus. The distal genes are required for processing of AI-2 following internalization (49). Adjacent to, but transcribed divergently from the lsr operon is lsrR, which encodes a repressor of lsr transcription, and lsrK, which encodes a kinase that phosphorylates intracellular AI-2 following import (49, 50). Phosphorylation of internalized AI-2 is required for induction of transcription of the lsr operon, suggesting that phospho-AI-2 is the inducer of this system. It is postulated that phospho-AI-2 binds to the LsrR repressor and inactivates it and that this results in derepression of lsr transcription (49) (Fig. 1A).
FIG. 1.
Model for AI-2 production and internalization in S. enterica serovar Typhimurium. (A) AI-2 (pentagons) is synthesized by LuxS and accumulates extracellularly. AI-2 is internalized by the Lsr ABC-type transporter, and internalized AI-2 is phosphorylated by the LsrK kinase. Phospho-AI-2 is the inducer of transcription of the lsr operon and is proposed to act by binding to LsrR, the repressor of the lsr operon, inactivating it. LsrF and LsrG are required for further processing of internalized AI-2. The dotted lines indicate hypothetical processes. (B) E. coli b1513 operon is homologous to the S. enterica serovar Typhimurium lsr operon. lsr gene designations are indicated under the annotations. b1516 (lsrB) encodes the periplasmic AI-2 binding protein. b1514 (lsrC) and b1515 (lsrD) encode the channel proteins, and b1513 (lsrA) encodes the ATPase that provides energy for AI-2 transport. b1517 (lsrF) is similar to genes specifying aldolases, and b1518 (lsrG) encodes a protein with an unknown function. There is no lsrE in the E. coli lsr operon. ydeV and ydeW encode proteins homologous to the AI-2 kinase LsrK and the lsr repressor LsrR, respectively.
E. coli has an operon that is homologous to the S. enterica serovar Typhimurium lsr operon and is annotated the b1513 operon (Fig. 1B). In the present work, we showed that the b1513 operon of E. coli also encodes an AI-2 transporter. Additionally, a genetic screen to identify E. coli mutants impaired in the ability to import AI-2 from culture fluids revealed that mutants blocked in glycerol or glycerol 3-phosphate (G3P) metabolism are unable to induce lsr transcription and thus cannot internalize AI-2. We propose that repression of lsr transcription is caused by the accumulation of cytoplasmic G3P and dihydroxyacetone phosphate (DHAP). G3P represses the lsr operon via catabolite repression, whereas DHAP causes repression by an LsrR-dependent and catabolite repression-independent route. We suggest that DHAP inhibits binding of phospho-AI-2 to LsrR and that this prevents induction of transcription of the lsr operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains used are listed in Table 1. Wild-type (WT) E. coli K-12 strain MG1655 (5) was used as the parental strain. Strains were grown in Luria-Bertani (LB) medium with shaking at 37°C. Where indicated below, medium was supplemented with carbon sources at a concentration of 0.4% (wt/vol) and/or with antibiotics at the following final concentrations: ampicillin, 100 mg liter−1; chloramphenicol, 25 mg liter−1; kanamycin, 50 mg liter−1; and tetracycline, 10 mg liter−1.
TABLE 1.
E. coli strains used in this study
| Strain | Relevant genotype | Parent strain | Source or strain construction (reference) |
|---|---|---|---|
| MG1655 | Wild type | ||
| KX11 | lsrK::Tn10Cm | MG1655 | See text |
| KX17 | glpD::Tn10Cm | MG1655 | See text |
| KX1200 | ΔluxS::Cm | MG1655 | Primers Ec39 and Ec40 for deletion |
| KX1108 | ΔlacZYA | MG1655 | Primers Ec42 and Ec43 for deletion, Cm removed by flip out (12) |
| KX1123 | lsr-lacZ | KX1108 | See text |
| KX1218 | lsr-lacZ ΔluxS::Cm | KX1123 | ΔluxS::Cm from strain KX1200 |
| KX1290 | lsr-lacZ ΔluxS | KX1218 | Cm removed by flip out (12) |
| KX1186 | lsr-lacZ lsrK::Tn10Cm | KX1123 | lsrK::Tn10Cm from strain KX11 |
| KX1372 | lsr-lacZ lsrK::Tn10Cm ΔluxS | KX1290 | lsrK::Tn10Cm from strain KX11 |
| KX1304 | lsr-lacZ glpD::Tn10Cm | KX1123 | glpD::Tn10Cm from strain KX17 |
| KX1306 | lsr-lacZ glpD::Tn10Cm ΔluxS | KX1290 | glpD::Tn10Cm from strain KX17 |
| KX1382 | ΔlsrCDB::Cm | MG1655 | Primers Ec51 and Ec52 for deletion |
| KX1310 | lsr-lacZ ΔglpR::Cm | KX1123 | Primers Ec63 and Ec64 for deletion |
| RJ70 | glpF::Tn10Tet glpK | (53) | |
| KX1420 | lsr-lacZ glpK | KX1123 | glpK from strain RJ70 |
| KX1328 | lsr-lacZ ΔlsrR::Kan | KX1123 | Primers Ec47 and Ec48 for deletion |
| RD14 | Δcya::Kan crp* | Winfried Boos laboratory collection | |
| KX1481 | lsr-lacZ Δcya::Kan | KX1123 | Δcya::Kan from strain RD14 |
| KX1468 | lsr-lacZ Δcya::Kan crp* | KX1481 | crp* from strain RD14 |
| KX1483 | lsr-lacZ Δcya::Kan crp* glpD::Tn10Cm | KX1468 | glpD::Tn10Cm from strain KX17 |
| KX1322 | lsr-lacZ ΔlsrR::Cm | KX1123 | Primers Ec47 and Ec48 for deletion |
| KX1374 | lsr-lacZ ΔlsrR::Kan glpD::Tn10Cm | KX1328 | glpD::Tn10Cm from strain KX17 |
| KX1536 | lsr-lacZ glpD::Tn10Cm glpK | KX1304 | glpK from strain RJ70 |
| KX1541 | lsr-lacZ glpD::Tn10Cm glpK Δcya::Kan crp* | KX1483 | glpK from strain RJ70 |
| DLT242 | gldA::Tn10Tet Δ(glpFKX) | MC4100 | (52) |
| KX1537 | lsr-lacZ gldA::Tn10Tet Δ(glpFKX) | KX1123 | gldA::Tn10Tet Δ(glpFKX) from strain DLT242 |
| KX1543 | lsr-lacZ Δcya::Kan crp* gldA::Tn10Tet Δ(glpFKX) | KX1468 | gldA::Tn10Tet Δ(glpFKX) from strain DLT242 |
| KX1547 | lsr-lacZ gldA::Tn10Tet ΔglpK glpD::Tn10Cm | KX1537 | glpD::Tn10Cm from strain KX17 |
| KX1549 | lsr-lacZ Δcya::Kan crp* gldA::Tn10Tet ΔglpK glpD::Tn10Cm | KX1543 | glpD::Tn10Cm from strain KX17 |
Genetic and molecular techniques.
Generalized transduction with bacteriophage P1 was performed as described previously (41). Plasmid preparation and transformation were performed using standard protocols (38). PCRs were performed using Taq DNA polymerase (Boehringer Mannheim Biochemicals) except when PCR products were used for cloning. In the latter cases, ExTaq DNA polymerase (Takara Biochemicals) was used. Sequencing reactions were performed by the Princeton University SynSeq facility.
Screening for E. coli mutants defective in AI-2 internalization.
To identify genes involved in AI-2 internalization, we screened for mutants with high levels of AI-2 activity in cell-free fluids from cultures in stationary phase. WT E. coli MG1655 was mutated with mini-Tn10Cm (28), and mutants were selected on LB agar plates containing chloramphenicol. Approximately 10,000 mutant colonies were ordered on grids on LB agar plates containing chloramphenicol, and following growth, aliquots were transferred to 96-well microtiter plates (Polysterene; Costar, Corning Incorporated) containing LB medium with 30% glycerol. The plates were frozen and stored at −80°C. For screening, frozen mutants were stamped onto 96-well 0.22-μm-pore-size filtration plates (Multiscreen-GV; MAGGV2210; Millipore) containing 150 μl of LB medium. The cultures were grown at 37°C with shaking for 14 h. Cell-free culture fluids were collected by vacuum filtration and assayed for AI-2 activity (as described below). Mutants that had detectable AI-2 activity in their culture fluids were selected for study, and this phenotype was verified by measuring AI-2 production throughout the growth curve. The mini-Tn10Cm insertion from each candidate mutant was backcrossed into MG1655 via P1 transduction, and the AI-2 internalization phenotype was verified. The location of each transposon insertion was identified by arbitrary primed PCR (36, 37). When necessary, PCR products were purified from 1% low-melting-point agarose (SeaPlaque; FMC Bioproducts) with β-agarase (New England Biolabs). The transposon-chromosome fusion junctions were sequenced with primer Ec67 (Table 2).
TABLE 2.
Primers used in this study
| Primer | Oligonucleotide sequence |
|---|---|
| Ec39 | TCAGAAAATTTTTAAAAAAATTACCGGAGGTGGCTAAATGGTGTAGGCTGGAGCTGCTTC |
| Ec40 | TCATTTGAACTGGCTTTTTTCAATTAATTGTGAAGATAGTTTACTGACTACATATGAATATCCTCCTTAGT |
| Ec42 | GAGCGCAACGCAATTAATGTGAGTTAGCTCACTCATTAGGCACCCCAGGCGTGTAGGCTGGAGCTGCTTC |
| Ec43 | GCTGAACTTGTAGGCCTGATAAGCGCAGCGTATCAGGCAATTTTTATAATCATATGAATATCCTCCTTAGT |
| Ec47 | GTGAAGAATGAATTATGACAATCAACGATTCGGCAATTTCAGAACAGGGAGTGTAGGCTGGAGCTGCTTC |
| Ec48 | CTCTATACGTTCTCCATCATTCCCGGTAATAAGGTCATGCAAATTTAACTCATATGAATATCCTCCTTAGT |
| Ec51 | CTGAAGTTTATTCAGAACAACCGTGAAATCACGGCACTGCTGGCGGTGGTGGTGTAGGCTGGAGCTGCTTC |
| Ec52 | GAAATCGTATTTGCCGATATTCTCTTTGTTGAATATCACGCGCTCCGGTAACATATGAATATCCTCCTTAGT |
| Ec63 | CCAGGGATTTATAAATGAAACAAACACAACGTCACAACGGTATTATCGAAGTGTAGGCTGGAGCTGCTTC |
| Ec64 | AAATACCTGGCGCGTTTTGGTCTGACGTGGGAAGCCGTGCAGGATCAGCACATATGAATATCCTCCTTAGT |
| Ec68 | GCGGAATTCGAGTTTCATATTCCAGACAGCCTTC |
| Ec69 | GCGGGATCCGAACTGGCGTTAATCTGACGTAG |
| Ec67 | CTGCCTCCCAGAGCCTG |
AI-2 activity assay.
The AI-2 activity in cell-free E. coli culture fluids was measured using the V. harveyi BB170 bioluminescence reporter assay, as described previously (1, 2). Cell-free culture fluids were prepared by filtration of liquid cultures (46, 47) or by filtration through 96-well filtration plates as described above. AI-2 activity is reported below as fold induction of light production compared with the background light obtained with the appropriate E. coli growth medium.
Time course of AI-2 production.
To measure AI-2 production in E. coli strains during growth, overnight cultures were diluted (1:100) into 200 ml of LB medium in 2-liter Erlenmeyer flasks. Aliquots were collected at various times and used for measurement of the optical density at 600 nm (OD600), preparation of cell-free culture fluids, and preparation of cell extracts for Western blot analysis when necessary (see below). To distinguish between mutants with reduced AI-2 internalization and mutants with growth defects, the AI-2 production and growth rates of the mutants were determined. Parallel 96-well filtration plates and standard 96-well microtiter plates containing 150 μl of LB medium and 2 μl of the overnight cultures of the candidate mutants were incubated for various times at 37°C with shaking. The cultures from the filter plates were used to prepare cell-free culture fluids for AI-2 activity assays, and the cultures in the standard plates were used for measurement of the optical density with a Wallac Victor2 model 1420 multilabel counter.
Western blot analysis.
To measure LuxS protein in E. coli, culture aliquots were collected throughout growth, and the OD600 was used to normalize the number of cells per milliliter. Culture volumes equivalent to 1 ml with an OD600 of 1 were harvested by centrifugation. The cells were resuspended in 250 μl of water and frozen at −80°C. To 100 μl of frozen cells, 50 μl of 3× sodium dodecyl sulfate-polyacrylamide electrophoresis sample buffer was added, and samples were boiled for 10 min. Identical samples (20 μl) were loaded into two separate sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis gels. One gel was used for protein visualization, and the other was used for Western transfer and analysis with anti-LuxS polyclonal antiserum (25). Anti-rabbit immunoglobulin G-alkaline phosphatase conjugate (Promega) was used for visualization.
Construction of a chromosomal single-copy lsr-lacZ transcriptional fusion.
An lsr-lacZ transcriptional fusion was constructed by the method described by Hand and Silhavy (23). The lsr promoter region, containing the lsrA (b1513)-lsrR (ydeW) intergenic region and about 200 nucleotides 5′ and 3′ of this region, was PCR amplified from the E. coli chromosome with primers Ec68 and Ec69 (Table 2). The resulting 721-bp PCR product was purified, digested with EcoRI and BamHI, and ligated to the EcoRI and BamHI sites located immediately upstream of the promoterless lacZ gene in pRS415 (23). The plasmid obtained was electroporated into KX1108 (ΔlacZYA), and subsequently the lsr-lacZ fusion was recombined onto λRS45 and integrated into the λ attachment (att) site of KX1108.
Construction of deletion and insertion mutants.
Deletions were constructed by methods described previously (12). Antibiotic resistance cassettes were amplified by PCR from plasmids pKD3 (chloramphenicol) and pKD4 (kanamycin), using primers with 20 bp of homology to the flanking regions of the antibiotic cassette and 50 bp of homology to the flanking regions of the gene to be deleted. The primers used for each deletion are listed in Table 2. glpK mutants were constructed by transducing the glpF::Tn10Tet insertion from strain RJ70 to the relevant strains. This glpF::Tn10Tet insertion is polar on glpK, and consequently, strains with this insertion are unable to phosphorylate glycerol and cannot grow on glycerol as the sole carbon source (53). Importantly, the insertion in glpF does not affect glycerol transport at the glycerol concentrations used in this work (31). P1 transduction from strain DLT242 [gldA::Tn10Tet Δ(glpFKX)] was used to construct the gldA and gldA glpK mutants (52). gldA is cotransducible with glpFKX. Therefore, gldA single mutants were obtained by selecting for tetracycline resistance followed by screening for growth on glycerol minimal medium, whereas gldA glpFKX double mutants were obtained by selection for tetracycline resistance followed by screening for the inability to grow on glycerol as the sole source of carbon.
Construction of the Δcya::Kan crp* strains.
To construct strains insensitive to catabolite repression, we used a P1 lysate from strain RD14 (Winfried Boos laboratory collection) which contains a cya deletion linked to kanamycin and the crp* mutation encoding a derivative of catabolite activator protein (CAP) that acts as a transcriptional activator in the absence of cyclic AMP (cAMP). These two mutations were transferred to the desired strains by two sequential P1 transductions. First, Δcya::Kan was transduced with P1 to KX1123 (lsr-lacZ) by using selection for Kanr (obtaining strain KX1481). Because a Δcya::Kan mutant cannot grow on glycerol, in a second step we used the P1 lysate from RD14 to transduce the crp* mutation into KX1481 (lsr-lacZ Δcya::Kan) by selecting for growth on M63 medium containing glycerol and kanamycin. This second step produced strain KX1468 (lsr-lacZ Δcya::Kan crp*). Whenever the glpD::Tn10Cm insertion was transduced into a strain containing the Δcya::Kan crp* mutations, we verified that the resulting strain retained the ability to grow on maltose minimal medium to ensure that the crp* mutation had been maintained.
β-Galactosidase assays.
Overnight cultures of E. coli were diluted 1:100 into fresh LB medium and grown with aeration at 37°C for 5 h or as indicated below. Cells from 1 ml of culture were harvested and resuspended in 1 ml of Z buffer for determination of the β-galactosidase activity as described previously (42). β-Galactosidase activity was calculated as follows: (OD420 minute−1 × dilution factor)/OD600. All assays were performed in triplicate. The error bars in the graphs below indicate the standard deviations.
RESULTS
Identification of genes involved in AI-2 internalization in E. coli.
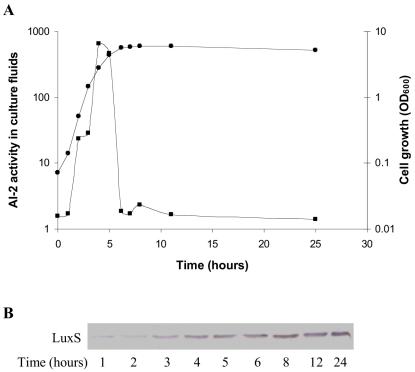
When E. coli was grown in LB medium, AI-2 activity increased during exponential growth and began to decline during the transition from exponential phase to stationary phase. By early stationary phase there was no detectable AI-2 in E. coli culture fluids (Fig. 2A). Antibodies to the LuxS protein showed that despite drastically reduced levels of extracellular AI-2, the AI-2 synthase LuxS was present even during late stationary phase (Fig. 2B). Thus, we could not account for the reduced AI-2 activity through obvious effects on LuxS production. Consistent with this finding, studies with S. enterica serovar Typhimurium have shown that transcription and translation of luxS remain constant throughout all phases of growth (3).
FIG. 2.
Extracellular AI-2 accumulation in E. coli. WT E. coli strain MG1655 was inoculated into LB medium at time zero, and at various times aliquots were taken. (A) Cell growth was monitored by measuring the optical density (•), and AI-2 activity in cell-free culture fluids was measured using the V. harveyi bioluminescence assay (▪). (B) LuxS production was determined by Western blotting using anti-LuxS antibodies.
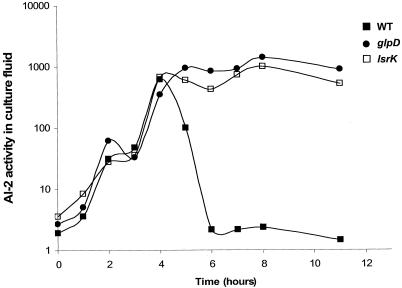
To identify genes that influence the levels of AI-2 in cell-free culture fluids, we constructed a library of random Tn10Cm transposon insertions in E. coli MG1655. Using an AI-2-specific reporter strain of V. harveyi, we screened the insertion mutants to identify those with AI-2 activity in the culture fluid in late stationary phase. Mutants with dramatic growth defects were discarded because we reasoned that slow growth would delay both extracellular AI-2 accumulation and disappearance. Ten thousand mutants were assayed, and two mutants were selected for study. The phenotypes of these two mutants are compared to that of the WT strain in Fig. 3. The transposon-chromosome fusion junctions of the selected mutants were amplified by PCR, and the insertion sites were identified by DNA sequence analysis coupled with BLAST database analysis.
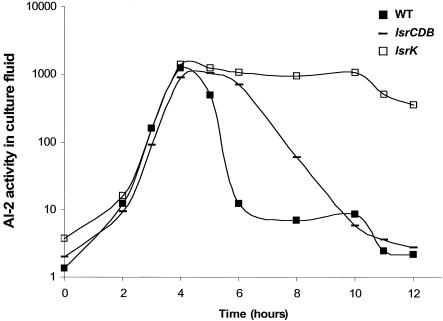
FIG. 3.
Extracellular AI-2 accumulation in E. coli mutants. AI-2 activity in cell-free culture fluids was measured using the V. harveyi bioluminescence bioassay. The following strains were analyzed: MG1655 (WT), KX17 (glpD), and KX11 (lsrK, annotated ydeV).
One transposon insertion that resulted in a mutant defective in AI-2 internalization was in glpD, the gene encoding the enzyme G3P dehydrogenase (GlpD), which catalyzes the aerobic oxidation of G3P to DHAP. This mutant displays a modest growth defect (data not shown). Nonetheless, it was chosen for further study because the defect in AI-2 internalization was more severe than would be expected to be due to the slightly lower growth rate. The second mutant had an insertion in ydeV, which encodes the homolog of the S. enterica serovar Typhimurium gene which we previously designated lsrK.
LsrK and the Lsr transporter are required for AI-2 internalization and processing in E. coli.
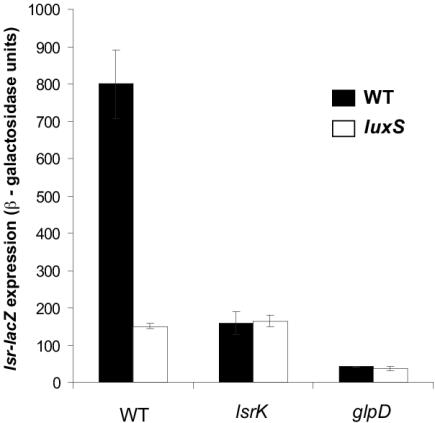
Finding ydeV in our screen of E. coli motivated us to examine the function and regulation of this gene, as well as the function and regulation of the other genes of the b1513 operon (homologous to the S. enterica serovar Typhimurium lsr operon) (Fig. 1B). To do this, we inserted a single copy of a b1513-lacZ promoter fusion at the att site of the E. coli chromosome and studied its regulation. Transcription of the b1513 operon in E. coli was induced in the WT strain but not in a ΔluxS mutant (Fig. 4). Addition of in vitro-synthesized AI-2 restored b1513 transcription in the ΔluxS mutant (data not shown). In the WT strain, expression of b1513-lacZ was maximal at 5 h of growth, which corresponded to maximal AI-2 accumulation in culture fluids, and mutants with mutations in transporter components (lsrCDB) were defective in internalization of AI-2 (Fig. 5).
FIG. 4.
Expression of the lsr operon in E. coli mutants in the presence and absence of luxS. The β-galactosidase activity of the lsr-lacZ (b1513-lacZ) fusion was determined in strains KX1123 (WT), KX1218 (luxS), KX1186 (lsrK), KX1372 (lsrK luxS), KX1304 (glpD), and KX1306 (glpD luxS) after 5 h of growth.
FIG. 5.
Extracellular AI-2 accumulation in E. coli Lsr transporter mutants. AI-2 activity in cell-free culture fluids was measured using the V. harveyi bioluminescence bioassay. The following strains were analyzed: MG1655 (WT), KX1382 (lsrCDB), and KX11 (lsrK).
The E. coli ydeV mutant displayed only low-level transcription of b1513 both in the presence and in the absence of luxS (Fig. 4). In the S. enterica serovar Typhimurium lsrK mutant, transcription of the lsr operon was similarly reduced, and this resulted in a low level of production of the Lsr transporter, which caused AI-2 accumulation and persistence in the extracellular medium. Figure 5 shows that this was true for the E. coli ydeV (lsrK) mutant. Therefore, with respect to regulation of expression of the operon and AI-2 uptake, the E. coli b1513 (lsrCDB) and ydeV (lsrK) mutants have phenotypes identical to those of the S. enterica serovar Typhimurium lsr operon and lsrK mutants, respectively. We concluded that these functions are analogous in E. coli and S. enterica serovar Typhimurium, and therefore we designated the b1513 operon of E. coli the lsr operon and the E. coli ydeV gene lsrK (Fig. 1B).
It was initially puzzlingly that of the lsr genes, only lsrK was identified in the present screen because the lsrACDB genes are also required to efficiently remove AI-2 from culture fluids. However, we determined that E. coli lsrCDB mutants but not lsrK mutants are capable of removing AI-2 from culture fluids, albeit significantly more slowly than the WT strain (Fig. 5). We propose that the slow AI-2 internalization occurs through some low-affinity transport system (49). Specifically, WT E. coli internalizes most of the AI-2 by 6 h, whereas an lsrCDB mutant requires 10 h for the equivalent internalization (Fig. 5) and the lsrK mutant internalizes little or no AI-2 even after 12 h (Fig. 3 and 5). Our determinations of AI-2 activity in the culture fluids of the transposon insertion mutants were made after 14 h growth. Therefore, we believe that the persistence of AI-2 for a longer period of time in culture fluids of the lsrK mutant than in culture fluids of the lsrCDB mutants accounted for our identification of lsrK but not lsrACDB in this experimental setup.
Interestingly, similar to the phenotype of an E. coli lsrK mutant, lsr-lacZ remained repressed in a glpD mutant irrespective of the presence of luxS (Fig. 4). We assumed that reduced lsr operon transcription in the glpD mutant resulted in an inability to assemble the Lsr transporter and to import AI-2, and this explains why AI-2 persisted in the cell-free culture fluids.
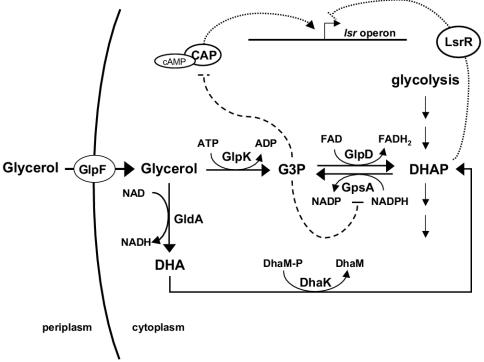
glp regulon and lsr repression.
GlpD is the enzyme responsible for funneling G3P to the glycolytic pathway, and it is essential for glycerol and G3P metabolism under aerobic conditions (Fig. 6) (30, 31). E. coli has another G3P dehydrogenase encoded by glpA, but this enzyme is functional only under anaerobic conditions (30, 31). glpD is a member of the glp regulon, which is under transcriptional control of the GlpR repressor. Specifically, the glp regulon is repressed by GlpR in the absence of glycerol. Growth on glycerol causes derepression of the regulon, and glycerol is imported via the GlpF permease. Internalized glycerol is phosphorylated to G3P by the glycerol kinase GlpK, and G3P is subsequently oxidized to DHAP by GlpD (Fig. 6). To understand why the lsr operon is repressed in a glpD mutant, we examined the expression of the lsr operon in E. coli in the presence of glycerol and G3P (Table 3). Addition of glycerol and G3P to the LB growth medium caused 21- and 5-fold repression of lsr-lacZ transcription, respectively. Glycerol probably caused greater lsr repression than G3P caused because glycerol is internalized more efficiently than G3P. Addition of either glycerol or G3P to the glpD mutant did not alter lsr expression. However, this result was anticipated because transcription of the lsr operon was already fully repressed in the glpD mutant (Table 3).
FIG. 6.
Aerobic glycerol and G3P metabolism in E. coli. Glycerol enters the cytoplasm through the glycerol facilitator (GlpF) and can be phosphorylated to G3P by the glycerol kinase (GlpK). In the presence of oxygen, G3P is oxidized by the G3P dehydrogenase (GlpD) to DHAP, which is further metabolized through the glycolytic pathway. Intracellular glycerol can also be oxidized to DHA by GldA. DHA is converted to DHAP by the DHA kinase (DhaK), which uses DhaM as a phosphoryl donor protein. G3P is required for phospholipid biosynthesis, and in the absence of extracellular glycerol, intracellular G3P is formed from DHAP by the G3P synthase (GpsA). In a glpD mutant G3P accumulates due to conversion of glycerol to G3P by GlpK. Intracellular G3P accumulation prevents cAMP formation by inhibiting the stimulation of adenylate cyclase via phospho-EIIAGlc (16). As a consequence, cAMP-CAP activation of the lsr operon is inhibited significantly. G3P accumulation also inhibits GpsA by a negative feedback mechanism, which leads to DHAP accumulation. We hypothesize that DHAP represses the lsr operon by a mechanism independent of cAMP-CAP that involves LsrR, the repressor of the lsr operon. The solid lines indicate enzymatic reactions, the dashed lines indicate regulatory interactions, and the dotted lines indicate the newly proposed regulatory interaction resulting from this work. FAD, flavin adenine dinucleotide; FADH2, reduced flavin adenine dinucleotide.
TABLE 3.
lsr-lacZ expression in E. coli grown in LB medium with glycerol or G3P
| Genotype | β-Galactosidase activity (u)a
|
Fold repression with:
|
|||
|---|---|---|---|---|---|
| LB medium | LB medium + glycerol | LB medium + G3P | Glycerol | G3P | |
| WT | 1,071 ± 66 | 50 ± 1 | 214 ± 2 | 21 | 5 |
| glpD | 41 ± 1 | 39 ± 1 | 49 ± 1 | 1 | 1 |
| glpR | 752 ± 68 | 26 ± 1 | 134 ± 4 | 29 | 6 |
| glpK | 885 ± 52 | 274 ± 2 | 171 ± 5 | 3 | 5 |
The β-galactosidase activities of the lsr-lacZ fusion were measured in strains KX1123 (WT), KX1304 (glpD), KX1310 (glpR), and KX1420 (glpK) following 5 h of growth in LB medium or LB medium containing 0.4% glycerol or 0.4% G3P.
Curiously, there is a putative GlpR binding site immediately downstream of the predicted lsrR promoter, encoding the repressor of the lsr operon. This observation suggested that GlpR could directly regulate the lsr operon by regulating lsrR expression. We found that this cannot be the case, however, because transcription of the lsr promoter, as measured by the lsr-lacZ fusion, was similar in WT E. coli and the ΔglpR mutant and, furthermore, lsr repression by glycerol and G3P occurred in the ΔglpR mutant (Table 3). In fact, repression of lsr expression by glycerol and G3P was slightly greater in the ΔglpR mutant than in WT E. coli. We reasoned that because the glp regulon was derepressed in the ΔglpR mutant, the glycerol and G3P transport systems were expressed at higher levels in the ΔglpR mutant than in the WT strain. Thus, compared to the WT strain, increased uptake of the two substrates occurred in the ΔglpR mutant, which in turn promoted increased repression of lsr expression.
Both glycerol and G3P repress lsr transcription.
Glycerol represses the maltose (mal) uptake and utilization regulon via catabolite repression (16, 17), and we suspected that glycerol could similarly repress lsr and, in turn, AI-2 uptake and utilization. The mal regulon of E. coli is a typical catabolite-sensitive regulon. Cyclic AMP and the catabolite activator protein (cAMP-CAP) control expression of several members of this regulon (6). Glycerol-mediated repression of the mal regulon occurs when glycerol is converted to G3P via phosphorylation by the glycerol kinase GlpK, but it does not occur in a glpK mutant (Fig. 6). Importantly, mal gene transcription is repressed following exogenous addition of G3P to either the glpK or glpD mutant (i.e., even when G3P cannot be converted to DHAP [Fig. 6]). Thus, G3P is the sole mediator of glycerol repression of the mal regulon, and neither glycerol, metabolites derived from glycerol, nor metabolites derived from G3P can substitute for G3P (16).
We investigated whether it was glycerol or G3P that was responsible for repression of lsr transcription by measuring lsr transcription in a glpK mutant. Repression by glycerol decreased from 21-fold in the WT strain to 3-fold in the glpK mutant (Table 3). Mutation of glpK did not alter the ability of G3P to repress transcription of the lsr operon, as fivefold repression occurred in the WT strain and the glpK mutant (Table 3). These results show that the majority of the repression of lsr transcription that occurs when E. coli is grown in glycerol is due to G3P and not to glycerol. Interestingly, while glycerol causes no repression of mal genes in the glpK mutant, glycerol consistently causes low-level (threefold) repression of lsr transcription in the glpK mutant. We verified that this low-level lsr repression was not due to the slight increase in pH that occurs during growth in LB medium by growing cells in buffered LB medium with or without glycerol (data not shown). Thus, we suggest that G3P causes repression of the lsr operon but, unlike regulation of mal gene expression, additionally, glycerol itself or some product derived from glycerol also causes partial repression of lsr expression.
G3P represses lsr transcription via catabolite repression.
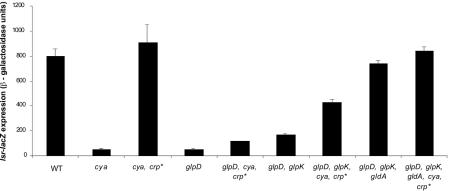
G3P-mediated repression of transcription of the mal genes in E. coli requires the cAMP-CAP system (16, 17). Double mutants harboring a deletion in the adenylate cyclase gene (cya) and a gain-of-function mutation in the CAP gene (crp*) are not sensitive to cAMP-CAP repression because the crp* mutation allows CAP to act as a transcriptional activator in the absence of cAMP. In a Δcya crp* genetic background, G3P does not repress transcription of mal regulon genes (16, 17). There is a CAP binding site upstream of the lsr operon, and consistent with this, lsr was repressed in the Δcya single mutant but not in the Δcya crp* double mutant (Fig. 7), showing that lsr transcription depends on cAMP and CAP. To examine whether G3P interferes with cAMP-CAP activation of the lsr operon, we added G3P to the Δcya crp* double mutant (Table 4). No G3P repression of lsr expression occurred in the Δcya crp* double mutant. Therefore, G3P repression occurs exclusively through prevention of cAMP-CAP-dependent activation of the lsr operon. Surprisingly, however, repression of the lsr operon did occur in a Δcya crp* mutant when glycerol was added (Table 4). Specifically, glycerol caused 21-fold repression in the WT strain and 8-fold repression in the Δcya crp* double mutant. Thus, repression by glycerol is partially independent of cAMP-CAP.
FIG. 7.
Effect of catabolite repression and the GldA pathway on the expression of lsr transcription. The β-galactosidase activity of the lsr-lacZ fusion was measured in strains KX1123 (WT), KX1481 (Δcya), KX1468 (Δcya crp*), KX1304 (glpD), KX1483 (glpD Δcya crp*), KX1536 (glpD glpK), KX1541 (glpD glpK cya crp*), KX1547 (glpD glpK gldA), and KX1549 (glpD glpK gldA cya crp*) after 5 h of growth in LB medium.
TABLE 4.
lsr-lacZ expression in E. coli mutants insensitive to catabolite repression grown in LB medium with G3P or glycerol
| Genotype | β-Galactosidase activity (u)a
|
Fold repression with:
|
|||
|---|---|---|---|---|---|
| LB medium | LB medium + G3P | LB medium + glycerol | G3P | Glycerol | |
| WT | 1,071 ± 66 | 226 ± 2 | 50 ± 1 | 5 | 21 |
| cya crp* | 1,094 ± 46 | 886 ± 12 | 137 ± 8 | 1 | 8 |
The β-galactosidase activities of the lsr-lacZ fusion were measured in strains KX1123 (WT) and KX1468 (Δcya crp*) following 5 h of growth in LB medium or LB medium containing 0.4% G3P or 0.4% glycerol.
Repression of lsr in a glpD mutant is caused by cAMP-CAP-dependent and -independent mechanisms.
We wondered if G3P-mediated inhibition of cAMP-CAP-dependent activation of lsr expression could explain our initial observation that the lsr operon is repressed in a glpD mutant. Specifically, in a glpD mutant, G3P metabolism is blocked and intracellular G3P accumulates (Fig. 6) (11, 56). We considered the possibility that in the glpD mutant increased intracellular G3P levels could lead to inhibition of cAMP-CAP-dependent activation of lsr. If this occurs, G3P repression of lsr should not occur in a glpD Δcya crp* triple mutant. Figure 7 shows that lsr expression was repressed 15-fold in the glpD mutant but only 7-fold in the glpD Δcya crp* triple mutant. These results show that G3P acting through cAMP-CAP accounts for a portion of the repression of the lsr operon observed in a glpD mutant, but an additional cAMP-CAP-independent mechanism must also be involved. To support this idea, we assayed the lsr expression phenotypes of the glpD glpK double mutant and the glpD glpK cya crp* quadruple mutant. In these mutants, G3P was not produced and glycerol accumulated (Fig. 6). The lsr operon remained repressed in a glpD glpK double mutant both in the presence and in the absence of a functional cAMP-CAP system (Fig. 7). These results, along with those shown in Table 4, verified that G3P accumulation cannot be the exclusive cause of lsr repression in a glpD mutant.
All of the results described above suggest that glycerol and/or a metabolite made from glycerol that is not G3P causes partial repression of lsr expression in the glpD mutant. A candidate for the metabolite is DHAP (Fig. 6). The GldA enzyme oxidizes glycerol to dihydroxyacetone (DHA) (26, 51, 52), which can subsequently be phosphorylated to DHAP by the DHA kinase, DhaK (22, 40) (Fig. 6). Although this route for glycerol metabolism is not sufficient to sustain growth on glycerol, it does allow conversion of glycerol to DHAP in a glpK mutant. To investigate the possibility that DHA or DHAP is involved in lsr repression, we transduced a gldA mutation into the glpD glpK double mutant and the glpD glpK Δcya crp* quadruple mutant. In both cases, inactivation of gldA restored expression of the lsr operon to WT levels (Fig. 7). We concluded that the lsr repression observed in glpD mutants unable to produce G3P is caused by the metabolism of glycerol via the GldA-DhaK pathway. We suggest that DHAP accumulates in the glpD single mutant and that it is the metabolite responsible for the cAMP-CAP-independent repression of the lsr operon. Accumulation of DHAP in a glpD mutant is expected because G3P accumulates and G3P feedback inhibits GpsA, the enzyme that catalyzes the conversion of DHAP to G3P (4, 10) (Fig. 6) (see Discussion).
We noted that in contrast to the results described above, the threefold repression of lsr expression that occurred in a glpK mutant grown on glycerol (Table 3) was likely not due to glycerol metabolism to DHAP through the GldA-DhaK pathway because this repression also occurred in a glpK gldA double mutant (data not shown). Rather, we believe that addition of extracellular glycerol does not induce the GldA-DhaK pathway when glpD is intact, and therefore, under the conditions used for the experiments whose results are shown in Table 3, glycerol itself appears to be able to cause a low level of lsr repression.
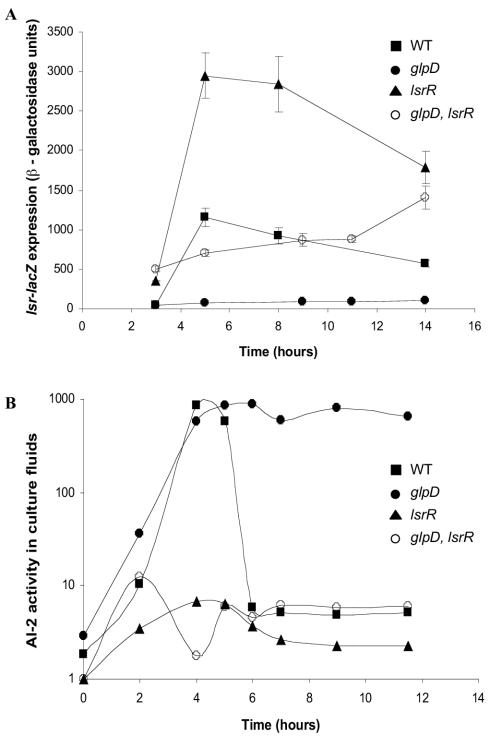
Repression of lsr expression in a glpD mutant requires LsrR.
We wondered whether repression of the lsr operon in the glpD mutant requires the known lsr regulator, LsrR. To test this, we measured the lsr expression and AI-2 activity phenotypes of the glpD and lsrR single mutants and the glpD lsrR double mutant. lsr operon expression was repressed in a glpD mutant (Fig. 8A), causing a reduction in AI-2 import, and consequently, AI-2 persisted in culture fluids (Fig. 8B). In contrast, inactivation of lsrR caused high-level expression of lsr (Fig. 8A), resulting in rapid AI-2 import and a dramatic loss of AI-2 activity from culture fluids (Fig. 8B). The repression of lsr transcription caused by mutation of glpD was partially relieved by mutation of lsrR (Fig. 8A). Importantly, this partial relief of transcriptional repression allowed AI-2 internalization that was as efficient as that in the lsrR single mutant (Fig. 8B). This result demonstrates that in terms of AI-2 transport, lsrR is epistatic to glpD, suggesting that LsrR is involved in the mechanism by which mutation of glpD causes lsr repression.
FIG. 8.
lsrR is epistatic to glpD. (A) β-Galactosidase activity of the lsr-lacZ fusion in strains KX1123 (WT), KX1304 (glpD), KX1328 (lsrR), and KX1374 (glpD lsrR) at different times during growth in LB medium. (B) AI-2 activity in cell-free culture fluids of strains MG1655 (WT), KX17 (glpD), KX1328 (lsrR), and KX1374 (glpD lsrR) as determined by the V. harveyi bioluminescence bioassay.
DISCUSSION
E. coli grown in LB medium releases and accumulates AI-2 in culture fluids during exponential growth. Maximal AI-2 activity is observed at the transition from exponential phase to stationary growth phase, after which the AI-2 activity rapidly disappears from the culture fluids. In S. enterica serovar Typhimurium, the disappearance of AI-2 from culture fluids is due to AI-2 uptake by the Lsr transport system, which is induced by the presence of AI-2. E. coli possesses an operon (previously designated the b1513 operon) homologous to the S. enterica serovar Typhimurium lsr operon. Transcriptional analysis of the E. coli lsr-lacZ fusion showed that AI-2 induces the expression of the lsr operon, and examination of mutants showed that the lsr operon is required for import and processing of AI-2. We concluded that the E. coli lsr operon functions analogously to the lsr operon of S. enterica serovar Typhimurium.
A screen for E. coli mutants defective in AI-2 internalization allowed us to identify lsrK and glpD. In these mutants, the lsr operon is uninducible, and so they display only low-level expression of the lsr operon both in the presence and in the absence of AI-2. We suspect that repression of transcription of the lsr operon in lsrK and glpD mutants results in their inability to assemble the Lsr transport apparatus, which in turn impairs their ability to internalize AI-2.
The E. coli lsrK homolog (annotated ydeV) is 79% identical to the S. enterica serovar Typhimurium lsrK gene. We previously showed that S. enterica serovar Typhimurium lsrK mutants do not internalize AI-2 like the WT does, and the lsr operon is not inducible by AI-2. Our characterization of the E. coli lsrK mutant showed it has phenotypes identical to those of the S. enterica serovar Typhimurium lsrK mutant (Fig. 3 and 4), suggesting that these mutants are functionally equivalent. We have shown explicitly that in S. enterica serovar Typhimurium LsrK phosphorylates AI-2. Therefore, lsrK mutants accumulate extracellular AI-2 because they cannot sequester AI-2 (as phospho-AI-2) in the cell. We propose that in E. coli, lsrK mutants do not induce lsr transcription in response to AI-2 because phospho-AI-2 is the antirepressor of lsr transcription, and this molecule is not produced in lsrK mutants.
A mutation in glpD causes a defect in AI-2 internalization, and, similar to the lsrK mutant, the glpD mutant does not induce transcription of lsr in response to AI-2. In a glpD mutant, G3P metabolism is blocked, and intracellular G3P accumulates as a consequence of phospholipid metabolism (11, 56). Our results show that the lsr operon is repressed by G3P via a cAMP-CAP-dependent mechanism. However, catabolite repression by G3P is not sufficient to explain lsr repression in the glpD mutant since this repression is not fully relieved by introduction of the Δcya crp* double mutation or by preventing G3P accumulation through introduction of a glpK mutation. Thus, an additional G3P-independent, cAMP-CAP-independent mechanism of lsr transcriptional repression must also be involved. When G3P metabolism is blocked, in addition to accumulation of G3P, DHAP can accumulate because G3P feedback inhibits GpsA-catalyzed conversion of DHAP to G3P (4, 10) (Fig. 6). Thus, increased G3P levels can promote increased DHAP levels. Therefore, the repression that we observed in the glpD mutant could have been due to DHAP or a metabolite derived from DHAP. Consistent with this hypothesis, when we eliminated glycerol metabolism to DHAP in the glpK glpD double mutant via inactivation of the GldA-DhaK pathway, repression of lsr was fully relieved.
Our results show that LsrR is required for the repression that we observed in the glpD mutant (Fig. 8). We propose that the cAMP-CAP-independent mechanism of lsr repression involves the interaction of DHAP (or possibly a metabolite derived from it) with the LsrR protein. Consistent with this, exogenous addition of DHA (which is converted to DHAP intracellularly) also causes LsrR-dependent lsr repression (data not shown). DHAP could act as an anti-inducer of the lsr operon by inhibiting the binding of phospho-AI-2 to LsrR, which could cause LsrR to remain locked in its active, repressing state. To validate this hypothesis, we are currently purifying LsrR for binding and competition assays with phospho-AI-2 and DHAP.
Understanding the physiology underlying the Glp-Lsr connection requires further analysis of the fate of internalized phospho-AI-2. In S. enterica serovar Typhimurium, the LsrF and LsrG proteins are involved in modifying phospho-AI-2, but the specific reactions that each carries out have not been characterized, nor are the products of these modifications known. It is possible that one of these products is DHAP since pentose phosphates are often converted to DHAP in order to be channeled to the glycolytic pathway for further metabolism. We are currently focusing on characterizing these biochemical reactions in both E. coli and S. enterica serovar Typhimurium.
Interestingly, the glpD mutant phenotype more closely mimics the lsrK mutant phenotype than the lsrCDB transporter mutant phenotypes. In transporter mutants, the presence of AI-2 in culture fluids is prolonged; however, AI-2 eventually disappears, presumably due to internalization by some low-affinity transporter. In contrast, AI-2 persists in culture fluids indefinitely in lsrK mutants, and we attribute this to a lack of phosphorylation or sequestration of internalized AI-2. Specifically, in an lsrK mutant, any AI-2 internalized by a secondary transporter does not get phosphorylated, and thus it cannot be sequestered. However, we do not believe that sequestration (i.e., AI-2 phosphorylation) is affected in the glpD mutant because in an lsrR glpD double mutant AI-2 is rapidly imported and remains sequestered (Fig. 8B), whereas in an lsrR lsrK double mutant extracellular AI-2 accumulates to wild-type levels (data not shown). We propose instead that the defect in AI-2 internalization is more severe in a glpD mutant than in the lsr transporter mutants because the secondary AI-2 transporter(s) is also subject to G3P catabolite repression in the glpD mutant. Consistent with this idea, in a glpD Δcya crp* triple mutant, although the lsr transporter is greatly repressed, most of the AI-2 is internalized by 10 h (data not shown).
Previous reports showed that high levels of extracellular AI-2 are detected when E. coli is grown on glucose, whereas no AI-2 can be detected in cell-free culture fluids when E. coli is grown in the absence of glucose (46). The present results explain both of these observations. First, in the presence of glucose, the lsr operon is not transcribed due to catabolite repression. Thus, AI-2 cannot be imported, and it accumulates in cell-free culture fluids. Second, in the absence of glucose, AI-2 is produced, but its presence is extremely transient due to rapid internalization by the Lsr transporter. DeLisa et al. (13) used DNA microarrays to identify genes controlled by AI-2 in E. coli. These experiments were performed with E. coli grown in the presence of glucose, and catabolite repression of transcription of the lsr operon by glucose could explain why none of the lsr genes was identified in this study. Similarly, it was reported that glucose, by an unknown mechanism, caused AI-2 to persist in cell-free culture fluids of E. coli (24). We show here that this mechanism is in fact cAMP-CAP-mediated repression of AI-2 import primarily through the Lsr apparatus.
At present, we do not understand the benefit that enteric bacteria derive from producing and releasing AI-2, only to internalize it later. Further work is necessary to determine if the physiological function of AI-2 as a signal in these bacteria is more significant under conditions in which AI-2 is imported and processed or under conditions in which the lsr transporter is not produced and AI-2 accumulates in the medium. In the first case, internalization of AI-2 could be used as a mechanism to terminate AI-2-controlled behaviors in E. coli or in other species in the vicinity. Alternatively, AI-2 internalization and modification could be used to transform the AI-2 signal into a different cytoplasmic signal. In the second case, in which the genes encoding the Lsr transport apparatus are repressed, cells that encounter AI-2 are exposed to this signal for a longer period of time than when the transporter is produced. Prolonged exposure to the signal could be useful for controlling other AI-2-dependent behaviors. There could be AI-2 receptors on the surface that, rather than internalize the signal, transduce the AI-2 sensory information to the cytoplasm to alter target gene expression. Studying the lsrK and lsrR mutants, in which the lsr operon is constitutively repressed and constitutively derepressed, respectively, should enable us to examine the role of AI-2 under these two conditions.
Acknowledgments
This work was supported by NIH grant 5RO1 GM065859 and by NSF grant MCB-0343821. K.B.X. was supported by Praxis XXI, Portugal award BPD-22064-99.
We are grateful to members of the Bassler lab for critical discussions of this work and to members of the Silhavy group for advice on experiments. We thank W. Boos for generously supplying strains and for insightful advice.
REFERENCES
- 1.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 3.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, R. M., and J. E. Cronan, Jr. 1975. Mutants of Escherichia coli defective in membrane phospholipid synthesis. Phenotypic suppression of sn-glycerol-3-phosphate acyltransferase Km mutants by loss of feedback inhibition of the biosynthetic sn-glycerol-3-phosphate dehydrogenase. J. Biol. Chem. 250:7153-7158. [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, J. Aduse-Opoku, M. A. Curtis, and M. Camara. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 9.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, D., V. Lightner, R. Edgar, P. Modrich, J. E. Cronan, Jr., and R. M. Bell. 1980. Regulation of phospholipid biosynthesis in Escherichia coli. Cloning of the structural gene for the biosynthetic sn-glycerol-3-phosphate dehydrogenase. J. Biol. Chem. 255:714-717. [PubMed] [Google Scholar]
- 11.Cronan, J. E., Jr., and C. O. Rock. 1996. Biosynthesis of membrane lipids, p. 612-636. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derzelle, S., E. Duchaud, F. Kunst, A. Danchin, and P. Bertin. 2002. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl. Environ. Microbiol. 68:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 16.Eppler, T., and W. Boos. 1999. Glycerol-3-phosphate-mediated repression of malT in Escherichia coli does not require metabolism, depends on enzyme IIAGlc and is mediated by cAMP levels. Mol. Microbiol. 33:1221-1231. [DOI] [PubMed] [Google Scholar]
- 17.Eppler, T., P. Postma, A. Schutz, U. Volker, and W. Boos. 2002. Glycerol-3-phosphate-induced catabolite repression in Escherichia coli. J. Bacteriol. 184:3044-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Federle, M. J., and B. L. Bassler. 2003. Interspecies communication in bacteria. J. Clin. Investig. 112:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 22.Gutknecht, R., R. Beutler, L. F. Garcia-Alles, U. Baumann, and B. Erni. 2001. The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 20:2480-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hand, N. J., and T. J. Silhavy. 2000. A practical guide to the construction and use of lac fusions in Escherichia coli. Methods Enzymol. 326:11-35. [DOI] [PubMed] [Google Scholar]
- 24.Hardie, K. R., C. Cooksley, A. D. Green, and K. Winzer. 2003. Autoinducer 2 activity in Escherichia coli culture supernatants can be actively reduced despite maintenance of an active synthase, LuxS. Microbiology 149:715-728. [DOI] [PubMed] [Google Scholar]
- 25.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, R. Z., J. C. Tang, and E. C. Lin. 1983. Experimental evolution of a novel pathway for glycerol dissimilation in Escherichia coli. J. Mol. Evol. 19:429-436. [DOI] [PubMed] [Google Scholar]
- 27.Kim, S. Y., S. E. Lee, Y. R. Kim, C. M. Kim, P. Y. Ryu, H. E. Choy, S. S. Chung, and J. H. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 28.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 29.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 30.Lin, E. C. 1976. Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 30:535-578. [DOI] [PubMed] [Google Scholar]
- 31.Lin, E. C. C. 1996. Dissimilatory pathways for sugars, polyols, and carboxylates, p. 307-342. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 32.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 33.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 35.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 37.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. N.Y.
- 39.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 40.Siebold, C., L. F. Garcia-Alles, B. Erni, and U. Baumann. 2003. A mechanism of covalent substrate binding in the X-ray structure of subunit K of the Escherichia coli dihydroxyacetone kinase. Proc. Natl. Acad. Sci. USA 100:8188-8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 42.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43809-821. [DOI] [PubMed] [Google Scholar]
- 46.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 48.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA. 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 50.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 51.Tang, J. C., E. J. St. Martin, and E. C. Lin. 1982. Derepression of an NAD-linked dehydrogenase that serves an Escherichia coli mutant for growth on glycerol. J. Bacteriol. 152:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Truniger, V., and W. Boos. 1994. Mapping and cloning of gldA, the structural gene of the Escherichia coli glycerol dehydrogenase. J. Bacteriol. 176:1796-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Truniger, V., W. Boos, and G. Sweet. 1992. Molecular analysis of the glpFKX regions of Escherichia coli and Shigella flexneri. J. Bacteriol. 174:6981-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 55.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 56.Yang, B., and T. J. Larson. 1998. Multiple promoters are responsible for transcription of the glpEGR operon of Escherichia coli K-12. Biochim. Biophys. Acta 1396:114-126. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]