Abstract
The amf gene cluster encodes a probable secretion system for a peptidic morphogen, AmfS, which induces aerial mycelium formation in Streptomyces griseus. Here we examined the transcriptional control mechanism for the promoter preceding amfT (PamfT) directing the transcription of the amfTSBA operon. High-resolution S1 analysis mapped a transcriptional start point at 31 nucleotides upstream of the translational start codon of amfT. Low-resolution analysis showed that PamfT is developmentally regulated in the wild type and completely abolished in an amfR mutant. The −35 region of PamfT contained the consensus sequence for the binding of BldD, a pleiotropic negative regulator for morphological and physiological development in Streptomyces coelicolor A3(2). The cloned bldD locus of S. griseus showed high sequence similarity to the S. coelicolor counterpart. Transcription of bldD occurred constitutively in both the wild type and an A-factor-deficient mutant of S. griseus, which suggests that the regulatory role of BldD is independent of A-factor. The gel retardation assay revealed that purified BldD and AmfR recombinant proteins specifically bind PamfT. Overproduction of BldD in the wild-type cell conferred a bald phenotype (defective in aerial growth and streptomycin production) and caused marked repression of PamfT activity. An amfT-depleted mutant also showed a bald phenotype but PamfT activity was not affected. Both the bldD-overproducing wild-type strain and the amfT mutant were unable to induce aerial growth of an amfS mutant in a cross-feeding assay, which indicates that these strains are defective in the production of an active AmfS peptide. The results overall suggests that two independent regulators, AmfR and BldD, control PamfT activity via direct binding to determine the transcriptional level of the amf operon responsible for the production and secretion of AmfS peptide, which induces the erection of aerial hyphae in S. griseus.
The gram-positive bacterial genus Streptomyces is characterized by the ability to perform complex morphological differentiation (3, 4). Early in the life cycle on solid media, the organism grows as branched multinucleoid substrate hyphae. In response to environmental and physiological signals, the older parts of the substrate hyphae produce aerial mycelium. After septa have formed at regular intervals along the hyphae, long chains of uninucleoid spores are formed. Streptomyces is also characterized by the ability to produce a wide variety of biologically active compounds, which have a number of applications in the medical, chemical, and agricultural industries (19). Studies in several species have demonstrated that the regulatory mechanisms for cellular differentiation and secondary metabolism are linked. For example, in Streptomyces griseus, both morphological differentiation and secondary metabolite formation are controlled by an autoregulatory substance, A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone). A-factor induces the transcription of specific regulatory genes for morphogenesis and secondary metabolism via the function of the receptor (ArpA) and global transcriptional activator (AdpA) (9).
We studied the regulatory role and function of the amf gene cluster in the onset of morphological development in S. griseus (25-27). The gene cluster consists of five coding sequences encoding a probable transmembrane protein (AmfT), a probable secreted peptide (AmfS), two HlyB-type ABC transporters (AmfB and AmfA), and a response regulator of a two-component regulatory system (AmfR) (Fig. 1A). The amf homologues of Streptomyces coelicolor A3(2) (2, 17) and Streptomyces avermitilis (11) are called ram and amf, respectively.
FIG. 1.
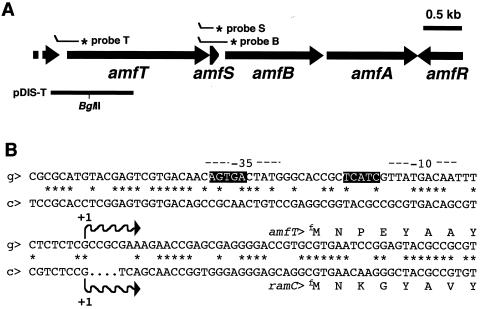
Schematic view of the amf gene cluster (A) and nucleotide sequence of PamfT (B). (A) The orientation and length of each amf coding sequence are shown. The positions of probes used for S1 mapping (T, B, and S) and the fragments cloned onto the disruption plasmid (pDIS-T) are also indicated. Asterisks show the radioactively labeled ends of the S1 probes. (B) The amfT promoter region of S. griseus (g) is shown with the corresponding sequence of the ramC promoter region of S. coelicolor A3(2) (c). The transcriptional start points assigned by high-resolution S1 protection assay and the sequences corresponding to the −35 and −10 regions are indicated by waved arrows and dashed lines, respectively. The dots represent sequence gaps between the two orthologous regions. The transcriptional start site of ramC has not been identified in S. coelicolor A3(2), and the site presented is that determined in S. lividans by Keiser et al. (12). The nucleotide sequences of the ramC promoter regions of S. coelicolor and S. lividans are identical. Boxed sequences correspond to the BldD-binding consensus of S. coelicolor A3(2) (7), which exists only in the amfT promoter.
Our previous study strongly suggested that AmfS acts as an extracellular morphogenic peptide that stimulates aerial growth (27). The AmfS precursor peptide expressed in substrate mycelium may be modified by an unknown mechanism and secreted by AmfA and AmfB to induce aerial growth. The activity of AmfS was observed in a cross-feeding experiment between the wild type and an amfS mutant, in which the latter colonies restore aerial growth upon receiving AmfS diffusing from the former colonies. The assay revealed that the amfR mutant is defective in the production of AmfS activity, which raised the possibility that the transcription of amfS and/or a related gene(s) is positively regulated by AmfR (27).
Recently, extensive studies by three research teams have shown that the ram gene cluster plays a significant regulatory role in developmental regulation in S. coelicolor A3(2), the best-studied model organism (12, 20). They revealed that promoter activities for ramR and ramCSAB, equivalent to amfR and amfTSBA, respectively, are developmentally regulated, and inactivation of either ramR or ramC abolishes aerial growth. The studies have also shown that RamR positively controls the promoter preceding ramC through direct binding. These reports of S. coelicolor studies support our previous observations on the amf genes of S. griseus and strongly reinforce the significance of the gene cluster in the control of Streptomyces development.
Here, we examined developmental regulation of the promoter preceding amfT, the main switch for expression of the amf genes. We find that the promoter is under dual regulation by AmfR and BldD, the central regulatory proteins for the onset of development in Streptomyces spp.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The wild-type strain S. griseus IFO13350 was obtained from the Institute of Fermentation Osaka, Japan. S. coelicolor A3(2) M145 was obtained from the John Innes Institute (Norwich, United Kingdom). Bacillus subtilis ATCC 6633 was used for the bioassay of streptomycin. Escherichia coli JM109 (18) and BL21(DE3) (Novagen) were used as hosts for DNA manipulation and expression of recombinant proteins. pUC19 (29) was used for general DNA manipulation. pT7Blue (Novagen) was used for TA cloning of PCR products. pGEX4T-2 (Pharmacia) and pET26b(+) (Novagen) were used as vectors for the effective expression of BldD and AmfR, respectively (see below). The conditions for genetic manipulation in E. coli and Streptomyces spp. were described by Maniatis et al. (18) and Kieser et al. (14), respectively. All Streptomyces plasmids used were described by Kieser et al. (14). S. griseus strains were grown in Bennett's sugar medium containing (in grams per liter) yeast extract (Difco Laboratories, Detroit, Mich.), 1; meat extract (Kyokuto, Tokyo, Japan), 1; NZ amine (Wako Pure Chemical Industries, Ltd., Tokyo, Japan), 2; and an appropriate sugar (Kokusan, Tokyo, Japan), 10 (pH 7.2), and YMP sugar medium, containing (in grams per liter) yeast extract (Difco), 2; meat extract (Kyokuto), 2; Bacto peptone (Difco), 4; NaCl, 5; MgSO4 · 7H2O, 2; and an appropriate sugar (Kokusan, Tokyo, Japan), 10 (pH 7.2). Agar (Kokusan) was supplied at 1.5% for solid media. For the selection of transformants, ampicillin (Wako) and kanamycin (Wako) at a final concentration of 50 μg ml−1 were used for E. coli. For S. griseus, thiostrepton (Sigma Chemical Company, St. Louis, Mo.) and kanamycin were added at a final concentration of 20 μg ml−1. Enzymes used for genetic manipulation were purchased from Takara-shuzo (Kyoto, Japan).
S1 nuclease mapping.
The transcriptional activities of the promoters preceding amfT (PamfT) and bldD (PbldD) were examined by an S1 protection assay. Methods for RNA preparation from cells grown on cellophane on the surface of agar medium and S1 nuclease mapping were as described by Kelemen et al. (13). Hybridization probes for PamfT (probe T; Fig. 1A) and PbldD were prepared by PCR with the oligonucleotide primer pairs TS1/TS2 and DS1/DS2 (Table 1), respectively. Probes for the intergenic region between amfT and amfB (probes S and B; Fig. 1A) were prepared as follows. The DNA fragments amplified with primers BS1/BS2 and BS1/BS3 were cloned onto pT7Blue, and the resultant plasmids were then used as templates for PCR with M13-RV (Takara)/BS2 and M13-RV/BS3 to generate probes S and B, respectively. Probes S and B contain a 5′-terminal mismatch region which distinguished the full-size protected fragments from unhybridized probe DNA.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequencea (5′-3′) | Restriction | Positions | Accession no. |
|---|---|---|---|---|
| TS1 | TCCGTGCTCTATTTCCGCACG | 10297-10317 | AB006206 | |
| TS2 | TGGGGTGCGTCGTAGAAGCG | 9841-9859 | AB006206 | |
| TD1 | CCGAATTCCTGTTCGTGCCCGTG | EcoRI | 10594-10616 | AB006206 |
| TD2 | GCCGAGACGTGAGATCTACCAGCCCTGG | Bg/II | 9673-9699 | AB006206 |
| TD3 | CCAGGGCTGGTAGATCTCACGTCTCGGC | Bg/II | 9673-9699 | AB006206 |
| TD4 | CGAAGCTTCTCCTCGTCGACCATG | HindIII | 8749-8766 | AB006206 |
| TG1 | TACTCCGAATTCACGCACGGT | EcoRI | 9886-9906 | AB006206 |
| TG2 | GGGAATTCAATACCCATCAGTACG | EcoRI | 10019-10042 | AB006206 |
| BS1 | AGCAGTTGATGCGCCTGTCGATGG | 7418-7441 | AB006206 | |
| BS2 | AGCTGTACTCACAGACCAGC | 7125-7144 | AB006206 | |
| BS3 | TGGAGCAGAGGAGGAGCCC | 6930-6949 | AB006206 | |
| DC1 | CGCGGATCCTCCAGCGAATACGCCAAAC | BamHI | 557-575 | AF045549 |
| DC2 | GTGCTCGAGGCTCAGAGCTCGTCGTG | XhoI | 1043-1060 | AF045549 |
| DC3 | ATGCATGCTCCCATACTAGG | SphI | 258-274 | U13854 |
| DC4 | CCGAGATCTGCATGTGTCAGAGG | Bg/II | 1020-1035 | U13854 |
| DS1 | TCGAAATTGCGTCATCCACG | 1250-1267 | AB114356 | |
| DS2 | AGTTTGGCCCCGAGCTGTTTGG | 1439-1461 | AB114356 | |
| DG2 | TCCACGCCGTGGAGGGAAAGG | 1485-1505 | AB114356 | |
| RC1 | AGACATATGACTACCGTGCTGCTGG | NdeI | 2561-2577 | AB006206 |
| RC2 | CGAAGGAAGCGGGCTGGATCCTCGAGCG | XhoI | 3142-3161 | AB006206 |
The restriction site is underlined.
For all probes, the downstream primers were labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. S1-protected fragments were analyzed on 6% polyacrylamide gels. The labeled primer TS2 was also used to generate dideoxy sequence ladders in the high-resolution analysis of PamfT. For high-resolution analysis of PbldD, Maxam-Gilbert sequence ladders prepared from the labeled hybridization probe were used as standards. Protected fragments were analyzed on a 6% polyacrylamide gel. The quality of RNA used for low-resolution analysis was checked by the control assay for hrdB, encoding a major sigma factor, with a probe described previously (16).
Cloning of S. griseus bldD.
The DNA fragment containing bldD was cloned by standard hybridization techniques from the chromosomal DNA of S. griseus. An internal 0.5-kb region of bldD was amplified from genomic DNA of S. coelicolor A3(2) by PCR with primers DC1 and DC2 (Table 1) and used as a probe for Southern hybridization with S. griseus chromosome. A 5-kb BamHI fragment that hybridized to the probe DNA was cloned at the BamHI site of pUC19 by the standard colony hybridization technique. The nucleotide sequence of the 2.4-kb region containing bldD was determined.
Preparation of recombinant BldD and AmfR proteins by E. coli host-vector systems.
For the preparation of recombinant proteins in E. coli, the coding sequences for bldD and amfR were amplified with primers DC1/DC2 and RC1/RC2 (Table 1) and cloned between the BamHI and XhoI sites of pGEX4T-2 and the NdeI and XhoI sites of pET-26b(+), respectively. The plasmid constructs directed the expression of BldD and AmfR as fusion proteins with an N-terminal glutathione S-transferase (GST) and a C-terminal hexameric histidine (6xHis) tag in E. coli JM109 and BL21(DE3), respectively. E. coli cells harboring the expression plasmids were cultured aerobically at 28°C in 100 ml of Luria broth (LB) liquid medium, to which was added 1 mM isopropylthiogalactopyranoside (IPTG) when the optical density at 600 nm reached 0.8. Cells were grown for 4 h after the addition of IPTG and harvested by centrifugation. The resultant cellular precipitate was suspended in an appropriate volume of phosphate-buffered saline (18) and disrupted by sonication. The soluble recombinant proteins were purified from the cell extract with appropriate affinity chromatographies following the method recommended by the manufacturer.
Gel mobility shift assay.
DNA-binding determinations by gel mobility shift assay followed the method described previously (25); 0.5 to 5.0 ng of 32P-labeled probe (10,000 to 20,000 cpm) was incubated with 1.0 to 20 μg of the recombinant proteins prepared as above at 30°C for 30 min in binding buffer containing 10 mM Tris-HCl (pH 7.0), 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10% (vol/vol) glycerol, 1 μg of poly(dI-dC), and 50 μg of bovine serum albumin per ml in a total volume of 50 μl. After incubation, complexes and free DNA were resolved on nondenaturing polyacrylamide gel containing 6% acrylamide. The gels were dried, and radioactive signals were detected with an image analyzer (Storm, Molecular Dynamics). For PamfT, the probe DNA was amplified with primers TG1 and TG2 (Table 1), digested at the restriction sites designed in the primer sequences, and labeled at the 5′ end with [α-32P]dATP with Klenow fragment. For PbldD, the DNA fragment amplified with primers DS1 and DG2 was labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase.
Disruption of amfT and overexpression of bldD.
amfT was disrupted by the standard homologous recombination technique, replacing the wild-type amfT allele with a mutated construct on a disruption plasmid by a double crossover event (pDIS-T; Fig. 1A). To construct pDIS-T, two 0.9-bp DNA fragments amplified by PCR with TD1/TD2 and TD3/TD4 (Table 1) were digested with EcoRI and BglII and with BglII and HindIII, respectively, and inserted between the EcoRI and HindIII sites of pUC19 by three-fragment ligation. The plasmid thus formed was cleaved with EcoRI and ligated to a 0.9-kb aphII (neomycin resistance) (1) cassette to generate pDIS-T. pDIS-T contains a nonsense codon and frameshift mutation in amfT at the position corresponding to the BglII site. pDIS-T was introduced into S. griseus wild-type cells by standard transformation, and neomycin-resistant segregants that carried an insertion of the whole pDIS-T region were selected. One of the neomycin-resistant strains thus obtained was then cultured in neomycin-free Bennett's liquid medium to promote the second crossover event that eliminates the neomycin resistance gene and one of the two amfT alleles. After checking by Southern hybridization, three true markerless disruptants that showed identical phenotypes were obtained. One of the recombinant strains was designated the amfT mutant.
For overexpression of BldD in S. griseus, the above gene cassette for the expression of GST-BldD was used as a template for PCR with primers DC3 and DC4 (Table 1), and the resultant amplicon was recovered as an SphI- and BglII-digested fragment. The fragment was then inserted between the SphI and BglII sites of pIJ702 to generate pIJ702-BldD. The plasmid thus formed carried the gene cassette downstream from the mel promoter and in the same orientation and directed the overexpression of GST-BldD driven by the promoter. The plasmid was introduced into wild-type S. griseus by the standard transformation technique.
Nucleotide sequence accession number.
The nucleotide sequence of bldD of S. griseus was submitted to DDBJ under accession no. AB114356.
RESULTS
S1 protection analysis of PamfT.
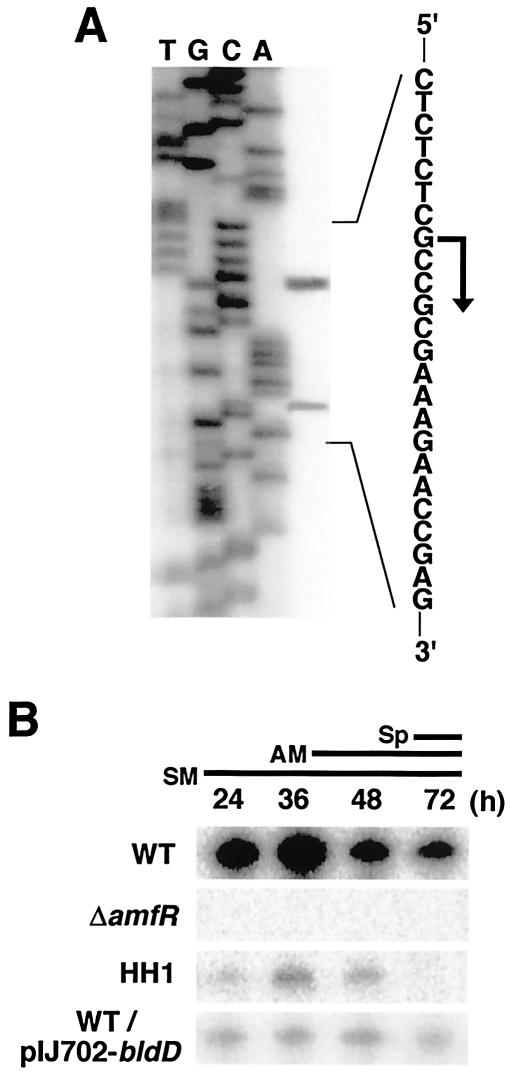
High-resolution S1 mapping assigned a transcriptional start point in PamfT 31 bp upstream from the translational initiation codon (GTG) of amfT (Fig. 1B and 2A). The minor signal that appeared 12 bp downstream from the start site is assumed to be a degradation product of the true protection fragment, judging from its slightly fluctuating signal intensity, although the signal also appeared when a different probe was used. We also examined transcription at the intergenic region between amfT and amfB and detected only full-size protection fragments derived from readthrough transcription from PamfT (data not shown). The result indicates that PamfT directs the major transcription of the amfTSBA operon.
FIG. 2.
S1 protection assay of the amfT promoter. The results of high-resolution (A) and low-resolution (B) analyses are shown. (A) The analysis assigned a transcriptional start point to the residue indicated by the bent arrow. RNA prepared from wild-type cells grown for 36 h on YMP/glucose solid medium was used for hybridization. Dideoxy sequencing ladders generated with primer TS2 (Table 1) were used as a reference. (B) RNA (10 μg) extracted from S. griseus cells grown for the indicated times on YMP/glucose solid medium was used for hybridization. The wild type grew as substrate mycelium (SM) on day 1, as a mixture of substrate and aerial mycelium (AM) on day 2, and as a mixture of aerial hyphae and spores (Sp) on day 3. WT, wild-type; ΔamfR, amfR mutant; HH1, A-factor-deficient; WT/pIJ702-BldD, wild-type strain harboring a plasmid directing overexpression of the GST-BldD fusion.
Low-resolution S1 analysis revealed that PamfT is developmentally regulated (Fig. 2B). In the wild-type strain, marked transcription occurred in the early growth phase and reached maximum in the transition phase. The activity was then reduced in the developmental phase. In contrast, promoter activity was completely abolished in a ΔamfR mutant (25) and markedly reduced in an A-factor biosynthesis mutant (HH1) (10). PamfT activity in ΔamfR and HH1 was restored to the wild-type level by introducing an intact amfR on a low-copy-number plasmid and by supplying synthetic A-factor, respectively (not shown). These results indicate that PamfT is positively controlled by A-factor and AmfR.
When the G residue corresponding to the major transcriptional start site is numbered +1, the sequences corresponding to −42 to −38 and −24 to −20 matched the consensus sequence for the binding of BldD of S. coelicolor A3(2), AGTgA(n)mTCACc (7). BldD is a transcriptional repressor globally regulating the expression of genes involved in morphological differentiation and secondary metabolite formation in S. coelicolor A3(2) (6-8). The presence of the consensus sequence suggested that BldD binds the promoter to control its activity in S. griseus. Interestingly, the consensus sequence was not present in the promoter region preceding ramC, the amfT ortholog of S. coelicolor, as previously described by O'Connor et al. (21) (Fig. 1B).
Cloning and transcriptional analysis of bldD of S. griseus.
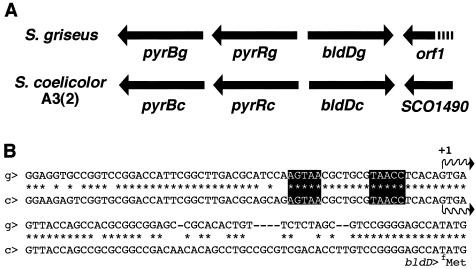
Since bldD from S. griseus has not been characterized, we cloned it by using bldD of S. coelicolor as a probe (see Materials and Methods). Nucleotide sequencing of the cloned DNA revealed the highly conserved gene organization of the bldD locus (Fig. 3A). All four coding sequences identified on the DNA fragment encoded proteins with marked sequence similarity (>90%) to their counterparts in S. coelicolor A3(2). BldD was especially well conserved, with extremely high sequence identity (164 of 167 amino acids). The locus is similarly highly conserved in Streptomyces avermitilis (11).
FIG. 3.
Comparison of bldD locus between S. griseus (g) and S. coelicolor A3(2) (c). (A) Schematic view of the bldD locus. In both organisms, bldD was flanked with coding sequences involved in pyrimidine biosynthesis (pyrB) and regulation (pyrR) and transcriptional antitermination mechanism (orf1 and SCO1490 for S. griseus and S. coelicolor, respectively). (B) Promoter regions preceding bldD. The transcriptional initiation site and the consensus sequence for BldD-binding are indicated by a waved arrow and solid boxes, respectively. Identical nucleotides are indicated by asterisks.
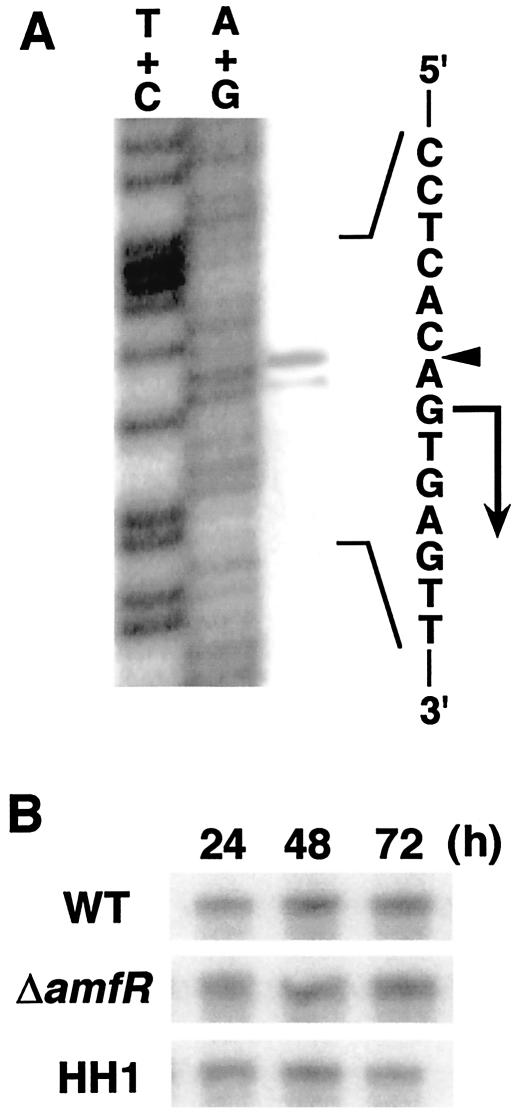
The promoter region of bldD (PbldD) of S. griseus also showed marked similarity to that of S. coelicolor A3(2) (Fig. 3B). High-resolution S1 protection analysis (Fig. 4A) assigned a transcriptional start site at the G residue 58 bp upstream from the deduced translational initiation site (ATG). In S. coelicolor A3(2), it is known that BldD regulates its own expression by binding to its promoter (6). The binding consensus was also present in S. griseus, AGTAA-7 bp-TAACC (Fig. 3B), which suggested that a similar regulatory mechanism functions in this organism. The binding of BldD to the promoter region of S. griseus was confirmed by gel mobility shift assay (see below) (Fig. 5B) and a DNase I footprinting experiment (not shown). The binding site was assigned to the same region as that of S. coelicolor.
FIG. 4.
S1 protection analysis of the bldD promoter of S. griseus. The results of high-resolution (A) and low-resolution (B) analyses are shown. (A) The arrowhead indicates the position of hybridized signal, and the transcriptional start site was assigned to the residue indicated by the bent arrow. Fragments generated by the chemical sequencing reactions are known to migrate 1.5 nucleotides further than the corresponding fragments generated by S1 nuclease digestion of DNA-RNA hybrids (22). RNA prepared from wild-type cells grown for 36 h on YMP/glucose solid medium was used for hybridization. Maxam-Gilbert sequence ladders were used as a standard. (B) Experimental conditions were those described in the legend to Fig. 2A.
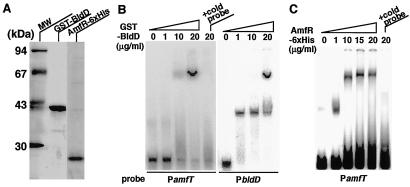
FIG. 5.
Gel mobility shift of PamfT and PbldD with purified BldD and AmfR fusions. (A) Purified GST-BldD and AmfR-6xHis proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were stained with Coomassie brilliant blue. Lane MW, molecular size standards. (B) Gel mobility shift by GST-BldD. The indicated amounts of purified GST-BldD were mixed with the probes for PamfT (157 bp) and PbldD (256 bp) and applied to a polyacrylamide gel. The probe for PbldD shows two different retardation patterns due to an unknown conformational transition depending on the concentration of BldD. A 100-fold molar excess of unlabeled probe was added to confirm the specificity of binding. (C) Gel mobility shift by AmfR. The indicated amounts of purified AmfR-6xHis protein were mixed with the probe for PamfT and applied to a polyacrylamide gel. A 100-fold molar excess of unlabeled probe was added to confirm the specificity of binding.
Low-resolution S1 analysis (Fig. 4B) showed that transcription from PbldD is constitutive throughout the vegetative and transition phases in the wild-type, HH1, and ΔamfR strains, which indicates that transcription of bldD is not affected by a deficiency in A-factor production or deletion of amfR.
Binding of BldD and AmfR to PamfT.
The above observations on PamfT activities suggested that the promoter is regulated positively by AmfR and negatively by BldD. Thus, we assessed the binding of the two regulatory proteins to PamfT in a gel mobility shift assay. BldD and AmfR were expressed and purified in E. coli expression systems as a GST-fused and a His-tagged recombinant protein, respectively (Fig. 5A). As shown in Fig. 5B and C, both the BldD and AmfR recombinant proteins caused a marked mobility shift of the probe DNA corresponding to −104 to +45 of PamfT (see Fig. 1B). Addition of a 100-fold excess of unlabeled probe DNA abolished the retardation. These results indicate that both the AmfR and BldD recombinant proteins can bind PamfT and suggest that their binding controls the initiation of transcription from the promoter in S. griseus.
Phenotypes conferred by overexpression of bldD and inactivation of amfT.
To assess the role of bldD as a negative regulator of transcription of the amf operon, we constructed a high-copy-number Streptomyces plasmid directing overexpression of the BldD recombinant protein and introduced it into the wild-type strain of S. griseus. The transformant was unable to form aerial mycelium and produced markedly reduced streptomycin on YMP/glucose agar medium (Fig. 6A). The bald phenotype was independent of the sugar supplied to the medium. The introduction of an empty vector did not affect the wild-type phenotypes, which confirms that the bald phenotype observed here is linked to overexpression of the BldD protein. Low-resolution S1 protection assays revealed that overproduction of bldD markedly represses PamfT activity (Fig. 2B).
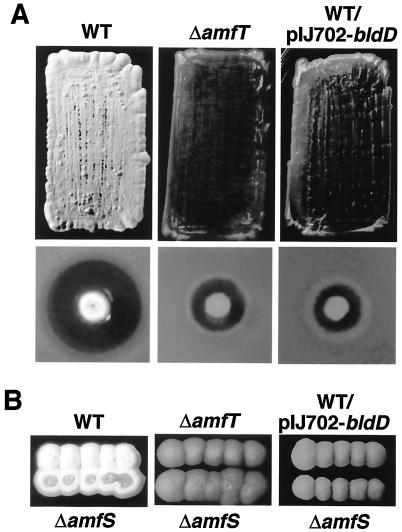
FIG. 6.
Phenotypes conferred by inactivation of amfT and overexpression of bldD in S. griseus. (A) For colony morphology (upper panels), patches were photographed after 5 days of growth at 28°C on YMP/glucose medium. The wild type shows a rough, white colony surface by growing aerial mycelium and spores, while the two other strains grow only substrate mycelium, and their colonies appear smooth and brown. For streptomycin production (lower panels), colonies grown for 5 days at 28°C on YMP/glucose medium were overlaid with soft agar containing spores of Bacillus subtilis and incubated overnight at 37°C. The amounts of streptomycin produced were estimated by growth inhibition of B. subtilis. (B) Cross-feeding assay with the amfS mutant. Each donor strain (upper colonies) was inoculated in close proximity to the amfS mutant (lower colonies) on YMP/glucose agar medium and assessed for its effect on the colony morphology of the amfS mutant. Patches were photographed after 5 days of growth at 28°C. WT, wild type; ΔamfT, amfT mutant; ΔamfS, amfS mutant; ΔamfR, amfR mutant; WT/pIJ702-bldD, wild type harboring a plasmid directing overexpression of GST-BldD.
We also generated a markerless amfT-depleted mutant by the standard homologous recombination technique. The mutant was unable to form aerial mycelium and produced markedly reduced streptomycin on YMP/glucose solid medium (Fig. 6A). The mutant formed aerial mycelium poorly on YMP/maltose medium. S1 analysis showed that PamfT activity in the amfT mutant is at the same level as in the wild type (not shown). The amfT mutant showed the wild-type phenotype when a pIJ922-derived low-copy-number plasmid that carried an intact amfT gene was introduced, which confirms that the mutant phenotype is linked to the inactivation of amfT.
The above two bald strains were subjected to a cross-feeding assay with the amfS mutant as a recipient (Fig. 6B). As reported previously (27), the amfS mutant grew aerial hyphae when the wild-type strain was inoculated in close proximity (Fig. 6B). The extracellular complementation phenomenon is ascribed to the activity of AmfS peptide secreted and supplied by the wild-type strain to the amfS mutant (27). The assay showed that both the amfT mutant and the wild-type strain overexpressing bldD are unable to induce aerial growth of the amfS mutant (Fig. 6B), which strongly suggests that both strains are defective in the production of AmfS peptide.
DISCUSSION
This study revealed that the two major regulators of Streptomyces development, AmfR and BldD, control the activity of PamfT, directing transcription of the amfTSBA operon of S. griseus. In S. griseus, A-factor regulates the onset of morphological and physiological development via the function of two transcriptional regulators, ArpA (A-factor receptor) and AdpA (central regulator). ArpA binds the promoter of adpA, repressing its transcription during vegetative growth. A-factor produced by the organism itself then binds ArpA and dissociates it from the promoter of adpA. The AdpA thus induced acts as a pleiotropic transcriptional activator of the downstream regulatory genes specific for morphogenesis and secondary metabolism (9). Yamazaki et al. recently revealed that AdpA binds the promoter of amfR to activate its transcription (28). Therefore, the dependence of PamfT on amfR shown in this study draws the conclusion that transcription of the amf operon is under the A-factor signaling cascade, predicted as A-factor < AdpA < AmfR < AmfS. This idea is consistent with the previous observation of the dependence of the AmfS activity on A-factor; an A-factor-deficient strain, S. griseus HH1, lacked the extracellular activity inducing aerial mycelium formation in the amfS mutant, and the activity was restored upon exogenous supply of A-factor (27).
This study also showed the presence of bldD in S. griseus. Although it has been clearly demonstrated that BldD plays an important role in developmental control in S. coelicolor A3(2) and that it functions as a transcriptional repressor (7), it is still not known why the mutation in bldD (which changes a Tyr at position 62 to Cys) (6) does not cause enhancement or acceleration of these phenotypes but confers a bald phenotype. It has been believed that a null mutation in bldD is lethal, but recently Elliot et al. reported that the gene is dispensable for viability and the null mutant also shows the bald phenotype (8). Our future studies should reveal the corresponding mutant phenotype in S. griseus.
The highly conserved nature of the bldD locus in Streptomyces spp. implies not only a general role but also marked dependence of BldD function on primary structure, which should have caused high selective pressure during evolution. Constitutive expression of bldD in S. griseus HH1, a mutant that grows only vegetative hyphae because of an A-factor deficiency, implies that BldD is expressed during the primary growth phase, repressing gene expression for the initiation of development in S. griseus. The wild-type transcriptional level of bldD in HH1 suggests that the role of BldD is independent of the A-factor cascade. The dependence of PamfT on both amfR and bldD therefore strongly suggests that the amf operon is an integration point for the two independent signaling networks, as pointed out in a recent review article (5).
The results of in vitro DNA-binding assays suggest that AmfR and BldD control PamfT activity through their direct binding. Although we need to confirm the direct interaction in vivo, it is partially supported by previous studies in S. coelicolor A3(2). O'Connor et al. (21) and Nguyen et al. (20) recently reported that RamR, the AmfR equivalent, binds the promoter of ramC, the amfT equivalent. Similar observations have been described by Keiser et al. (12) for the ram locus of Streptomyces lividans, a close relative of S. coelicolor A3(2). O'Connor et al. (21) and Keiser et al. (12) also showed that a bldD mutant of S. coelicolor is defective in RamC production or transcription, although there are no candidate sites for BldD binding in the promoter preceding ramC. The sequence heterogeneity between the amfT and ramC promoter regions (Fig. 1B) is in marked contrast to the high similarity in the bldD promoters (Fig. 3B) and of interest in terms of the evolution and diversity of the genus Streptomyces. There could be another regulatory target(s) for BldD in S. coelicolor that affects RamC production.
Another example of the difference in BldD recognition between S. griseus and S. coelicolor A3(2) is the promoter of sigH, a stress response sigma factor gene. Kelemen et al. previously reported that BldD serves as a connection channel between stress response and morphological development in S. coelicolor by showing that one of the promoters of the sigH operon is a binding target of BldD (13). On the other hand, the sigH operon of S. griseus, which we characterized recently (23), has a similar sequence in the corresponding promoter region, but it was not bound by the BldD recombinant protein used in this study (our unpublished result). Thus, we assume that there is a certain diversity in the constituents of the BldD regulon among Streptomyces spp., while the BldD protein itself is highly conserved.
The phenotype of the amfT mutant suggests that amfT plays a crucial role in the onset of morphological and physiological development in S. griseus. Furthermore, the mutation in amfT abolished the secretion of a substance(s) that induces aerial growth in the amfS mutant (Fig. 6B) without affecting the transcriptional activity of PamfT. Our previous study strongly suggested that the secreted substance contains a modified form of the amfS gene product (27). Since the markerless mutational construction in amfT does not affect the transcription of amfS, we assume that the mutant is blocked in the process of translation, modification, or secretion of AmfS.
During the review of the manuscript for this article, Kodani et al. (15) published a high-impact paper which reports that the gene product of ramS, the amfS counterpart of S. coelicolor A3(2), is identical to the extracellular surfactant peptide long known as SapB that is essential for the erection of aerial hyphae. SapB production is assumed to be the final event in the regulatory cascade leading to the onset of aerial mycelium formation, such that it has been an important marker to order the bld gene hierarchy in S. coelicolor studies (24). They find that SapB is derived from the C-terminal half of RamS and transformed into a lantibiotic-like structure by an unknown modification process. In the paper, they also suggest that the modification is mediated by RamC, based on its sequence similarity to proteins involved in lantibiotic biosynthesis. Our study on the chemical structure of the AmfS peptide revealed the same properties as RamS/SapB, which strongly suggests that AmfS is the SapB equivalent of S. griseus (our unpublished results). The evidence also supports the idea that AmfT is involved in the modification of AmfS, although AmfT does not show distinct similarity to enzymes related to lantibiotic synthesis. We believe that recent progress in this area should contribute much to our understanding of the molecular mechanisms that control the onset of morphological development in Streptomyces spp.
Acknowledgments
We thank S. Horinouchi and Y. Ohnishi for helpful discussion.
This study was supported by a Grant-in-Aid for scientific research (no. 15380066) and the 21st Century COE program of MEXT, Japan.
REFERENCES
- 1.Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327-336. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 4.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 6.Elliot, M., F. Damji, R. Passantino, K. Chater, and B. Leskiw. 1998. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 180:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliot, M. A., M. J. Bibb, M. J. Buttner, and B. K. Leskiw. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:257-269. [DOI] [PubMed] [Google Scholar]
- 8.Elliot, M. A., T. R. Locke, C. M. Galibois, and B. K. Leskiw. 2003. BldD from Streptomyces coelicolor is a non-essential global regulator that binds its own promoter as a dimer. FEMS Microbiol. Lett. 225:35-40. [DOI] [PubMed] [Google Scholar]
- 9.Horinouchi, S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:d2045-d2057. [DOI] [PubMed] [Google Scholar]
- 10.Horinouchi, S., Y. Kumada, and T. Beppu. 1984. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J. Bacteriol. 158:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 12.Keijser, B. J., G. P. van Wezel, G. W. Canters, and E. Vijgenboom. 2002. Developmental regulation of the Streptomyces lividans ram genes: involvement of RamR in regulation of the ramCSAB operon. J. Bacteriol. 184:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelemen, G. H., P. H. Viollier, J. Tenor, L. Marri, M. J. Buttner, and C. J. Thompson. 2001. A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol. Microbiol. 40:804-814. [DOI] [PubMed] [Google Scholar]
- 14.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 15.Kodani, S., M. E. Hudson, M. C. Durrant, M. J. Buttner, J. R. Nodwell, and J. M. Willey. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 101:11448-11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komatsu, M., Y. Kuwahara, A. Hiroishi, K. Hosono, T. Beppu, and K. Ueda. 2003. Cloning of the conserved regulatory operon by its aerial mycelium-inducing activity in an amfR mutant of Streptomyces griseus. Gene 306:79-89. [DOI] [PubMed] [Google Scholar]
- 17.Ma, H., and K. Kendall. 1994. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176:3800-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Miyadoh, S. 1993. Research on antibiotic screening in Japan over the last decade: a producing microorganisms approach. Actinomycetologica 7:100-106. [Google Scholar]
- 20.Nguyen, K. T., J. M. Willey, L. D. Nguyen, L. T. Nguyen, P. H. Viollier, and C. J. Thompson. 2002. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol. Microbiol. 46:1223-1238. [DOI] [PubMed] [Google Scholar]
- 21.O'Connor, T. J., P. Kanellis, and J. R. Nodwell. 2002. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 45:45-57. [DOI] [PubMed] [Google Scholar]
- 22.Sollner-Webb, B., and R. H. Reeder. 1979. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell 18:485-499. [DOI] [PubMed] [Google Scholar]
- 23.Takano, H., K. Hosono, T. Beppu, and K. Ueda. 2003. Involvement of σH and related sigma factors in glucose-dependent initiation of morphological and physiological development of Streptomyces griseus. Gene 320:127-135. [DOI] [PubMed] [Google Scholar]
- 24.Tillotson, R. D., H. A. Wosten, M. Richter, and J. M. Willey. 1998. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30:595-602. [DOI] [PubMed] [Google Scholar]
- 25.Ueda, K., C. W. Hsheh, T. Tosaki, H. Shinkawa, T. Beppu, and S. Horinouchi. 1998. Characterization of an A-factor-responsive repressor for amfR essential for onset of aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 180:5085-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueda, K., K. Miyake, S. Horinouchi, and T. Beppu. 1993. A gene cluster involved in aerial mycelium formation in Streptomyces griseus encodes proteins similar to the response regulators of two-component regulatory systems and membrane translocators. J. Bacteriol. 175:2006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueda, K., K. Oinuma, G. Ikeda, K. Hosono, Y. Ohnishi, S. Horinouchi, and T. Beppu. 2002. AmfS, an extracellular peptidic morphogen in Streptomyces griseus. J. Bacteriol. 184:1488-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki, H., Y. Takano, Y. Ohnishi, and S. Horinouchi. 2003. amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regualtory cascade in Streptomyces griseus. Mol. Microbiol. 50:1173-1187. [DOI] [PubMed] [Google Scholar]
- 29.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]